Abstract
The Notch signaling pathway is a key regulator of stem cells during development, and its deregulated activity is linked to developmental defects and cancer. Transcriptional activation of Notch target genes requires cleavage of the Notch receptor in response to ligand binding, production of the Notch intracellular domain (NICD1), NICD1 migration into the nucleus, and assembly of a transcriptional complex. Post-translational modifications of Notch regulate its trafficking, turnover, and transcriptional activity. Here, we show that NICD1 is modified by small ubiquitin-like modifier (SUMO) in a stress-inducible manner. Sumoylation occurs in the nucleus where NICD1 is sumoylated in the RBPJ-associated molecule (RAM) domain. Although stress and sumoylation enhance nuclear localization of NICD1, its transcriptional activity is attenuated. Molecular modeling indicates that sumoylation can occur within the DNA-bound ternary transcriptional complex, consisting of NICD1, the transcription factor Suppressor of Hairless (CSL), and the co-activator Mastermind-like (MAML) without its disruption. Mechanistically, sumoylation of NICD1 facilitates the recruitment of histone deacetylase 4 (HDAC4) to the Notch transcriptional complex to suppress Notch target gene expression. Stress-induced sumoylation decreases the NICD1-mediated induction of Notch target genes, which was abrogated by expressing a sumoylation-defected mutant in cells and in the developing central nervous system of the chick in vivo. Our findings of the stress-inducible sumoylation of NICD1 reveal a novel context-dependent regulatory mechanism of Notch target gene expression.
Subject terms: DNA, Nuclear receptors
Introduction
The Notch pathway regulates development of most tissues. The output of Notch signaling is strictly dose-dependent [1, 2], and Notch target gene expression needs to be fine-tuned for the cells to meet changes in their physiological environment [3–5]. Various forms of stress conditions such as hypoxia, irradiation, inflammation, and oxidative stress influence Notch activity [4, 6–8], but the mechanisms regulating context-specific Notch target gene expression at the level of the transcriptional complex are, however, unclear.
Notch signaling is activated by direct cell–cell contact with Notch ligand-expressing cells. Upon ligand binding the Notch receptor is cleaved, resulting in the release and translocation of the Notch intracellular domain (NICD) to the nucleus to activate target genes [9], such as bHLH genes of the Hes and Hey families, which function as transcriptional repressors [10, 11]. In the nucleus, the RBPJ-associated molecule (RAM) domain of NICD binds to the transcription factor Suppressor of Hairless (CSL), which is followed by the binding of a secondary low-affinity ankyrin repeat (ANK) on NICD to CSL [12]. The interaction between NICD and CSL leads to an allosteric change in CSL causing displacement of co-repressors, which activates CSL, which then recruits the transcriptional co-activator protein Mastermind-like (MAML) to activate target genes [12, 13].
Post-translational modifications (PTMs) regulate Notch activity [2]. PTMs influence nuclear translocation, target gene expression, and half-life of NICD [1, 2]. NICD1 is methylated by co-activator-associated arginine methyltransferase 1, which regulates NICD1 stability and the expression of specific Notch target genes [14]. PIM kinases phosphorylate NICD1 and regulate its nuclear localization and transcriptional activity [15]. In addition, NICD1 is subjected to hydroxylation [16] and acetylation [17], and inhibition of global sumoylation increases Notch target gene expression [18], but no direct role of sumoylation in the regulation of Notch1 has been reported.
The functional consequences of the modification of proteins by small ubiquitin-like modifiers (SUMO) vary depending on the target and range from regulating transcription, cytoplasmic-nuclear transport, and DNA repair to altering protein–protein interactions [19]. Sumoylation has been implicated to regulate cell fate specification during development [20].
The binding of SUMO to its substrate occurs stepwise involving an E1-activating enzyme, an E2 ubiquitin enzyme 9 (Ubc9), and, in most cases, E3 ligases [21]. Only a small fraction of most SUMO substrates are sumoylated at steady state, challenging the detection of sumoylated proteins [22]. In addition to the SUMO consensus target sequence ψKxE (ψ is a bulky hydrophobic amino-acid residue, K is the target lysine, x is any residue, and E represents glutamate) [23], atypical sites with little similarity to the consensus sequences exist [24]. Sentrin-specific proteases (SENPs) regulate the conjugation/deconjugation balance by desumoylating the SUMO target proteins [25].
The genomic DNA is wrapped around histones. Histones undergo constant acetylation and deacetylation, which impacts chromatin landscape and regulates gene expression including Notch target genes [[56]26]. Histone deacetylases (HDACs) are divided into four classes based on function and DNA sequence similarity: class I (HDACs 1, 2, 3, and 8), class II (HDACs 4, 5, 6, 7, 9, and 10), sirtuin class III, and class IV (HDAC11) [27]. In addition, HDACs target non-histone proteins, including transcriptional factors, which may represent general regulatory mechanisms in biological signaling. Class II HDACs, including HDAC4, have been reported to act as SUMO E3 ligases [28]. HDAC4 is also recruited by sumoylated LAP1, a member of the CEBP family of transcription factors, thereby attenuating the binding of HDAC4 on the cyclooxygenase 2 promoter and repressing its transcription [29].
Here, we addressed the key question of how transcriptional tuning of Notch target genes by sumoylation occurs during cell stress. We demonstrate that NICD1 is sumoylated in the nucleus in the RAM domain upon heat stress, with consequent suppression of Notch target genes. We show by biochemical assays and molecular modeling that NICD1 can be sumoylated within the ternary transcriptional complex. Sumoylation leads to the recruitment of HDAC4 to the transcriptional complex, and represses the expression of specific classical Notch1 target genes Hes and Hey. In vivo in the developing CNS of chicken embryos Notch target genes are repressed upon stress by WT Notch, whereas the target gene activation capacity is retained by a sumo-deficient Notch mutant. Thus, stress-inducible sumoylation provides a regulatory mechanism for dynamic Notch target gene expression in vivo.
Results
The intracellular domain of Notch1 is sumoylated
We have previously developed screening approaches to purify and identify novel SUMO2 substrates in heat-shocked cells [30, 31]. The screens identified Notch1 as a putative SUMO substrate and indicated that the sumoylation of Notch1 occurs within a sequence of the RAM domain containing four lysine residues (K1774, K1780, K1781, and K1782). Therefore, we wanted to determine whether Notch is indeed subject to sumoylation. The lysines are non-consensus sumoylation target sites located in NICD1 (Fig. 1a). K1774, K1780, K1781, and K1782 are unique to Notch1 and are conserved in several species including chicken, mouse, rat, and human (Fig. 1b).
Fig. 1.

Notch as a sumoylation target protein. a The sequence around the putative sumoylation site K1774/K1780/K1781/K1782 is located in the intracellular domain of Notch. b Sequence alignment of the Notch1 region surrounding the putative sumoylation site K1780 using the Clustal Omega web-based tool. The putative sumoylation site is highlighted in gray
To study whether NICD1 is a SUMO substrate, we first expressed GFP-Flag-NICD1 and Histidine-tagged SUMO1 (His-SUMO1) in HeLa cells. Whole-cell extracts were collected, and samples were immunoprecipitated with His beads and immunoblotted with a GFP-specific antibody. Sumoylation of NICD1 was detected in a band between 150 and 250 kDa (Fig. 2a). Furthermore, overexpressed ΔENotch1 (ΔEN1), a construct coding for a membrane-tethered form of Notch1, which is constitutively cleaved by γ-secretase to generate NICD1, demonstrated, although weakly, sumoylation of NICD1 as detected with an antibody (Val-1744) that recognizes activated cleaved Notch1 (Fig. 2b).
Fig. 2.
Notch1 is modified by SUMO. a GFP-Flag-NICD1 is modified by Histidine-SUMO1 (His-SUMO1) in HeLa cells. n = 4. b Activated Notch1 (ΔEN) is modified by transfected His-SUMO1 in HeLa cells. The samples were blotted with α-Val1744 recognizing NICD1. n = 4. c Heat shock increases the modification of Notch1 by SUMO1 and SUMO3. HeLa cells were transfected with GFP-Flag-NICD1 and His-SUMO1 (S1) or His-SUMO3 (S3). The samples were left untreated or heat-shocked for 1 h at 42 °C followed by a 1-h recovery at 37 °C (HS + recovery). n = 4. d Proteotoxic stress enhances Notch sumoylation. Transfected HeLa cells were left untreated, or were treated with the proteasome inhibitor Bortezomib at 37 °C for 8 or 16 h (h). n = 3. e Notch1 is modified by endogenous SUMO2/3. HeLa cells were heat-shocked and recovered as in (c). Hemagglutinin-SUMO2 (HA-SUMO2) was transfected as a positive control and pCMVKM empty vector (pCMV) as a negative control. n = 3. f Modification of Notch1 by endogenous SUMO1 is not detectable. The indicated samples were treated as in (e), but blotted with α-SUMO1. His-SUMO1 + GFP-Flag-NICD1 was transfected as a positive control. n = 3. g SENP1 and SENP2 deconjugate SUMO1 and SUMO2 from Notch1. COS7 cells were heat-shocked and recovered as in (c), lysed, and immunoblotted with α-GFP antibody. The sumoylated form of Notch1 is indicated with an arrow or a bracket and the unmodified form with an asterisk. n = 3
Various environmental stresses can increase/decrease sumoylation, and stress stimuli have been shown to boost the interaction between SUMO and several of its target proteins to provide an important post-translational regulatory mechanism [31].
To examine whether stress has an effect on NICD1 sumoylation, we transfected HeLa cells with GFP-Flag-NICD1 together with either His-SUMO1 or His-SUMO3 and heat-shocked the cells. Heat shock significantly enhanced modification of NICD1 by both His-SUMO1 and His-SUMO3 (Fig. 2c). Heat-shocked cells with transfected SUMO displayed several bands between 150 and 250 kDa, indicating that NICD1 potentially contains multiple sumoylation sites (Fig. 2c). Ligand-activated Full-Length Notch1 (FLN; Supplementary Fig. 1A) and endogenous Notch (Supplementary Fig. 1B) were also shown to be sumoylated as NICD1. Without overexpressed SUMO, sumoylation of NICD1 decreased significantly, supporting the notion that conjugation of endogenous SUMO to Notch is challenging to detect (Supplementary Fig. 2A).
Acute heat shock enhanced transcription of Notch target genes (Supplementary Fig. 2B). In contrast, we observed the most efficient sumoylation of NICD1 upon recovery from acute stress in cells exposed to a 1-h heat shock at 42 °C, followed by a 1-h recovery at 37 °C (Supplementary Fig. 2C). To determine if other stress conditions induce Notch sumoylation, we exposed HeLa cells to proteotoxic stress by treating the cells with the proteasome inhibitor Bortezomib, and detected increased sumoylation of Notch (Fig. 2d). To establish which endogenous SUMO isoforms are capable of modifying Notch1 we transfected GFP-Flag-NICD1 into HeLa cells, immunoprecipitated Notch1, and immunoblotted with an antibody recognizing either SUMO2/3 or SUMO1 (Fig. 2e, f and Supplementary Fig. 2D). Only endogenous SUMO2/3 was able to modify NICD1. Next, we detected that the de-conjugating enzymes SENP1 and SENP2 were able to remove His-SUMO1 and His-SUMO2 from NICD1 (Fig. 2g).
Stress-inducible sumoylation of Notch occurs in the nucleus
Sumoylation is known to regulate a variety of cellular processes including nuclear transport and expression of genes [32]. We assessed by immunofluorescence confocal microscopy whether the nuclear localization of NICD1 is altered in response to stress and sumoylation. A prominent portion of NICD1 generated from ΔEN1 was detected in the nucleus in cells overexpressing SUMO. Since heat stress increased the modification of NICD1 by SUMO (Fig. 2c), it was of interest to determine whether overexpressed SUMO, in combination with heat shock, further enhanced the nuclear accumulation of NICD1. The strongest nuclear localization was observed when heat shock was combined with overexpression of Notch and SUMO (Fig. 3a, b). In average, 58% of those cells displayed strong nuclear staining of Notch1, whereas only 41% of the cells expressing only endogenous SUMO displayed strong nuclear staining (Fig. 3b, Supplementary Figs. 3A, B).
Fig. 3.
SUMO enhances the nuclear localization of Notch1, interacts with Notch1 in the nucleus, and increases the levels of Notch1. a Sumoylation of Notch1 increases the nuclear localization of Notch1. COS7 cells were transfected with activated Notch1 (ΔEN) and/or His-SUMO1 (S1), or a control vector. Twenty-four hours after transfection the cells were left untreated or heat-shocked for 1 h at 42 °C followed by a 1-h recovery at 37 °C (HS). Images were taken with a Zeiss LSM510 META confocal microscope. n = 3. b Heat shock combined with transfected SUMO increases the nuclear localization of Notch1 most efficiently. Values are statistically significant at **p < 0.01 and represent averages from three independent experiments. Hundred cells in average were counted for each sample. c Notch1 is sumoylated in the nucleus. COS7 cells were heat-shocked and recovered as in (a). Nuclear fractions (NE) and cytoplasmic fractions (Cyt) were extracted and the samples were immunoblotted. GFP-Flag-NICD1 (NICD1) shows strong sumoylation in the nuclear fractions, whereas the cytoplasmic fractions show no conjugation of His-SUMO1 (S1) to NICD1. n = 3. d Consistent with (c), NICD1 cleaved from overexpressed ΔEN does not interact with His-SUMO1 (S1) in the cytoplasm, but only in the nucleus. The samples were treated as in (c), but immunoblotted with α-Notch1 C20. e COS7 cells were transfected as indicated, and left untreated or treated with the protein translation inhibitor cycloheximide (CHX). The cells were heat-shocked at 42 °C followed by a 1-h recovery at 37 °C (HS + rec.), or left untreated, and the samples were lysed and immunoblotted. f The levels of Notch1 are increased with increasing amounts of transfected SUMO1. COS7 cells were transfected with GFP-Flag-NICD (NICD) and different amounts of His-SUMO1 (S1), heat-shocked and recovered, lysed, and immunoprecipitated against Notch1 with protein G-sepharose beads and α-Notch1 C20, and analyzed with immunoblotting. The sumoylated form of Notch1 is indicated with an arrow or a bracket and the unmodified form with an asterisk. The lysates showed less difference in the levels of NICD, presumably due to decreased sumoylation of Notch1 in the presence of desumoylating isopeptidases. n = 3
SUMO interacts with many of its substrates in the nucleus [33]. Although SUMO1, SUMO2/3, SAE1/SAE2, and Ubc9 are predominantly located in the nucleus [34], SUMO modifications have also been observed to occur in the cytoplasm, plasma membrane, ER membrane, and mitochondria [35]. To determine the cellular localization of NICD1 sumoylation, we extracted cytoplasmic and nuclear fractions and observed NICD1 to be sumoylated only in the nuclear fractions (Fig. 3c, d). Therefore, it was important to investigate the functional outcome of NICD1 sumoylation in the nucleus. Alterations in protein levels have been documented for a variety of SUMO target proteins [36]. To determine whether sumoylation affects NICD levels, we inhibited de novo protein synthesis by cycloheximide treatment for up to 6 h and monitored the amount of NICD with and without heat shock and overexpressed SUMO (Fig. 3e and Supplementary Figs. 3C and D). Immunoblotting displayed higher levels of NICD by overexpression of SUMO both with and without heat shock. In a SUMO dose response experiment SUMO enhanced NICD levels in a dose-dependent manner (Fig. 3f and Supplementary Fig. 3E).
Sumoylation of NICD1 decreases expression of Notch target genes
Dynamic expression of Notch target genes Hes and Hey is crucial for a proper timing of Notch-driven fate-determining steps during development and to elicit proper cellular responses in a changing environment. Since the mechanisms of dynamic control of Notch target gene expression during different cellular contexts have remained elusive, we investigated a putative effect of stress-induced sumoylation on Notch-mediated transcription. We first used molecular modeling to assess whether sumoylation of NICD1 within the Notch1 transcriptional complex is possible. Secondary structure prediction for NICD1 follows the secondary structure pattern of the crystallized Notch1 transcription complex [37]. K1774 is located in the last helix (F1773-S1779) of the crystallized RAM peptide, which is followed by a predicted loop region (K1780-L1794) with K1780, K1781, and K1782. The secondary structure prediction for residues 1780–1794 agrees with the notion that sumoylation often occurs on residues located in loop regions or in intrinsically disordered regions [32]. On the basis of our three-dimensional model of the sumoylated Notch1 transcription complex, the loop with K1780, K1781, and K1782 is on the surface of the complex (Fig. 4 and Supplementary Fig. 4). The RAM peptide interacts with the CSL subunit of the Notch1 transcription complex, but K1780, K1781, and K1782 are not involved in these interactions. Instead, they are solvent-exposed and hence easily accessible to sumoylation without having an impact on the interaction with CSL or any other component of the Notch1 transcriptional complex (Fig. 4 and Supplementary Fig. 4).
Fig. 4.
Three-dimensional model of sumoylated Notch1 transcription complex. CSL (light green), MAML1 (purple), DNA (orange), ANK repeat domain (cyan), and the N-terminal part (until E1777) of the RAM peptide (cyan) are from the crystal structure of the Notch1 transcription complex (PDB 3V79; 56). The C-terminal part of the RAM peptide (A1778-D1790) containing the sumoylated K1780 (shown as a representative for sumoylation with cyan spheres), K1781 and K1782 was modeled, along with SUMO (green) and the E2-conjugating enzyme Ubc9 (wheat), using the crystal structure of sumoylated Ran GTPase-activating protein 1 (PDB 3UIP; 80) as a template for modeling
We generated a tetra-SUMO mutant (TSM) of GFP-Flag-NICD1, where the four conserved lysines in the RAM domain were mutated to arginine, to generate K1774/1780/1781/1782R. The TSM could not be sumoylated as efficiently as wild-type NICD1, demonstrating that the RAM domain is indeed a bona fide sumoylation site (Fig. 5a). Since stress-induced sumoylation increased the nuclear localization of NICD1 (Fig. 3a, b), we studied by immunofluorescence confocal microscopy how the TSM localizes in the cell. In response to heat shock and overexpressed SUMO, the nuclear accumulation of the TSM was significantly weaker compared to the wild type (Fig. 5b). Thereafter, Notch1 activity was analyzed using a 12×CSL luciferase reporter gene and, interestingly, the TSM displayed higher activity than the wild-type NICD1 (Fig. 5c), suggesting that sumoylation decreases Notch transcriptional activity. Next, we investigated whether the repressive effect of sumoylation also applies to target gene expression. Using quantitative RT-PCR analysis, we found that the expression of the classical Notch1 target genes Hes1, Hey1, and Hey2 upon heat stress decreased by SUMO. The results showed that addition of SUMO decreased the expression of each target gene regardless of the recovery time after heat shock (Figs. 5d–f). The TSM increased the expression levels of Hes1, Hey1, and Hey2, which further supports our finding that the RAM domain is a fundamental SUMO site and a key regulator of Notch1 transcriptional activity. All three target genes were most repressed by heat shock-induced sumoylation of wild-type NICD1. Since the TSM increased the expression of Hey1 and Hey2, but not Hes1 during heat stress, albeit all these target genes showed the same response to sumoylation (Figs. 5g–i), stress-induced repression of Hes1 is likely to involve additional mechanisms besides sumoylation of NICD1. In addition, Notch sumoylation clearly repressed the expression of the three mentioned Notch target genes during proteotoxic stress. This applied both to ligand-activated FLN in HEK 293 FLN cells (Fig. 5j) and overexpressed NICD in HeLa cells (Fig. 5k). As with heat shock (Figs. 5h–i) the TSM increased target gene expression during proteotoxic stress (Fig. 5k).
Fig. 5.
Sumoylation decreases Notch1 activity and represses the expression of Notch1 target genes. a K1774/1780/1781/1782R GFP-Flag-NICD1 (TSM) cannot be sumoylated as efficiently as wild-type GFP-Flag-NICD1 (WT). The sumoylated form of Notch1 is indicated with an arrow and the unmodified form with an asterisk. n = 3. b Wild-type NICD1 localizes more potently in the nucleus than TSM. COS7 cells were transfected with GFP-Flag-NICD1 (WT) or TSM, and some samples with His-SUMO1 (S1) as indicated. Images were taken with a Zeiss LSM510 META confocal microscope. c TSM increases the activity of the Notch1 signaling pathway. Notch1 activity was measured from HeLa cells by the 12×CSL luciferase reporter reflecting the activity of the Notch1 signaling pathway. The values shown are normalized luciferase units. n = 3. d–f Sumoylation (S1) decreases the expression of Notch (N1) target genes Hes1, Hey1, and Hey2 during heat shock. The relative expression of mRNA was analyzed by quantitative RT-PCR. n = 3. g–i Sumoylation (S1) decreases expression of Notch1 (WT) target genes Hes1, Hey1, and Hey1, whereas TSM increases the expression of Hes1, Hey1, and Hey2. The relative expression of mRNA was analyzed by quantitative RT-PCR. n = 3. j, k SUMO (S1) represses the expression of the indicated Notch target genes during proteotoxic stress in the form of Bortezomib (Brz). The relative expression of mRNA was analyzed by quantitative RT-PCR n = 3
To examine whether mutated NICD1 (NOTCH1-TSM) lacking the domains necessary for proper sumoylation retains a capacity to activate downstream target genes in vivo, we electroporated chick embryos with vectors either encoding wild-type NICD1 (NOTCH1-WT) or the mutant NOTCH1-TSM. When electroporated, both constructs inhibited the upregulation of the neuronal marker TUJ1 and induced expression of the Notch1 target gene Hes5 (Figs. 6a–d). However, when heat-shocked at 55 °C for 1 h prior to retrieval there was a pronounced reduction in RNA levels of Hes5 in the embryos electroporated with NOTCH1-WT (Figs. 6e, f). In contrast, in embryos electroporated with NOTCH1-TSM there was still a robust increase in Hes5 expression despite the heat shock (Fig. 6g, h). The effects of heat shock on Hes1 expression in NOTCH1-WT- and NOTCH1-TSM-electroporated embryos were not as pronounced as those on Hes5 (Supplementary Fig. 5), in line with the data on the effect of TSM on Hes1 expression during stress in vitro (Fig. 5g). To test whether also other stresses influence Notch sumoylation and target gene expression in vivo we treated NOTCH1-WT and NOTCH1-TSM electroporated embryos with Bortezomib to induce proteotoxic stress (Supplementary Fig. 6). Bortezomib reduced Hes5 in embryos expressing NOTCH1-WT (Supplementary Fig. 6). In embryos exposed to 50 μM Bortezomib there was a stronger reduction in Hes5 levels in NOTCH1-WT than in NOTCH1-TSM electroporated embryos (Supplementary Fig. 6). Taken together, both heat stress and proteotoxic stress affected Notch target gene expression in vivo through NICD sumoylation.
Fig. 6.
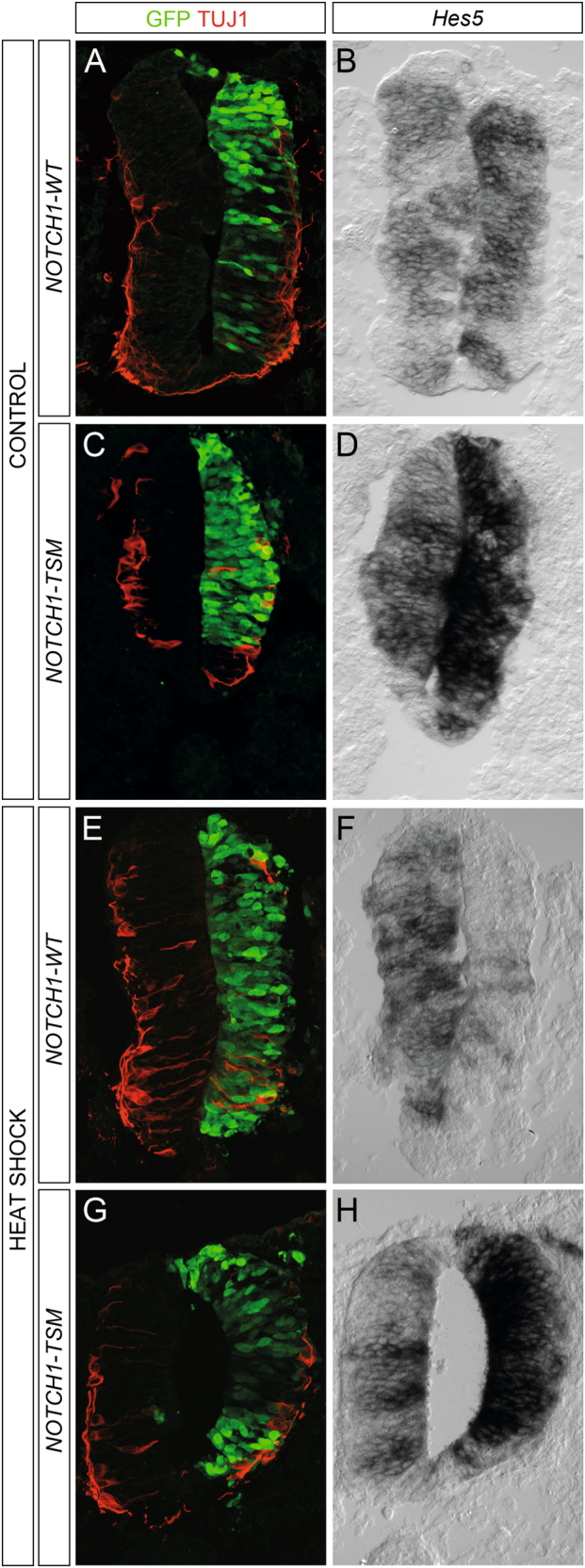
Sumoylation-resistant Notch1 mutant retains capacity to activate Hes5 after heat shock. a–d In embryos not exposed to heat shock (CONTROL) there is a reduction in TUJ1 immunoreactivity and an increase in Hes5 expression both when electroporated with NOTCH1-WT (a, b) and NOTCH1-TSM (c, d). n = 3. e–h In embryos exposed to heat shock for 1 h at 55 °C (HEAT SHOCK) there is a reduction in Hes5 levels in embryos electroporated with NOTCH1-WT (e, f), whereas NOTCH1-TSM electroporation induced high levels of the Hes5 transcript (g, h). n = 3
Sumoylated NICD1 interacts with the transcription factor CSL and the deacetylase HDAC4 to repress gene expression
To reveal by which mechanism sumoylation of NICD1 suppressed the activity of Notch1 (Fig. 5c) and expression of its target genes Hes1, Hey1, and Hey2 (Figs. 5d–k), we addressed the effect of sumoylation on the interaction between NICD1 and CSL. For this purpose, we performed a modified ChIP assay where we crosslinked NICD1 and CSL on DNA in order to maintain their interaction within the transcriptional complex on DNA (Fig. 7a). SUMO did not impair the NICD1–CSL–DNA interaction. This is in accordance with the three-dimensional model of the sumoylated Notch1 transcriptional complex, in which the K1780/K1781/K1782 are easily available for sumoylation, and neither SUMO nor the conjugating enzyme Ubc9 interferes with the NICD1–CSL–DNA interactions (Fig. 4, Supplementary Fig. 4). The modeled complex suggests that Ubc9 interacts with CSL, but the RAM peptide and K1780/K1781/K1782 mediate the majority of the interactions with SUMO and Ubc9. Therefore, we conclude that sumoylation mediates its repressive effect on Notch1 target genes mainly through some other mechanism(s) than by decreasing the interaction with CSL.
Fig. 7.
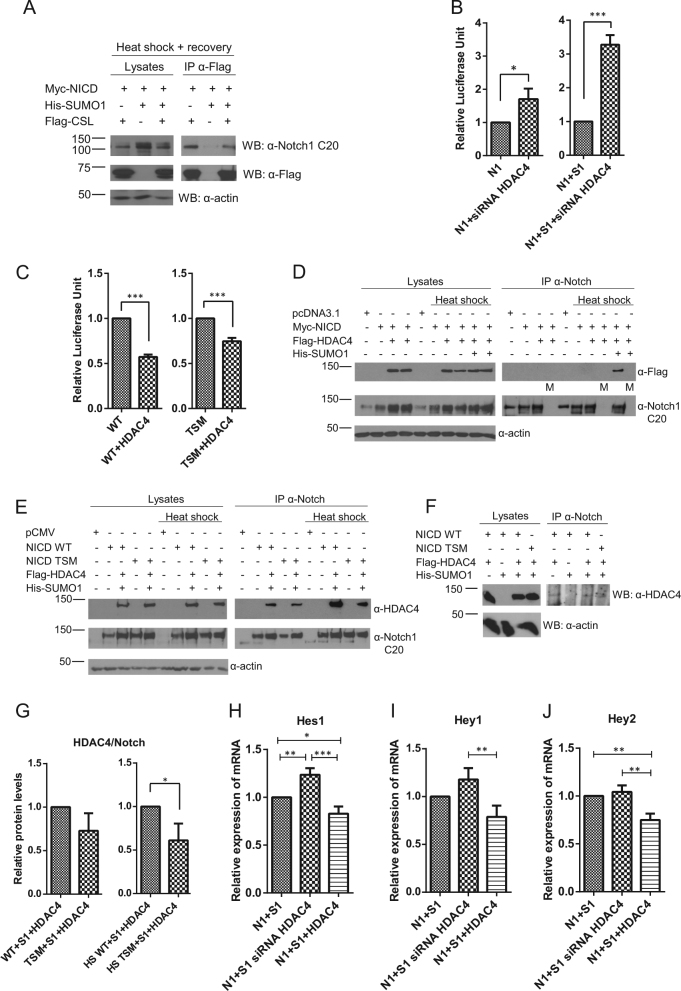
HDAC4 decreases Notch1 activity in the presence of SUMO. a Sumoylated Notch1 binds to CSL and remains bound to DNA. n = 4. b Blocking the expression of HDAC4 by siRNA increases Notch1 activity. Notch1 activity was measured from HeLa cells by the 12×CSL luciferase reporter reflecting the activity of the Notch1 signaling pathway. The values shown are normalized luciferase units. The difference in Notch1 activity is greater in samples with overexpressed SUMO1 (S1) n = 3. c HDAC4 represses the activity of wild-type GFP-Flag-NICD1 (WT) more than it represses the activity of the K1774/1780/1781/1782R GFP-Flag-NICD1 (TSM). n = 3. d SUMO increases Notch1–HDAC4 interaction. Control samples without antibody during IP are indicated with an “M” referring to mock. n = 3. e Consistent with the results in (c), K1774/1780/1781/1782R GFP-Flag-NICD1 (NICD TSM) decreases NICD1–HDAC4 interaction compared to wildtype NICD1 (NICD WT). f Notch interacts with HDAC4 on the DNA. n = 3. g The levels of HDAC4 as related to Notch in the immunoprecipitates in (e) were quantified. Values indicate the average of three independent experiments. n = 3. h–j Inhibition of HDAC4 expression by siRNA increases, and HDAC4 overexpression decreases the expression of Notch1 (N1) target genes Hes1, Hey1, and Hey2 in the presence of SUMO1 (S1). The relative expression of mRNA was analyzed by quantitative RT-PCR. n = 3
An important mechanism by which sumoylation mediates repression of gene expression is association of sumoylated proteins with HDACs [38]. HDACs negatively regulate transcription by causing local structural changes in the chromatin, which stems from HDACs deacetylating histones and in some cases by HDACs recruiting and modifying additional repressors or co-repressors [29]. Intriguingly, when screening for protein modifiers interacting with Notch under stress, we observed an interaction between HDAC4 and Notch (data not shown). To investigate a potential role of HDAC4 in the SUMO-mediated effect on Notch1 activity we downregulated HDAC4 with small interfering RNAs (siRNAs). Downregulation increased the stress-induced Notch1 activity and the increase was more pronounced in the presence of SUMO (Fig. 7b, right and left graph). We found that HDAC4 repressed the activity of wild-type Notch1 more than that of TSM in the presence of heat shock (Fig. 7c, left and right graph). We confirmed that HDAC4 interacted with NICD1 and demonstrated that the interaction was enhanced in the presence of SUMO and heat shock (Fig. 7d). We next analyzed the binding of the TSM to HDAC4 and observed a reduced interaction (Fig. 7e), demonstrating that sumoylation of NICD1 enhances the binding to HDAC4. To find out whether NICD1 and HDAC4 interact on the chromatin, we crosslinked the proteins on DNA in a modified ChIP assay, pulled down Notch, and immunoblotted the pulled down samples with an HDAC4-specific antibody, which showed the presence of HDAC4 (Fig. 7f).
Since HDAC4 lowered Notch activity in the presence of SUMO1 (Fig. 7b), HDAC4 may have a repressive role in the expression of specific Notch1 target genes as a consequence of NICD1 sumoylation. Thus, we performed overexpression and siRNA-mediated knockdown of HDAC4 and analyzed the expression of the Notch1 target genes Hes1, Hey1, and Hey2 in the presence of heat shock and SUMO. Silencing of HDAC4 activated Notch target genes, whereas overexpression of HDAC4 had an opposite effect (Figs. 7h–j). A model of the stress-inducible repression of Notch target gene expression and the regulatory mechanisms involved is presented in Fig. 8. Taken together, these results support the repressive role of HDAC4 in the regulation of Notch1 target genes as a result of NICD1 being conjugated by SUMO.
Fig. 8.
A model of the involvement of SUMO and HDAC4 in the repression of Notch1 target gene expression. In the absence of SUMO, NICD binds to CSL leading to the recruitment of MAML and consequent activation of specific Notch target genes. Recruitment of HDAC4 to the NICD/CSL/MAML transcriptional complex by sumoylated NICD promotes HDAC4-mediated transcriptional repression
Discussion
Here we show that stress-inducible conjugation of SUMO1 to NICD1 regulates Notch target gene expression. Specifically, sumoylation of NICD1 represses the expression of Notch target genes during stress conditions. Sumoylation of NICD1 does not interfere with the formation of the transcriptional complex, and sumoylated Notch1 interacts with CSL on chromatin. This finding is supported by our three-dimensional model of the Notch1 transcription complex demonstrating that K1780/K1781/K1782 are accessible for sumoylation, and neither SUMO nor the presence of the SUMO-conjugating enzyme Ubc9 interferes with the binding of NICD1 to CSL. The repressive effect of SUMO appears to be mediated by the recruitment of HDAC4 to the transcriptional complex, which is in line with several studies reporting that SUMO promotes HDAC-mediated transcriptional repression [29, 39–41]. In agreement, we show that wild-type Notch1 interacts more potently with HDAC4 than the TSM, and silencing of HDAC4 enhances Notch target gene expression. Sumoylation enhanced the nuclear levels of NICD1, but reduced its transcriptional activity. As sumoylation is highly dynamic and reversible, this provides a mechanism for transcriptional switching and Notch can be reactivated upon desumoylation. Notch activity is regulated at several levels of the pathway and PTMs have been shown to modulate receptor trafficking, proteolytic processing, NICD1 nuclear translocation, and stability [1, 2, 14, 16]. Our data reveal a new regulatory mechanism and show that NICD1 is sumoylated in the nucleus to control the activity of the Notch transcriptional complex.
It has previously been shown that sumoylation regulates other proteins in the Notch signaling pathway [42]. For example, MAML1 undergoes sumoylation, which causes enhanced interaction between MAML1 and HDAC7 leading to decreased MAML1 transcriptional activity, and oxidative stress-induced sumoylation of Hes1 represses the expression of GADD45α [39, 43]. Sumoylation has been shown to be a mechanism for coordinated regulation of signaling pathways. In some cases a protein group of the same pathway can be simultaneously multisumoylated, stabilizing the interaction between the modified proteins [44]. Furthermore, protein complexes may be held together by several SUMO–SUMO interacting motifs (SIMs) [45], and elimination of one SUMO site may thus have no major physiological consequences. Proteins engaged in interrelated complexes have been demonstrated to be subjects to synchronous SUMO2 modification in heat-shocked cells [46]. Since SUMO-mediated regulation operates through signaling network control, the physiological significance of sumoylation of a single target protein may become more difficult to substantiate [45]. Such network control is feasible in the regulation of the Notch1 pathway, given that both MAML1 in the NICD1 transcriptional complex and the target gene Hes1 are sumoylated [39, 42, 43]. Coordinated control of the activity of different components in the Notch pathway would constitute a dynamic, but robust cell-intrinsic system to regulate cellular responses to Notch activation.
The Hes family of repressors controlled by Notch signaling display cyclic expression patterns, mainly controlled through negative feedback, to coordinate cell fate decisions during development. Sumoylation has been implicated to influence cell fate specification during development [20], and sumoylation of NICD1 provides an additional regulatory mechanism for dynamic expression of Notch target genes. Since sumoylation is stress-inducible, modification of NICD1 by SUMO provides a cell autonomous mechanism for regulating Notch1 in response to the cellular context. We observed that stress reduced WT Notch-mediated expression of Hes5 in the developing chick CNS, but expression was not affected in cells expressing the mutated Notch, indicating that the stress-induced sumoylation of Notch represses target genes in vivo. Hence, sumoylation provides a mechanism for fine-tuning Notch-mediated cell functions in response to changes in the physiological state. Due to the reversible nature of sumoylation, SUMO conjugation allows for transcriptional switching at the level of chromatin without the requirement of reactivation of the pathway by interaction with ligand-expressing cells.
To date, only a few PTMs of Notch have been reported. Sumoylation presents a mechanism for cell autonomous, context-specific modulation of Notch target gene expression. Since dysregulation of Notch signaling is linked to a wide range of pathologies [2], mechanistic insight into SUMO-mediated Notch regulation provides a novel aspect of the pathway, which can be amendable to therapeutic intervention.
Materials and methods
Cell culture, transfection, immunoprecipitation, and immunoblotting
African green monkey COS7 cells (generous gift from J. Eriksson group, Turku Centre for Biotechnology), human cervical cancer HeLa cells (ATCC), and human HEK 293 cells expressing FLN (generous gift from U.Lendahl group, Karolinska Institut, Stockholm) were cultured in DMEM (Dulbecco’s Modified Eagle’s Medium, Sigma-Aldrich) supplemented with 10% fetal calf serum, 2 mM L-glutamine, 100 U/ml penicillin, and 100 μg/ml streptomycin. For HEK 293 FLN cells, 10 μg/ml puromycin was added to DMEM. The cells were grown at 37 °C in an atmosphere containing 5% CO2. Transfection was performed by resuspending 10 μg of plasmid DNA, and HeLa or COS7 or HEK 293 FLN cells in 400 μl Opti-MEM (Invitrogen) in BTX cuvettes. Cells were subjected to electroporation at 220 V and 975 μF (Gene Pulser Transfection Apparatus, Bio-Rad). Prior to treatments, cells were left to recover for 24 h. For blocking the expression of HDAC4 by siRNA, Genesolution siRNA pools for HDAC4 (cat.no. 1027416) and control siRNAs (cat.no. 1027415) were purchased from Qiagen. The siRNAs were prepared according to the instructions of the manufacturer. HeLa cells were separately transfected with siRNAs by using Lipofectamine 2000 (Invitrogen). The cells were then cultured for 48–72 h prior to transfection of vector plasmids into the cells followed by harvesting of the cells for luciferase assay or quantitative RT-PCR. The TSM K1774/1780/1781/1782R was generated from human wild-type GFP-Flag-NICD1 by using the QuikChange® Site-Directed Mutagenesis kit (Stratagene) and sequenced to ensure successful mutagenesis.
The immunoprecipitations were performed on transfected and treated HeLa or COS7 cells. For harvesting, cells were washed twice with cold PBS (phosphate-buffered saline). For immunoprecipitating polyhistidine-tagged proteins, Talon magnetic beads (Clontech) were used. The cells were resuspended in 1% SDS (sodium dodecyl sulfate) in PBS, and the lysates were boiled for 5 min. A concentration of 8 M urea/PBS/20 mM N-Ethylmaleimide (NEM, Sigma-Aldrich) was added into each sample. The DNA was sheared into smaller fragments by sonication, which was followed by a centrifugation for 10 min at 15,000g. Lysates were taken from the supernatants, which were incubated with 40 μl of washed Talon magnetic beads under rotation for 1 h at 4 °C, and beads were washed six times with 8 M urea/PBS/850 mM NaCl and once with 8 M urea/PBS, and eluted with 20 μl of 200 mM imidazole/8 M urea/PBS. Proteins were run on an 8% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane (Protran nitrocellulose, Schleicher & Shuell), and immunoblottted with α-GFP (Invitrogen) or α-cleaved Notch1 (Val-1744, Cell Signaling). α-HSC70 (Stressgen) was used as a loading control.
For immunoprecipitating Myc-NICD1-, GFP-Flag-NICD1-, or the TSM-transfected and -treated (either control treatment or 1 h heat shock at 42 °C water bath + 1 h recovery at 37 °C) HeLa cells were resuspended in 1% SDS in PBS, and the lysates were boiled for 5 min. Triton X-100 (1%)/20 mM NEM in PBS was added into each sample, and the samples were sonicated followed by a centrifugation for 10 min at 15,000g. Lysates were taken from the supernatants, which were incubated with either Flag M2 beads (Sigma-Aldrich), or with 40 μl of protein G-sepharose (GE Healthcare) and 3 μl of α-Notch1 C20 (Santa Cruz Biotechnology) under rotation for 1 h at 4 °C. The beads were washed six times with 1% Triton X-100/20 mM NEM in PBS, and the proteins were eluted with 3× Laemmli sample buffer sample buffer. The proteins were run on an 8% SDS-polyacrylamide gel, and immunoblottted with α-SUMO1 (Zymed Laboratories), α-SUMO2/3 (Zymed Laboratories), α-Notch1 C20 (Santa Cruz Biotechnology), Flag (Sigma-Aldrich), or HDAC4 (Abcam). α-actin (Cell Signaling) was used as a loading control.
To study Notch sumoylation in HEK 293 FLN cells in which FLN was activated with Notch-specific ligands, cell culture plates were coated with Protein G (Invitrogen) 50 μg/ml in PBS at room temperature overnight. The plates were washed three times with PBS and blocked with 10 mg/ml of bovine serum albumin (BSA) at PBS for 1 h at room temperature. The plates were washed three times with PBS and incubated with recombinant Jagged1-FC (R&D Systems) with a final concentration of 0.5 μg/ml in 0.1% BSA/PBS for 2 h at room temperature. During the last incubation, HEK 293 FLN cells were transfected. After incubation, the plates were washed three times with PBS and once with DMEM, and immediately plated with the transfected cells. Cells in which Notch target gene expression was studied were treated 24 h after transfection as described in section (Quantitative RT-PCR). To study whether ligand-activated FLN can be sumoylated as NICD1 in HEK 293 FLN cells, the plates were 24 h after transfection left untreated or treated with 1 h heat shock at 42 °C water bath + 1 h recovery at 37 °C), and the lysates were collected with 3× Laemmli sample buffer. The proteins were run on an SDS-polyacrylamide gel and immunoblottted with α-Notch1 C20 (Santa Cruz Biotechnology). α-actin (Cell Signaling) was used as a loading control.
To study the effect of Bortezomib-induced sumoylation, HEK 293 FLN cells or HeLa cells were 24 h after transfection exposed to Bortezomib in dimethyl sulfoxide (DMSO) at a final concentration of 100 nm for 8 h at 37 °C, and in Fig. 2f also for 16 h. The HeLa cells were either lysed in 3× Laemmli sample buffer and analyzed by immunoblotting, or treated as described in section (Quantitative RT-PCR).
To study desumoylation of Notch, transfected COS7 cells were heat-shocked for 1 h at 42 °C followed by a recovery for 1 h at 37 °C. The cells were lysed in 3× Laemmli sample buffer and the proteins were analyzed by western blotting with an antibody targeted against GFP.
To study whether sumoylation enhances the levels of Notch, COS7 cells were transfected with GFP-Flag-NICD and some samples in addition with His-SUMO1. Twenty-four hours after transfection, the cells were control-treated or treated with the protein translation inhibitor cycloheximide (Sigma-Aldrich) at a final concentration of 10 µg/ml in ethanol for 0.5, 1, 2, 4, or 6 h. The cells were left untreated or heat-shocked for 1 h at 42 °C followed by a recovery for 1 h at 37 °C, and the samples were lysed in 3× Laemmli sample buffer and immunoblotted with α-GFP (Invitrogen). To study Notch sumoylation in a SUMO dose-dependent manner, increasing amounts of SUMO was transfected into COS7 cells. After 24 h, for heat shocking the plates were kept for 1 h at 42 °C water bath followed by a 1-h recovery at 37 °C. The cells were resuspended in 1% SDS in PBS and the lysates were boiled for 5 min. Triton X-100 (1%)/20 mM NEM in PBS was added into each sample and the samples were sonicated followed by a centrifugation for 10 min at 15,000g. Lysates were taken from the supernatants, and the supernatants were incubated with 40 μl of protein G-sepharose (GE Healthcare) and 3 μl of α-Notch1 C20 (Santa Cruz Biotechnology) under rotation for 1 h at 4 °C. The beads were washed six times with 1% Triton X-100/20 mM NEM in PBS and the proteins were eluted with 3× Laemmli sample buffer. The proteins were run on an 8% SDS-polyacrylamide gel and immunoblotted with α-Notch1 C20 (Santa Cruz Biotechnology).
Immunocytochemistry and confocal microscopy
Transfected COS7 cells were plated on coverslips and left to recover for 24 h. Cells were left untreated or heat-shocked for 1 h at 42 °C followed by a 1-h recovery at 37 °C. Cells were fixed in 3% PFA, washed with PBS, permeabilized for 5 min in 0.1% Triton X-100 in PBS, blocked for 1 h with 3% BSA/0.05% Triton X-100 in PBS, and incubated overnight with primary antibody rabbit α-Notch1 C20 (Santa Cruz Biotechnology), which had been diluted 1:200 in blocking solution. The cells were washed three times with PBS, and incubated for 1 h with secondary antibody donkey α-rabbit Alexa 488 (Molecular Probes), which had been diluted 1:1000 in blocking solution. The cells were washed three times with PBS, and for visualization of nuclei the cells were incubated for 2 min with 0.5 mg/ml DAPI in 0.1 M PBS and mounted to slides with ProLong Gold mounting medium (Invitrogen). Immunostaining was visualized with a Zeiss LSM510 META confocal microscope. Cells transfected with GFP-Flag-NICD1 WT or the TSM were visualized by GFP. Values shown in the figures are statistically significant at ***P < 0.001 or **P < 0.01, and are averages from three independent experiments. Hundred cells in average were counted for each sample.
Extraction of nuclear and cytoplasmic fractions
For extracting nuclear fractions of transfected and treated (1 h heat shock at 42 °C + 1 h recovery at 37 °C) COS7 cells, 1–1.5×107 cells were resuspended in buffer I (10 mM Tris pH 7.4, 10 mM NaCl, 3 mM MgCl2, 1× Complete Mini EDTA-free Protease Inhibitor Cocktail Tablets, 2 mM dithiothreitol (DTT)) and incubated on ice for 10 min. For breaking the nuclear membrane, 10% NP40 was added to a final concentration of 0.2%. The tube was flipped 10 times to achieve homogenization. The sample was incubated for 1 min on ice and centrifuged for 4 min at 400g, and the pellet was resuspended in buffer I. The sample was centrifuged for 4 min at 400g, and the pellet was resuspended in buffer II (20 mM Tris pH 7.4, 40 mM Na4P2O7, 5 mM MgCl2, 50 mM NaF, 100 mM Na3VO4, 10 mM EDTA, 1% Triton, and 1% SDS). NP40 (10%) was added to each sample, incubated 10 min on ice, and centrifuged for 10 min at 20,000g. Laemmli sample buffer (3×) was added to the collected supernatant and the samples were boiled for 5 min.
For extracting cytoplasmic fractions, COS7 cells were resuspended in buffer I and incubated on ice for 10 min. For breaking the nuclear membrane, 10% NP40 was added to a final concentration of 0.2%. The tube was flipped 10 times to achieve homogenization. The sample was incubated for 2 min on ice and centrifuged for 2 min at 10,000g. For elimination of nuclear residue, the supernatant was centrifuged for 2 min at 10,000g. Laemmli sample buffer (3×) was added to the collected supernatant and the samples were boiled for 5 min. The proteins from nuclear and cytoplasmic extracts were separated in SDS-PAGE and immunoblotted with α-GFP (Invitrogen) and α-Notch1 C20 (Santa Cruz Biotechnology). α-HSC70 (Stressgen) was used as a loading control.
Quantitative RT-PCR
The RNeasy kit (Qiagen) was used to isolate RNA from transfected and control-treated/heat-shocked/Bortezomib-treated HeLa cells or HEK 293 FLN cells. DNase I treatment of 1 μg of each RNA sample was carried out by using RQ1 DNase (Promega M610A). The DNase-treated samples were reverse-transcribed to cDNA by using the M-MLV Reverse transcriptase protocol (Promega). Kapa Probe Fast qPCR Master Mix (Kapa Biosystems) was utilized and Taqman quantitative RT-PCR was performed with 7900HT Fast Real-Time PCR System (Applied Biosystems). Primers for human Hes1, Hey1, and Hey2 were designed by using Universal Probe Library Assay Design Center (Roche Applied Biosciences). The primers for Hes1, Hey1, and Hey2 were purchased from Oligomer and were as follows: Hes1 forward 5′-GTGAAGCACCTCCGGAAC-3′, Hes1 reverse 5′-GTCACCTCGTTCATGCACTC-3′, Hey1 forward 5′-CGAGCTGGACGAGACCAT-3′, Hey1 reverse 5′-GAGCCGAACTCAAGTTTCCA-3′, Hey2 forward 5′-CCCGCCCTTGTCAGTATC-3′, and Hey2 reverse 5′-TTGTTTGTTCCACTGCTGGT-3′. Taqman probes from Roche were used, no. 60 for Hes1, no. 39 for Hey1, and no. 73 for Hey2. Primers and probes for human GAPDH have been described earlier [47]. Relative quantities of Hes1, Hey1, and Hey2 transcripts were normalized against their own GADPH. Experiments were repeated at least three times. The results were analyzed with SDS 2.3 and RQ manager software (Applied Biosystems) and statistics was made with GraphPad Prism 5. Values shown in the figures are statistically significant at ***P < 0.001, **P < 0.01, or *P < 0.05 as indicated. Values indicate the average of three independent experiments.
Notch activity assay
Transfected and heat-shocked HeLa cells were washed with PBS and lysed with passive lysis buffer (Promega), which had been diluted to 1× with mq H2O. For measuring luciferase activity from cells expressing the 12×CSL luciferase reporter gene, 2 µl of sample was mixed with 18 µl of passive lysis buffer, and the luciferase activity was measured from triplicates with a luminometer (Thermo Scientific). The 12×CSL luciferase reporter has been described earlier [48, 49]. For measuring β-galactosidase activity, a mixture of 10 µl of sample and 240 µl of ONPG (ortho-nitrophenyl-β-galactoside) buffer was incubated for 30 min at 37 °C, and the absorbance was measured from triplicates with a Multiskan Ascent photometer (Thermo Scientific) at 420 nm. Notch activity was calculated by relating the luciferase expression levels to the expression levels of β-galactosidase. To verify equal expression levels of Notch, lysates for immunoblotting were taken prior to adding passive lysis buffer. The lysates were immunoblotted against Notch with α-Notch1 C20 (Santa Cruz Technology), and α-actin (Cell Signaling) was used as a loading control. Values shown in the figures are statistically significant at ***P < 0.001 or *P < 0.05 as indicated. Values indicate the average of at three independent experiments.
Chromatin immunoprecipitation
For studying the impact of sumoylation on the interaction between Notch and CSL, or Notch and HDAC4, transfected COS7 cells were heat-shocked for 1 h at 42 °C followed by a recovery for 1 h at 37 °C. For crosslinking of protein–DNA interactions, 1 ml of 11% formaldehyde containing 100 M NaCl, 1 mM EDTA, 0.5 mM EGTA, and 50 mM HEPES was added into the cell plate containing 10 ml of DMEM. The plates were rotated for 5 min after which 660 μl of 2.5 M glycine were added dropwise and the plates were rotated for another 5 min. The formaldehyde was removed and the plates were washed once with cold PBS inside a hood. The cells were harvested with 5 ml of PBS into polystyrene falcon tubes in which the cells were washed twice by centrifugation for 5 min, 1000g at 4 °C. Two milliliters of Joost’s lysis buffer (1% SDS, 10 mM EDTA pH 8, 50 mM Tris-HCL pH 8)/1× protease inhibitor (Roche) was added/sample and let to stand for 10 min. The DNA was sheared into smaller fragments with a Bioruptor sonicator. Lysates were taken, and 700 μl of sample was mixed with 700 μl of ChIP buffer (150 mM NaCl, 20 mM Tris-HCl pH 8, 1% Triton X-100)/1× protease inhibitor (Roche), and was incubated under rotation for 1 h at 4 °C with 20 μl of Flag M2 beads (Sigma-Aldrich) for studying Notch-CSL interaction, or 40 μl of protein G-sepharose (GE Healthcare) and 3 μl of α-Notch1 C20 (Santa Cruz Biotechnology) for studying Notch–HDAC4 interaction. The samples were centrifuged for 1 min, 2000g at 4 °C. The beads were washed twice with wash buffer 1 (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 150 mM NaCl), twice with wash buffer 2 (0.1% SDS, 1% Triton X-100, 2 mM EDTA pH 8, 500 mM NaCl, 20 mM Tris-HCl pH 8), and twice with wash buffer 3 (2 mM EDTA pH 8, 20 mM Tris-HCl pH 8, 10% glycerol), and the proteins were eluted with 3× Laemmli sample buffer. The proteins were run on an 8% SDS-polyacrylamide gel and immunoblottted with α-Notch1 C20 (Santa Cruz Biotechnology), α-Flag (Sigma-Aldrich), or α-HDAC4 (Abcam). α-actin (Cell Signaling) was used as a loading control.
In ovo electroporation
Fertilized chicken eggs were incubated at 38 °C for 42–45 h to reach stage HH11. Albumen (5 ml) was withdrawn with a syringe and the top of the shell was opened in order to expose the embryo. NICD1-WT and NICD1-TSM (K1774/1780/1781/1782R), previously subcloned from a pCMV vector into a pCIG vector, were injected in the neural tube using pulled glass microcapillaries (1.5 μg/μl in PBS; 1 mM MgCl2; fast green). Five electric pulses of 21 V for 50 ms each were discharged at intervals of 1 s. The opening of the eggs was then taped in order to avoid the embryos to dry. Eggs were incubated at 38 °C for 16 h. Heat shock: control eggs were incubated for one more hour at 38 °C, while heat-shocked eggs were incubated at 55 °C for 1 h and then dissected in PBS. Bortezomib treatment: 20 h after electroporation, the eggs were opened again in order to check survival and to have access to the developing embryo. Bortezomib (5 or 50 μM) were injected in the amniotic cavity where the developing embryo lays. The eggs were then taped again to avoid the embryos from drying and incubated for two more hours at 38 °C. After the stress treatment, embryos were retrieved and dissected in PBS. After dissection, neural tubes were incubated in 4% paraformaldehyde (PFA) for 1 h. After washing with PBS, neural tubes were transferred in 30% sucrose solution overnight and then cryosectioned (10 μm) onto Superfrost Plus slides (Thermo Scientific). Immunohistochemistry was performed with Tuj1 (MMS-435P, Covance; 1:2000) and GFP (ab6673, Abcam; 1:500) antibodies. In situ hybridization was performed as described [50] using chick probes for Hes1 and Hes5, cDNA encoding Hes1 and Hes5 was obtained from MRC Geneservice (chEST356J15 and chEST382I21).
Structural modeling of sumoylated Notch1 transcription complex
The crystal structure of the Notch1 transcription complex (PDB 3V79) [37] was used as a basis for modeling the sumoylation of NICD1 into the complex. According to the amino-acid sequence of human NICD1 (UniProtKB P46531), the crystallized RAM peptide (residues K1759 – E1777) ends three amino acids before the sumoylated K1780. Because of the lack of sumoylated homologs with known structure, we used the crystal structure of sumoylated Ran GTPase-activating protein 1 (PDB 3UIP) [51] as a template to model the sumoylated loop (A1778 – D1790) of the RAM peptide in complex with SUMO1 and Ubc9. In the BODIL modeling environment [52], the last helix before the sumoylated K524 in Ran GTPase-activating protein 1 was first manually superimposed on top of the last helix of the crystallized RAM peptide in the Notch 1 transcription complex. Thereafter, the coordinates for the RAM peptide were saved together with the superimposed coordinates of the sumoylated loop (residues G521 – N524) of Ran GTPase-activating protein 1, SUMO1, and Ubc9. On the basis of these superimposed coordinates, amino acids A1778 – D1790 in NICD1 were modeled in complex with SUMO1 and Ubc9 with the program MODELLER [53]. Hence, the resulting model of the sumoylated Notch1 transcription complex is a hybrid between the crystallized structure of the complex and a modeled structure of the sumoylated loop (A1778 – D1790), SUMO1 and Ubc9. Secondary structure prediction was performed with Jpred [54], sequence alignments with the program MALIGN [55] in the BODIL modeling environment [52], and pictures were prepared with PyMOL (version 1.6; Schrödinger, LLC).
Statistical analyses
Cell culture and chicken egg experiments were repeated three or more times. One-way ANOVA with Bonferronis’ post hoc test or two-tailed Student’s t-test (GraphPad Prism Software, CA, USA) were used to determine the statistical difference. p values of *p < 0.05, **p < 0.01, ***p < 0.001 were considered as significant with 95% confidence interval.
Electronic supplementary material
Acknowledgments
This work was supported by the Academy of Finland grant 218062 (C.S.) and Sigrid Juselius Foundation (C.S., L.S., T.A.S., and K.M.D.) and Cancer Society of Finland (C.S. and L.S.). We are grateful to Helena Saarento for technical assistance, the cell imaging core at the Turku Center of Biotechnology is thanked for access to infrastructure. Use of Biocenter Finland infrastructure at Åbo Akademi University (bioinformatics, structural biology, and translational activities) is acknowledged, along with the National Doctoral Programme in Informational and Structural Biology.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing financial interest.
Footnotes
Edited by D. Aberdam
Electronic supplementary material
The online version of this article (10.1038/s41418-017-0002-6) contains supplementary material, which is available to authorized users.
References
- 1.Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- 2.Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov. 2014;13:357–78. doi: 10.1038/nrd4252. [DOI] [PubMed] [Google Scholar]
- 3.Main H, et al. Interactions between Notch- and hypoxia-induced transcriptomes in embryonic stem cells. Exp Cell Res. 2010;316:1610–24. doi: 10.1016/j.yexcr.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 4.Gustafsson MV, et al. Hypoxia requires notch signaling to maintain the undifferentiated cell state. Dev PNAS Cell. 2005;9:617–28. doi: 10.1016/j.devcel.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 5.Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci USA. 2008;105:6392–7. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao L, et al. Notch-1 signaling regulates microglia activation via NF-ÎoB pathway after hypoxic exposure in vivo and in vitro. PLoS ONE. 2013;8:e78439. doi: 10.1371/journal.pone.0078439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frías A, et al. A switch in Akt isoforms is required for notch-induced snail1 expression and protection from cell death. Mol Cell Biol. 2016;36:923–40. doi: 10.1128/MCB.01074-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh P, et al. In vivo mapping of notch pathway activity in normal and stress hematopoiesis. Cell Stem Cell. 2013;13:190–204. doi: 10.1016/j.stem.2013.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pézeron G, Millen K, Boukhatmi H, Bray S. Notch directly regulates the cell morphogenesis genes Reck, talin and trio in adult muscle progenitors. J Cell Sci. 2014;127:4634–44. doi: 10.1242/jcs.151787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischer A, Gessler M. Delta-notch--and then? Protein interactions and proposed modes of repression by Hes and Hey bHLH factors. Nucleic Acids Res. 2007;35:4583–96. doi: 10.1093/nar/gkm477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bray S, Bernard F. Notch targets and their regulation. Curr Top Dev Biol. 2010;92:253–75. doi: 10.1016/S0070-2153(10)92008-5. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Zang C, Liu XS, Aster JC. The role of notch receptors in transcriptional regulation. J Cell Physiol. 2015;230:982–8. doi: 10.1002/jcp.24872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kopan R, Ilagan MXG. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hein K, et al. Site-specific methylation of Notch1 controls the amplitude and duration of the Notch1 response. Sci Signal. 2015;8:ra30. doi: 10.1126/scisignal.2005892. [DOI] [PubMed] [Google Scholar]
- 15.Santio NM, et al. Phosphorylation of Notch1 by Pim kinases promotes oncogenic signaling in breast and prostate cancer cells. Oncotarget. 2016 doi: 10.18632/oncotarget.9215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng X, et al. Interaction with factor inhibiting HIF-1 defines an additional mode of cross-coupling between the Notch and hypoxia signaling pathways. Proc Natl Acad Sci USA. 2008;105:3368–73. doi: 10.1073/pnas.0711591105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guarani V, et al. Acetylation-dependent regulation of endothelial Notch signalling by the SIRT1 deacetylase. Nature. 2011;473:234–8. doi: 10.1038/nature09917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Licciardello MP, et al. NOTCH1 activation in breast cancer confers sensitivity to inhibition of SUMOylation. Oncogene. 2014 doi: 10.1038/onc.2014.319. [DOI] [PubMed] [Google Scholar]
- 19.Droescher M, Chaugule VK, Pichler A. SUMO rules: regulatory concepts and their implication in neurologic functions. Neuromol Med. 2013;15:639–60. doi: 10.1007/s12017-013-8258-6. [DOI] [PubMed] [Google Scholar]
- 20.Lomelí H, Vázquez M. Emerging roles of the SUMO pathway in development. Cell Mol Life Sci. 2011;68:4045–64. doi: 10.1007/s00018-011-0792-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watts FZ. Starting and stopping SUMOylation. What regulates the regulator? Chromosoma. 2013;122:451–63. doi: 10.1007/s00412-013-0422-0. [DOI] [PubMed] [Google Scholar]
- 22.Hwang J, Kalejta RF. In vivo analysis of protein sumoylation induced by a viral protein: detection of HCMV pp71-induced Daxx sumoylation. Methods. 2011;55:160–5. doi: 10.1016/j.ymeth.2011.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sampson DA, Wang M, Matunis MJ. The small ubiquitin-like modifier-1 (SUMO-1) consensus sequence mediates Ubc9 binding and is essential for SUMO-1 modification. J Biol Chem. 2001;276:21664–9. doi: 10.1074/jbc.M100006200. [DOI] [PubMed] [Google Scholar]
- 24.Yang X-J, Chiang C-M. Sumoylation in gene regulation, human disease, and therapeutic action. F1000Prime Rep. 2013;5:45. doi: 10.12703/P5-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henley JM, Craig TJ, Wilkinson KA. Neuronal SUMOylation: mechanisms, physiology, and roles in neuronal dysfunction. Physiol Rev. 2014;94:1249–85. doi: 10.1152/physrev.00008.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abend A, Kehat I. Histone deacetylases as therapeutic targets - from cancer to cardiac disease. Pharmacol Ther. 2015;147:55–62. doi: 10.1016/j.pharmthera.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 27.Foti SB, Chou A, Moll AD, Roskams AJ. HDAC inhibitors dysregulate neural stem cell activity in the postnatal mouse brain. Int J Dev Neurosci. 2013;31:434–47. doi: 10.1016/j.ijdevneu.2013.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Sando R, et al. HDAC4 governs a transcriptional program essential for synaptic plasticity and memory. Cell. 2012;151:821–34. doi: 10.1016/j.cell.2012.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang W-L, Lee Y-C, Yang W-M, Chang W-C, Wang J-M. Sumoylation of LAP1 is involved in the HDAC4-mediated repression of COX-2 transcription. Nucleic Acids Res. 2008;36:6066–79. doi: 10.1093/nar/gkn607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blomster HA, et al. Novel proteomics strategy brings insight into the prevalence of SUMO-2 target sites. Mol Cell Proteomics. 2009;8:1382–90. doi: 10.1074/mcp.M800551-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo C, Henley JM. Wrestling with stress: roles of protein SUMOylation and deSUMOylation in cell stress response. IUBMB Life. 2014;66:71–7. doi: 10.1002/iub.1244. [DOI] [PubMed] [Google Scholar]
- 32.Gareau JR, Lima CD. The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Mol Cell Biol. 2010;11:861–71. doi: 10.1038/nrm3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi Y, Iwase M, Strunnikov AV, Kikuchi Y. Cytoplasmic sumoylation by PIAS-type Siz1-SUMO ligase. Cell Cycle. 2008;7:1738–44. doi: 10.4161/cc.7.12.6156. [DOI] [PubMed] [Google Scholar]
- 34.Sekhri P, Tao T, Kaplan F, Zhang X-D. Characterization of amino acid residues within the N-terminal region of Ubc9 that play a role in Ubc9 nuclear localization. Biochem Biophys Res Commun. 2015;458:128–33. doi: 10.1016/j.bbrc.2015.01.081. [DOI] [PubMed] [Google Scholar]
- 35.Kaminsky R, et al. SUMO regulates the assembly and function of a cytoplasmic intermediate filament protein in C. elegans. Dev Cell. 2009;17:724–35. doi: 10.1016/j.devcel.2009.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947–56. doi: 10.1038/nrm2293. [DOI] [PubMed] [Google Scholar]
- 37.Choi SH, et al. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–9. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia-Dominguez M, Reyes JC. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta. 2009;1789:451–9. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 39.Lindberg MJ, Popko-Scibor AE, Hansson ML, Wallberg AE. SUMO modification regulates the transcriptional activity of MAML1. FASEB J. 2010;24:2396–404. doi: 10.1096/fj.09-149401. [DOI] [PubMed] [Google Scholar]
- 40.Yang S-H, Sharrocks AD. SUMO promotes HDAC-mediated transcriptional repression. Mol Cell. 2004;13:611–7. doi: 10.1016/S1097-2765(04)00060-7. [DOI] [PubMed] [Google Scholar]
- 41.Girdwood D, et al. P300 transcriptional repression is mediated by SUMO modification. Mol Cell. 2003;11:1043–54. doi: 10.1016/S1097-2765(03)00141-2. [DOI] [PubMed] [Google Scholar]
- 42.Poulin G, Dong Y, Fraser AG, Hopper NA, Ahringer J. Chromatin regulation and sumoylation in the inhibition of Ras-induced vulval development in Caenorhabditis elegans. EMBO J. 2005;24:2613–23. doi: 10.1038/sj.emboj.7600726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chiou H-YC, Liu S-Y, Lin C-H, Lee EH. Hes-1 SUMOylation by protein inhibitor of activated STAT1 enhances the suppressing effect of Hes-1 on GADD45α expression to increase cell survival. J Biomed Sci. 2014;21:53. doi: 10.1186/1423-0127-21-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Psakhye I, Jentsch S. Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell. 2012;151:807–20. doi: 10.1016/j.cell.2012.10.021. [DOI] [PubMed] [Google Scholar]
- 45.Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–86. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- 46.Tammsalu T, et al. Proteome-wide identification of SUMO2 modification sites. Sci Signal. 2014;7:rs2. doi: 10.1126/scisignal.2005146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ostling P, Björk JK, Roos-Mattjus P, Mezger V, Sistonen L. Heat shock factor 2 (HSF2) contributes to inducible expression of hsp genes through interplay with HSF1. J Biol Chem. 2007;282:7077–86. doi: 10.1074/jbc.M607556200. [DOI] [PubMed] [Google Scholar]
- 48.Jarriault S, et al. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 49.Kato H, et al. Involvement of RBP-J in biological functions of mouse Notch1 and its derivatives. Development. 1997;124:4133–41. doi: 10.1242/dev.124.20.4133. [DOI] [PubMed] [Google Scholar]
- 50.Tsuchida T, et al. Topographic organization of embryonic motor neurons defined by expression of LIM homeobox genes. Cell. 1994;79:957–70. doi: 10.1016/0092-8674(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 51.Gareau JR, Reverter D, Lima CD. Determinants of small ubiquitin-like modifier 1 (SUMO1) protein specificity, E3 ligase, and SUMO-RanGAP1 binding activities of nucleoporin RanBP2. J Biol Chem. 2012;287:4740–51. doi: 10.1074/jbc.M111.321141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehtonen JV, et al. BODIL: a molecular modeling environment for structure-function analysis and drug design. J Comput Aided Mol Des. 2004;18:401–19. doi: 10.1007/s10822-004-3752-4. [DOI] [PubMed] [Google Scholar]
- 53.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 54.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:197–201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson MS, May AC, Rodionov MA, Overington JP. Discrimination of common protein folds: application of protein structure to sequence/structure comparisons. Methods Enzymol. 1996;266:575–98. doi: 10.1016/S0076-6879(96)66036-4. [DOI] [PubMed] [Google Scholar]
- 56.Giaimo BD, Oswald F, Borggrefe T. Dynamic chromatin regulation at Notch target genes. Transcription. 2016;8:61-66 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.