Abstract
Spatial structure impacts microbial growth and interactions, with ecological and evolutionary consequences. It is therefore important to quantitatively understand how spatial proximity affects interactions in different environments. We tested how proximity influences colony size when either Escherichia coli or Salmonella enterica are grown on various carbon sources. The importance of colony location changed with species and carbon source. Spatially explicit, genome-scale metabolic modeling recapitulated observed colony size variation. Competitors that determine territory size, according to Voronoi diagrams, were the most important drivers of variation in colony size. However, the relative importance of different competitors changed through time. Further, the effect of location increased when colonies took up resources quickly relative to the diffusion of limiting resources. These analyses made it apparent that the importance of location was smaller than expected for experiments with S. enterica growing on glucose. The accumulation of toxic byproducts appeared to limit the growth of large colonies and reduced variation in colony size. Our work provides an experimentally and theoretically grounded understanding of how location interacts with metabolism and diffusion to influence microbial interactions.
Introduction
Microbial interactions help determine functions from nutrient cycling to human health [1, 2]. Spatial structure mediates microbial interactions [3]; however, the relationship between proximity and interaction strength remains unclear [4]. Quantifying, and being able to predict, the effect of location on microbial interactions is critical for understanding the function of microbial systems as well as their ecological and evolutionary dynamics.
Spatial structure modulates the resource competition that shapes microbial communities [5–7]. Competition influences community assembly [8] and stability [9], and influences selection on microbial traits [10]. Spatial structure alters the scope of competition [6]. In agitated liquid environments, all cells tend to have equal access to resources and interactions are global. In contrast, in structured environments, cells interact more strongly with neighbors than with distant individuals. This localizing effect of spatial structure has been repeatedly shown to influence the outcomes of microbial evo-ecological experiments [10–23].
The specific location of bacteria in spatially structured environments matters. Within a biofilm or colony, bacteria at the edge have lower local density and grow faster than those in the center [4, 24–27], which can segregate competing genotypes [21, 28–31]. Between-colony interactions are also influenced by colonies’ locations. A competing colony’s effect is magnified if it is located between a focal colony and a nutrient source [17]. Furthermore, the coexistence of competing genotypes can be highly sensitive to inter-colony distances [10, 20]. Finally, hints of the importance of location are also being detected in complex natural ecosystems. For example, changes in the spacing between Aggregatibacter actinomycetemcomitans and Streptococcus gordonii determine virulence in oral abscesses [32].
While it is known that location matters, we lack a rigorous framework for understanding and predicting the impact of location on interactions. Interaction strength is likely a function of distance, but by what distance-based measure: the distance to the closest competitor, a function of all competitor distances, or a measurement of how competitors divide the available territory? Ecologists often use distance metrics to explain variance in plant growth [33], and a linearly-weighted distance model captured a decline in bacterial colony size due to crowding [34]. In contrast, Voronoi diagrams, which measure the territory that is closer to a focal colony than any other colonies [35], have been used to investigate pattern formation as bacteria cover a surface [36]. To date, there has not been a rigorous test of the ability of different geometric models to explain variance in colony size.
In addition to this geometric description, the question arises what minimal biophysical model can predict the location-based effects on colony growth in variable environments. Microbes typically interact through chemicals that they consume and excrete [37, 38]. Does accounting for metabolism and diffusion suffice to predict the variation in colony growth? Genome-scale metabolic models and flux-balance analysis can quantitatively predict the metabolites that microbes consume and excrete, and therefore can predict the ecological interactions that emerge from intracellular mechanisms [17, 39, 40]. Diffusion can be incorporated to predict system dynamics in structured environments [17]. We therefore can test to what extent colony variation is purely a function of metabolism and diffusion by comparing computational predictions against experimental observations. If factors such as toxicity, signals, or stochastic differences in lag time drive colony variation, then the model, which does not take these effects into account, will do a poor job. Determining the extent to which metabolic mechanisms drive spatial effects will be critical for predicting growth in complex natural settings.
Here, we investigated how location influences interactions in arguably the simplest scenario—monocultures grown on homogeneous surfaces. We plated monocultures of either Escherichia coli or Salmonella enterica on various media and used high-resolution scanners to investigate the size of colonies and the associated variance within each plate. We then used simulations and geometric descriptions to determine how much colony variation is explained by metabolic mechanisms, what aspect of location best explained variation in growth, and how variation was influenced by nutrient uptake, diffusion, and duration of growth. Finally, we investigated one case in which variation differed from expectation and suggest that this deviation was caused by byproduct toxicity. Our work provides a quantitative framework for understanding and predicting the effect of location on microbial competition.
Methods
Strains and media
We used cells of either Salmonella enterica serovar Typhimurium LT2 or Escherichia coli K12-MG1655. In the genome-scale metabolic modeling, these strains were represented by the iRR_1083 model [41] and the iJO_1366 model [42], respectively. The Petri dish experiments either used Luria–Bertani (LB) media (10 g/L tryptone, 10 g/L NaCl, 5 g/L yeast extract) or a modified Hypho minimal media (7.26 mM K2HPO4, 0.88 mM NaH2PO4, 3.78 mM [NH4]2SO4, 0.41 mM MgSO4, 1 mL of a metal mix [43]). The minimal media contained glucose (16.6 mM), citrate (10.2 mM), lactose (8.33 mM), or acetate (12.5 mM) as the limiting resource. The glucose concentration in the low and medium glucose treatments was 4.15 and 8.33 mM. Experiments where acetate was added to glucose plates had concentrations of 16.6 mM glucose and 12.5 mM acetate. All Petri dishes contained 25 mL of media with 1% agar. Experiments to visualize acidification of the media used bromothymol blue at a concentration of 0.08 g/L. To reduce condensation dishes were left open for 30 min as the agar solidified. UV lights were used to maintain sterility as plates cooled.
For Petri dish experiments, after spreading approximately 60 cells onto a Petri dish, a piece of matte black Kydex Plastic Sheet (0.08 in. thick) was placed within the upper lid of the dish to improve contrast and reduce reflections. Petri dishes were placed onto a Canon Perfection V600 scanner agar side down and a 600 dpi image was scanned every 20 min for almost 150 h. We housed scanners in a 30 °C incubator. Each treatment (a unique combination of a species and a media type) was repeated in separate Petri dishes 3–8 times. The replicates per treatment are in Supplementary Table 2. We tracked colony areas over time, and quantified acidity, using custom image analysis software (Supplementary Material).
Computational modeling
Simulations were run in COMETS, a platform developed to model growth and interactions in structured environments using genome-scale metabolic networks [17]. Biomass and resources are distributed on a lattice. Then dynamic flux-balance analysis determines optimal metabolic activity and growth for all biomass based on the local environment at each time step. Biomass and resources each diffuse with specific diffusion coefficients. Michaelis–Menten kinetics constrain resource uptake.
In addition, we used simplified models in which the genome-scale metabolic model was replaced with a set of differential equations. The simplified model describes a reaction-diffusion system in which bacteria grow under Monod kinetics. Bacteria and resource spread via diffusion, with the diffusion coefficient for the bacteria, DB, being much smaller than that for the resource, DR (Eqs. 1):
| 1a |
| 1b |
| 1c |
The first term in the differential equations describes diffusion ( is the two-dimensional Laplace operator), the second term describes conversion of the resource into biomass. γ sets the resources used per biomass produced, and was equal to 1 unless otherwise stated. The maximum growth rate, μmax, is approached as the resource concentration R increases. The saturation concentration is set by km.
For the simple Monod simulations the “world” was a 5 cm × 5 cm square, into which 60 colonies were seeded at random locations. Resources were distributed uniformly at a concentration of 1e−6 mmol per box. These simulations were run until resources were fully consumed unless otherwise stated. The genome-scale metabolic model simulations were conducted in circular environments that were 90 mm in diameter and seeded with biomass and resources to mimic the experimental conditions. Genome-scale simulations were run for equal lengths of time as the laboratory experiments unless otherwise stated. Other simulation parameters are provided in Supplementary Table 1. Simulations were carried out using the University of Minnesota Supercomputing Institute’s Mesabi cluster.
To simulate toxicity and resource competition, we modified our simplified model to include a toxin (A), which diffuses like the resource and which is produced as biomass grows with a conversion of λ (Eqs. 2). The toxicity of A on growth changes with A*, where higher values are less toxic:
| 2a |
| 2b |
| 2c |
| 2d |
| 2e |
To test whether toxicity improved the fit to experimental data for S. enterica grown on glucose, we parameterized the simplified model to match the genome-scale model in the absence of toxins (Supplementary Fig. 4B–E). To test the effect of toxicity on Voronoi response, we ran simulations of model 2 for 150 h in an environment mimicking experiments with S. enterica grown on a glucose Petri dish using λ = 0.2 mmol/g, which was conservatively set at approximately 10x less than produced by E. coli during growth on glucose [44]. The model used to generate the data in Fig. 5d used A* = 0.01 mM.
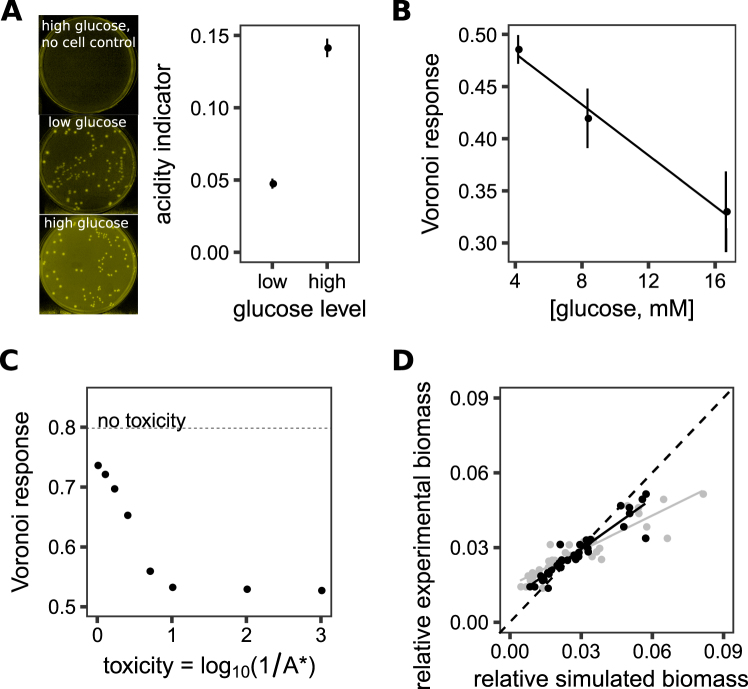
Fig. 5.
Generation of toxic waste reduces the Voronoi response. a S. enterica colonies grown on Petri dishes with glucose concentration below (low) or above (high) the Crabtree threshold, which is the glucose concentration when acetic acid is produced during growth in the presence of oxygen. The Petri dishes contained the pH dye bromothymol blue, which is dark green at neutral pH and becomes yellow as it acidifies. High glucose dishes had higher acidity despite less total growth (see Supplementary Methods for image analysis of yellow intensity, Supplementary Figure 3F for yield data). b The Voronoi response as a function of glucose concentration. Both the 8 and 16.6 mM concentrations are at or above the Crabtree threshold. c The Voronoi response after 150 h in the simplified models modified so that toxic metabolites are generated as biomass grows. The model parameter A* is a measure for the effect of toxicity on growth (see Methods/main text). The dotted line indicates the Voronoi response when toxicity is removed from the model. d Scatter plots of a simulation mimicking S. enterica grown on a glucose Petri dish, without (gray) or with (black) toxicity in the model. The x-axis is the relative biomass in simulation, and the y-axis is the relative biomass measured in the experimental Petri dish. Adding toxicity significantly improved the fit, by bringing the relationship between the x and y data closer to the line with slope 1 passing through the origin (shown with the dashed line)
Statistics
In the programming language R, we used the spatstat package to find Voronoi areas with the dirichletArea function [45]. After calculation of any distance metric, colonies <5 mm from a Petri dish edge were excluded. We also used R to carry out analysis of variance (ANOVA), analysis of covariance (ANCOVA), t tests, and linear regressions as described throughout the results.
Results
Variance in colony size is context-dependent
We tested whether species and resource identity influenced the variance in colony size within monoculture plates. Approximately 60 S. enterica or E. coli cells were grown on Petri dishes with different carbon sources, and colony areas were measured using flatbed scanners and custom software (Fig. 1a, see Methods for more detail). Within every plate/replicate and treatment we found a range of colony sizes, as seen in the example density plots of the final colony areas in Fig. 1b. Because the average colony size differed substantially across treatments (see, for example, Fig. 1b), we used the coefficient of variation of colony area at the end of the experiment (standard deviation over mean) within a plate to compare variation in colony size between treatments. Differences in media and species caused large differences in the coefficient of variation across treatment (ANOVA, F(7,32) = 18.9, p = 1.07e−9, Fig. 1c), suggesting that spatial effects were highly context-dependent.
Fig. 1.
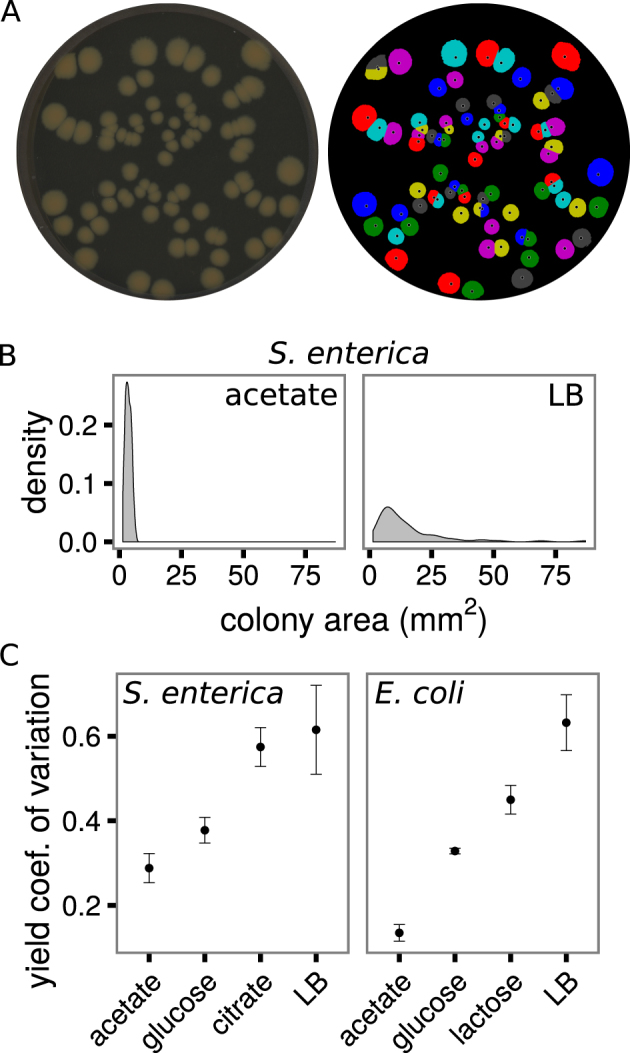
The variance in colony yields depends on the species and environment. a A snapshot of S. enterica colonies on LB media (left) and the yields (areas) of those colonies determined by automated image analysis (right). b Kernel density distributions of S. enterica colony yields grown on acetate or LB. c The coefficient of variation of colony yields for each species on each carbon source. Error bars are standard error of mean. Four to eight Petri dishes are included in each point
Variance in colony size can be predicted with models that pair metabolism and diffusion
We tested whether the observed variations in colony size could be predicted from the interplay of intracellular metabolic mechanisms, diffusion, and colony location, by running simulations that combine genome-scale metabolic modeling with diffusion calculations. Our computational platform, COMETS, uses dynamic flux-balance analysis to predict the growth and metabolic activity of bacteria by identifying the metabolic strategy that maximizes biomass production at each time step [17, 39]. Biomass and metabolites diffuse to simulate growing colonies and the resource gradients that arise as a result of microbial metabolism. Note that colony expansion is the result of both increasing biomass and diffusion [46].
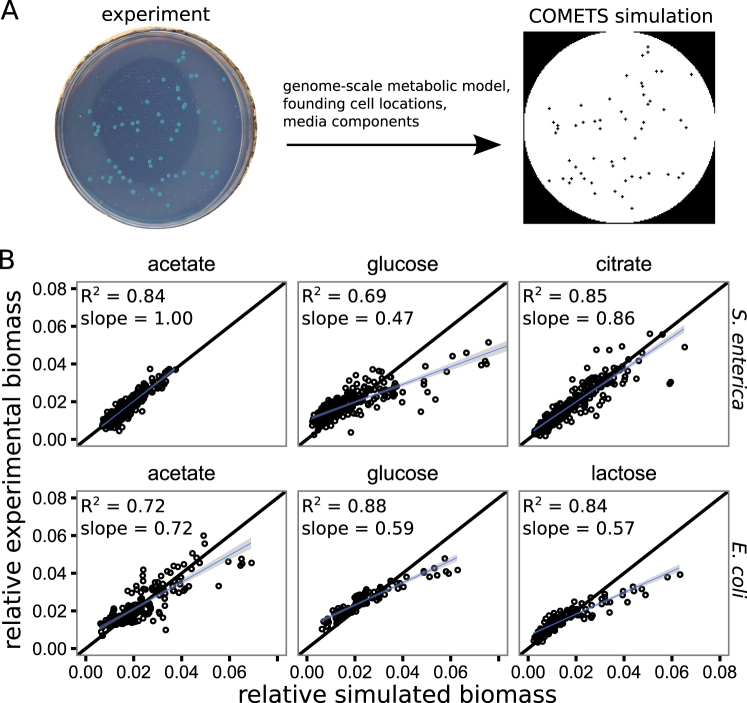
Simulations were initiated with resources and colony locations that matched each experimental plate (Fig. 2a). We plotted the relative yields (yield of a colony/total yield on a Petri dish) for simulations against those for experiments (Fig. 2b). We used relative yields because the measurements of interest were the relative differences between colonies on a plate, which can be compared with relative numbers even if the specific yield measurement (area vs. biomass) differs. The relative colony sizes in simulations were well correlated with the relative colony sizes in experiments, although the predictive ability of the simulations depended on the treatment (mixed-effects linear regression with subsequent F tests, main effect of simulated yields: F(1,1479) = 2027, p < 2.2e−16, main effect of treatment: F(5,24) = 11, p = 1.3e−5, interaction: F(5,1470) = 85, p < 2.2e−16). Deviations from simulated predictions had a slope <1, meaning there was more variability in colony size in simulations than in experiments. These over-predictions were most pronounced when the carbon resource was a sugar (i.e., glucose or lactose). Below, we further explore the deviations caused by S. enterica growth on glucose.
Fig. 2.
Genome-scale metabolic modeling recapitulates the variance in colony yields. a We used genome-scale metabolic modeling in the COMETS platform to test the mechanisms generating the observed variance in colony yields. The relevant genome-scale metabolic model was seeded into an environment at the sites from which colonies initiated in experiments. Dynamic flux-balance analysis calculations and subsequent metabolite and biomass diffusion were carried out in discrete time steps for a duration mimicking the laboratory experiments. b A comparison of the relative colony yields measured in experiments (y-axis) to the relative colony yields predicted by the COMETS simulations (x-axis) for the treatments accessible with COMETs (defined media, i.e., not LB). Each facet contains data from four to eight Petri dish experiments/simulations. The black line has slope = 1 and intercept = 0, while the blue lines and surrounding gray are linear regression lines with standard error. A high R2 suggests that the relative spatial effects are captured by the model, while a slope close to 1 suggests an accurate prediction of amount of variance in colony yield
Relative colony size is driven by the location of adjacent competitors
To understand how location determines colony size, we tested the explanatory power of different metrics that varied in the influence assigned to potentially competing colonies. We focused on metrics that had previously been used in the forestry or microbiology literature [33, 34, 36]. These metrics tested whether colony size could be best predicted by (i) the distance to the nearest neighbor (Fig. 3a), (ii) the sum of the inverse distances to all neighbors (Fig. 3b), (iii) the log of the sum of the squared inverse distances (Fig. 3c), (iv) or a colony’s Voronoi area, which is the area on a Petri dish which is closer to a focal colony than to any other colony (Fig. 3d, see also Fig. 3e) [35].
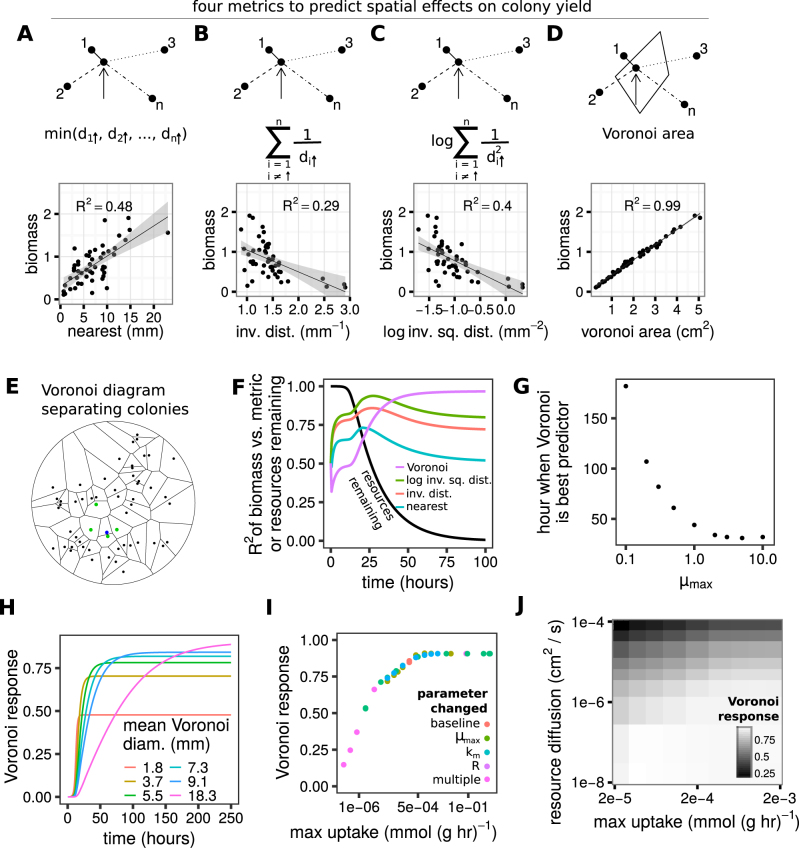
Fig. 3.
Voronoi diagrams capture the effect of location on yield better than other distance metrics. Using the simplified differential equations model, four metrics were tested to determine which colonies interact to generate variation in colony size and to what extent. a–d show a cartoon of the measurement and the metric plotted against simulated colony yield (biomass). a The distance to the closest colony, such that the yield of the focal colony (indicated by the arrow) would be predicted from the distance to colony 1, which is closest, but no other colony would be considered. b The sum of the inverse linear distances to every colony, such that the yield of the focal colony would be predicted by the distance to every colony, with each colony’s influence inversely proportional to its distance. c Like b, but colonies become quadratically less important as distance increases. d The territory closest to a colony, described by a Voronoi diagram. Here, the focal colony’s Voronoi area is shown (solid line polygon). A Voronoi diagram divides a plane into areas around colony initiation sites such that all the space in a territory is closer to its enclosed colony than to any other colony, which is accomplished by drawing perpendicular lines half-way through lines connecting a focal colony to Voronoi neighbors. e A Voronoi diagram drawn for all colony initiation sites on a Petri dish. For a focal colony (blue), its Voronoi neighbors are the green colonies. f The change in R2 over time for the different metrics vs. simulated colony biomass. The black line shows the proportion of resources left. g The hour when the R2 for Voronoi areas vs. simulated colony biomass surpasses the R2 for all other metrics, as a function of the max growth rate (μmax) used in the simulation. h The magnitude of the spatial effect (the “Voronoi response”) as a function of time. Different lines are from simulations with different average Voronoi areas, which is inversely proportional to initial cell density. i The Voronoi response as a function of the maximum potential per-mass uptake. Changes from the default (baseline) values (Supplementary Table 1) in maximum growth rate, km, starting resource concentration, or any combination of these parameters (multiple) all have similar effects j The Voronoi response is determined by the balance between the maximum uptake rate of a colony (x-axis) and the rate of resource diffusion (y-axis)
To abstract away species/environment-specific intracellular metabolism, we ran these tests with a simplified model that simulated biomass growth on an explicit limiting resource using Monod kinetics paired with diffusion (see Methods). We simulated conditions with a high maximum growth rate (μmax=1/h) until all resources were used and asked which metric correlated best with colony size. While all metrics were somewhat predictive, the Voronoi areas had almost perfect prediction. The high R2 of Voronoi areas suggested that under these parameters removing non-Voronoi neighbors (Fig. 3e) would have negligible effects on the size of a focal colony; this prediction was confirmed with “colony dropout” simulations (Supplementary Figure 1A).
We next tested how the predictive power of Voronoi diagrams (the R2 of Voronoi area vs. colony biomass) changed over the course of growth. At the start of simulations all colonies were the same size, and then variance between colonies increased as colonies grew and drew down the resources available to them. The correlation between Voronoi area and colony size increased through time, as did the performance of Voronoi relative to other metrics (Fig. 3f, see Supplementary Figure 1B for analysis with non-parametric Spearman’s ρ). Voronoi area became the best predictor of colony size once the majority of resources were consumed (Fig. 3f) and was the best predictor across multiple resource diffusion coefficients (Supplementary Figure 1C). Consistently, as maximum growth rate increased, resources were consumed faster, and the time required for Voronoi to be the best predictor decreased (Fig. 3g).
In addition to understanding which competitors caused differential growth (i.e., the relative performance of different metrics), we were also interested in the strength of spatial effects (i.e., the magnitude of the best metric). We defined the “Voronoi response” to measure the extent to which colonies have monopolized resources that started in their respective territories. The Voronoi response is the slope of the line that is generated when plotting against (Supplementary Figure 1D). Note that the Voronoi response, the R2 of Voronoi area vs. colony biomass, and the yield coefficient of variation are correlated (Supplementary Figure 1E). Voronoi response declines from a max of 1 towards 0 as colony size becomes less dependent on Voronoi area, which can occur because some colonies are accessing resources that originated in other territories, or because resources have not yet been fully consumed. As with the R2, the Voronoi response increased through time (Fig. 3h). When the size of the average Voronoi area is increased, it takes longer to reach the maximum Voronoi response, but the final Voronoi response is larger meaning colonies are able to monopolize more of the resources that start in their territory. In the other direction, extremely dense environments have small final Voronoi responses, which will approach zero as areas become extremely small (Fig. 3h).
Finally, we tested how the maximum Voronoi response (when no nutrients remain) is influenced by the balance between resource uptake and diffusion. Increasing maximum resource uptake rate through a variety of parameters all increased the Voronoi response until saturating (Fig. 3i). Increasing resource diffusion (DR) reduced the Voronoi response (Supplementary Figure 1G). The final magnitude of the Voronoi response was determined by the balance of nutrient uptake to diffusion (Fig. 3j). While the ratio of uptake to diffusion changed the maximum Voronoi response, it did not change the relative performance: Voronoi outperformed other metrics if all resources were consumed (Supplementary Fig. 1C).
The Voronoi response varied in laboratory experiments
Voronoi area was a good predictor of colony size variation across many laboratory treatments (Fig. 4a, Supplementary Figure 2A). Voronoi area was significantly better than other metrics on rich LB media (ANOVA followed by Tukey's multiple comparisons: LB, p < 5e−4), and as good as other metrics in most other treatments. Voronoi area fared significantly worse than the inverse distance metrics for both species on acetate, and for S. enterica on glucose (ANOVA followed by Tukey's multiple comparisons: acetate, p < 1e−7; glucose p < 1e−7).
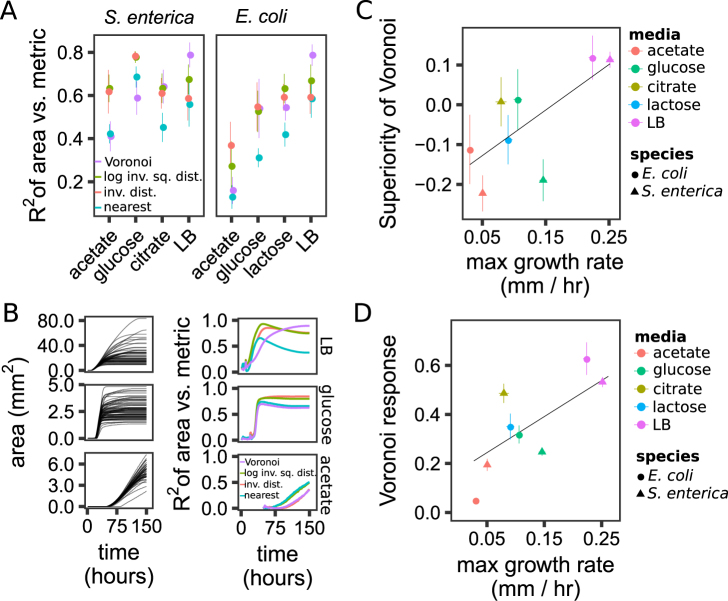
Fig. 4.
The Voronoi response changes between experimental treatments, but generally increases with the maximum colony growth rate. a The performance of the different metrics on the experimental data (R2 of the metric vs. colony areas at 150 h). b The growth curves of individual colonies (left) and change in R2 of the metrics over time (right) for representative S. enterica Petri dishes on three different media. c The superiority of the Voronoi metric (the R2 of the Voronoi metric minus the R2 of the log inverse-squared distance metric) plotted over the maximum growth rates measured in experiments, with a significant least-squares linear fit. d The Voronoi response plotted over the maximum growth rate, with a significant least-squares linear fit. The bars in a, c, d are standard error of the means calculated over Petri dishes, with four to eight Petri dishes per treatment
The relative performance of spatial metrics in different treatments is consistent with our simulation-based findings on the effects of resource depletion (Fig. 4b and Supplementary Figure 2B,C). For example, on LB plates S. enterica colonies reached the carrying capacity and Voronoi areas rose to outperform other metrics (Fig. 4b, compare to Fig. 3f). Conversely, on acetate S. enterica grows slowly and appeared to still be growing. S. enterica on glucose violated our expectations: colony growth had plateaued, but Voronoi did not outperform other metrics (Fig. 4b). We hypothesize why this was below.
The connection between resource depletion and relative metric performance is further supported by an analysis of growth rate. Consistent with simulations (Fig. 3f), in laboratory experiments faster growth rate correlated strongly with the relative superiority of Voronoi (the R2 of Voronoi minus the R2 of the log. sum Inv. dist. metric) (Fig. 4c, linear regression, slope = 1.15, p = 5.4e−4). This correlation suggests that the variable performance of Voronoi in different treatments was at least partially explained by differences in the proportion of resource consumed by the end of the experiment. Additionally, genome-scale simulations showed incomplete resource utilization after 150 h for the substrates that caused slow growth (Supplementary Figure 3A, B, C).
Experimental data also support the computational prediction that the Voronoi response (i.e., the magnitude of spatial effects) is influenced by the growth rate. The Voronoi response in laboratory experiments increased as maximum growth rate increased (Fig. 4d, linear regression, slope = 1.7, F(1,43) = 31.3, p = 1.4e−6). This is in agreement with simulations, although at least part of the small Voronoi response for low growth rate treatments might be due to residual nutrients in these treatments (see above).
Deviations from expected spatial patterns suggest production of toxic waste
S. enterica grown on glucose deviated from expected spatial patterns. In this treatment (i) colony size variation was poorly explained by Voronoi areas (relative to other metrics) despite rapid initial growth and cessation of growth before the end of the experiment (Fig. 4a–c, Supplementary Figure 2B, C) and (ii) the treatment was the most poorly predicted by the genome-scale metabolic modeling (Fig. 2b). This led us to hypothesize that a mechanism in addition to competition for diffusing resources was occurring in this treatment. S. enterica can generate potentially toxic acetate during growth on glucose, so we hypothesized that acetate accumulation arrested growth of large colonies [47–49]. This is particularly likely as we realized that our glucose concentration was in a range in which bacteria are prone to the Crabtree effect. The Crabtree effect causes fermentation to be preferred over respiration above glucose concentrations of ~8 mM, resulting in secretion of high levels of acetate even in the presence of oxygen [44].
Several lines of experimental evidence are consistent with toxic byproducts reducing colony size variation when S. enterica is grown on glucose. First, the pH indicator bromothymol blue was used to demonstrate that glucose plates became acidified by growth of S. enterica (Fig. 5a). Second, if glucose concentration was dropped below the Crabtree threshold (~8 mM, [44]), then less acidification was detected (Fig. 5a) despite more biomass being produced (Supplementary Figure 4F). Reducing the amount of glucose and thus acidification also increased the response of colony size to Voronoi area (Fig. 5b, linear regression, slope = −0.012, p = 4.9e−4). Conversely, if acetate was added to glucose plates, the maximum colony size, and the Voronoi response, decreased (Supplementary Figure 4G, H).
Finally, incorporating production of toxic waste into the differential equation model decreased the Voronoi response observed in simulations and improved the fit between experiment and simulation. Incorporating toxins into our simulations reduced the Voronoi response (Fig. 5c). We additionally tested whether toxins improved the ability of simulations to predict colony size. Incorporating toxin production into a parameterized version of the simplified model significantly improved the ability of simulations to predict observed colony sizes for S. enterica on glucose (Fig. 5d, ANCOVA, interaction term between toxicity and simulated biomass in predicting experimental biomass, p = 7.2e−4).
Discussion
Understanding the quantitative way that spatial proximity affects interactions between bacterial colonies will allow us to better understand and manage microbial ecosystems. We found that the impact of location on bacterial colony size was context-dependent and strongly influenced by both species and resource identity. Encouragingly, spatially explicit, genome-scale metabolic models were able to predict much of the context-dependent variation in colony size by modeling the interaction between diffusion and intracellular metabolism. A simplified model of differential equations demonstrated that variation in colony size is driven by the size of Voronoi areas, though the relative performance of metrics changes over the course of growth. Furthermore, differences in Voronoi response are primarily driven by differences in the rate at which colonies grow and consume resources. Faster consumption causes size variation between colonies to be larger and mitigates diffusion’s tendency to reduce colony size variation. These general ecological relationships serve as a useful null model from which to predict spatial effects caused by resource competition. We demonstrated the utility of this null model by identifying a toxin-mediated interaction that we did not anticipate. In summary, we provide an experimentally and theoretically grounded understanding of how location interacts with metabolism and diffusion to influence microbial interactions.
Voronoi diagrams identified the competitors that drove colony variance once resources were depleted. The relative performance of Voronoi suggests that the arrangement of competing colonies (i.e., colony geometry) is an important determinant of relative growth. This is an intuitive result as diffusing resources are most likely to be consumed by the closest colony and Voronoi diagrams demarcate the resource-containing area closest to each colony. Interestingly, the superiority of this metric is time-dependent, however, and Voronoi diagrams only outperform other metrics once most resources have been consumed. Much of the variation in relative performance across experimental treatments is consistent with growth rate-specific differences in the extent of resource depletion by the end of the experiment. Simulations demonstrated that after resource depletion Voronoi diagrams outperform other metrics at explaining how the location of competitors determines relative microbial growth across treatments. The lack of resource depletion in experiments can explain situations where genome-scale simulations are good predictors of relative colony size, but Voronoi diagrams are not, for example, with E. coli grown on acetate.
Metabolism influenced the importance of location by altering the balance of nutrient uptake to diffusion. Nutrients that generated rapid growth increased the variance in final colony size, and led to a larger response to location (i.e., Voronoi response). The Voronoi response also became larger as colonies were further apart on average. It is important to note that decreasing the magnitude of spatial effects is not equivalent to decreasing competition. The average colony size and total biomass on a plate are equivalent whether competition is local or global (assuming all resources are consumed). However, if the balance of uptake and diffusion causes interactions to be local, spatial location matters, and some colonies will grow much larger than others.
The success of genome-scale models to predict the effects of competition suggests that it will be possible to quantitatively predict microbial metabolic interactions in complex, spatially structured environments. Genome-scale metabolic models can be generated directly from sequence data using known gene–protein reaction associations [40, 42]. High-throughput methods to generate models from sequence data are improving [50–55], and therefore spatially explicit tools such as COMETS may be increasingly useful to generate quantitative predictions of the effect of location on growth and microbial interactions. However, it should be noted that COMETS did not perfectly predict all observed variation even in our simplified system, and incorporating non-metabolic interactions such as toxicity into genome-scale modeling frameworks will likely be important.
Voronoi areas and genome-scale simulations provide null models for size variance between competing colonies, and departures from the null suggest additional interactions are occurring. S. enterica deviated from model expectations when growing on glucose, leading us to suspect that toxins were altering interactions. The production and response of S. enterica to organic acids are certainly well established [48]; however, we did not anticipate that they would be sufficient to halt growth on our plates. Indeed, E. coli also produces acidic waste, but did not deviate from expectations on glucose. This lack of deviation is likely because our E. coli is less sensitive to acetate than S. enterica (Supplementary Figure 4A). More broadly, the detection of toxicity in our system serves as an example of how quantitative analysis can aid in the identification of species interactions. Different biological phenomena likely cause specific departures from the null expectations, akin to the reduced variance caused by waste accumulation. Further research will be aimed at finding spatial signatures of biological phenomena in microbial systems.
A quantitative understanding of how location mediates microbial interactions has important consequences for understanding and harnessing microbial evolutionary ecology. It is well established that spatial structure can alter the interactions between microbes [4] and plays a critical role in determining health outcomes [7]. Quantifying how space mediates interactions will allow for more rigorous understanding of community composition, and improve prediction of dynamics such as competitive exclusion. Additionally, understanding organisms’ interaction strengths is critical for understanding the evolution of microbial traits. For example, it was recently demonstrated that the level of antibiotic secretion can be explained by the relative strength of interaction with sensitive and resistant competitors [10]. As technology which allows for fine-scale placement of cells matures [56–58], we can create spatial arrangements that maximize selection of competitive phenotypes of interest. As we strive to move beyond descriptions of microbial diversity to explanations and management of diversity it will be critical to develop quantitative understanding of microbial interactions.
Electronic supplementary material
Acknowledgements
The authors thank S Zhuang for advice on the scanning technique, B Adamowicz for help with experiments, and the UMN theory group for useful discussions. J Chacón was funded by the Biocatalysis Initiative through UMN.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0038-0) contains supplementary material, which is available to authorized users.
References
- 1.Arrigo KR. Marine microorganisms and global nutrient cycles. Nature. 2005;437:343–8. doi: 10.1038/nature04159. [DOI] [PubMed] [Google Scholar]
- 2.Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nat Rev Genet. 2012;13:260–70. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connell JL, Kim J, Shear JB, Bard AJ, Whiteley M. Real-time monitoring of quorum sensing in 3D-printed bacterial aggregates using scanning electrochemical microscopy. Proc Natl Acad Sci USA. 2014;111:18255–60. doi: 10.1073/pnas.1421211111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nadell CD, Drescher K, Foster KR. Spatial structure, cooperation, and competition in biofilms. Nat Rev Microbiol. 2016;14:589–600. doi: 10.1038/nrmicro.2016.84. [DOI] [PubMed] [Google Scholar]
- 5.Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–50. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- 6.Mitri S, Foster KR. The genotypic view of social interactions in microbial communities. Annu Rev Genet. 2013;47:247–73. doi: 10.1146/annurev-genet-111212-133307. [DOI] [PubMed] [Google Scholar]
- 7.Stacy A, McNally L, Darch SE, Brown SP, Whiteley M. The biogeography of polymicrobial infection. Nat Rev Microbiol. 2016;14:93–105. doi: 10.1038/nrmicro.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.David LA, Weil A, Ryan ET, Calderwood SB, Harris JB, Chowdhury F, et al. Gut microbial succession follows acute secretory diarrhea in humans. MBio. 2015;6:1–14. doi: 10.3391/mbi.2015.6.1.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shade A, Peter H, Allison SD, Baho DL, Berga M, Bürgmann H, et al. Fundamentals of microbial community resistance and resilience. Front Microbiol. 2012;3:1–19. doi: 10.3389/fmicb.2012.00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gerardin Y, Springer M, Kishony R. A competitive trade-off limits the selective advantage of increased antibiotic production. Nat Microbiol. 2016;1:16175. doi: 10.1038/nmicrobiol.2016.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dechesne A, Or D, Smets BF. Limited diffusive fluxes of substrate facilitate coexistence of two competing bacterial strains. FEMS Microbiol Ecol. 2008;64:1–8. doi: 10.1111/j.1574-6941.2008.00446.x. [DOI] [PubMed] [Google Scholar]
- 12.Allen B, Gore J, Nowak MA. Spatial dilemmas of diffusible public goods. Elife. 2013;2013:1–11. doi: 10.7554/eLife.01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gralka M, Stiewe F, Farrell F, Möbius W, Waclaw B, Hallatschek O, et al. Allele surfing promotes microbial adaptation from standing variation. Ecol Lett. 2016;19:889–98. doi: 10.1111/ele.12625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greig D, Travisano M. Density-dependent effects on allelopathic interactions in yeast. Evolution. 2008;62:521–7. doi: 10.1111/j.1558-5646.2007.00292.x. [DOI] [PubMed] [Google Scholar]
- 15.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–6. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 16.Harcombe H, Betts A, Shapiro JW, Marx CJ. Adding biotic complexity alters the metabolic benefits of mutualism. Evolution. 2016;70:1871–81. doi: 10.1111/evo.12973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harcombe WR, Riehl WJ, Dukovski I, Granger BR, Betts A, Lang AH, et al. Metabolic resource allocation in individual microbes determines ecosystem interactions and spatial dynamics. Cell Rep. 2014;7:1104–15. doi: 10.1016/j.celrep.2014.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Allison SD. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett. 2005;8:626–35. doi: 10.1111/j.1461-0248.2005.00756.x. [DOI] [Google Scholar]
- 19.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–4. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 20.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–93. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitri S, Clarke E, Foster KR. Resource limitation drives spatial organization in microbial groups. ISME J. 2015;10:1–12. doi: 10.1038/ismej.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Penn AS, Conibear TCR, Watson RA, Kraaijeveld AR, Webb JS. Can Simpson’s paradox explain cooperation in Pseudomonas aeruginosa biofilms? FEMS Immunol Med Microbiol. 2012;65:226–35. doi: 10.1111/j.1574-695X.2012.00970.x. [DOI] [PubMed] [Google Scholar]
- 23.Chao L, Levin BR. Structured habitats and the evolution of anticompetitor toxins in bacteria. Proc Natl Acad Sci USA. 1981;78:6324–8. doi: 10.1073/pnas.78.10.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gandhi SR, Yurtsev EA, Korolev KS, Gore J. Range expansions transition from pulled to pushed waves as growth becomes more cooperative in an experimental microbial population. Proc Natl Acad Sci USA. 2016;113:6922–7. doi: 10.1073/pnas.1521056113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Persat A, Nadell CD, Kim MK, Ingremeau F, Siryaporn A, Drescher K, et al. The mechanical world of bacteria. Cell. 2015;161:988–97. doi: 10.1016/j.cell.2015.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pirt SJ. A kinetic study of the mode of growth of surface colonies of bacteria and fungi. J Gen Microbiol. 1967;47:181–97. doi: 10.1099/00221287-47-2-181. [DOI] [PubMed] [Google Scholar]
- 27.Cole JA, Kohler L, Hedhli J, Luthey-Schulten Z. Spatially-resolved metabolic cooperativity within dense bacterial colonies. BMC Syst Biol. 2015;9:15. doi: 10.1186/s12918-015-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci USA. 2007;104:19926–30. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hallatschek O, Nelson DR. Life at the front of an expanding population. Evolution. 2010;64:193–206. doi: 10.1111/j.1558-5646.2009.00809.x. [DOI] [PubMed] [Google Scholar]
- 30.Korolev KS, Muller MJ, Karahan N, Murray AW, Hallatschek O, Nelson DR. Selective sweeps in growing microbial colonies. Phys Biol. 2012;9:026008. doi: 10.1088/1478-3975/9/2/026008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Momeni B, Brileya KA, Fields MW, Shou W. Strong inter-population cooperation leads to partner intermixing in microbial communities. Elife. 2014;3:e02945. doi: 10.7554/eLife.02945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc Natl Acad Sci USA. 2014;111:7819–24. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tome M, Burkhart HE. Distance-dependent competition measures for predicting growth of individual trees. Forest Sci. 1989;35:816–31. [Google Scholar]
- 34.Guillier L, Pardon P, Augustin JC. Automated image analysis of bacterial colony growth as a tool to study individual lag time distributions of immobilized cells. J Microbiol Methods. 2006;65:324–34. doi: 10.1016/j.mimet.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Okabe A, Boots B, Sugihara K, Nok Chiu S. Spatial tesselations: concepts and applications of Voronoi diagrams. 2nd ed. New York, NY: Wiley; 2000. [Google Scholar]
- 36.Lloyd DP, Allen RJ. Competition for space during bacterial colonization of a surface. J R Soc Interface. 2015;12:20150608. doi: 10.1098/rsif.2015.0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Germerodt S, Bohl K, Lück A, Pande S, Schröter A, Kaleta C, et al. Pervasive selection for cooperative cross-feeding in bacterial communities. PLoS Comput Biol. 2016;12:1–21. doi: 10.1371/journal.pcbi.1004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mahadevan R, Edwards JS, Doyle FJ., III Dynamic flux balance analysis of diauxic growth in Escherichia coli. Biophys J. 2002;83:1331–40. doi: 10.1016/S0006-3495(02)73903-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Orth JD, Thiele I, Palsson BOslash. What is flux balance analysis? Nat Biotechnol. 2010;28:245–8. doi: 10.1038/nbt.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Raghunathan A, Reed J, Shin S, Palsson B, Daefler S. Constraint-based analysis of metabolic capacity of Salmonella typhimurium during host-pathogen interaction. BMC Syst Biol. 2009;3:38. doi: 10.1186/1752-0509-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Orth JD, Conrad TM, Na J, Lerman JA, Nam H, Feist AM, et al. A comprehensive genome-scale reconstruction of Escherichia coli metabolism. Mol Syst Biol. 2011;7:535. doi: 10.1038/msb.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Delaney NF, Kaczmarek ME, Ward LM, Swanson PK, Lee MC, Marx CJ. Development of an optimized medium, strain and high-throughput culturing methods for Methylobacterium extorquens. PLoS ONE. 2013;8:e62957. doi: 10.1371/journal.pone.0062957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luli GW, Strohl WR. Comparison of growth, acetate production, and acetate inhibition of Escherichia coli strains in batch and fed-batch fermentations. Appl Environ Microbiol. 1990;56:1004–11. doi: 10.1128/aem.56.4.1004-1011.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baddeley A, Rubak E, Turner R. Spatial point patterns: methodology and applications with {R} London: Chapman & Hall/CRC Press; 2015. [Google Scholar]
- 46.Murray JD. Mathematical biology, I: An introduction. 3rd ed. New York: Springer-Verlag; 2002.
- 47.Rhee M, Lee S, Dougherty RH, Kang D. Antimicrobial effects of mustard flour and acetic acid against Escherichia coli O157: H7, Listeria monocytogenes, and Salmonella ent. Appl Environ Microbiol. 2003;69:2959–63. doi: 10.1128/AEM.69.5.2959-2963.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cappuyns AM, Bernaerts K, Vanderleyden J, Van Impe JF. A dynamic model for diauxic growth, overflow metabolism, and AI-2-mediated cell-cell communication of Salmonella typhimurium based on systems biology concepts. Biotechnol Bioeng. 2009;102:280–93. doi: 10.1002/bit.22044. [DOI] [PubMed] [Google Scholar]
- 50.Agren R, Liu L, Shoaie S, Vongsangnak W, Nookaew I, Nielsen J. The RAVEN Toolbox and its use for generating a genome-scale metabolic model for Penicillium chrysogenum. PLoS Comput Biol. 2013;9:e1002980. doi: 10.1371/journal.pcbi.1002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feist AM, Herrgård MJ, Thiele I, Reed JL, Palsson BO. Reconstruction of biochemical networks in microbial organisms. Nat Rev Microbiol. 2009;7:129–43. doi: 10.1038/nrmicro1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henry CS, Dejongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol. 2010;28:969–74. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]
- 53.Krumholz EW, Libourel IGL. Sequence-based network completion reveals the integrality of missing reactions in metabolic networks. J Biol Chem. 2015;290:19197–207. doi: 10.1074/jbc.M114.634121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:206–14. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thiele I, Palsson BOslash. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc. 2010;5:93–121. doi: 10.1038/nprot.2009.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Connell JL, Ritschdorff ET, Whiteley M, Shear JB. 3D printing of microscopic bacterial communities. Proc Natl Acad Sci USA. 2013;110:18380–5. doi: 10.1073/pnas.1309729110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ferris CJ, Gilmore KG, Wallace GG, In Het Panhuis M. Biofabrication: an overview of the approaches used for printing of living cells. Appl Microbiol Biotechnol. 2013;97:4243–58. doi: 10.1007/s00253-013-4853-6. [DOI] [PubMed] [Google Scholar]
- 58.Xu T, Petridou S, Lee EH, Roth EA, Vyavahare NR, Hickman JJ, et al. Construction of high-density bacterial colony arrays and patterns by the ink-jet method. Biotechnol Bioeng. 2004;85:29–33. doi: 10.1002/bit.10768. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.