Abstract
Changes in frequency and amplitude of rain events, that is, precipitation patterns, result in different water conditions with soil depth, and likely affect plant growth and shape plant and soil microbial activity. Here, we used 18O stable isotope probing (SIP) to investigate bacterial and fungal communities that actively grew or not upon rewetting, at three different depths in soil mesocosms previously subjected to frequent or infrequent watering for 12 weeks (equal total water input). Phylogenetic marker genes for bacteria and fungi were sequenced after rewetting, and plant-soil microbial coupling documented by plant 13C-CO2 labeling. Soil depth, rather than precipitation pattern, was most influential in shaping microbial response to rewetting, and had differential effects on active and inactive bacterial and fungal communities. After rewetting, active bacterial communities were less rich, more even and phylogenetically related than the inactive, and reactivated throughout the soil profile. Active fungal communities after rewetting were less abundant and rich than the inactive. The coupling between plants and soil microbes decreased under infrequent watering in the top soil layer. We suggest that differences in fungal and bacterial abundance and relative activity could result in large effects on subsequent soil biogeochemical cycling.
Introduction
Water availability is a key regulator of ecosystem functioning, directly controlling plant and soil microbial activity. The predicted large changes in precipitation brought on by climate change include periods of increased water limitation followed by larger magnitude rain events for many parts of the world [1]. Changes in the total amount of precipitation constrain ecosystem functioning, and their effects have been documented in many water input reduction experiments (e.g., [2–8]). However, climate change is predicted to affect not only the amount but also the temporal distribution of rain. Changes in frequency and amplitude of rain events, that is, precipitation patterns, likely shape the activity of plants and soil microorganisms [9–11].
Microbial reactivation upon rewetting is a key moment in ecosystem functioning. Substrate becomes readily available to soil microorganisms, triggering the microbial activity that drives soil biogeochemical cycles. However, not all microbes respond similarly to rewetting events, indicating adaptation in life strategies that may be phylogenetically conserved ecological traits [12, 13]. Furthermore, microbial communities having been exposed to a history of erratic moisture fluctuations are adapted to these conditions and show smaller changes in response to rewetting events [54].
Within soil microbial communities, bacteria and fungi differ in their resistance to desiccation, as well as in their response to rewetting [14–16]. Fungal populations have been shown to be more resistant to water limitation, likely due to their ability to access water from distant micropores with their extensive hyphal network [17]. Bacteria typically respond faster than fungi to changes in water availability, albeit with a wide range of responses in the bacterial community [12, 13]. Soil microorganisms drive biogeochemical cycles in soil, such that changes in the relative contribution of bacteria and fungi may affect ecosystem functioning. For example, increased fungal:bacterial ratio resulting from dry–wet cycles improved soil nutrient retention [18]. It is therefore crucial to consider both bacterial and fungal responses when evaluating effects of changes in precipitation on the soil microbial community.
Dry–wet cycles are expected to affect soil differentially at different depths, since the top soil layer experiences more fluctuating water conditions, is wet more often, and dries out quicker than the deeper layers, likely shaping microbial community composition and function [19, 20]. Decreased bacterial diversity upon hydration is supported by physical modeling approaches that indicate increased competition when soil rehydration restores connectivity [20–22]. Moreover, as most roots are usually developed in the top soil layers and plants’ root activity is expected to respond to precipitation patterns (e.g. [23]), we expect the strongest microbial response in the top soil layers. Under mesic conditions, plants generally remain alive between rain events and their interaction with soil microorganisms may drastically impact how the system responds to precipitation changes. The large amounts of plant carbon allocated to the soil by rhizodeposition is crucial for heterotrophic microorganisms, particularly when considering the carbon costs of microbial water resistance strategies, such as active osmoregulation and exopolysaccharide production (Canarini 2015). The presence of plants can therefore increase microbial resistance and resilience to water stress through sustained C inputs [24, 25]. However, if drought conditions persist, photosynthesis is reduced and becomes less coupled with below-ground processes [3]. Despite the intricate link between soil microbes and plants, many drying–rewetting studies focusing on microbial community composition or activity have been performed on systems devoid of live plants, as incubations in soil alone or because plants died during the dry period preceding rewetting.
The present study investigated the response to a rewetting event of bacterial and fungal populations that were actively growing or not (referred to as “active” and “inactive, respectively), at different soil depths in a plant-soil system with a history of contrasting precipitation patterns, that is, its precipitation legacy. Using 18O stable isotope probing (18O-SIP), 18O-labeled water was applied upon rewetting in order to discriminate microbial communities, which are actively growing from those that were not. We hypothesized that (1) rewetting would result in growth of only a small fraction of soil microbes, which closely track soil moisture fluctuations, (2) the microbial community response to rewetting would vary with soil depth, (3) precipitation legacy would affect both plants and microbes, and (4) the precipitation legacy effect would be more pronounced at shallow soil depth.
Materials and methods
Experimental set-up
Mesocosms (56 cm high, 36 cm wide, and 2 cm deep, Fig. S1) were filled (uniform bulk density 1.2 g cm−3) with sandy soil collected from 0 to 25 cm depth in an ungrazed grassland (Varenne-Saint-Germain, France). Sieving (2 mm mesh) ensured the soil and its associated microbial community was homogeneous throughout the mesocosms. Soil texture was 92% sand, 4% silt, 4% clay, pH was 5.9, cation exchange capacity was 4.0 cmol kg−1, organic matter was 2.6%, total N 0.12%, and total C 1.51%. Winter wheat (Triticum aestivum cv Soissons) was germinated on the same grassland soil, then 72 h after their germination plantlets were transferred to mesocosms (14 plants per mesocosm, 2.5 cm apart) grown under controlled conditions (20/15 °C daytime/nighttime temperature, 18-h photoperiod). After a 2-week establishment period during which all mesocosms were watered daily (ensuring that the plants were properly developed before initiating the experimental treatments), two watering frequency treatments were applied, with the same total amount of water given to each treatment. The high-frequency treatment (i.e., frequent water input) consisted of daily water inputs with the minimum volume of water required to avoid wilting (from 1.4 to 6.3 mm as plant water requirement grew with their size). The low-frequency treatment (infrequent water input) consisted of one input event every 2–3 days for 2 weeks, until the plants were strong enough to survive one input every 5–6 days. The volume equaled the sum total of water delivered over the same period in the high-frequency treatments (from 2.8 to 37.5 mm). The mesocosms were weighed daily to monitor soil water content. Forty mesocosms were used for the experiment. Before wet-up, 10 were used for soil and plant sampling and 10 for 13C-CO2 labeling (5 replicates per treatment). After wet-up, 10 were used for H218O labeling and 10 for the unlabeled control (5 replicates per treatment).
Before final wet-up: soil and plant sampling
Three target soil layers were determined at 0–5, 10–15, and 30–35 cm depth, based on the wetting fronts recorded during the experiment. Following a watering event, the 0–5 cm layer was uniformly wet in both treatments, the 10–15 cm layer was uniformly wet in the low-frequency treatment but infrequently wet in the high-frequency treatment, and the 30–35 cm layer was infrequently wet in the low-frequency treatment but uniformly dry in the high-frequency treatment. Twelve weeks after germination (i.e., after 18 low-frequency watering cycles), leaf gas exchange was measured on the youngest, fully developed leaf of three plants per mesocosm (Li-6400 XT portable photosynthesis system, Li-Cor, Lincoln, USA) in five mesocosms per treatment, then all plants were harvested and measured for leaf surface area (Li-3100C, Li-Cor), dry biomass (including root biomass), root length density (WinRHIZO software, Regent Instruments Inc., Canada), and C and N content (NC 2500 elemental analyzer, Carlo Erba, Milan, Italy).
Before final wet-up: plant-soil coupling (13C-CO2 labeling)
Fourteen weeks after germination, five mesocosms per treatment were labeled for 1.5 h with 13C-CO2 as follows. An airtight transparent plastic tent was closed around the mesocosms and CO2 concentration within the tent was allowed to be drawn down by plant photosynthesis to approximately 300 ppm. Fans inside the tent ensured good mixing of air. Labeling was performed by dissolving CaCO3 powder (50% 13C, 50% 12CO2) with HCl 1 M and pumping the resulting gas through the tent. 12CO2 concentration inside the tent was monitored (Walz GFS 3000 infra-red gas analyzer, Heinz Walz GmbH, Effeltrich, Germany), and averaged 964.2 μmol mol−1 (range: 579.3–1215.3 μmol mol−1) over the labeling period. As we added a 50%:50% mixture of 12CO2 and 13CO2, total CO2 concentration was approximately twice the measured values. Temperature during labeling period averaged 23.0 °C. After labeling, the tent was removed and the greenhouse flushed with outside air. Repeated measurements of root, leaf, and microbial biomass isotopic signature were made in each mesocosm for 5 days as follows, while precipitation treatments were maintained. Five 4.6 cm wide vertical strips were marked out, each comprising two plant individuals and 2.3 cm away from the next strip. Each day for 5 days, one side of the mesocosms was opened, three soil layers (0–5, 10–15, and 30–35 cm depth) in one random vertical strip per mesocom were sampled and replaced by sand before the mesocosms were closed again. Each layer was subsampled: one subsample was used for microbial biomass and isotopic signature, the other was washed and the roots dried (48 h at 65 °C) and ground for 13C signature measurement. The youngest fully developed leaf of two plants was taken on the first and last sampling day, dried, and ground for 13C signature measurement. Soil microbial biomass C was determined by chloroform fumigation extraction [26]. One 10 g subsample was fumigated for 24 h with chloroform vapor, whereas another was not. Microbial C was extracted by vigorous shaking in K2SO4 0.5 M. Organic C concentration and its isotopic signature were determined by oxidizing extractable carbon to CO2 [27]. In all, 1 mL of extracted C plus 1 mL of the oxidizing agent (supersaturated potassium persulfate oxidizing solution :100 mL H2O+ 4.0 g K2S2O8+ 200 mL of 85% H3PO4) were added to 12 mL vials, then flushed for 5 min with helium to remove atmospheric CO2. To complete the oxidation, vials were heated to 100° for 1 h. Finally, soil microbial biomass C was calculated as follows ((total C in fumigated soil)–(total C in unfumigated soil))/0.45 [26]. C concentrations and 13C signature were analyzed with a GasBench II system coupled to an isotope ratio mass spectrometer (Delta Plus XL, Thermo Finnigan Mat, Bremen, Germany). Plant biomass 13C signature was measured by carbon isotope analysis (precision of 0.1‰): combustion in an elemental analyzer (EA1110 CHN, Carlo Erba) coupled via a Conflo II interface to an isotope ratio mass spectrometer (Delta S, Finnigan MAT). 13C/12C ratio is expressed in δ notation (‰) relative to the Vienna Pee Dee Belemnite (VPDB) standard.
Final wet-up: H218O application
At the end of the experiment, the short-term response of the active microbial community to a large-scale rain event was assessed with a SIP experiment. Rewetting soil with 18O-labeled water results in the heavy 18O stable isotope being assimilated into the DNA of actively growing and replicating communities, which can then be separated and sequenced [28]. No water was added to the mesocosms for 48 h prior to rewetting. The rewetting SIP was performed in situ, by opening the sides of the mesocosms and applying H218O (5 18O-labeled mesocosms, 98.7 atom% 18O, Eurisotop, Saint-Aubin, France) or molecular grade unlabeled H2O (five control mesocosms) to a 2 cm diameter area in each of the three target soil layers. Molecular grade water was applied to the rest of the soil volume, except a non-watered 4 cm wide buffer zone that was maintained between the labeled and unlabeled water application zones to avoid mixing. To ensure a long enough exposure of the microbial community to the 18O label and compensate plant water uptake, H218O (and its H2O counterpart in the control mesocosms) was applied over 5 days as daily 2 ml applications for 2 days then daily 0.5 ml applications for the next 3 days. Following each application, the mesocosms were closed, returned to their original vertical position in the greenhouse, and the plants bagged in clear plastic to decrease soil moisture loss from evapotranspiration. Six days after initial wet-up, the labeled areas and their control counterparts were sampled and the soil kept at −20 °C prior to SIP processing.
DNA extraction and fractionation
For each sample, DNA was extracted separately from three 0.5 g subsamples (FastDNA kit, MP Biomedicals, Solon, OH, USA), and quantified (Quantus Fluorometer, Promega, Sunnyvale, USA) before pooling the subsamples. Enriched DNA was separated from unenriched DNA through isopycnic centrifugation using a CsCl gradient. In all, 5 µg of extracted DNA was combined with 3.5 ml of CsCl (1.89 g ml−1), 0.3 ml of gradient buffer (200 mM Tris 8.0, 200 mM KCl, 2 mM EDTA), 0.9 ml TE buffer and added to a 4.7 ml centrifuge tube (Beckmann-Coulter, Fullerton, USA) and ultracentrifuged (60,000 rpm—i.e., 149,723 × g at the average radius r(av)-, 18 °C, 115 h).
DNA from a H218O-labeled soil sample and its natural abundance control were always processed in the same ultracentifuge run. Following centrifugation, each tube was divided into 70 µl fractions and their density determined (AR200 refractometer, Reichert, Depew, USA). DNA was purified by adding 300 µl of molecular grade H2O, 10 µl of glycogen (20 mg ml−1) and 400 µl of isopropanol to each fraction, before overnight incubation at 5 °C. The fractions were centrifuged (13,400 × g, 15 min), the precipitate washed with filter-sterilized 70% ethanol, suspended in 50 µl of TE buffer and kept at −80 °C. DNA concentration in each fraction was quantified by fluorometry (Quant-iT PicoGreen dsDNA assay kit, Invitrogen, Cergy-Pontoise, France). The fractions constituting each sample were binned into four groups based on their density: 1.668 < light ≤ 1.708, 1.708 < mid-light ≤ 1.714, 1.714 < mid-heavy ≤ 1.722, 1.722 < heavy<1.740 g cm−3. The DNA of the active microbial community was defined as the DNA present in the heavy fraction of H218O sample when no DNA was present in the heavy fraction of the natural abundance control. The inactive microbial community was defined based on the DNA in the light fraction of the H218O sample. The bacterial and fungal communities were quantified and the DNA in the binned fractions sequenced as described below.
Quantification of the bacterial and fungal communities
The abundance of the soil bacterial and fungal communities was assessed by quantitative polymerase chain reaction (qPCR), using bacterial 16S ribosomal RNA (rRNA) encoding gene primers 341F 5ʹ-CCTACGGGAGGCAGCAG-3ʹ/534R 5ʹ-ATTACCGCGGCTGCTGGCA-3ʹ [29] and fungal internal transcribed spacer (ITS) region primers ITS3 5ʹ-GCATCGATGAAGAACGCAGC-3ʹ/ITS4 5ʹ-TCCTCCGCTTATTGATATGC-3ʹ [30], respectively. Quantification was based on SYBR Green dye increasing fluorescence intensity during amplification, in a ViiA7 (Life Technologies, Carlsbad, CA, USA). Real-time PCR assays were carried out in triplicate 15 µl reactions containing SYBR green PCR Master Mix (Takyon Low ROX SYBR 2 × MasterMix blue dTTP, Eurogentec, France), 1 µM of each primer, 250 ng of T4 gene 32 (QBiogene, France) and 1 ng of DNA. Standard curves were obtained using serial dilutions of linearized plasmids containing the cloned genes (efficiency: 89–99%). Template-free controls gave negligible values. No inhibition was detected.
Amplicon generation and MiSeq sequencing
Illumina next-generation amplicon sequencing was used to sequence 240 samples. Amplicons were generated in two steps [31]. In the first step, the bacterial 16S rRNA gene V3–V4 hypervariable region was amplified by PCR using the following fusion primers including overhang adapters to allow subsequent addition of multiplexing index sequences [32]: Pro341F (5ʹ-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGRSGCAGCAG-3ʹ) and Pro805R (5ʹ-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG GACTACCAGGGTATCTAAT-3ʹ). PCR was carried out in duplicate 15 µL reactions containing 7.5 µL Phusion High-Fidelity PCR Master Mix (Thermo Scientific), 0.25 µM of each primer, 250 ng T4 gp32 (MPBio) and 1 ng template DNA. Thermal cycling conditions were 98 °C for 3 min followed by 25 cycles of 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, with a final extension at 72 °C for 10 min. Duplicate first step PCR products were pooled then used as template for the second step PCR. In the second step, PCR amplification added multiplexing index sequences to the overhang adapters using a unique multiplex primer pair combination for each sample, in duplicate 30 µL reactions containing 15 µL Phusion High-Fidelity PCR Master Mix (Thermo Scientific), 1 µM of one forward and one reverse multiplex primer and 6 µL of first step PCR product. Thermal cycling conditions were 98 °C for 3 min, 8 cycles of 98 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, final extension at 72 °C for 10 min. PCR products were pooled, cleaned-up, and purified using AMPure XP beads (Beckman-Coulter), quantified with picogreen (Thermo Scientific), followed by equimolar pooling and gel purification. Sequencing was performed on MiSeq (Illumina, 2 × 250 bp, MiSeq reagent kit v2, 500 cycles). Demultiplexing and trimming of Illumina adaptors and barcodes was done with Illumina MiSeq Reporter software (version 2.5.1.3). Fungal ITS rRNA region was amplified similarly, using the primers ITS3F (5ʹ-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGNNNNGCATCGATGAAGAACGCAGC-3ʹ) and ITS4R (5ʹ-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGNNNNTCCTCSSCTTATTGATATGC-3ʹ), modified from White et al. [30], with 30 cycles for the first step PCR and 10 cycles for the second step PCR.
Bioinformatic analysis of the 16S rRNA gene and ITSr RNA region amplicons
Sequences were assembled using PEAR [33]. Further quality checks were conducted using the QIIME pipeline [34] and short sequences were removed (<330 bp for 16S rRNA genes and <230 bp for ITS region). Reference-based and de-novo chimera detection, as well as operational taxonomic unit (OTU) clustering were performed using VSEARCH [35] and the adequate reference databases (Greengenes for 16S, UNITE for ITS region). Identity thresholds were set at 94% for 16S rRNA gene data, based on replicate sequencings of a bacterial mock community containing 40 bacterial species, and 97% for ITS region data for which we did not have a mock community. Representative 16S rRNA genes sequences for each OTU were aligned using PyNAST [36] and a phylogenetic tree constructed using FastTree [37]. Taxonomy was assigned using UCLUST [38] and the latest released Greengenes database (v.05/2013 [39]) for 16S rRNA gene, and BLAST [40] and the UNITE reference database (v.7-08/2016, [41] for ITS region.
Bacterial and fungal α-diversity metrics were calculated in QIIME based on rarefied OTU tables (10,000 sequences per sample for both 16S and ITS region). UniFrac distance matrices [42] and Bray–Curtis dissimilarity matrices were computed for 16S rRNA genes and ITS region, respectively. Net relatedness index (NRI) of the bacterial communities was calculated based on mean phylogenetic distance [43, 44], using a null model of random community phylogenetic relationships (picante package [45]; 999 runs, not abundance weighted).
Statistical analyses
Statistical analyses were performed using R 3.1.2 [46] on n = 5 replicate mesocosms per treatment. Data measured at the overall mesocosm level were assessed by analysis of variance, using precipitation regime as fixed effect variable. The precipitation pattern treatment, applied at the mesocosm level, generated a depth gradient within the mesocosms, therefore this nested design was accounted for in the statistical analysis and allowed the deconvolution of precipitation pattern, depth, and their interaction. Data measured at different soil depths were analyzed by analysis of variance using a linear mixed-effects model that accounted for the experimental design by including precipitation regime, soil depth, activity (when relevant, based on communities present in heavy vs. light DNA fractions), and their interaction as fixed effects variables and mesocosm as the random effect variable. Bacterial UniFrac distances and fungal Bray–Curtis dissimilarities were used for principal coordinate analysis, and analyzed by non-parametric permutational multivariate analysis of variance [47]. The OTUs responding significantly to experimental treatment were detected using a linear mixed-effects model followed by a test to account for false discovery rates [48]. The OTUs that responded significantly were hierarchically clustered into groups, and the significance of the clustering verified against random clustering.
Results
Precipitation patterns and plant performance
After an initial drop in soil water content, the treatments were stabilized at relatively dry conditions (Fig. 1). The theoretical soil water retention curve for our soil, based on soil texture, bulk density, and horizon [49], shows that our soils dried down close to the theoretical wilting point, and never reached field capacity when watered (Fig. S2). Frequent water input significantly increased live above-ground biomass and decreased dead above-ground biomass, but left root biomass unchanged (Fig. S3). However, root biomass distribution was significantly affected: frequent water inputs significantly increased root biomass in the top soil layer, and decreased it in the middle and bottom soil layers (Fig. 2). Root length density mirrored root biomass pattern (data not shown). We found no significant effect of precipitation regime on stomatal conductance or photosynthetic rate scaled by plant leaf area to account for treatment effect on leaf development, integrated over 4 days at the end of the treatment period (see Fig. S4 for photosynthesis).
Fig. 1.
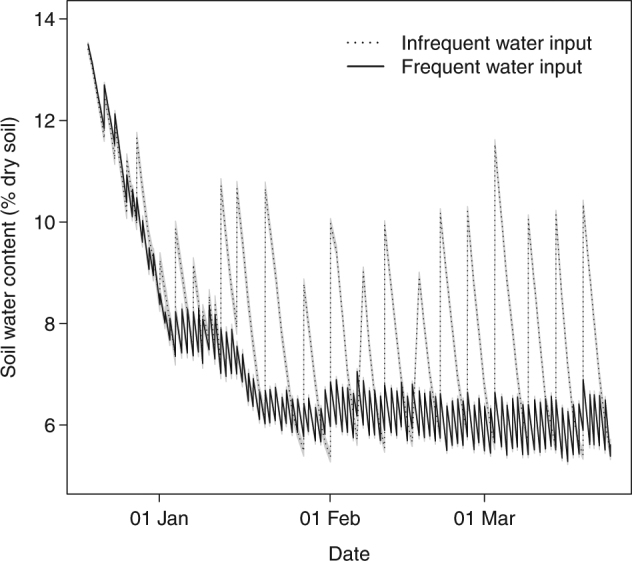
Dynamics of soil water content in the experimental treatments (infrequent and frequent water input, dotted and full curves, respectively) over the duration of the experiment. Lines and shaded polygons around them indicate mean ± standard error (n = 5)
Fig. 2.
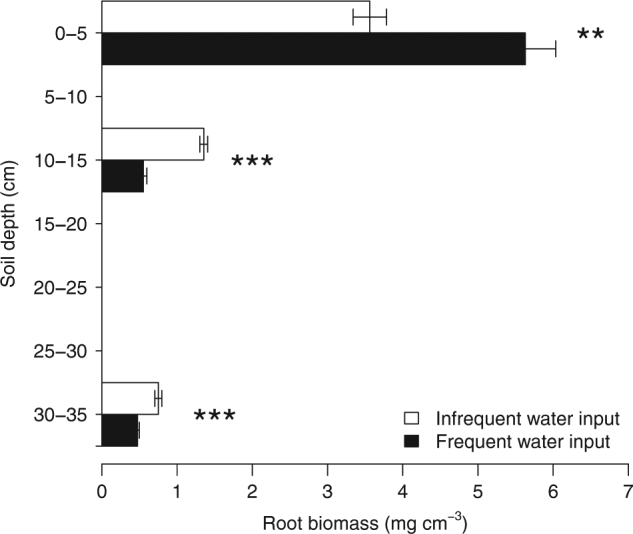
Root biomass after 12 weeks under contrasting precipitation pattern treatments (open bars: infrequent water input, closed bars: frequent water input), in three soil depth layers (top: 0–5 cm, middle: 10–15 cm, bottom: 30–35 cm). Bars indicate mean ± standard error (n = 5). Significance between treatments in each layer: * 0.05>p>0.01, ** 0.01>p>0.001, *** 0.001>p.
Plant-microbial coupling
Bulk root 13C signature was not significantly affected by treatment, soil layer or time (Fig. S5). Soil microbial biomass C significantly decreased with depth (p = 0.023; 222.1 ± 37.4, 140.4 ± 15.4, 131.6 ± 19.8 μg C g−1 dry soil in the top, middle, and bottom soil layers, respectively), but was left unchanged by precipitation pattern. Soil microbial biomass 13C signature was significantly higher under frequent water input in the top soil layer, compared with all other treatments and soil layers, and relatively stable over the 5-day measurement period (Fig. S6). The average 5-day microbial biomass 13C signature was significantly related to root biomass under frequent precipitation inputs (R2 = 0.80, p = 0.011), but not under infrequent inputs (Fig. S7).
Microbial community abundance, composition, and diversity
Both bacterial 16S rRNA gene and fungal ITS region abundances were significantly higher (p < 0.001) in the inactive (4.8 × 109 ± 4.6 × 108 and 6.1 × 108 ± 7.2 × 107 copies g−1 soil, respectively) than in the active (9.7 × 107 ± 2.8 × 107 and 5.4 × 106 ± 9.9 × 105 copies g−1 soil, respectively) communities, and was left unchanged by precipitation pattern or depth.
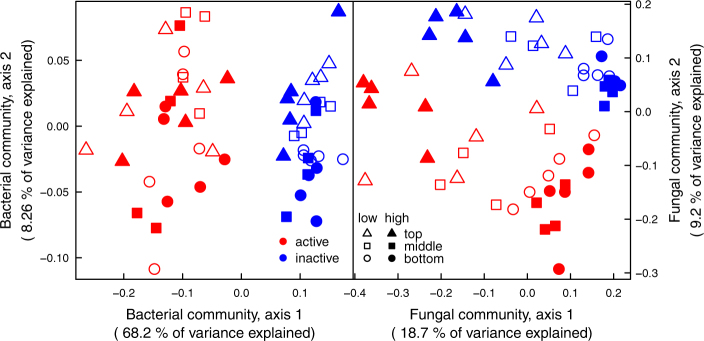
The 18O-SIP allowed us to discriminate the active from the inactive soil microbial community after rewetting, accounting for 61.1% and 9.3% of the variability in bacterial and fungal community composition, respectively (Fig. 3, Table S1). Differences among microbial communities with depth accounted for 4.0% and 14.3% of the overall bacterial and fungal data variability, respectively (Table S1). When considered separately, active and inactive bacterial and fungal communities differed with depth, which explained between 11% and 20% of the variance (Table S2). Precipitation pattern left soil bacterial community composition unchanged, accounted at most for 5% of the variability in overall, active, and inactive fungal community composition (Table S2), and affected overall and inactive fungal community composition differently at different depths (significant precipitation pattern treatment × depth interaction).
Fig. 3.
Principal coordinates analysis (PCoA) of the UniFrac pairwise dissimilarity of the relative abundance of bacterial sequences based on 16S rRNA gene a and of the Bray–Curtis distance of the relative abundance of fungal sequences based on ITS rRNA region b. The active (red symbols) and the inactive (blue symbols) communities were determined in the infrequent (open symbols) and frequent (closed symbols) water input treatments, in three soil depth layers (top: 0–5 cm, triangles; middle: 10–15 cm, squares, bottom: 30–35 cm, circles)
The active bacterial and fungal communities after rewetting were significantly less rich than the inactive (Fig. 4), but more even and less phylogenetically diverse for bacteria, whereas their evenness remained unchanged for fungi. Consistently, NRI was significantly higher in the active than in the inactive bacterial community, indicating stronger phylogenetic clustering (12.04 ± 0.38 vs. 8.43 ± 0.33, respectively). We found no significant effect of precipitation pattern on the α-diversity of microbial communities. We detected no significant effects of depth on the α-diversity of the active bacterial community after rewetting. In the inactive bacterial community, all indices decreased significantly with depth, except NRI, which increased (indicating increased phylogenetic clustering) with depth. Evenness-related indexes responded differently to precipitation pattern at different depths, driven by decreased evenness in the top soil layer under infrequent water inputs, which was not detected in the other soil layers (Fig. 4a, Fig. S8). In both the active and inactive fungal communities, evenness significantly increased with soil depth, driven by a large decrease in the top soil layer under frequent water inputs (Fig. 4b, Fig. S9).
Fig. 4.
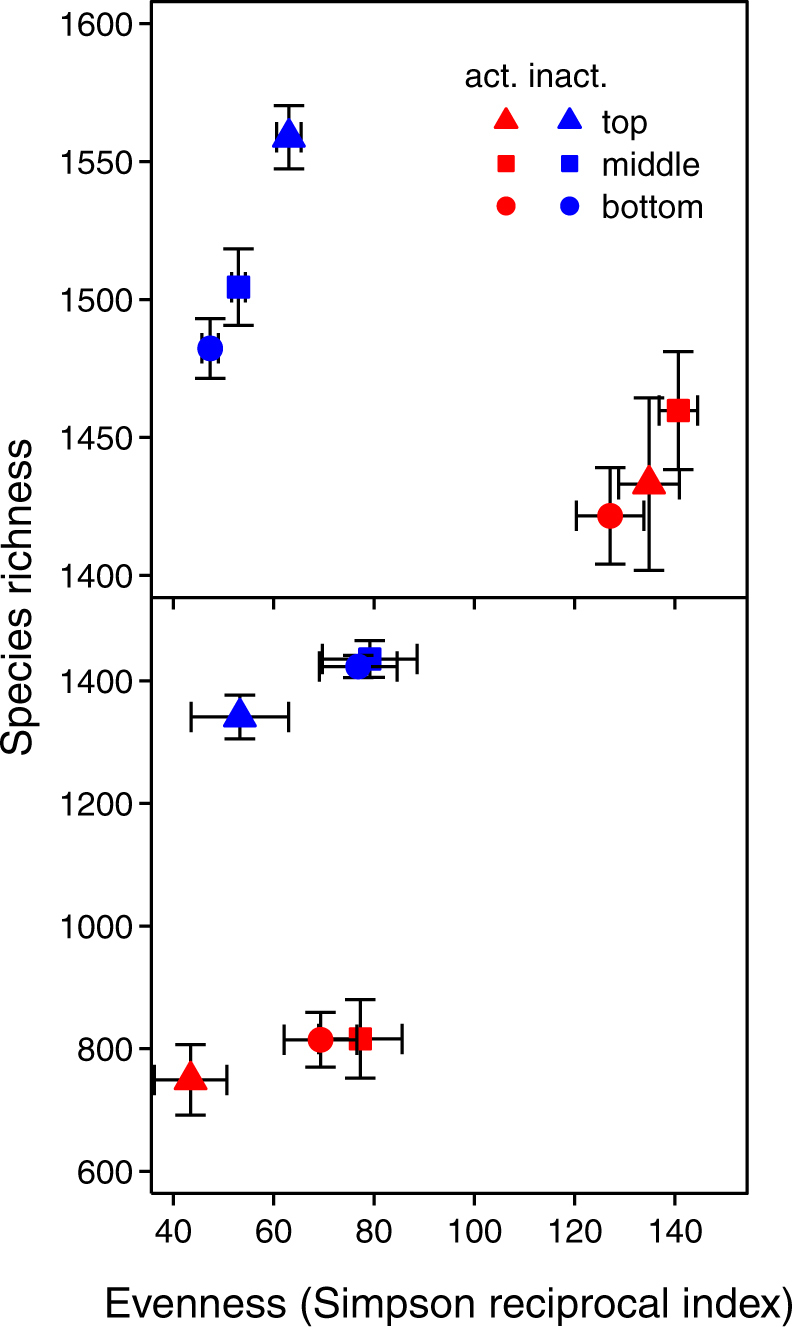
Bacterial a and fungal b observed species richness and evenness (determined by the Simpson reciprocal index) in the active (red symbols) and inactive (blue symbols) communities at three soil depths (top: 0–5 cm, triangles; middle: 10–15 cm, squares, bottom: 30–35 cm, circles). Bars indicate mean ± standard error
Significantly responsive groups
No microbial OTU responded significantly to precipitation pattern. In the active and inactive bacterial and fungal communities, OTUs responded significantly to soil depth, significantly clustering by soil depth into two groups (Fig. S10, S11, S12, S13) comprising the OTUs that were relatively more present in the top and middle soil layers (“top group”) or in the bottom and middle soil layers (“bottom group”). The active bacterial OTUs, which responded significantly to depth belonged predominantly to the Proteobacteria (mostly Alphaproteobacteria, driven by Sphingomonads in the top group and Betaproteobacteria and Actinobacteria phyla in the bottom group, Fig. 5a). The depth-significant inactive bacterial OTUs were dominated by Alphaproteobacteria and Actinobacteria in the top and bottom group, respectively (Fig. 5b). In the active fungal community, the significant OTUs belonged predominantly to the Ascomycota phyla (Fig. 6a): the top group was dominated by Chaetosphaeriales and Xylariales orders of Sordariomycetes, the bottom group by Hypocreales (Sordariomycetes) and Eurotiales (Eurotiomycetes). In the inactive fungal community, no taxonomic orders dominated clearly in the top group, which included mostly Helotiales and Chaetothyriales (Leothimycetes and Eurotiomycetes classes of Ascomycota, respectively), and Agaricomycetes (Agaricomycetes class of Basidiomycota), whereas the bottom group was dominated by Hypocreales and Eurotiales (Sordariomycetes class of Ascomycota, Fig. 6b).
Fig. 5.
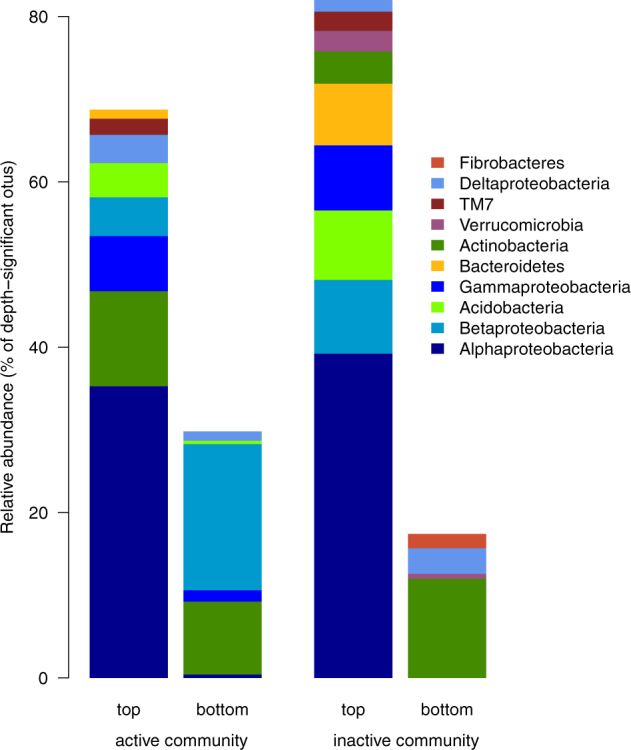
Relative abundance of the OTUs that responded significantly to soil depth in the active a and inactive b bacterial communities (phylum assignment provided, class provided for Proteobacteria). Based on their relative abundance, the OTUs clustered by top and bottom groups
Fig. 6.
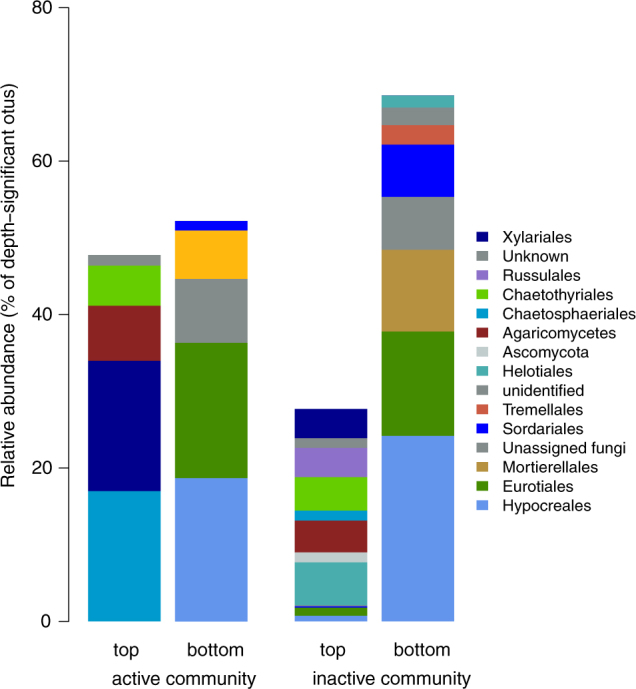
Relative abundance of the OTUs that responded significantly to soil depth in the active a and inactive b fungal community (order provided). Based on their relative abundance, the OTUs clustered by top and bottom groups
Discussion
The root biomass of live plants responded differentially to precipitation pattern, in addition to the expected decreasing root density gradient with depth (Fig. 2). Root distribution response is expected to determine C rhizodeposition, thus affecting microbial community function under dry–wet cycles [9]. Under frequent water inputs, the coupling between plants and soil microbes was most apparent by the top soil layer, where most roots were located. However, under infrequent water inputs, this coupling in the top soil layer broke down, with no additional plant C transferred to microbial biomass despite larger root biomass. These results are consistent with reports of reduced coupling between plants and soil microbes under drought [3, 50], which can even extend to ulterior drought events [7]. We found no effect of precipitation pattern on the amount of C photosynthesized, and no differences in 13C label in root biomass among treatments, soil depth, or over time, suggesting that the coupling between plants and soil microbes was affected through changes in rhizodeposition or microbial access to rhizodeposits [20, 50–52].
Precipitation pattern effects in our system were limited, both at the community level and the OTU level. Several studies have also found little response of the present bacterial community composition to altered precipitation in the field, even after a year or more of treatment [53–55]. However, altered precipitation has been shown to affect the functional response of soil microbes to subsequent rewetting events [54, 56], suggesting that some response related to microbial activity could be expected. Our results suggest that both soil depth and microbial activity may have to be considered: at the community level, we found a significant effect of precipitation patterns on the soil microbial community that was inactive upon rewetting and located in the 0–5 cm soil layer. These effects were opposite for bacteria and fungi: infrequent precipitation regime decreased bacterial evenness and relatedness but increased fungal evenness. Our results are consistent with reduced bacterial relatedness under drier conditions [57], which prevail in the uppermost soil layer under infrequent water inputs as it dries out first and remains dry for several days. Thus, in our system, precipitation pattern legacy effects upon rewetting had a larger influence on the microbial seedbank than on the active players. This points toward the importance of phylogenetic differences in persistence and mortality as drivers of community change under changing water regimes.
Within a plant-soil system, we were able to characterize the effects of precipitation pattern and soil depth on the active and inactive soil microbial communities. Bacteria responded to rewetting with a large offset in community structure between the active and the inactive community, whereas fungi showed a less contrasted response (Fig. 3). The soil bacterial community tracked soil moisture conditions more closely than the soil fungal community did, which is consistent with the generally higher resistance to dry periods of fungi compared with bacteria [14, 16, 58–60], as well as the more stable properties of fungal-based food webs [18, 61]. The active fungal community was much less abundant (one-tenth) and less rich than the inactive fungal community (Fig. 4), indicating that only a few fungal groups were poised for quick growth upon rewetting, in contrast with the rapid reactivation of many phylogenetically clustered bacterial groups that is consistent with earlier studies [12, 13]. Indeed, the active bacterial community was only moderately less abundant and rich, and more phylogenetically clustered than the inactive community. Furthermore, the phylogenetic clustering of active bacteria in response to wetting lends support to the idea that phylogeny may have ecological significance [62]. We hypothesize that the life strategy of the active bacteria was based on dynamic adjustment to transient water and nutrient availability whenever water film properties allowed it, whereas the inactive bacterial community relied more on maintaining functionality under drier conditions. In systems that are structured by water availability, such coexisting strategies likely drive the present bacterial communities (i.e., DNA-based), reflecting spatial patterns, whereas the potentially active (i.e., RNA-based communities) or actively growing communities track soil water availability [13, 63].
Soil depth was the main factor shaping the differences among soil bacterial and fungal communities and in our system after rewetting. The inactive bacterial community was more diverse and even in the top soil layer than in the deeper layers (Fig. 4), in line with the expected depth pattern of soil bacterial diversity [64]. In contrast, the inactive fungal community was less diverse and even in the top soil. Due to their filamentous life-form, fungi are less dependent than bacteria on water film continuity to access substrates. Increased fungal evenness and richness is often measured in the top soil [65, 66]. Nevertheless, the strong coupling between plants and soil microbes in the top soil under frequent water inputs may have favored the activity of fungi that are tightly involved with roots, resulting in increased dominance. The dominant depth-significant bacterial OTUs shifted with depth (Fig. 5), from Alphaproteobacteria in the top group to Betaproteobacteria and Acidobacteria in the active and inactive communities (respectively) in the bottom group, suggesting that a change in the identity of the dominant depth-responsive bacteria was involved in the depth effect measured at the community level. This change was clear in the active bacterial community, despite no apparent depth response of its α-diversity. Similarly, despite similar α-diversity patterns in the active and inactive fungal communities, the depth-responsive fungal OTUs in the inactive community were evenly distributed in the top group and dominated by Hypocreales (an order that includes many parasitic fungi) in the bottom group, whereas in the active community they shifted with depth (Fig. 6). These shifts in the identity of the dominant species, despite no change in α-diversity, suggests that different functional responses may have occurred at different depths.
Top soil is a dynamic nexus: where most root biomass is, where the legacy of precipitation pattern over time impacts the diversity of inactive fungal and bacterial communities, where plant-soil microbial coupling is tightest. Deeper soil layers show a different picture: different microbial diversity patterns, as well as different microbial groups that are actively growing or not upon rewetting. Our results indicate that the contrasting response between bacteria and fungi transcends soil depth. Changes in fungal:bacterial ratio have often been observed as a consequence of climate change, and are projected to have potential biogeochemical cycling, as well as for food web stability. Here, we extend this view and show that not only bacterial and fungal abundances respond differently to environmental drivers but that their relative activity is also affected. Thus, predicted changes in precipitation patterns may affect the activity patterns of microbial populations, based on their life strategy, which could result in large-scale effects on soil biogeochemical processes, including soil C budgets and nutrient cycling.
Electronic supplementary material
Acknowledgements
This research was supported by funding from the ANR (project INCITE, ANR-13-JSV7-0004), the European Commission (Career Integration Grant FP7-MC-CIG #618010) and the Conseil Régional de Bourgogne to RB. IE was supported in part by an INRA Ph.D. fellowship (Département Environnement et Agronomie). AG acknowledges support by an SNF grant (31003A_159866). We thank François Nuge and Xavier Buisson for kindly letting us use their soil, Michel Laderach (Dijon Céréales) for providing the seeds, Karine Palavioux, Céline Bernard, Franck Zenk, Damien Gironde, and Noureddine El Mjiyad for help in the greenhouse, Jérôme Fromentin for the ultracentrifuge, Samuel Jacquiod for sharing on functional response groups, Livio Antonielli for bioinformatics discussions, Virginie Bourion for root scans, Florian Bizouard for grinding, Marielle Adrian and Marie-Claire Héloir for the Li-6400, Arnaud Coffin and Marjorie Ubertosi for soil retention data. Matthieu Barret kindly provided the bacterial mock community, assembled within the MetaBAR project funded by the INRA MEM Metaprogramme.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
These authors contributed equally: Ilonka C. Engelhardt, Amy Welty.
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0079-z) contains supplementary material, which is available to authorized users.
References
- 1.IPCC. Climate change 2007: impacts, adaptation and vulnerability. Contribution of Working Group II to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, UK: Cambridge University Press; 2007. [Google Scholar]
- 2.Sowerby A, Emmett BA, Tietema A, Beier C. Contrasting effects of repeated summer drought on soil carbon efflux in hydric and mesic heathland soils. Glob Change Biol. 2008;14:2388–404. doi: 10.1111/j.1365-2486.2008.01643.x. [DOI] [Google Scholar]
- 3.Ruehr NK, Offerman C, Gessler A, Winkler BJ, Ferrio JP, Buchmann N, et al. Effects of drought on allocation of recent carbon: from beech leaves to soil respiration. New Phytol. 2009;184:950–61. doi: 10.1111/j.1469-8137.2009.03044.x. [DOI] [PubMed] [Google Scholar]
- 4.Bloor JMG, Bardgett RD. Stability of above-ground and below-ground processes to extreme drought in model grassland ecosystems: interactions with plant species diversity and soil nitrogen availability. Perspect Plant Ecol Evol Syst. 2012;14:193–204. doi: 10.1016/j.ppees.2011.12.001. [DOI] [Google Scholar]
- 5.Hartmann AA, Niklaus PA. Effects of simulated drought and nitrogen fertilizer on plant productivity and nitrous oxide (N2O) emissions of two pastures. Plant Soil. 2012;361:411–26. doi: 10.1007/s11104-012-1248-x. [DOI] [Google Scholar]
- 6.Bimüller C, Dannenmann M, Tejedor J, von Lützow M, Buegger F, Meier R, et al. Prolonged summer droughts retard soil N processing and stabilization in organo-mineral fractions. Soil Biol Biochem. 2014;68:241–51. doi: 10.1016/j.soilbio.2013.10.003. [DOI] [Google Scholar]
- 7.Fuchslueger L, Bahn M, Hasibeder R, Kienzl S, Fritz K, Schmitt M, et al. Drought history affects grassland plant and microbial carbon turnover during and after a subsequent drought event. J Ecol. 2016;104:1453–65. doi: 10.1111/1365-2745.12593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hoover DL, Knapp AK, Smith MD. The immediate and prolonged effects of climate extremes on soil respiration in a mesic grassland. J Geophys Res Biogeosci. 2016;121:1034–44. doi: 10.1002/2015JG003256. [DOI] [Google Scholar]
- 9.Canarini A, Dijkstra FA. Dry-rewetting cycles regulate wheat carbon rhizodeposition, stabilization and nitrogen cycling. Soil Biol Biochem. 2015;81:195–203. doi: 10.1016/j.soilbio.2014.11.014. [DOI] [Google Scholar]
- 10.Chou WW, Silver WL, Jackson RD, Thompson AW, Allen-Diaz B. The sensitivity of annual grassland carbon cycling to the quantity and timing of rainfall. Glob Change Biol. 2008;14:1382–94. doi: 10.1111/j.1365-2486.2008.01572.x. [DOI] [Google Scholar]
- 11.Nielsen UN, Ball BA. Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob Chang Biol. 2015;21:1407–21. doi: 10.1111/gcb.12789. [DOI] [PubMed] [Google Scholar]
- 12.Placella SA, Brodie EL, Firestone MK. Rainfall-induced carbon dioxide pulses result from sequential resuscitation of phylogenetically clustered microbial groups. Proc Natl Acad Sci USA. 2012;109:10931–6. doi: 10.1073/pnas.1204306109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barnard RL, Osborne CA, Firestone MK. Responses of soil bacterial and fungal communities to extreme desiccation and rewetting. ISME J. 2013;7:2229–41. doi: 10.1038/ismej.2013.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries FT, Shade A. Controls on soil microbial community stability under climate change. Front Microbiol. 2013;4:265. doi: 10.3389/fmicb.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Landesman WJ, Dighton J. Shifts in microbial biomass and the bacteria: fungi ratio occur under field conditions within 3 h after rainfall. Microb Ecol. 2011;62:228–36. doi: 10.1007/s00248-011-9811-1. [DOI] [PubMed] [Google Scholar]
- 16.Barnard RL, Osborne CA, Firestone MK. Changing precipitation pattern alters soil microbial community response to wet-up under a Mediterranean-type climate. ISME J. 2015;9:947–57. doi: 10.1038/ismej.2014.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Boer W, Folman LB, Summerbell RC, Boddy L. Living in a fungal world: impact of fungi on soil bacterial niche development. FEMS Microbiol Rev. 2005;29:795–811. doi: 10.1016/j.femsre.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Gordon H, Haygarth PM, Bardgett RD. Drying and rewetting effects on soil microbial community composition and nutrient leaching. Soil Biol Biochem. 2008;40:302–11. doi: 10.1016/j.soilbio.2007.08.008. [DOI] [Google Scholar]
- 19.Ebrahimi A, Or D. Hydration and diffusion processes shape microbial community organization and function in model soil aggregates. Water Resour Res. 2015;51:9804–27. doi: 10.1002/2015WR017565. [DOI] [Google Scholar]
- 20.Tecon R, Or D. Bacterial flagellar motility on hydrated rough surfaces controlled by aqueous film thickness and connectedness. Sci Rep. 2016;6:19409. doi: 10.1038/srep19409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dechesne A, Wang G, Gülez G, Or D, Smets BF. Hydration-controlled bacterial motility and dispersal on surfaces. Proc Natl Acad Sci USA. 2010;107:14369–72. doi: 10.1073/pnas.1008392107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang G, Or D. Hydration dynamics promote bacterial coexistence on rough surfaces. ISME J. 2013;7:395–404. doi: 10.1038/ismej.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Volkmann TH, Haberer K, Gessler A, Weiler M. High-resolution isotope measurements resolve rapid ecohydrological dynamics at the soil-plant interface. New Phytol. 2016;210:839–49. doi: 10.1111/nph.13868. [DOI] [PubMed] [Google Scholar]
- 24.Hasibeder R, Fuchslueger L, Richter A, Bahn M. Summer drought alters carbon allocation to roots and root respiration in mountain grassland. New Phytol. 2015;205:1117–27. doi: 10.1111/nph.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.von Rein I, Gessler A, Premke K, Keitel C, Ulrich A, Kayler ZE. Forest understory plant and soil microbial response to an experimentally induced drought and heat-pulse event: the importance of maintaining the continuum. Glob Chang Biol. 2016;22:2861–74. doi: 10.1111/gcb.13270. [DOI] [PubMed] [Google Scholar]
- 26.Vance ED, Brookes PC, Jenkinson DS. An extraction method for measuring soil microbial biomass C. Soil Biol Biochem. 1987;19:703–7. doi: 10.1016/0038-0717(87)90052-6. [DOI] [Google Scholar]
- 27.Lang SQ, Bernasconi SM, Früh-Green GL. Stable isotope analysis of organic carbon in small (μg C) samples and dissolved organic matter using a GasBench preparation device. Rapid Commun Mass Spectrom. 2012;6:9–16. doi: 10.1002/rcm.5287. [DOI] [PubMed] [Google Scholar]
- 28.Blazewicz SJ, Schwartz E. Dynamics of 18O incorporation from H218O into soil microbial DNA. Microb Ecol. 2011;61:911–6. doi: 10.1007/s00248-011-9826-7. [DOI] [PubMed] [Google Scholar]
- 29.Muyzer G, de Waal EC, Uitierlinden AG. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl Environ Microbiol. 1993;59:695–700. doi: 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White TJ, Bruns T, Lee S, Taylor JW. PCR protocols: a guide to methods and applications. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. New York: Academic Press Inc.; 1990. pp. 315–22. [Google Scholar]
- 31.Berry D, Ben Mahfoudh K, Wagner M, Loy A. Barcoded primers used in multiplex amplicon pyrosequencing bias amplification. Appl Environ Microbiol. 2011;77:7846–9. doi: 10.1128/AEM.05220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Takahashi S, Tomita J, Nishioka K, Hisada T, Nishijima M. Development of a prokaryotic universal primer for simultaneous analysis of Bacteria and Archaea using next-generation sequencing. PLoS ONE. 2014;9:e105592. doi: 10.1371/journal.pone.0105592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang J, Kobert K, Flouri T, Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–20. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rognes T, Flouri T, Nichols B, Quince C, Mahe F. VSEARCH: a versatile open source tool for metagenomics. PeerJ. 2016;4:e2584. doi: 10.7717/peerj.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics. 2010;26:266–7. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum-evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26:1641–50. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–1. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 39.McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–8. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Webb M, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403-10. [DOI] [PubMed]
- 41.Abarenkov K, Nilsson RH, Larsson KH, Alexander IJ, Eberhardt U, Erland S, et al. The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytologist. 2010;186:281–5. [DOI] [PubMed]
- 42.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Webb CO. Exploring the phylogenetic structure of ecological communities: an example for rain forest trees. Am Nat. 2000;156:145–55. doi: 10.1086/303378. [DOI] [PubMed] [Google Scholar]
- 44.Webb CO, Ackerly DD, M MA, Donoghue MJ. Phylogenies and community ecology. Annu Rev Ecol Syst. 2002;33:475–505. doi: 10.1146/annurev.ecolsys.33.010802.150448. [DOI] [Google Scholar]
- 45.Kembel SW, Cowan PD, Helmus MR, Cornwell WK, Morlon H, Ackerly DD, et al. Picante: R tools for integrating phylogenies and ecology. Bioinformatics. 2010;26:1463–4. doi: 10.1093/bioinformatics/btq166. [DOI] [PubMed] [Google Scholar]
- 46.R Core Team (2014). R: a language and environment for statistical computing. Vol. ISBN 3-900051-07-0: Vienna, Austria.
- 47.Anderson MJ. A new method for non-parametric multivariate analysis of variance. Austral Ecol. 2001;26:32–46. [Google Scholar]
- 48.Strimmer K. fdrtool: a versatile R package for estimating local and tail area-based false discovery rates. Bioinformatics. 2008;24:1461–2. doi: 10.1093/bioinformatics/btn209. [DOI] [PubMed] [Google Scholar]
- 49.Al Majou H, Bruand A, Duval O, Le Bas C, Vautier A. Prediction of soil water retention properties after stratification by combining texture, bulk density and the type of horizon. Soil Use Manag. 2008;24:383–91. doi: 10.1111/j.1475-2743.2008.00180.x. [DOI] [Google Scholar]
- 50.Fuchslueger L, Bahn M, Fritz K, Hasibeder R, Richter A. Experimental drought reduces the transfer of recently fixed plant carbon to soil microbes and alters the bacterial community composition in a mountain meadow. New Phytol. 2014;201:916–27. doi: 10.1111/nph.12569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moyano FE, Manzoni S, Chenu C. Responses of soil heterotrophic respiration to moisture availability: an exploration of processes and models. Soil Biol Biochem. 2013;59:72–85. doi: 10.1016/j.soilbio.2013.01.002. [DOI] [Google Scholar]
- 52.von Rein I, Kayler ZE, Premke K, Gessler A. Desiccation of sediments affects assimilate transport within aquatic plants and carbon transfer to microorganisms. Plant Biol (Stuttg) 2016;18:947–61. doi: 10.1111/plb.12486. [DOI] [PubMed] [Google Scholar]
- 53.Cruz-Martinez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF. Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 2009;3:738–44. doi: 10.1038/ismej.2009.16. [DOI] [PubMed] [Google Scholar]
- 54.Evans SE, Wallenstein MD. Soil microbial community response to drying and rewetting stress: does historical precipitation regime matter? Biogeochemistry. 2012;109:101–16. doi: 10.1007/s10533-011-9638-3. [DOI] [Google Scholar]
- 55.Slaughter LC, Weintraub MN, McCulley RL. Seasonal effects stronger than three-year climate manipulation on grassland soil microbial community. Soil Sci Soc Am J. 2015;79:1352. doi: 10.2136/sssaj2014.10.0431. [DOI] [Google Scholar]
- 56.Zeglin LH, Bottomley PJ, Jumpponen A, Rice CW, Arango M, Lindsley A, et al. Altered precipitation regime affects the function and composition of soil microbial communities on multiple time scales. Ecology. 2013;94:2334–45. doi: 10.1890/12-2018.1. [DOI] [PubMed] [Google Scholar]
- 57.Taketani RG, Lanconi MD, Kavamura VN, Durrer A, Andreote FD, Melo IS. Dry season constrains bacterial phylogenetic diversity in a semi-arid rhizosphere system. Microb Ecol. 2017;73:153–61. doi: 10.1007/s00248-016-0835-4. [DOI] [PubMed] [Google Scholar]
- 58.Jumpponen A, Jones KL. Tallgrass prairie soil fungal communities are resilient to climate change. Fungal Ecol. 2014;10:44–57. doi: 10.1016/j.funeco.2013.11.003. [DOI] [Google Scholar]
- 59.Bapiri A, Bååth E, Rousk J. Drying–rewetting cycles affect fungal and bacterial growth differently in an arable soil. Microb Ecol. 2010;60:419–28. doi: 10.1007/s00248-010-9723-5. [DOI] [PubMed] [Google Scholar]
- 60.Yuste JC, Penuelas J, Estiarte M, Garcia-Mas J, Mattana S, Ogaya R, et al. Drought-resistant fungi control soil organic matter decomposition and its response to temperature. Glob Change Biol. 2011;17:1475–86. doi: 10.1111/j.1365-2486.2010.02300.x. [DOI] [Google Scholar]
- 61.de Vries FT, Liiri ME, Bjornlund L, Bowker MA, Christensen S, Setala HM, et al. Land use alters the resistance and resilience of soil food webs to drought. Nat Clim Change. 2012;2:276–80. doi: 10.1038/nclimate1368. [DOI] [Google Scholar]
- 62.Morrissey EM, Mau RL, Schwartz E, Caporaso JG, Dijkstra P, van Gestel N, et al. Phylogenetic organization of bacterial activity. ISME J. 2016;10:2336–40. [DOI] [PMC free article] [PubMed]
- 63.Angel R, Pasternak Z, Soares MI, Conrad R, Gillor O. Active and total prokaryotic communities in dryland soils. FEMS Microbiol Ecol. 2013;86:130–8. doi: 10.1111/1574-6941.12155. [DOI] [PubMed] [Google Scholar]
- 64.Eilers KG, Debenport S, Anderson S, Fierer N. Digging deeper to find unique microbial communities: the strong effect of depth on the structure of bacterial and archaeal communities in soil. Soil Biol Biochem. 2012;50:58–65. doi: 10.1016/j.soilbio.2012.03.011. [DOI] [Google Scholar]
- 65.Jumpponen A, Jones KL, Blair J. Vertical distribution of fungal communities in tallgrass prairie soil. Mycologia. 2010;102:1027–41. doi: 10.3852/09-316. [DOI] [PubMed] [Google Scholar]
- 66.Vargas-Gastelum L, Romero-Olivares AL, Escalante AE, Rocha-Olivares A, Brizuela C, Riquelme M. Impact of seasonal changes on fungal diversity of a semi-arid ecosystem revealed by 454 pyrosequencing. FEMS Microbiol Ecol. 2015;91:fiv044. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.