Abstract
Bacteriophages are central members and potential modulators of the gut microbiome; however, the ecological and evolutionary relationships of gut bacteria and phages are poorly understood. Here we investigated the abundance and diversity of lysogenic bacteria (lysogens) in the bacterial community of C57BL/6J mice by detecting integrated prophages in genomes reconstructed from the metagenome of commensal bacteria. For the activities of lysogens and prophages, we compared the prophage genomes with the metagenome of free phages. The majority of commensal bacteria in different taxa were identified as lysogens. More lysogens were found among Firmicutes and Proteobacteria, than among Bacteroidetes and Actinobacteria. The prophage genomes shared high sequence similarity with the metagenome of free phages, indicating that most lysogens appeared to be active, and that prophages are spontaneously induced as active phages; dietary interventions changed the composition of the induced prophages. By contrast, CRISPR-Cas systems were present in few commensal bacteria, and were rarely active against gut phages. The structure of the bacteria-phage infection networks was “nested-modular”, with modularity emerging across taxonomic scales, indicating that temperate phage features have developed over a long phylogenetic timescale. We concluded that phage generalists contribute to the prevalence of lysogeny in the gut ecosystem.
Introduction
The gut microbiota is recognized as a hidden modifier of host physiology and metabolism [1]. Although the microbiome-mediated host phenotypes are attributed to the interplay between different types of microorganisms [2, 3], most microbiome studies have focused on bacteria. Viruses are central members of the gut microbiota; most of them are bacterial viruses (bacteriophages or phages) [1]. The bacteriophage community fraction constitutes up to 17% of the human fecal metagenome [4], and bacteriophages are as numerous as commensal bacteria [5]. Metagenomic studies revealed that temperate bacteriophages are dominant and adaptive members of the human and murine gut viromes [6, 7], and alterations in their composition are associated with microbiome-associated diseases [8–11], giving rise to potential implications of gut bacteriophages in the host health and disease. Thus, it is expected that the temperate behavior of bacteriophages significantly affects the composition and function of the gut microbiome; to verify this, we first need to assess the extent of lysogeny, and determine how bacteria and phages interact in the gut ecosystem.
Bacteria–phage interactions are central to the bacterial physiology and metabolism, promoting genetic diversity and evolution of bacterial communities [12]. Bacteriophages destroy the host cells by lysis, transfer genes between hosts, and modify host phenotypes through lysogenic conversion. Temperate phages have been much less studied than lytic phages because of the different outcomes of the infection modes [13]. E.g., the outcome of lytic infections is virion progeny and lysed host cells, while the lysogenic infections leads to no apparent cellular changes in microbial communities. Lysogeny is estimated by counting the virions and lysogenic cells after prophage induction by mitomycin C treatment [14, 15]; however, this method is limited to cultivable bacteria, and has low accuracy because of an unsuccessful induction of prophages [16, 17]. Hence, the identification and quantification of lysogenic bacteria (lysogens) in environmental communities remains challenging [13]. The recovery of genomes from metagenomic data is an emerging technique that produces dozens to hundreds of draft-quality genomes from heterogeneous metagenomic sequences [18], and that enables not only the linking of functional and taxonomic data for inherent microbes [19, 20], but allowing the assembly of in situ genomes of uncultured microbial lineages [21, 22]. Since temperate phages are part of the host genome, the in situ recovery of the individual genomes of resident bacteria from metagenomic data provides an opportunity for studying lysogeny in natural communities, including the gut microbiome.
In the current study, we examined the abundance and diversity of commensal lysogens and their prophages in the gut bacterial and phage communities. We classified lysogens based on prophage detection in bacterial genomes reconstructed from the fecal bacterial metagenome, and evaluated the predominance of commensal lysogens in situ in a bacterial community. We used specific pathogen-free C57BL/6 mice maintained on sterilized water and animal diets to minimize exogenous contamination of resident microbial communities. We obtained two genomic sets of the gut phage community: a set of latent phages, integrated in host bacterial genomes (integrated prophages); and a set of active phages, existing freely outside the host bacteria (free phages). We established lysogen fractions of the bacterial metagenome, and identified active lysogens and prophages by performing sequence comparisons of the free and integrated phage datasets. We exploited the phage-prophage connections to predict bacteria-phage infection patterns in the predominantly lysogenized gut ecosystem. We believe that the current study elucidates the ecology and evolution of bacteria and phages in the gut ecosystem in relation to host health, and presents an alternative tool for the study of environmental lysogeny.
Materials and Methods
Animal experiment
Animal experiments were approved by the Institutional Animal Care and Use Committee of Kyung Hee University (KHPASP(SE)-16-133). Specific-pathogen free C57BL/6J male mice at 4-week of age (n = 6, Japan SLC, Japan) were maintained in ventilated plastic isolators (25 ± 1 °C temperature, 48 ± 6% relative humidity and a 14-h light/10-h dark cycle), and received sterilized food and water (pH 7.75) ad libitum. Since diet is a strong driving force of the gut bacterial and phage communities [23, 24], C57BL/6 mice were subjected to consecutive dietary shifts (Supplementary Figure S1a). The mice were fed a low-fat diet (LFD, TD06416, Harlan Laboratories, USA) first for 3 weeks, after which they were separated into two groups (n = 3 per group); one group (FORTH) received a high-fat, high-sucrose diet (HFHS, TD06415, Harlan Laboratories) for 3 weeks and the other group (BACK) received a low-fat, high-plant-polysaccharide diet (LFPP, 2918C, Harlan Laboratories) for 3 weeks. Diet was switched again to either the LFPP or HFHS diet for another 3 weeks. Last, diet was returned to the LFD diet for 3 weeks. Approximately 0.5 g feces per individual were collected in the third week of each phase, and were stored immediately at –80 °C.
DNA extraction and whole-genome shotgun sequencing
The fecal sample was suspended in a 0.02-μm filtered saline magnesium buffer [25]. The suspension was divided into pellet and supernatant by centrifugation at 2500 × g for 5 min at 4 °C. This step was repeated four times, and then the sample was centrifuged again at 5000 × g for 10 min at 4 °C to obtain complete separation (Supplementary Figure S1b). The pellet was used for DNA extraction of bacterial metagenome as described by Zoetendal et al. [26]. The supernatant was used for virus-like particle purification (modification: 0.45-μm filtration) and DNA extraction of viral metagenome, as described previously [27]. Bacterial contamination of viral DNAs was evaluated by quantitative PCR with bacterial 16S rRNA gene-specific primers using iQ SYBR Green Supermix (Bio-Rad, USA). The viral DNAs were amplified using the Illustra Genomiphi V2 DNA Amplification Kit (GE Healthcare, USA), according to the manufacturer’s instructions. The bacterial and viral DNAs were sequenced on an Illumina HiSeq4000 sequencer (2 × 101 bp), according to manufacturer’s instructions (Illumina, USA). The accession number of European Nucleotide Archive for the bacterial and phage metagenomes is PRJEB22007.
De novo assembly, binning and refinement of bacterial metagenome
Raw reads were quality-filtered, and high-quality reads with a mean (±SD) of 4.0 ± 0.5 Gb per bacterial metagenome (n = 24) were retained (Supplementary Table S1 and Figure S1b). The paired-end reads were co-assembled using IDBA-UD [28], and were binned to 482 draft-quality genomes (bacterial bins) based on tetranucleotide frequency and read abundance using MetaBAT [29]. The bacterial bins were refined using “merge”, “outliers” and “modify” commands of CheckM [30], resulting in 456 bins (Supplementary Table S1 and Figure S2a). The refined bins were re-assembled to scaffolds with the reads extracted from a single sample whose reads were mapped the most to the refined bins. Putative completeness and contamination of bacterial bins were estimated using the lineage-specific single-copy-gene workflow of CheckM. The bacterial bins that were <30% completed and >10% contaminated were filtered out; consequently, 181 bacterial bins were obtained (Supplementary Figure S2a and Table S2). The detailed procedures for de novo assembly, binning, refinement and taxonomic identification are described in Supplementary Materials and Methods.
Detection of putative prophages and metagenome assembly of free phages
Prophage sequences were predicted in the scaffolds (>1 kb) of bacterial bins using VirSorter [31]. To minimize a risk of false-positives, we only retained viral predictions presenting at least one viral hallmark gene, or enrichment in viral-like genes or non-Caudovirales genes (category 1 and 2 in entirely viral; category 4 and 5 in prophages). Raw reads of the phage metagenome (n = 24) were quality-filtered to get high-quality, paired-end reads as described in the bacterial metagenome. Using IDBA-UD, high-quality reads with a mean (±SD) of 0.7 ± 0.2 Gb were assembled separately into 443 ± 517 contigs (>1 kb) encoding 2625 ± 2349 genes per sample (Supplementary Table S1).
Protein and viral clusters
We collected the following two references: 4485 RefSeq DNA viral genomes (RefSeq_viral); and 12,498 prophage genomes from 5492 RefSeq microbial genomes (RefSeq_proph) [32]. The proteins of the phage metagenome, the prophage genomes and two references were clustered at a minimum of 60% sequence identity and 80% alignment [33]. Viral clusters were created using a shared gene content-based network analysis [34], which generates approximate genus-level viral populations. This analysis was started using 1361 and 287 non-redundant, long contigs (≥3 kb) of the phage metagenome and the prophage genomes, respectively. Protein sequence similarities were calculated based on reciprocal BLAST hits, and protein clusters were defined using the Markov Clustering algorithm. The protein clusters were used for gene content-based viral clusters using vConTACT [34]. The detailed procedures for protein and viral clusters are described in Supplementary Materials and Methods.
CRISPR-Cas array identification
The CRISPR arrays of bacterial bins were predicted using PILER-CR [35]. Spacers were compared with the contigs of the phage metagenome using CRISPRTarget parameters [36]. Complete or nearly-complete matches (at most two mismatches) over the whole length of the spacers were selected for accurate matches to protospacers [37].
Viral tRNA identification
The sequences of tRNA genes were identified in the contigs of the phage metagenome using ARAGON [38]. The tRNA sequences were compared against the scaffolds of bacterial bins, and perfect matches were considered significant using BLASTn analysis.
Bacteria-phage infection network analysis
The nestedness temperature calculator (NTC) [39] was used for nestedness using FALCON [40], and the Bipartite, Recursively Induced Modules (BRIM) method [41] was used for modularity using “lpbrim” in R package (https://github.com/biometry/bipartite). The detailed procedure for the network analysis is described in Supplementary Materials and Methods.
Statistical analysis
A P-value of 0.05 was considered significant. Using Prism 5 for Windows (GraphPad Software, USA), differences in the abundance of bacterial bins were determined using unpaired two-tailed Student’s t-tests. Correlation analyses of the genome completeness and size of bacterial bins with lysogen percentage and abundance were calculated using Spearman’s rank correlation. Using R package “vegan”, the principal coordinate analysis (PCoA) of active prophages was performed with Jaccard dissimilarity, and the PCoA of lysogens and non-lysogens was performed with Bray-Curtis dissimilarity. Statistical significance of beta-diversity was assessed using function “adonis” with 999 permutations. Comparisons in Jaccard dissimilarities were defined using One-way ANOVA and Tukey’s post-hoc test.
Results
The recovery of bacterial genomes from metagenomic data
Using metagenome assembly and binning processes, a bacterial community set was obtained, comprising 181 bacterial bins (>30% genome completeness and <10% contamination) of commensal bacteria (Fig. 1a). A mean (±SD) of 37.9 ± 7.9% reads per sample were mapped to 181 bacterial bins. According to the phylogenetic analysis of the conserved marker genes [30], the bacterial bins were assigned to the phyla Firmicutes (133 bins), Bacteroidetes (36 bins), Actinobacteria (4 bins), Proteobacteria (4 bins), Tenericutes (1 bin), Deferribacteres (1 bin), Verrucomicrobia (1 bin) and unknown (1 bin) (Fig. 1a). The bacterial bins of the Firmicutes and Bacteroidetes phyla were dominant, followed by Actinobacteria, Deferribacteres, Tenericutes, Proteobacteria and Verrucomicrobia (Supplementary Figure S2b). Since many gut bacteria are currently not cultivable [42], 91 bacterial bins were taxonomically distant from known cultured genera, with the bacterial bins distributed over 34 known genera (Supplementary Figure S2c). Of these, 14 and 12 genera represented the top 20 core genera of the mouse and human gut microbiomes, respectively [42]. Nine bins were closely related to three species of the altered Schaedler flora [43], and 10 bins were closely related to Lachnospiraceae bacteria isolated from murine cecal contents [44]. Altogether, a community of 181 bacterial bins representing the dominant commensal gut bacteria was captured by genome reconstruction from metagenomic data.
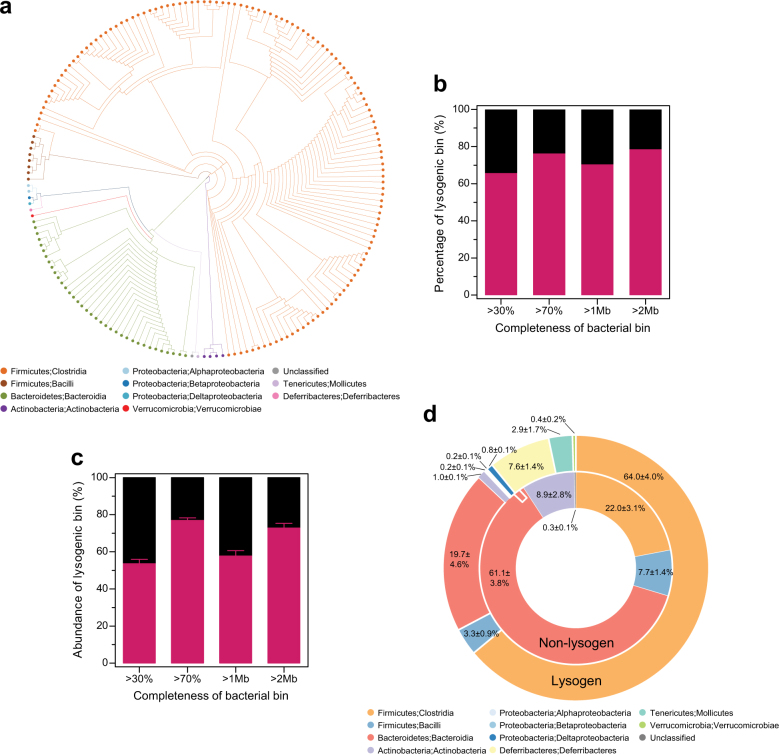
Fig. 1.
The distribution and abundance of commensal lysogens in the gut bacterial community. a The phylogenetic tree of 181 bacterial bins that were generated based on 43 conserved marker genes, and designated at the class taxon level by different colors. b The percentage and c abundance of lysogenic bins (n = 24) per genome completeness (>30 and >70%) and size (>1 and >2 Mb). d Bacterial taxonomic profiles of lysogenic and non-lysogenic bins (n = 24) shown at the class-level. All data are presented as the mean ± SEM
The predominance of commensal lysogens
Since nucleotide composition of the viral genome is polarized toward the host genome during co-evolution [45], we anticipated that prophage elements would be included in 181 bacterial bins. The prophage sequences were computationally predicted based on the presence of viral “hallmark” genes [31]. In total, 336 genomic fragments (12,914 ± 12,460 bp, mean ± SD) of putative prophages were predicted for 119 bacterial bins (65.8%); these were regarded as lysogenic bins (Fig. 1b). The abundance of lysogenic bins (53.6 ± 2.4%) was higher than that of non-lysogenic bins (46.4 ± 2.4%) (Fig. 1c). This number increased to 76.3% in bacterial bins with >70% completeness, and 78.6% in bacterial bins with >2 Mb genome size in the percentage of lysogenic bins (Fig. 1b), and it increased to a mean of 76.9 ± 1.4% in bacterial bins with >70% completeness and 72.9± 2.5% in bacterial bins with >2 Mb genome size in the abundance of lysogenic bins (Fig. 1c). The observed proportion of lysogens is much larger than that in the publicly available bacterial genomes, where half of the genomes (46–54%) are lysogens [46], suggesting that prophages are widely distributed in commensal bacteria. The percentage and abundance of lysogenic bins were positively correlated with genome completeness and size (P < 0.01) (Supplementary Figure S3), indicating that the dataset provided a minimal estimation of the actual number of lysogens in commensal bacteria.
We examined the fitness consequences of commensal lysogens by comparing the mean abundance of individual bins in lysogens and non-lysogens because the carriage of temperate phages can be costly for their host [47]. The mean abundance of total bins was significantly lower in lysogenic than in non-lysogenic bins, but the difference disappeared or was reversed for bacterial bins with >70% completeness and with >2 Mb genome size, although lysogen proportion was increased (Supplementary Figure S4a). This was verified in Firmicutes and Bacteroidetes where a sufficient number of bacterial bins were available, but no differences were observed between the lysogenic and non-lysogenic bins (Supplementary Figure S4b), showing that lysogeny might result in no detectable fitness cost for the bacterial host in the gut.
Taxonomic preference and dietary response of the commensal lysogens
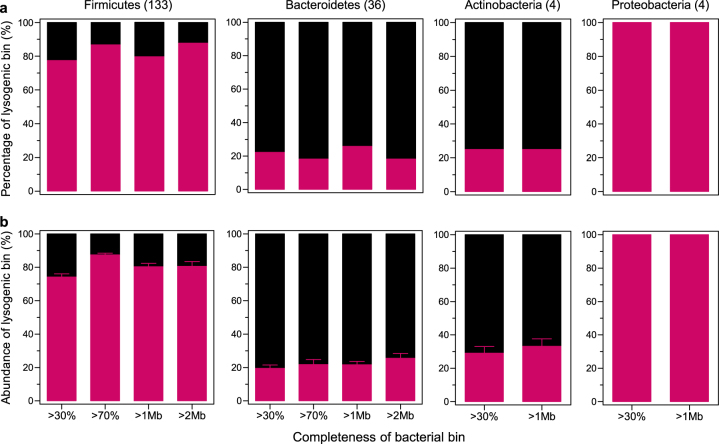
While the lysogenic bins were widely distributed over all the observed bacterial taxa, the non-lysogenic bins were limited to a few bacterial taxa (Fig. 1d). To determine the bacterial taxa that were preferentially lysogenized, we examined the distribution of lysogenic bins in different bacterial phyla. The lysogenic bins were distributed in all bacterial phyla, but were fewer in Bacteroidetes (18.2–25.8%) and Actinobacteria (25.0%) than in Firmicutes (77.4–87.7%) and Proteobacteria (100%) (Fig. 2a). The same patterns were observed for the abundance of the lysogenic bins; the lysogenic bins were more abundant in Firmicutes (74.1 ± 1.9–87.4 ± 1.0%) and Proteobacteria (100%) than in Bacteroidetes (19.4 ± 2.0–25.5 ± 2.9%) and Actinobacteria (29.1 ± 4.0–33.1 ± 4.5%) (Fig. 2b). At genus-level, the lysogenic bins were found in all of the detected bacterial genera, except for Alistipes, Bifidobacterium, and Lachnospiraceae bacterium M18-1 (Supplementary Figure S5). Since the activities of temperate phages in the gut-associated Bifidobacterium have been extensively studied [48], it needs to be further assessed with a sufficient number of bacterial bins. However, lysogenic bins were less frequent in all Bacteroidetes genera, including Alistipes (Supplementary Table S2). Altogether, these results suggest that lysogeny may be favorable in particular bacterial taxa, which occurs within genus- or at a lower level.
Fig. 2.
The taxonomic distribution of commensal lysogens. a The percentage and b abundance of lysogenic bins (n = 24) shown by phylum-level bacterial taxa according to genome completeness (>30 and >70%) and size (>1 and >2 Mb). The number of bacterial bins per bacterial phylum is indicated in parentheses. All data are presented as the mean ± SEM
We next determined dietary variations in the abundance of the lysogenic and non-lysogenic bins. According to the results of Bray-Curtis dissimilarity-based PCoA, abundance of lysogenic and non-lysogenic bins was affected by diet type (P = 0.001). Changes in abundance were associated more with the LFPP diet than with the LFD or the HFHS diets (Supplementary Figure S6a and b). Similar to a previous study [23], the HFHS diet increased the abundance of Firmicutes (Clostridia and Bacilli), while the LFPP diet increased that of Bacteroidetes (Supplementary Figure S6c, d and e). Interestingly, the lysogenic bins were more abundant in mice fed the HFHS diet, whereas non-lysogenic bins were more abundant in mice fed the LFPP diet (Supplementary Figure S6a and b). The type of diet made a larger contribution to variations in lysogenic bins of Clostridia than to changes in non-lysogenic bins (Supplementary Figure S6c), whereas the type of diet made a similar contribution to variations in both lysogenic and non-lysogenic bins in Bacilli and Bacteroidetes (Supplementary Figure S6d and e). These data suggested that gut lysogeny may occur differently in response to different diets in the commensal bacteria of diverse bacterial taxa.
The prophages induced from commensal lysogens form a subset of the phage community
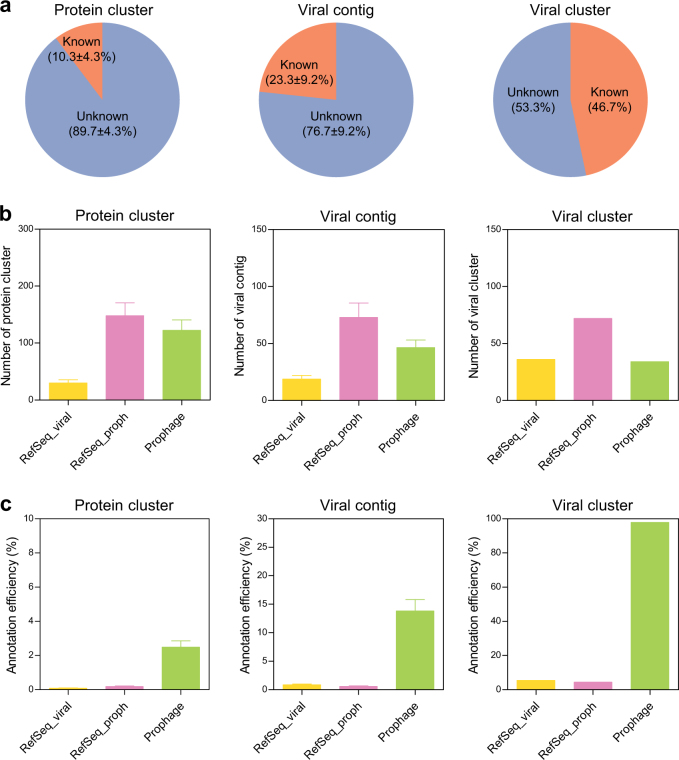
We compared the prophage genomes of the commensal lysogens with the metagenome of free phages to estimate the proportion of prophages in the active phage assemblage. The proteins encoded by the phage metagenome were first clustered using sequence similarity-based comparisons [33]; this resulted in 2348 ± 2106 protein clusters with 455 ± 1411 singletons. The representative sequences were then compared with protein sequences from the prophage genomes and two reference data sets (Refseq_viral and RefSeq_proph). Most protein clusters (89.7 ± 4.3%) shared no significant similarity with the proteins from the three databases (Fig. 3a). The annotated protein clusters were assigned to Refseq_proph (55.2 ± 2.6%), the integrated prophages (51.1 ± 3.0%) and Refseq_viral (12.9 ± 1.8%) (Fig. 3b). Then, the viral contigs were classified as “known” if they encoded at least one annotated protein cluster. A large number of viral contigs (76.7 ± 9.2%) encoded no proteins in the three databases (Fig. 3a). The identified contigs were annotated to Refseq_proph (72.4 ± 2.3%), the integrated prophages (52.6 ± 2.5%) and Refseq_viral (18.5 ± 1.5%) (Fig. 3b). These data indicated that the genetic content of the gut phage community is dominated by prophage-associated genes largely uncharacterized.
Fig. 3.
High sequence similarities between the integrated prophages and the free phages. a The overall percentage of phage metagenome annotations in the reference databases shown for protein clusters (n = 24), viral contigs (n = 24) and viral clusters. b The percentage of “known” phage metagenome annotated to three reference databases (the integrated prophages, RefSeq_viral and RefSeq_proph) shown for protein clusters, viral contigs and viral clusters. c The annotation efficiency of the phage metagenome to three reference databases shown for protein clusters, viral contigs and viral clusters. All data are presented as the mean ± SEM
Next, viral clusters were created using a shared gene content-based network analysis [34], resulting in 184 viral clusters with 589 singletons (Supplementary Table S3). From these, 98 viral clusters had no connection to any genomes in the three databases (Fig. 3a), suggesting that a large fraction of the gut phage populations remains unexplored. The 86 known viral clusters were grouped with phage genomes from Refseq_proph (72 clusters), the integrated prophages (34 clusters) and Refseq_viral (37 clusters) (Fig. 3b). However, the databases were highly biased toward Refseq_viral and Refseq_proph with respect to the number of genomes. Indeed, the annotation efficiency (%ratio of the number of sequences homologous with the sequences of free phages to the total number of reference sequences) of the prophage genomes was at least 14-fold higher than that of the Refseq_viral and Refseq_proph (Fig. 3c), speculating that the prophages of diverse commensal bacteria induced in situ may contribute in large part to the gut phage community.
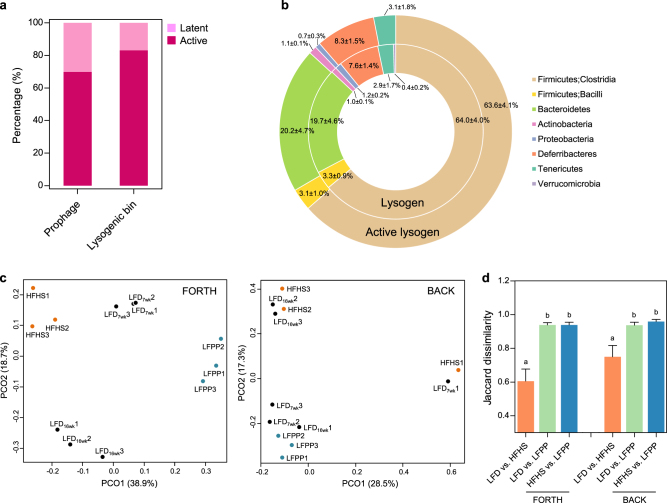
To verify this, we aligned the metagenomic reads of free phages with the prophage fragments (BLASTn, >95% identity and >90% alignment), regarding prophages as induced and bacterial bins as active, if they aligned. Consequently, 234 prophages (69.6%, 38,856 ± 13,121 reads per phage metagenome) of 99 lysogenic bins (83.2%) were active (Fig. 4a), and these active prophages were distributed in all taxa of the commensal lysogens, except Verrucomicrobia (Fig. 4b). They were not quantitatively evaluated because of the amplification bias toward single-stranded DNA viruses [49, 50]. The network plot of viral clusters also provided a community-wide overview of phage-prophage connections (Fig. 5). These connections were not restricted to a certain viral cluster or bacterial taxon, and were rarely observed across a class or higher level of bacterial classification, indicating that lysogeny occurs under the tight constraints of bacteria-phage specificity. Interestingly, the composition of the active prophages was affected by diet type, as shown for the bacterial hosts. According to the Jaccard dissimilarity-based PCoA, the composition of the active prophages was different among the three types of diet (P = 0.001 in FORTH, and P = 0.057 in BACK) (Fig. 4c); notably, these differences were more pronounced for the LFPP diet than for the LFD and HFHS diets (P < 0.01) (Fig. 4d). Of the active prophages, 37 prophages of 27 lysogenic bins were constantly detected regardless of diet type. Taken together, these data suggested that the majority of the prophages in the commensal lysogens are active prophages, not defective or cryptic prophages, and prophage induction from the commensal lysogens in diverse bacterial taxa is spontaneous and responsive to diet.
Fig. 4.
Prophages are spontaneously induced in active commensal lysogens. a The percentage of induced prophages and active lysogenic bins estimated by mapping phage metagenomic reads to the prophage sequences. b The taxonomic profiles of lysogenic and active lysogenic bins compared at the phylum level (n = 24). The composition of induced prophages for different diets determined using c the Jaccard dissimilarity-based PCoA (Adonis; FORTH, P = 0.001; BACK, P = 0.057) and d Jaccard distance comparisons (one-way ANOVA; FORTH, P < 0.001; BACK, P = 0.006). All data are presented as the mean ± SEM. LFD low-fat diet, HFHS high-fat, high-sucrose diet, and LFPP low-fat, high-plant-polysaccharide diet
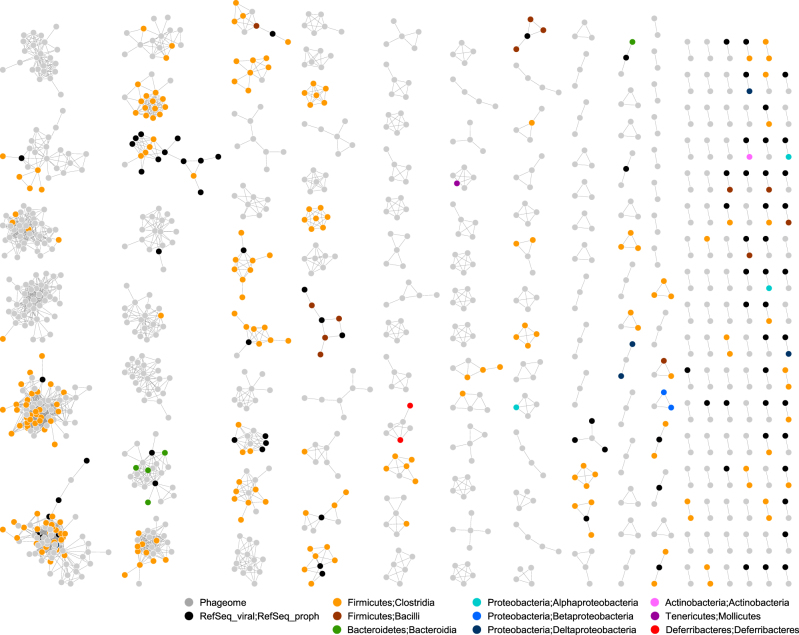
Fig. 5.
The network visualization of viral clusters comprising genomic sequences of the free phages and the integrated prophages. The nodes for different bacterial bins are designated by different colors
The taxonomic diversity of free phages and integrated prophages
Overall, 113 clusters (61.4%) of the free phages and 46 clusters (65.9%) of the integrated prophages remained unassigned at the family or lower level (Supplementary Figure S7a). In the free phages, the families Siphoviridae (42.7%) and Myoviridae (40.4%), contributing to the temporal stability of the communities [51], were abundant, whereas Podoviridae (9.2%) and Microviridae (3.9%), responsible for genetic variation of the communities [51], were not much (Supplementary Figure S7b). In the integrated prophages, Siphoviridae (42.7%) and Myoviridae (40.4%) were dominant in different bacterial taxa, while Podoviridae were detected only in Proteobacteria (Supplementary Figure S7c). Beyond the family level, 18 phage genera were detected (Supplementary Figure S7d). Most of these were reported in the human gut metagenome [52], but we identified 10 additional taxa that had not been previously reported. Phi29virus was only found in the free phages, while Lambdavirus was found only in the integrated prophages. Muvirus, thought to be a spontaneously induced phage [53], was consistently detected in the free phages and integrated prophages [52]. These data indicated that the gut phage community comprises phage taxa known to have a temperate life cycle, many of which are novel.
The associations between CRISPR-Cas immune systems and lysogeny
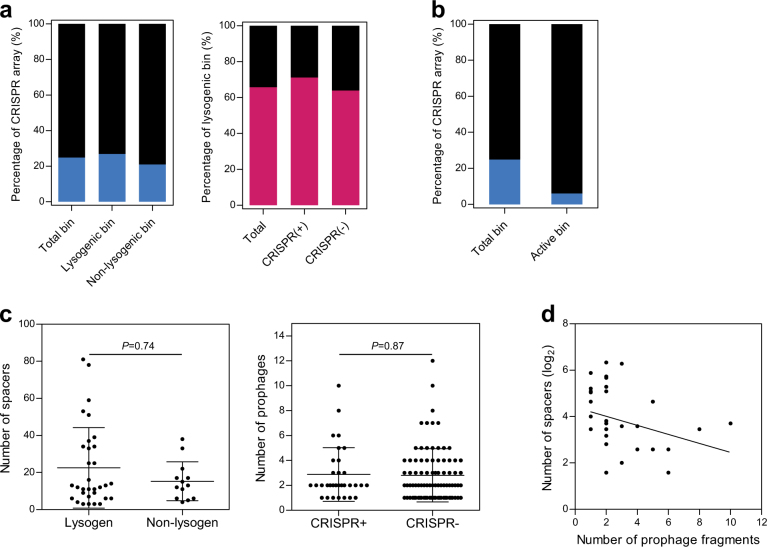
The CRISPR-Cas prokaryotic immune system records the sequence information of virulent phages that had previously infected the cell, in a genomic array [54]. To analyze the association between lysogeny and CRISPR-Cas systems, we searched for CRISPR arrays in bacterial bins using CRISPR repeat identification (Supplementary Table S4). In contrast to the prophage distribution, CRISPR arrays were present in only 45 bacterial bins (24.9%) (Fig. 6a), which is approximately half as many than what has been reported for the publicly available bacterial genomes (47%) [46]. We next explored the active CRISPR arrays, by searching for protospacer sequences in the phage metagenome; we found that only 6.5% of all spacers (60/920 spacers) in 11 bacterial bins showed exact matches to viral sequences (Fig. 6b), implying that the gut phages had not been subjected to the CRISPR-Cas systems of commensal bacteria. Although the presence of CRISPR arrays or prophages was not associated with the number of prophage fragments or CRISPR spacers, respectively (Fig. 6c), a negative correlation between the number of CRISPR spacers and prophage fragments were apparent (P = 0.030) (Fig. 6d). This was in agreement with the tendency of lysogens to have few spacers in CRISPR arrays, with the non-lysogens more likely to have many spacers in CRISPR arrays [46]. Altogether, the contrasting distribution of prophages and CRISPR-Cas systems enabled us to hypothesize that symbiotic relationships exist between commensal bacteria and phages.
Fig. 6.
The distribution of CRISPR-Cas systems in bacterial bins. a The percentage of CRISPR arrays in bacterial bins according to the identification of lysogen and the presence of CRISPR arrays. b The percentage of bacterial bins encoding spacers homologous to the phage metagenomic sequences in CRISPR arrays. c Comparison of the spacer numbers in lysogenic (n = 32) and non-lysogenic bins (n = 13) (two-tailed Student t-test), and comparison of the prophage numbers in the presence (n = 32) and absence (n = 87) of CRISPR-Cas systems (two-tailed Student t-test). d Correlation between the number of spacers in the CRISPR arrays of bacterial bins encoding CRISPR-Cas systems and the number of prophages in lysogenic bins (n = 32, Spearman’s rank correlation, r = −0.38, P = 0.03). All data are presented as the mean ± SEM
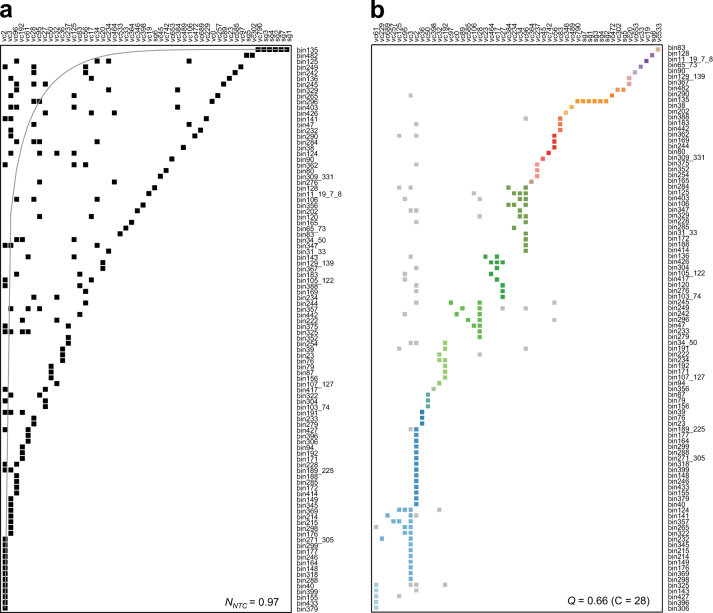
The “nested-modular” bacteria-phage infection networks
The pattern of bacteria-phage infection networks can be predictive of their underlying ecological and evolutionary processes [55]. The phage-prophage connections of the viral clusters allowed us to define bacteria-phage infections, resulting in the host assignment for 34 viral clusters. A search for viral transfer RNA genes of the phage metagenome originating from their hosts [56], resulted in the host assignment for 10 viral clusters and five singletons (Supplementary Table S5). The addition of protospacer-spacer pairs resulted in the host assignment for eight viral clusters and two singletons. Using a binary adjacency matrix composed of 50 columns for viral clusters and 98 rows for bacterial bins, we examined the nestedness and modularity of the bacteria-phage infection networks. These network analyses revealed the nestedness (NNTC = 0.97) and modularity (Q = 0.66) of the patterns of bacteria-phage infections (Fig. 7a, b). The “nested-modular” infection patterns were similar to the antagonistic co-evolution patterns of bacteria and phages [55], such that phages have evolved from specialists to generalists, whose interactions diversified into multi-scale structures under modular constraints. The local modules exhibited viral cluster-centric nested structures (Supplementary Figure S8a), indicating that few generalist phages interact with a broad range of bacterial hosts. Indeed, notably, many viral clusters were linked to multiple hosts from different phyla (VC3), classes (VC20 and 27), families (13 VCs) and genera (6 VCs) (Supplementary Figure S8b). This trend is congruent with results of global virome [57] and transduction [58] studies. Thus, the “nested-modular” infection patterns reflect the notion that gut phages readily interact with commensal bacteria that share similar genetic modules outlined by allelic variation; most of those interactions result in lysogeny.
Fig. 7.
The bacteria–phage interaction networks between viral clusters and bacterial bins. The matrix is composed of virus clusters (vc; rows) and bacterial bins (columns). These bacteria–phage networks are described as a nested by NTC (NNTC = 0.97); and b modular by lp-Brim (Q = 0.66, c = 28). The gray curve is an isocline of perfect nestedness. The different modules are highlighted in color
Discussion
The current study was designed to measure active commensal lysogens and prophages in the gut bacterial and phage metagenomes using genome-resolved metagenomic analyses. By capturing prophage genomes from the reconstructed genomes of bacterial species from the metagenomic data, we found that the majority of commensal bacteria in a variety of bacterial taxa are lysogens. By comparing two metagenome datasets of integrated prophages and free phages, we determined the lysogen activity and prophage induction in the gut environment, providing evidence that commensal bacteria may serve as a genetic reservoir of the gut phage community. The prophages of active lysogens were induced differently in response to different diets, suggesting that diet might be a potential regulator of lytic-lysogenic switches in the commensal lysogens. Further, these prophage-mediated bacteria-phage connections enabled the disentanglement of ecological and evolutionary relationships of the symbiotic interactions that develop in the gut ecosystem using infection network analysis, revealing “nested-modular” evolutionary structures in lysogenic infections similar to the antagonistic interactions of marine bacteria and phages [59].
The “nested-modular” structure of bacteria-phage infections implies that temperate phages have evolved from specialists to generalists via module-based evolution at multiple levels in the gut. Notably, the observation that many of the generalist phages infect multiple hosts representing various taxonomic backgrounds implies that gut-associated lysogenic interactions have evolved over the phylogenetic time of commensal bacteria. This long-term co-evolution may have induced phage-mediated gene transfers among commensal bacteria, resulting in taxonomic heterogeneity of the gut bacteria, particularly in the Clostridia class. This is corroborated by the low distribution and activity of CRISPR-Cas systems of commensal bacteria. Experimentally, using a gnotobiotic mouse model, fecal bacterial isolates were shown to have low susceptibility to fecal phage infections in vivo [60]. In this context, module-based generalist temperate phages most likely represent keystone phages in the assembly and functions of the gut phage community, which is consistent with the results of a study of the core bacteriophage groups in the healthy gut microbiome [61, 62].
The high distribution of active lysogens and prophages predicts widespread lysogenic infections which regulate host gene expression by disrupting functional and regulatory genes through phage integration and excision [63]. The inverse distribution of prophages and CRISPR-Cas systems suggests that commensal lysogens exhibit prophage-mediated resistance to superinfection by related phages [64]. No differences in the abundance of lysogenic and non-lysogenic bins indicate that the fitness energy costs of lysogeny would be insignificant in the rich-nutrient environment of the gut [65], thereby promoting the spread of lysogeny among commensal bacteria. In addition, the observed active prophages whose induction was independent of diet type indicated spontaneous prophage induction [53], enhancing competitive fitness by eliminating competitor bacterial strains [66]. This also promotes the spread of phages over the surrounding, resulting in an increase of lysogens by lysogenic conversion [53]. Widespread lysogeny contributes to the phage-mediated immunity predicted to occur on the surface of and inside the mucosal layer [67, 68]. In this regard, viral dysbiosis observed in colitis [9, 10], diet-induced obesity [8] and Clostridium difficile infection [69] may be consequences of abnormal signaling in the phage-mediated immunity because these diseases are associated with defects in the mucus layer [70–72].
Intriguingly, a large fraction of the commensal lysogens was identified in Firmicutes and Proteobacteria, compared to Bacteroidetes and Actinobacteria. According to the Piggyback-the-Winner model [73], lysogeny is favored in rapidly growing and dense communities. This was supported by a large-scale genome study reporting that minimum doubling time is a biological trait that is highly correlated with prophage prevalence [46]. The Firmicutes, particularly Clostrdia, constitute a more active, fast-growing population of the gut microbiota than Bacteroidetes in response to dietary and antibiotics interventions [23, 74]. The Proteobacteria population is also characterized by a high replication rate in the infant gut [75], and rapidly expands in response to diverse stimuli [76]. The different growth rates among the four phyla may be attributed to lysogeny occurring more frequently in Firmicutes and Proteobacteria, although this needs to be further evaluated in a large-scale study.
The Microviridae and Inoviridae families are thought to undergo lysogeny [77], but were not observed among commensal lysogens in this study. Because nucleotide composition patterns of these families were weakly similar to those of their host genome [32], our detection strategy was unlikely to capture genomic sequences of single-stranded DNA phages. Particularly, Microviridae are largely detected in the mammalian digestive system [5], contributing to community changes of the human gut virome [51]. Considering that members of Bacteroidetes are the candidate hosts of Microviridae [5], the actual fraction of Bacteroidetes lysogens is thought to be underestimated.
The current study revealed that active lysogens and prophages predominate in the gut microbial community, and strengthens the notion from the point of view of bacterial host that lysogeny is the main form of phage infection among commensal bacteria in the mammalian gut. Furthermore, this study showed that the bacteria-phage infection networks of lytic models such as Kill-the-Winner dynamics [55, 78] are coupled with the high-density/high-growth lysogeny of the Piggyback-the-Winner model [73] to confer broad availability of bacterial hosts as well as the benefits of lysogeny [64, 79, 80]. Using genome recovery from metagenomic data, our findings expanded the knowledge of lysogenic interactions between gut bacteria and phages as well as unknown diversity of gut phages. Also, our study elucidated that the activity of temperate phages is not optional, but essential for the stability and maintenance of bacterial populations in the gut. Since these relationships have been conserved for a long evolutionary time, information about the lysogenic infections of commensal bacteria can refine our knowledge of the underlying functions of the gut microbiome, and help characterize what constitutes a healthy gut microbiome. Future studies are needed to address the ecological role of commensal lysogens and phage generalists in gut microbial functions, and to assess whether lysogenic infections can be used to manipulate gut microbial homeostasis.
Electronic supplementary material
Acknowledgements
This work was supported by grants from the Basic Science Research Program (2015R1D1A4A01019807) funded by the Ministry of Education, the Mid-Career Researcher Program (2016R1E1A1A02921587) funded by the Ministry of Science, ICT & Future Planning through the National Research Foundation of Korea, and the Agricultural Microbiome R&D Program (grant number: 914006-4) funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA) through Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry (IPET). We thank Dr. Donovan H. Parks, Dr. Simon Roux, Dr. Ben Bolduc and Dr. Ho Bin Jang for technical advice in computational analyses.
Compliance with Ethical Standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-018-0061-9) contains supplementary material, which is available to authorized users.
References
- 1.Mirzaei MK, Maurice CF. Menage a trois in the human gut: interactions between host, bacteria and phages. Nat Rev Microbiol. 2017;15:397–408. doi: 10.1038/nrmicro.2017.30. [DOI] [PubMed] [Google Scholar]
- 2.Borrel G, McCann A, Deane J, Neto MC, Lynch DB, Brugere JF, et al. Genomics and metagenomics of trimethylamine-utilizing Archaea in the human gut microbiome. ISME J. 2017;11:2059–74. doi: 10.1038/ismej.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atarashi K, Tanoue T, Oshima K, Suda W, Nagano Y, Nishikawa H, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–6. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 4.Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21:1616–25. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim MS, Park EJ, Roh SW, Bae JW. Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol. 2011;77:8062–70. doi: 10.1128/AEM.06331-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lim ES, Zhou Y, Zhao G, Bauer IK, Droit L, Ndao IM, et al. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat Med. 2015;21:1228–34. doi: 10.1038/nm.3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, Rohwer F, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466:334–8. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim MS, Bae JW. Spatial disturbances in altered mucosal and luminal gut viromes of diet-induced obese mice. Environ Microbiol. 2016;18:1498–510. doi: 10.1111/1462-2920.13182. [DOI] [PubMed] [Google Scholar]
- 9.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–60. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang JY, Kim MS, Kim E, Cheon JH, Lee YS, Kim Y, et al. Enteric viruses ameliorate gut inflammation via Toll-like receptor 3 and Toll-like receptor 7-mediated interferon-b production. Immunity. 2016;44:889–900. doi: 10.1016/j.immuni.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 11.Zhao G, Vatanen T, Droit L, Park A, Kostic AD, Poon TW, et al. Intestinal virome changes precede autoimmunity in type I diabetes-susceptible children. Proc Natl Acad Sci USA. 2017;114:E6166–E6175. doi: 10.1073/pnas.1706359114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinbauer MG. Ecology of prokaryotic viruses. FEMS Microbiol Rev. 2004;28:127–81. doi: 10.1016/j.femsre.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 13.Howard-Varona C, Hargreaves KR, Abedon ST, Sullivan MB. Lysogeny in nature: mechanisms, impact and ecology of temperate phages. ISME J. 2017;11:1511–20. doi: 10.1038/ismej.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang SC, Paul JH. Seasonal and diel abundance of viruses and occurrence of lysogeny/bacteriocinogeny in the marine-environment. Mar Ecol Progress Ser. 1994;104:163–72. doi: 10.3354/meps104163. [DOI] [Google Scholar]
- 15.Jiang SC, Paul JH. Occurrence of lysogenic bacteria in marine microbial communities as determined by prophage induction. Mar Ecol Progress Ser. 1996;142:27–38. doi: 10.3354/meps142027. [DOI] [Google Scholar]
- 16.Knowles B, Bailey B, Boling L, Breitbart M, Cobian-Guemes A, Del Campo J, et al. Variability and host density independence in inductions-based estimates of environmental lysogeny. Nat Microbiol. 2017;2:17064. doi: 10.1038/nmicrobiol.2017.64. [DOI] [PubMed] [Google Scholar]
- 17.Paul JH, Weinbauer M. Detection of lysogeny in marine environments. Man Aquat Viral Ecol. 2010;4:30–33. doi: 10.4319/mave.2010.978-0-9845591-0-7.30. [DOI] [Google Scholar]
- 18.Sangwan N, Xia F, Gilbert JA. Recovering complete and draft population genomes from metagenome datasets. Microbiome. 2016;4:8. doi: 10.1186/s40168-016-0154-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nielsen HB, Almeida M, Juncker AS, Rasmussen S, Li J, Sunagawa S, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–8. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 20.Sharon I, Morowitz MJ, Thomas BC, Costello EK, Relman DA, Banfield JF. Time series community genomics analysis reveals rapid shifts in bacterial species, strains, and phage during infant gut colonization. Genome Res. 2013;23:111–20. doi: 10.1101/gr.142315.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol. 2013;31:533–8. doi: 10.1038/nbt.2579. [DOI] [PubMed] [Google Scholar]
- 22.Ormerod KL, Wood DL, Lachner N, Gellatly SL, Daly JN, Parsons JD, et al. Genomic characterization of the uncultured Bacteroidales family S24-7 inhabiting the guts of homeothermic animals. Microbiome. 2016;4:36. doi: 10.1186/s40168-016-0181-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carmody RN, Gerber GK, Luevano JM, Jr., Gatti DM, Somes L, Svenson KL, et al. Diet dominates host genotype in shaping the murine gut microbiota. Cell Host Microbe. 2015;17:72–84. doi: 10.1016/j.chom.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Howe A, Ringus DL, Williams RJ, Choo ZN, Greenwald SM, Owens SM, et al. Divergent responses of viral and bacterial communities in the gut microbiome to dietary disturbances in mice. ISME J. 2016;10:1217–27. doi: 10.1038/ismej.2015.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thurber RV, Haynes M, Breitbart M, Wegley L, Rohwer F. Laboratory procedures to generate viral metagenomes. Nat Protoc. 2009;4:470–83. doi: 10.1038/nprot.2009.10. [DOI] [PubMed] [Google Scholar]
- 26.Zoetendal EG, Heilig HG, Klaassens ES, Booijink CC, Kleerebezem M, Smidt H, et al. Isolation of DNA from bacterial samples of the human gastrointestinal tract. Nat Protoc. 2006;1:870–3. doi: 10.1038/nprot.2006.142. [DOI] [PubMed] [Google Scholar]
- 27.Reyes A, Blanton LV, Cao S, Zhao G, Manary M, Trehan I, et al. Gut DNA viromes of Malawian twins discordant for severe acute malnutrition. Proc Natl Acad Sci USA. 2015;112:11941–6. doi: 10.1073/pnas.1514285112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–8. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 29.Kang DD, Froula J, Egan R, Wang Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. Peer J. 2015;3:e1165. doi: 10.7717/peerj.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roux S, Enault F, Hurwitz BL, Sullivan MB. VirSorter: mining viral signal from microbial genomic data. Peer J. 2015;3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roux S, Hallam SJ, Woyke T, Sullivan MB. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. 2015;4:e08490. doi: 10.7554/eLife.08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): a marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE. 2013;8:e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bolduc B, Jang HB, Doulcier G, You ZQ, Roux S, Sullivan MB. vConTACT: an iVirus tool to classify double-stranded DNA viruses that infect Archaea and Bacteria. Peer J. 2017;5:e3243. doi: 10.7717/peerj.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. PILER-CR: fast and accurate identification of CRISPR repeats. BMC Bioinf. 2007;8:18. doi: 10.1186/1471-2105-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Biswas A, Gagnon JN, Brouns SJ, Fineran PC, Brown CM. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013;10:817–27. doi: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Edwards RA, McNair K, Faust K, Raes J, Dutilh BE. Computational approaches to predict bacteriophage-host relationships. FEMS Microbiol Rev. 2016;40:258–72. doi: 10.1093/femsre/fuv048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laslett D, Canback B. ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res. 2004;32:11–16. doi: 10.1093/nar/gkh152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Almeida-Neto M, Guimaraes P, Guimaraes PR, Loyola RD, Ulrich W. A consistent metric for nestedness analysis in ecological systems: reconciling concept and measurement. Oikos. 2008;117:1227–39. doi: 10.1111/j.0030-1299.2008.16644.x. [DOI] [Google Scholar]
- 40.Beckett SJ, Boulton CA, Williams HT. FALCON: a software package for analysis of nestedness in bipartite networks. F1000 Res. 2014;3:185. doi: 10.12688/f1000research.4831.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barber MJ. Modularity and community detection in bipartite networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2007;76:066102. doi: 10.1103/PhysRevE.76.066102. [DOI] [PubMed] [Google Scholar]
- 42.Xiao L, Feng Q, Liang S, Sonne SB, Xia Z, Qiu X, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–8. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 43.Wannemuehler MJ, Overstreet AM, Ward DV, Phillips GJ. Draft genome sequences of the altered schaedler flora, a defined bacterial community from gnotobiotic mice. Genome Announc. 2014;2:e00287–14. doi: 10.1128/genomeA.00287-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Duck LW, Walter MR, Novak J, Kelly D, Tomasi M, Cong Y, et al. Isolation of flagellated bacteria implicated in Crohn’s disease. Inflamm Bowel Dis. 2007;13:1191–201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 45.Pride DT, Meinersmann RJ, Wassenaar TM, Blaser MJ. Evolutionary implications of microbial genome tetranucleotide frequency biases. Genome Res. 2003;13:145–58. doi: 10.1101/gr.335003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Touchon M, Bernheim A, Rocha EP. Genetic and life-history traits associated with the distribution of prophages in bacteria. ISME J. 2016;10:2744–54. doi: 10.1038/ismej.2016.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Paepe M, Tournier L, Moncaut E, Son O, Langella P, Petit MA. Carriage of λ latent virus is costly for its bacterial host due to frequent reactivation in monoxenic mouse intestine. PLoS Genet. 2016;12:e1005861. doi: 10.1371/journal.pgen.1005861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lugli GA, Milani C, Turroni F, Tremblay D, Ferrario C, Mancabelli L, et al. Prophages of the genus Bifidobacterium as modulating agents of the infant gut microbiota. Environ Microbiol. 2016;18:2196–213. doi: 10.1111/1462-2920.13154. [DOI] [PubMed] [Google Scholar]
- 49.Kim KH, Bae JW. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl Environ Microbiol. 2011;77:7663–8. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roux S, Solonenko NE, Dang VT, Poulos BT, Schwenk SM, Goldsmith DB, et al. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. Peer J. 2016;4:e2777. doi: 10.7717/peerj.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Minot S, Bryson A, Chehoud C, Wu GD, Lewis JD, Bushman FD. Rapid evolution of the human gut virome. Proc Natl Acad Sci USA. 2013;110:12450–5. doi: 10.1073/pnas.1300833110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waller AS, Yamada T, Kristensen DM, Kultima JR, Sunagawa S, Koonin EV, et al. Classification and quantification of bacteriophage taxa in human gut metagenomes. ISME J. 2014;8:1391–402. doi: 10.1038/ismej.2014.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nanda AM, Thormann K, Frunzke J. Impact of spontaneous prophage induction on the fitness of bacterial populations and host-microbe interactions. J Bacteriol. 2015;197:410–9. doi: 10.1128/JB.02230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–70. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 55.Weitz JS, Poisot T, Meyer JR, Flores CO, Valverde S, Sullivan MB, et al. Phage-bacteria infection networks. Trends Microbiol. 2013;21:82–91. doi: 10.1016/j.tim.2012.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Bailly-Bechet M, Vergassola M, Rocha E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007;17:1486–95. doi: 10.1101/gr.6649807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, et al. Uncovering Earth’s virome. Nature. 2016;536:425–30. doi: 10.1038/nature19094. [DOI] [PubMed] [Google Scholar]
- 58.Popa O, Landan G, Dagan T. Phylogenomic networks reveal limited phylogenetic range of lateral gene transfer by transduction. ISME J. 2017;11:543–54. doi: 10.1038/ismej.2016.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Flores CO, Valverde S, Weitz JS. Multi-scale structure and geographic drivers of cross-infection within marine bacteria and phages. ISME J. 2013;7:520–32. doi: 10.1038/ismej.2012.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Reyes A, Wu M, McNulty NP, Rohwer FL, Gordon JI. Gnotobiotic mouse model of phage-bacterial host dynamics in the human gut. Proc Natl Acad Sci USA. 2013;110:20236–41. doi: 10.1073/pnas.1319470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, Boling L, et al. A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun. 2014;5:4498. doi: 10.1038/ncomms5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manrique P, Bolduc B, Walk ST, van der Oost J, de Vos WM, Young MJ. Healthy human gut phageome. Proc Natl Acad Sci USA. 2016;113:10400–5. doi: 10.1073/pnas.1601060113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Feiner R, Argov T, Rabinovich L, Sigal N, Borovok I, Herskovits AA. A new perspective on lysogeny: prophages as active regulatory switches of bacteria. Nat Rev Microbiol. 2015;13:641–50. doi: 10.1038/nrmicro3527. [DOI] [PubMed] [Google Scholar]
- 64.Bondy-Denomy J, Qian J, Westra ER, Buckling A, Guttman DS, Davidson AR, et al. Prophages mediate defense against phage infection through diverse mechanisms. ISME J. 2016;10:2854–66. doi: 10.1038/ismej.2016.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Obeng N, Pratama AA, Elsas JD. The significance of mutualistic phages for bacterial ecology and evolution. Trends Microbiol. 2016;24:440–9. doi: 10.1016/j.tim.2015.12.009. [DOI] [PubMed] [Google Scholar]
- 66.Duerkop BA, Clements CV, Rollins D, Rodrigues JL, Hooper LV. A composite bacteriophage alters colonization by an intestinal commensal bacterium. Proc Natl Acad Sci USA. 2012;109:17621–6. doi: 10.1073/pnas.1206136109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barr JJ, Auro R, Furlan M, Whiteson KL, Erb ML, Pogliano J, et al. Bacteriophage adhering to mucus provide a non-host-derived immunity. Proc Natl Acad Sci USA. 2013;110:10771–6. doi: 10.1073/pnas.1305923110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Silveira CB, Rohwer FL. Piggyback-the-Winner in host-associated microbial communities. NPJ Biofilms Micro. 2016;2:16010. doi: 10.1038/npjbiofilms.2016.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zuo T, Wong SH, Lam K, Lui R, Cheung K, Tang W et al. Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome. Gut. 2017. 10.1136/gutjnl-2017-313952. [DOI] [PMC free article] [PubMed]
- 70.Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci USA. 2013;110:9066–71. doi: 10.1073/pnas.1219451110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Johansson ME, Gustafsson JK, Holmen-Larsson J, Jabbar KS, Xia L, Xu H, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–91. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McGuckin MA, Linden SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011;9:265–78. doi: 10.1038/nrmicro2538. [DOI] [PubMed] [Google Scholar]
- 73.Knowles B, Silveira CB, Bailey BA, Barott K, Cantu VA, Cobian-Guemes AG, et al. Lytic to temperate switching of viral communities. Nature. 2016;531:466–70. doi: 10.1038/nature17193. [DOI] [PubMed] [Google Scholar]
- 74.Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell. 2013;152:39–50. doi: 10.1016/j.cell.2012.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown CT, Olm MR, Thomas BC, Banfield JF. Measurement of bacterial replication rates in microbial communities. Nat Biotechnol. 2016;34:1256–63. doi: 10.1038/nbt.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33:496–503. doi: 10.1016/j.tibtech.2015.06.011. [DOI] [PubMed] [Google Scholar]
- 77.Krupovic M, Forterre P. Single-stranded DNA viruses employ a variety of mechanisms for integration into host genomes. Ann N Y Acad Sci. 2015;1341:41–53. doi: 10.1111/nyas.12675. [DOI] [PubMed] [Google Scholar]
- 78.Thingstad TF. Elements of a theory for the mechanisms controlling abundance, diversity, and biogeochemical role of lytic bacterial viruses in aquatic systems. Limnol Oceanogr. 2000;45:1320–8. doi: 10.4319/lo.2000.45.6.1320. [DOI] [Google Scholar]
- 79.Paul JH. Prophages in marine bacteria: dangerous molecular time bombs or the key to survival in the seas? ISME J. 2008;2:579–89. doi: 10.1038/ismej.2008.35. [DOI] [PubMed] [Google Scholar]
- 80.Weitz JS, Beckett SJ, Brum JR, Cael BB, Dushoff J. Lysis, lysogeny and virus-microbe ratios. Nature. 2017;549:E1–E3. doi: 10.1038/nature23295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.