Abstract
Although amyotrophic lateral sclerosis (ALS), also known as Lou Gehrig’s disease, was first described in 1874, a flurry of genetic discoveries in the last 10 years has markedly increased our understanding of this disease. These findings have not only enhanced our knowledge of mechanisms leading to ALS, but also have revealed that ALS shares many genetic causes with another neurodegenerative disease, frontotemporal lobar dementia (FTLD). In this review, we survey how recent genetic studies have bridged our mechanistic understanding of these two related diseases and how the genetics behind ALS and FTLD point to complex disorders, implicating non-neuronal cell types in disease pathophysiology. The involvement of non-neuronal cell types is consistent with a non-cell autonomous component in these diseases. This is further supported by studies that identified a critical role of immune-associated genes within ALS/FTLD and other neurodegenerative disorders. The molecular functions of these genes support an emerging concept that various non-autonomous functions are involved in neurodegeneration. Further insights into such a mechanism(s) will ultimately lead to a better understanding of potential routes of therapeutic intervention.
Facts
ALS and FTLD are severe neurodegenerative disorders on the same disease spectrum.
Multiple cellular processes including dysregulation of RNA homeostasis, imbalance of proteostasis, contribute to ALS/FTLD pathogenesis.
Aberrant function in non-neuronal cell types, including microglia, contributes to ALS/FTLD.
Strong neuroimmune and neuroinflammatory components are associated with ALS/FTLD patients.
Open Questions
Why can patients with similar mutations have different disease manifestations, i.e., why do C9ORF72 mutations lead to motor neuron loss in some patients while others exhibit loss of neurons in the frontotemporal lobe?
Do ALS causal mutations result in microglial dysfunction and contribute to ALS/FTLD pathology? How do microglia normally act to mitigate neurodegeneration in ALS/FTLD?
To what extent do cellular signaling pathways mediate non-cell autonomous communications between distinct central nervous system (CNS) cell types during disease?
Is it possible to therapeutically target specific cell types in the CNS?
Introduction
Amyotrophic lateral sclerosis (ALS) is a neurodegenerative disorder affecting motor neurons [1], with an incidence of 1–2/100,000 per year [2]. Mean survival time is 3–5 years after onset, with no current cure [3]. Riluzole and Radicava are the two ALS therapies currently available, each displaying a modest impact on disease outcome [4–6]. In contrast to ALS, frontotemporal lobar degeneration (FTLD) pathology, is characterized by degeneration of the frontal and temporal lobes. FTLD is a major cause of early-onset (<65 years) dementia [7]. FTLD prevalence is somewhat higher than ALS and varies among different studies, ranging from 4–15 per 100,000 [8, 9]. Unlike ALS, there are no approved disease-modifying drugs; patients are treated with drugs that mitigate symptoms. These two diseases were once considered distinct disorders, as ALS patients typical present with loss of voluntary motor strength while FTLD patients exhibit behavioral and/or speech impairments. However, because of an increased understanding of the genetics underlying ALS and FTLD, they are now considered to be part of a continuum. Several lines of evidence support this notion: different individuals within the same family may present with either ALS or FTLD, or a single affected individual may bear symptoms of both syndromes, while mutations of the same genes have been found in both ALS and FTLD patients [10–13]. One relatively underappreciated aspect of the genetics is the contribution of the immune response in both diseases. This is consistent with observations in other neurodegenerative disease genome-wide association studies (GWAS) that identified immune-genes as disease risk factors [14–16]. In both Alzheimer’s disease (AD) and Parkinson’s disease (PD) there is a distinct immune signature defined by GWAS variants and transcriptomic analysis [14–16]. These data suggest the immune system is a strong modulator of neurodegeneration by acting in a non-cell autonomous fashion to affect neuronal function and survival. In this review, we examine how ALS/FTLD-associated genes can be examined in the context of different cell types of the central nervous system (CNS) (i.e., the neuro-immune axis) to more comprehensively understand ALS/FTLD pathology.
Clinical features of ALS/FTLD: patient symptoms, progression, and pathology
ALS is characterized by the rapid development of progressive muscle weakness and atrophy throughout the body [17]. The disease affects both upper motor neurons (UMNs), in the motor cortex, and lower motor neurons (LMNs), in the brainstem and spinal cord. ALS often presents in an isolated region, affecting speech (dysarthria), swallowing (dysphagia), or arm or leg strength that spreads to the entire neuroaxis [18]. Typical ALS diagnosis occurs at age 55 with life expectancy of 3 years after diagnosis [19]. Rare, juvenile forms of ALS with onset before 25 years old were described, with extremely aggressive clinical progression due to certain mutations in FUS [20] and slow progression due to mutations in Alsin (ALS2), Senataxin (SETX), or Spasticsin (SPG11) [21]. Not all motor neurons (MNs) are susceptible to degeneration, but nearly all ALS patients succumb to respiratory failure [19].
Generally, ALS is divided into sporadic (sALS, 90–95%) and familial (fALS, 5–10%) forms. Further classification includes initial symptom presentation site (bulbar vs. spinal) and progression rate (fast vs. slow progressors), with spinal onset as the most common (~70%). Approximately half of ALS patients display minor changes in cognition and behavior, while ~15% may eventually develop frontotemporal dementia [22]. ALS-FTLD is an ALS subtype with signs of cognitive and/or behavioral changes observed in ~5–15% of cases [23]. This disease manifestation is enriched in genetic mutations including Chromosome 9 Open Reading Frame 72 (C9ORF72) and TANK Binding Kinase 1 (TBK1) variants of ALS.
Despite these distinct categories, sALS and fALS are clinically and pathologically similar. The primary post-mortem neuropathological features of ALS patients are atrophy of anterior horn cells and gliosis in anterior and lateral columns of the spinal cord [24]. In the brain, there is motor cortex atrophy which may also be accompanied by ubiquitin-positive inclusions in neurons of the frontal and temporal lobes, hippocampus and striatum with accompanying dementia [25–29]. TAR DNA binding protein 43 (TDP-43) constitutes the main component of these inclusions in the vast majority of ALS/FTLD patients [30, 31]. Interestingly, TDP-43 brain inclusions are absent from ALS patients with Super Oxide Dismutase 1 (SOD1) or fused in sarcoma (FUS) mutations. Peripherally, there is myelin atrophy and axonal degeneration [18].
FTLD is characterized by pathological frontal and temporal lobe atrophy [32, 33]. There are three major FTLD subtypes, depending on specific protein aggregates observed in post-mortem tissue: FTLD-U (ubiquitin and TDP-43), FTLD-tau, and FTLD-FUS [31, 34, 35], with approximately half of all FTLD patients exhibiting TDP-43 pathology while the remainder have tau or FUS pathology [36]. That TDP-43 pathology is also found in nearly all ALS patients suggests a shared pathophysiology between these two diseases.
Looking at ALS/FTLD through a genetic perspective
Mutations in SOD1, the first causative ALS gene identified in 1993 [37], are amongst the most penetrant ALS associated alleles [38–40]. It is thought that mutant SOD1 acts as a gain of function, acquiring toxicity through protein misfolding and aggregation [41]. Because MN loss is a disease hallmark, early research focused on a neuron-centric view of ALS. The identification of hyper-phosphorylated, ubiquitylated, and cleaved TDP-43 as a pathological hallmark of ALS/FTLD [31] was a breakthrough, suggesting a common mechanism among the different genetic cases. Additionally, post-mortem studies in sALS, ALS with dementia, and SOD1-negative fALS revealed TDP-43 immunoreactivity [42], whereas SOD1 pathology is observed only in the presence of SOD1 mutations. Collectively, the data imply SOD1-mediated ALS arises from a distinct mechanism separate from most sALS and fALS cases that feature TDP-43 cytoplasmic accumulation [31]. Furthermore, clinically, SOD1-ALS does not associate with concomitant FTLD and patients are less likely to display significant cognitive changes [43].
In 2008–2009, mutations in Transactive Response DNA Binding Protein 43 kDa (TARDBP), encoding the TDP-43 protein, and FUS were identified in ALS patients [44–46]. Mutations in the TARDBP C-terminal glycine-rich region and the FUS C-terminal region promote cytoplasmic aggregation of these proteins. For TDP-43, the glycine-rich regions encode low-complexity domains that promote aggregation and potential transcellular propagation [47]. These two RNA-binding proteins (RBPs) play roles in mRNA transcript processing, splicing, stability, and RNA-protein stress granules (SGs). Although TDP-43 and FUS protein aggregates are found in ALS patients carrying their respective mutations, TDP-43 pathology does not seem to overlap with FUS or SOD1 pathology.
TDP-43 and FUS interact with the proteins hnRNPA1, hnRNPA2B1 (Heterogeneous Nuclear Ribonucleoprotein A1 and A2B1), and MATR3 (Matrin 3) [48, 49] in their RNA metabolic functions. Mutations affecting the protein structure of the prion-like domain of hnRNPA1 and hnRNPA2B1 also cause multi-system proteinopathy and ALS [48], while mutations in MATR3 cause fALS whereby patients often exhibit myopathy [49]. These findings transformed the view of ALS, suggesting ALS is not limited to protein aggregation but includes dysregulation of multiple pathways; thus its etiology is more complex than originally believed. One common theme is that mutant forms of these proteins lead to dysregulation of cellular homeostasis in the CNS. These proteins contain RNA-binding domains and/or aggregation-prone protein domains, which increase the tendency of these molecules to form aggregates [50–52].
Regardless of the cause/effect relationship between these aggregated proteins and disease, one hypothesis suggests the mechanism(s) involved in regulating aggregates, e.g., protein turnover/autophagy, might also become compromised in aggregate bearing cells and in turn contribute to disease. Consistent with this notion are numerous studies linking several important autophagy and proteostasis genes to ALS: Sequestosome 1 (SQSTM1)/p62, Optineurin (OPTN), TBK1, Ubiquilin 2 (UBQLN2), Valosin Containing Protein (VCP), and cyclin F (CCNF) [53–60] (Table 1). While the link between SOD1, TDP-43, and FUS with autophagy-related genes may be the ability to maintain cellular homeostasis, it remains unclear why this pathology affects only a subpopulation of MNs. Perhaps mutant proteins possess inherent cellular toxicities, overwhelm the response mechanism, and compromise their ability to handle the initial insult; thus, a second insult arises in the response mechanism, further disrupting homeostasis, leading to additional dysfunction and death (Fig. 1). As many ALS causal genes have multiple cellular functions, such mutations may disrupt more than one homeostatic pathway in neurons and/or non-neuronal cells. Cells that require fast and efficient mechanisms to maintain normal homeostasis, such as neurons, are likely to be more susceptible to situations where multiple housekeeping functions are dysregulated. One intriguing finding that supports a combination of cellular pathways leading to a common pathology is the presence of multiple disease gene variants in more than one ALS-associated gene in a single patient. A recent study showed that having more than one causal mutation is linked to earlier disease onset, but not other clinical features such as duration or site of onset [61].
Table 1.
RNA expression of ALS/FTLD-related genes in different cell types of the CNS
| Gene name | ALS/FTLD related | Function | Microglia expression | Neuron expression | Gene name | ALS/FTLD related | Function | Microglia expression | Neuron expression |
|---|---|---|---|---|---|---|---|---|---|
| ALS2 | ALS | Vesicle trafficking | Low | High | NEK1 | ALS | Cytoskeletal dynamics | Low | Medium |
| ATXN2 | ALS/FTLD | Endocytosis/RNA metabolism | Low | High | OPTN | ALS/FTLD | Autophagy–lysosome pathway | Low | High |
| ANG | ALS/FTLD | Angiogenesis | Medium | Low | PFN1 | ALS | Cytoskeletal dynamics | High | Low |
| C9ORF72 | ALS/FTLD | Protein/RNA aggregates, autophagy–lysosomal pathway, cytoskeletal dynamics | High | High | SCFD1 | ALS | Vesicle trafficking | High | High |
| C21ORF2 | ALS | Mitochondrial dysfunction, cytoskeletal dynamics | Low | Low | SETX | ALS | DNA/RNA metabolism | High | Medium |
| CCNF | ALS/FTLD | Protein degradation | Low | Low | SIGMAR1 | ALS/FTLD | Protein degradation | Low | High |
| CHCHD10 | ALS/FTLD | Mitochondrial dysfunction | Low | Low | SOD1 | ALS | Protein/RNA aggregates | Low | High |
| CHMP2B | ALS/FTLD | Protein degradation | High | High | SPG11 | ALS | DNA damage repair | High | High |
| DCTN1 | ALS/FTLD | Cytoskeletal dynamics | Low | High | SQSTM1 | ALS/FTLD | Autophagy–lysosome pathway | High | Medium |
| FIG4 | ALS | Protein degradation | Medium | High | TAF15 | ALS | RNA metabolism | High | Medium |
| FUS | ALS/FTLD | Protein/RNA aggregates | High | Medium | TBK1 | ALS/FTLD | Autophagy–lysosome pathway | High | Medium |
| HNRNPA1 | ALS | Protein/RNA aggregates | High | Medium | TDP-43 | ALS/FTLD | Protein/RNA aggregates | Medium | High |
| HNRNPA2B1 | ALS | Protein/RNA aggregates | High | Medium | TIA1 | ALS/FTLD | Protein/RNA aggregates | Medium | High |
| MATR3 | ALS/FTLD | RNA metabolism | Medium | High | TUBA4A | ALS | Cytoskeletal dynamics | Low | High |
| MAPT | ALS/FTLD | Cytoskeletal dynamics | Low | High | UBQLN2 | ALS/FTLD | Protein degradation | Low | High |
| MOBP | ALS | Oligodendrocytes | Low | Low | VAPB | ALS | Vesicle trafficking | Low | Low |
| NEFH | ALS | Cytoskeletal dynamics | Medium | High | VCP | ALS/FTLD | Protein degradation | High | High |
This table lists data from the Barres Lab [111] showing some of the major ALS/FTLD-related genes and their associated cellular functions, together with their RNA expression from human tissue derived from different CNS cell types. For a more comprehensive list of the genetic landscape of ALS/FTLD refer to Guerreiro et al. [13].
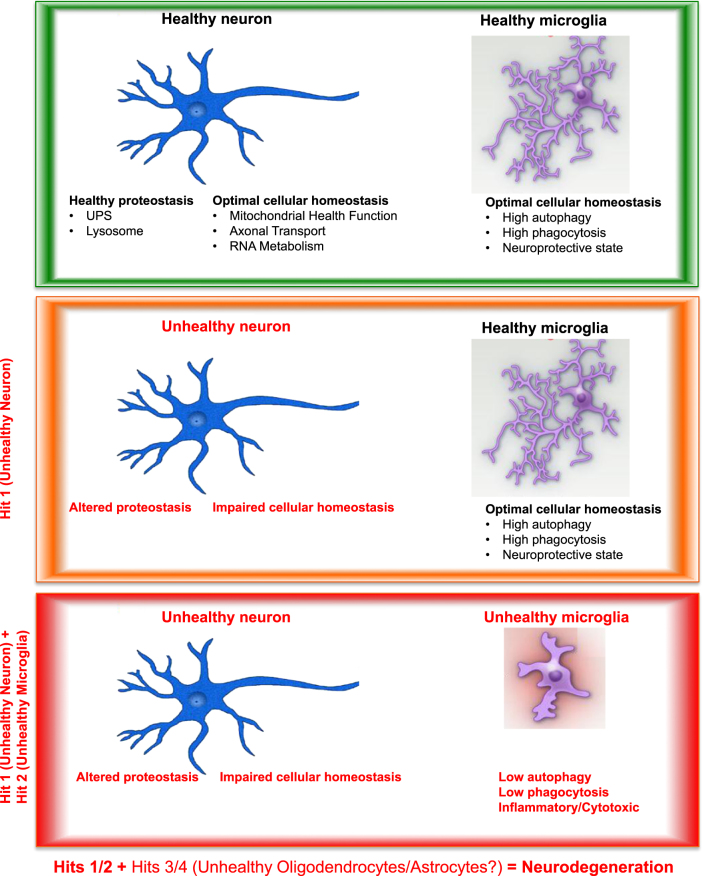
Fig. 1.
A model of neuron–microglia interplay in disease progression. In a healthy individual, all cells within the CNS are equipped with a myriad of functionalities to maintain optimal homeostasis as well as an ability to respond to acute insults. This process is regulated in both an autonomous and non-cell autonomous manner. A healthy neuron undergoes normal mitochondrial fission/fusion and elimination of damaged mitochondria, efficient axonal transport, and RNA metabolism. In healthy microglia, key functionalities actively maintain CNS homeostasis by directly interacting with the extracellular space via both phagocytosis- and lysosomal-mediated flux. During the aging process, neurons can become damaged due to an ALS causal mutation, sustained head trauma, environmental stresses, or some combination thereof; in ALS, this leads to motor neuron dysfunction (Hit 1). This may lead to alterations in neuronal proteostasis and/or basic cellular functions, e.g., in mitochondria biogenesis/dynamic, axonal transport, or RNA metabolism. Under such conditions, the genetics as well as the mechanistic data indicate that microglial function also becomes impaired (Hit 2). This microglial impairment is manifested via dysfunction in autophagy, phagocytosis and/or other homeostatic cellular pathways in these cells, resulting in a shift in their secretory/inflammatory profiles. Thus, it is these multi-cell and multi-pathway failures (Hit 1 + Hit 2) that we hypothesize lead to the initiation of ALS/FTLD. As other cells (e.g., astrocytes, oligodendrocytes) in the affected micro-environment respond, they too become functionally aberrant in a similar fashion, causing further damage and progressive degeneration (Hits 1/2 + Hits 3/4)
An example of homeostasis is the formation and dissociation of SGs, cytoplasmic membrane-free organelles that arise as a physiological response to cellular stress and subsequently cleared by autophagy. SGs comprised various proteins involved in RNA metabolism, transport, and transcription, and they also contain several RBPs such as TDP-43, FUS, and hnRNPA1 [62]. Other proteins found in SGs include Senataxin, mutations in which cause a rare form of juvenile ALS [63], and Ataxin-2 (ATXN-2), where intermediate-length polyQ protein expansions are associated with ALS [64]. In disease conditions, aberrant homeostasis leads to SG accumulation and fibril formations containing RBPs and other associated proteins [65, 66]. The low-complexity RBPs are more predisposed to a phenomenon known as liquid-liquid phase separation (LLPS), which may initiate or exacerbate disease pathology by facilitating the formation and continued presence of SGs due to impaired dissociation [65, 66]. Recently, it was reported that mutations in the low-complexity domain of T cell-restricted intracellular antigen-1 (TIA1) cause ALS/FTLD [67]. The TIA1 mutations impair SG dynamics, and SG recruited TDP-43 protein becomes insoluble. The data suggest altered SG formation impacts ALS pathology through aberrant LLPS function by affecting clearance of SG-associated proteins such as TDP-43.
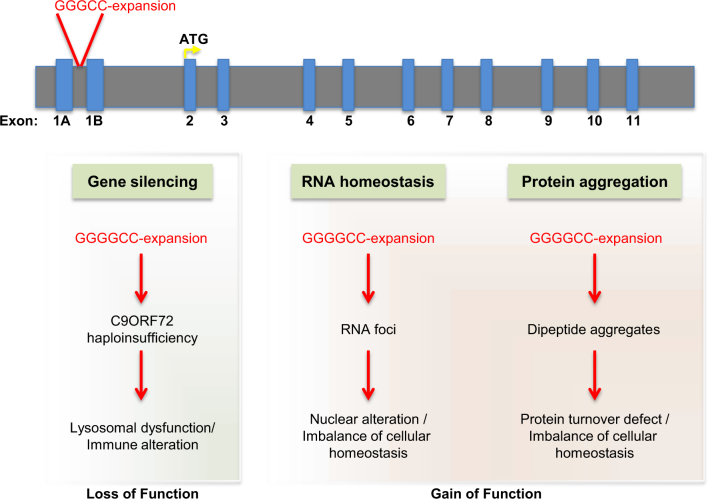
Another class of genes identified as ALS/FTLD risk factors are those involved in cytoskeletal dynamics. The identification of Microtubule Associated Protein Tau (MAPT), Dynactin Subunit 1 (DCTN1), Neurofilament Heavy Polypeptide (NEFH), Profilin (PFN1), NIMA Related Kinase 1 (NEK1), and Tubulin Alpha 4a (TUBA4A) [68–74] as ALS causal genes (Table 1) support this notion. It should be noted that most of the genes described thus far are rare genetic occurrences in the totality of ALS. The expansion of a hexanucleotide (G4C2) repeat in an intronic region of the C9ORF72 gene was identified as the most common mutation in familial ALS/FTLD patients [75–79]. This expansion occurs in the intronic region between two alternative non-coding first exons and leads to three observable phenotypes (Fig. 2). First, C9ORF72 intronic expansion allows for RNA foci formation that can sequester a subset of nuclear RBPs [80–85]. Second, C9ORF72-associated dipeptide repeat (DPR) proteins are generated by Repeat Associated Non-AUG (RAN) translation of the (G4C2) repeats. Depending on direction and frame of translation, these DPRs comprised poly-GP, GA, GR, PA, and PR. Poly-GA and GR appear to be more aggregation-prone, whereas poly-GA, GR, and PR appear to be more cytotoxic [86, 87]. It was recently proposed that arginine-rich DPRs such as PR may induce SG formation and alter SG dynamics [66]. Patients harboring expanded (G4C2) repeats also exhibit pathological cytoplasmic TDP-43 aggregates in neurons. Finally, hexanucleotide expansion reduces endogenous C9ORF72 levels. In this regard, studies suggest a C9ORF72 role in early formation of autophagosome and lysosome function [88–90]. Furthermore, C9ORF72 could interact with Cofilin, an actin-binding protein, to take part in cytoskeletal dynamics [91]. Thus, hexanucleotide expansion-mediated loss of C9ORF72 could be detrimental via numerous mechanisms. Given its involvement in many of the pathologies known to be affected in ALS/FTLD, further investigation of C9ORF72 functions may help elucidate a common pathway combining aggregated proteins, autophagy/lysosome, and cytoskeletal dynamics.
Fig. 2.
C9ORF72 associates with multiple cellular pathways relevant to ALS/FTLD. A schematic representation of the genomic structure of C9ORF72, showing the location of the (GGGGCC) hexanucleotide expansion in the intronic region between two alternative non-coding first exons. This expansion leads to three observable phenotypes. One is C9ORF72 loss-of-function leading to a blockade of lysosomal function and an alteration in immune function. Based on the RNA expression profile of this gene, this haploinsufficiency phenotype primarily affects microglia in the CNS. In addition, there are two other phenotypes observed in both C9ORF72 ALS/FTLD patients and in animal models of the disease. The (GGGGCC) hexanucleotide expansion leads to the formation on RNA foci, which results in the mislocalization and sequestration of RNA-binding proteins causing nuclear and cellular dysfunction. Finally, through repeat-associated non-ATG-dependent translation, (GGGGCC) hexanucleotide repeats can produce five different types of dipeptide repeat sequences, depending on the direction (sense and anti-sense) and frame of translation (GA, GR, GP, PR, and PA). These aberrant species lead to protein aggregation, toxicity, lysosomal blockade, and overall cellular dysfunction. This most common genetic cause of ALS/FTLD illustrates the potential contribution of disruptions to multiple cellular pathways in various cell types to disease
Non-cell autonomous pathology and the role of glia in ALS/FTLD
Because MN loss is a disease hallmark, much of the early research focused on a neuron-centric view of ALS/FTLD, yet recent studies have implicated non-neuronal cell types in disease, including astrocytes, oligodendrocytes, and microglia. These observations were made after histogical assessments and transcriptomic profiling of disease tissue, which identified a strong non-neuronal signature in ALS and other neurodegenerative diseases [92–94]. While early views of glial activation in disease emphasized a secondary role for microglia and astrocytes as neuronal injury responders, more recently these cells are thought to be more actively involved in disease [95]. Such interpretations align with expression patterns of several ALS causal genes, which are limited to non-neuronal cell types (Table 1). The prevailing view is that these cells act in a non-cell autonomous manner to affect ALS/FTLD pathology.
Consistent with the idea that neuro-inflammation is relevant to disease, the microglial transcriptional signature changes during disease progression in SOD1G93A mice [95, 96]. Moreover, SOD1G93A-derived microglia upregulate factors linked to neurotoxicity, indicating an ability of microglia to influence surrounding cells [95, 96]. While the functional consequence of the microglial changes are not fully understood, this dramatic transcriptional reorganization suggests a role for microglia in pathogenesis. A seminal study from the Cleveland lab established that in SOD1G93A mice, microglia-specific transgene ablation prolongs survival [97]. Introduction of wild-type donor-derived microglia into microglial-deficient SOD1G93A/PU.1−/− mice slowed MN loss and extended survival compared to mice receiving SOD1G93A microglia, providing further evidence of a non-cell autonomous microglial role [98]. Furthermore, Gerber et al. [99] described a role for microglia in the initiation of ALS in a hSODG93A mouse model while others have shown that during disease progression microglia switch to an activated neurodegenerative state [100, 101]. Altered microglia function is also observed in an animal model of FTLD [102]. Interestingly, MAPT, a protein whose misfolding, mislocalization, and aggregation are hallmarks of AD, is mutated in FTLD [103]. The FTLD-associated mutation P301S MAPT was utilized in various mouse models of tauopathies, where robust microglia activation is observed prior to tau tangle formation [104]. This study also showed tau pathology was attenuated by treating with an immunosuppressive agent, FK506. A recent study also showed that microglia play an active role in propagating tau pathology [105]. These data underscore the role that microglia play in ALS/FTLD—not only as responders to pathology but also as mediators of disease progression.
While microglia clearly play a role in ALS/FTLD, some toxicity leading to MN loss may occur through a detrimental effect mediated by astrocytes. Consistent with this idea, MN death was observed in neurons co-cultured with astrocytes derived from induced pluripotent stem (iPS) cells from sALS patients, but not from control patient-derived astrocytes [106]. Additional studies implicating a non-cell autonomous role for astrocytes includes one in which mutant SOD1 astrocytes selectively kill primary spinal and embryonic mouse stem cell-derived MNs, an effect believed to be mediated by soluble toxic factors released from astrocytes [107]. The mechanism of this non-cell autonomous toxicity was shown to be RIPK1-dependent necroptosis in MNs [106]. Interestingly, an in vivo study reducing expression of mutant SOD1 in astrocytes did not affect disease onset in a SOD1G37R transgenic mouse model, but slowed disease progression [108].
Additionally, there is significant evidence that oligodendrocytes contribute to ALS pathologies in mouse models. Removal of mutant SOD1, specifically from oligodendrocytes, extends survival of SOD1G93A ALS mice [109]. Finally, a recent GWAS identified mutations in the Myelin-Associated Oligodendrocyte Basic Protein (MOBP), a gene whose expression is thought to be limited to oligodendrocytes, as a risk locus for ALS [110]. Given its restricted expression pattern, it seems likely that oligodendrocytes harboring MOBP mutations fail to communicate properly with MNs, and that this defective signaling contributes to disease.
Based on these findings, it is evident that ALS/FTLD pathology is mediated by multiple cell types and that intracellular communication between different CNS cell types contribute to pathology. Many ALS/FTLD-associated genes are highly expressed in microglia, as compared to other CNS cell types (Table 1), including some recently discovered ALS/FTLD causal genes—C9ORF72, OPTN, TBK1, and SQSTM1/p62 [111]. These observations, while somewhat unexpected, underscore the importance of non-cell autonomous processes in ALS pathophysiology.
Microglia function in development and homeostasis
While microglia were first identified many years ago, the complexity of their origin and function has only recently been elucidated [112, 113]. As sentinels of the CNS, microglia play a vital role in both physiological and pathological conditions. Below, we briefly mention some critical microglia functions and speculate as to how disruption of microglia homeostasis might contribute to ALS/FTLD pathology and progression.
During embryonic and postnatal development, microglia are critical for proper CNS formation by promoting neuronal survival through release of neurotrophic factors. Examples of microglia involvement during development include: secretion of Insulin-like Growth Factor-1 (IGF-1) to support cortical neuron survival [114], microglial–neuronal interactions to promote survival via CX3CL1/CX3CR1 fractalkine signaling [115, 116], cellular pruning via nerve growth factor-induced cell death in retinal neurons [117], removal of cellular debris through phagocytosis resulting from apoptosis [118] and elimination of neurons undergoing regulated cell death via phagocytosis and lysosome degradation in zebrafish [119, 120]. Additional roles include clearance of myelin debris during development and aging [121–123] and activity-dependent synaptic pruning during development and disease via complement receptor CR3 pathway in a mouse AD model [124].
Microglia rely on their ability to phagocytose and eliminate substrates through lysosomal degradation during normal and disease conditions. Supporting this notion, inefficient clearance of myelin debris in CX3CR1-deficient mice impairs remyelination [125]. Furthermore, impaired phagocytosis in Triggering Receptor Expressed On Myeloid Cells 2 (TREM2)-deficient microglia exacerbates ischemic damage in experimental stroke [126]. TREM2, which has been genetically linked to AD [127, 128], was identified as an ALS/FTLD risk allele [129, 130]. Thus, we hypothesize that in pathological conditions normal microglial function is compromised due to excess cellular debris resulting from dying or damaged MNs. Furthermore, in the context of aging and/or neurodegeneration, their dysregulation leads to deleterious consequences, though exactly how they are involved in ALS/FTLD pathology is less well understood.
It is noteworthy that defects in pathways known to be involved in ALS pathophysiology are not limited to MNs, but are also observed in microglia. Given their essential non-cell autonomous supportive CNS role, altered microglia functions may also impact MN physiology and viability. Thus, we propose that homeostatic non-cell autonomous interactions are disrupted in disease, leading to further CNS dysfunction and damage.
A role for microglia in ALS/FTLD
Neuroinflammation, characterized by astrogliosis, microglial activation, and peripheral immune cell invasion, is one of the hallmarks of ALS patients [131–134]. In fact, the presence and morphological changes of these cells, as well as the manifestation of self-recognizing antibodies in patients, suggested that ALS was an autoimmune disease [135]. While this idea may not accurately reflect the nature of ALS, that this hypothesis was presented underscores the importance of immune modulation in disease. In post-mortem ALS patient tissue there is increased microgliosis in numerous regions which exhibit MN degeneration including the motor cortex, portions along the corticospinal tract, and in the ventral horn of the spinal cord [131, 132]. In fact, the level of microglial activation in post-mortem tissue correlates with end-stage clinical measures of executive function [136]. These studies detail the presence of microglia and other immune cells at end stage; however, the role of these cells during initiation and progression has not been fully elucidated.
To address this, different imaging approaches examined the dynamic nature of microglia in ALS. In vivo positron emission tomography (PET) imaging identified microglial activation in several different regions in ALS patients. Using either 11C-PK11195 or PBR-28, translocator protein radio-ligands that image microglial activation, robust PET signal was observed in regions corresponding to MN injury in ALS [137, 138] and FTLD patients [139]. These data indicate that immune-associated clinical markers are disease relevant as higher PBR-28 signal correlates with worse ALS clinical function [137]. This is also consistent with the finding that increased microglial pathology associated with worse clinical outcomes [140]. These data show a distinct correlation between microglial activation and damage to UMNs [141] and that there is a clear role for microglial activation at various disease stages [142]. Based on our improved understanding of microglial physiology, accumulating evidence suggests they help regulate CNS homeostasis and contribute to degeneration in ALS.
Microglia have been implicated in mediating pathology in SOD1G93A mice; however, whether this is due to a gain or loss of microglial function is unclear. Given its central role as the immune homeostatic surveillance system in the brain, one intriguing possibility is that loss-of-function mutations in genes implicated in microglial function alters the ability of these cells to efficiently maintain tissue homeostasis. To illustrate this point, a recent study showed that C9ORF72 gene is highly expressed in myeloid cells and required for proper immune and microglial function [90]. C9ORF72 null mice present an altered immune profile and an accumulation of lysosomes in microglia, suggesting microglia dysregulation may contribute to ALS/FTLD pathology through dysfunction of the microglial clearance system [90]. While C9ORF72 loss-of-function mice display observable phenotypes, they do not recapitulate ALS/FTLD patient pathology or other more aggressive ALS mouse models. This suggests the C9ORF72 intronic expansion contributes to patient pathology. In fact, several laboratories introduced human disease causing forms of C9ORF72 into mice via BAC transgenics or adeno-associated virus (AAV) technology to recapitulate gain-of-function effects [143–146]. While these gain-of-function models exhibit features of ALS with varying degrees of neurodegeneration and behavioral deficits, they do not completely recapitulate the severity of ALS or other ALS mouse models. These studies suggest that both C9ORF72 gain- and loss-of-function play a role in patients harboring the (G4C2) intronic expansion. These observations are important because much of the therapeutic effort for this target focus on attenuating the gain-of-function of mutant C9ORF72. Thus, this strategy may not prove sufficiently efficacious if loss of C9ORF72 also contributes to disease due to improper immune and microglial function. One approach is to examine the C9ORF72 BAC or AAV models in the context of C9ORF72 null background to test the hypothesis that lysosomal function and protein degradation will be exacerbated with C9ORF72 haploinsufficiency.
These studies highlight a pathological role for microglia in disease; however, the cause/effect relationship between microglial activity and disease remains unclear. Do signals from degenerating MNs trigger microglial responses? Alternatively, do changes observed in microglia cause MN degeneration or both? Regardless of the sequence of events, such scenarios are indicative of non-cell autonomous interactions, and these interactions may serve as key drivers of disease progression. Thus, given their role in supporting neuronal physiology and overall health, we consider evidence of defects in microglia as having deleterious impacts on neurons due to a failure in its sentinel functions, and per definition, occur through cell-to-cell interactions.
Microglial-mediated autophagy and protein turnover in ALS/FTLD
ALS mutations affect genes involved in protein degradation (UBQLN2, OPTN, SQSTM1/p62, VCP, charged multivesicular body protein 2B (CHMP2B), FIG4 phosphoinositide 5-phosphatase (FIG4)), or genes that result in the formation of protein aggregates (TDP-43, FUS, C9ORF72, SOD1); this suggests that disruptions to pathways that clear aggregated or misfolded proteins are likely to contribute to disease onset and/or progression. These aggregates and/or their oligomeric precursors induce cellular stress and therefore negatively affect homeostasis [147, 148], ultimately contributing to axonal degeneration and cell death. Thus, identifying and investigating specific protein degradation pathways that contribute to ALS pathology is necessary to better understand disease. One such pathway is the autophagy/lysosomal system, which, upon its induction, enhanced TDP-43 turnover and improved survival in ALS models [149].
Autophagy is an evolutionarily conserved process handling the degradation of cytoplasmic cargo to regulate cellular metabolism and protein homeostasis [150–152] and is critically important to normal brain homeostasis as autophagy-deficient mice exhibit neurodegeneration and neuronal accumulation of aggregate-prone proteins [153, 154].
With the emergence of flux into the lysosome as an important modality to clear intracellular aggregates and damaged organelles, the autophagic machinery has emerged as a vital defense against neurodegeneration. Genetic studies provide a direct connection between ALS/FTLD-related genes and the autophagy–lysosome pathways. For example, TBK1 [1, 58] controls autophagosome maturation during autophagy-mediated antimicrobial defense [155] and autophagosomal engulfment of polyubiquitylated mitochondria [156]. Other studies showed a requirement for TBK1 and OPTN for efficient mitophagy [157, 158]. The notion that lysosomal-mediated clearance of damaged mitochondria is important for CNS homeostasis is consistent with current PD research, which indicates that PINK1 and PARKIN2 are mitophagy regulators [159–161]. Additionally, SQSTM1/p62 facilitates degradation of ubiquitylated protein aggregates by autophagy [162] and also induces mitophagic clearance after NLR Family Pyrin Domain Containing 3 (NLRP3) agonists that cause mitochondria damage [163]. Finally, C9ORF72 may regulate autophagy and endocytic transport as C9ORF72 co-localizes with Rab proteins in neuronal cell lines and ALS patient MNs [164].
To date, significant research focused on aggregate and organelle clearance within susceptible neuronal populations. However, TBK1, OPTN, and SQSTM1/p62 also have strong immune system connections, suggesting that autophagy dysregulation in cells responsible for the immune response may contribute to neurodegeneration. Furthermore, C9ORF72 loss causes lysosome accumulation in macrophages and microglia, which is observed in C9ORF72-ALS patients [90]. An additional direct link between microglial lysosome function and ALS/FTLD is the FTLD-associated gene Progranulin [165, 166]. This gene, which is highly expressed in microglia, is linked to FTLD and Gaucher’s disease, a severe lysosomal storage disorder [167]. Furthermore, Progranulin loss leads to a defect in microglial synaptic phagocytosis in a complement- and presumably, lysosomal-dependent manner [102]. These observations strongly suggest that these ALS- and FTLD-associated proteins involved in autophagy and lysosomal function also perform an important role in CNS immune homeostasis. Altered lysosomal function may shift microglial homeostasis in the CNS, resulting in a highly compromised state and leading to disease susceptibility and/or progression. Possible outcomes may be a failure of normal microglia support functions, disrupted intercellular signaling and/or release of soluble neuro-toxic factors.
We hypothesize that lysosomal flux is critical to microglia function; when flux is dysregulated in microglia, their cellular output (cytokine, cell–cell interactions, etc.) is altered. This idea is consistent with findings in various OPTN mutant mouse models [168]. OPTN depletion in microglia affects myelin homeostasis, which in turn, results in an impairment of oligodendrocyte myelination. Loss of OPTN in oligodendrocytes and myeloid cells (not in astrocytes or MNs) produced subtle deficits in axonal myelination, features observed in ALS/FTLD patients and in SOD1G93A mice [168]. Interestingly, the microglial transcriptional profile derived from OPTN-deficient mice was also altered, consistent with microglia dysregulation. We speculate this effect acts as a mechanism driving ALS pathology, as OPTN is associated with autophagy and endocytosis/phagocytosis. From work in the OPTN mouse model, it is hypothesized that microglia have a vital role in maintaining CNS homeostasis and thereby preventing axonal pathology [168]. This supports the possibility that non-cell autonomous activity of axonal maintenance is critical in preventing ALS/FTLD pathology.
While ALS/FTLD genetics point to a prominent role for the autophagy/lysosomal pathway in disease, the precise role of the ubiquitin–proteasomal pathway in etiology is unclear. Interestingly, ubiquitin-positive inclusions are a hallmark of ALS [30, 31], implicating this degradative pathway in disease. However, to date, GWAS have not linked Ubiquitin–Proteasome System (UPS) machinery components with ALS/FTLD. However, missense mutations in the UBQLN2 gene were identified in X-linked dominant ALS/FTLD [55]. While this protein has been implicated in delivery of ubiquitylated cargo to the proteasome [169], its function is not fully understood and it may be more relevant for lysosomal function [170]. Thus, the role of the UPS pathway needs to be further explored and in particular, focusing on how this pathway is regulated in microglia in neurodegeneration. Furthermore, the balance between the UPS and the autophagy/lysosomal pathway in these cells needs closer examination. Future experiments examining how homeostasis and protein turnover is altered in the autophagy/lysosomal and ubiquitin/proteasomal pathways in ALS/FTLD will rely on animal models to determine the degree of crosstalk within and between various cell types in disease.
As many of ALS causal genes fall into very distinct cellular functions or pathways, one question is whether simultaneous dysfunction of multiple pathways contribute to disease. For example, TARDBP mutations disrupt at least two pathways (e.g., RNA processing and aggregation clearance) in all cells, yet ultimately causes MN-specific cell death. Likewise, disease causing C9ORF72 mutations also disrupt numerous pathways including endosomal trafficking, lysosomal function, and the immune system [90, 164]. These impairments, in parallel with the appearance of mutant C9ORF72-depedent nuclear RNA foci and dipeptide accumulation, suggest a multi-system dysregulation that leads to ALS. An intriguing hypothesis for the prevalence of C9ORF72 pathology in ALS/FTLD is that C9ORF72 hexanucleotide expansion mutations simultaneously affect more than one vulnerable process on the path to ALS/FTLD (Fig. 1).
Conclusions and future directions—multi-pathway dysfunction as a cause for neurodegeneration?
This review provides a broad summary of ALS/FTLD in the context of clinical presentations, genetics, and non-cell autonomous contributions of microglia, astrocytes and oligodendrocytes to disease. In particular, we describe how disruptions to ALS-related pathways in these cells, with particular emphasis on the autophagy/lysosomal and ubiquitin pathways, may impact MN physiology and viability. The review also highlights the growing consensus that microglia are essential in maintaining proper neuronal homeostasis, and that dysregulation of microglia-mediated pathways are also involved in neurodegeneration. Thus, the genetics of ALS/FTLD as well as data generated from pre-clinical disease models indicate disrupted CNS microglial function and suggest that these aberrant activities contribute to onset or progression. Such a hypothesis is in line with pathological features observed in patients. With the ability to understand that ALS/FTLD may in part be caused by a dysfunction in microglia, an important step in developing a therapy is to determine the best way to target a specific cell type in the CNS. In this context, targeting a phagocytic cell in the CNS may in fact be advantageous. One emerging approach to address the role of ALS/FTLD-related pathways is to utilize a single-cell sequencing approach in animal models, patient-derived tissue and human post-mortem brain to elucidate shared transcriptional or epigenetic signatures [171]. This technology will aid in simultaneously identifying conserved, dominant and/or disrupted pathways in multiple cell types in the diseased CNS.
Such efforts may help define a common mechanism central to ALS/FTLD neuropathophysiology or point to multiple independent mechanisms within each of the different cell types. One can imagine expanding the therapeutic strategy to include a combinatorial approach whereby two or more independent cellular processes will need to be targeted in different cell types. Additionally, these approaches may help inform biomarker strategies ultimately aiding in diagnosis and progression, as well as providing a means to assess treatment efficacy, in ALS/FTLD patients.
In summary, ALS/FTLD patients present with a large degree of varying clinical symptoms and subtypes, underscoring the complexity and heterogeneity of disease etiology. A more comprehensive pathogenesis of ALS/FTLD might be explained by the disruption of several different pathways and non-neuronal cell types affecting non-autonomous signaling. Given these observations, it is tempting to speculate that perhaps one preferentially affected pathway may give rise to a particular type of ALS/FTLD onset. Viewing this disease from a broader perspective will help elucidate additional mechanisms that may apply more broadly to other neurodegenerative diseases, and serve as a compelling basis for investigators to devise novel strategies that target specific cell types and pathways.
Acknowledgements
We thank Dr. Junying Yuan for helpful discussion and support.
Compliance with ethical standards
Conflict of interest
MWK, SCS are employees of Biogen. SSWH was employed and Biogen and is now an employee of GSK. DO was employed and Biogen and is now an employee of Sanofi. HC has no conflict of interest.
Footnotes
These authors contributed equally: Hongbo Chen and Mark W. Kankel.
Edited by E. Baerecke
References
- 1.Cirulli ET, Lasseigne BN, Petrovski S, Sapp PC, Dion PA, Leblond CS, et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science. 2015;347:1436–41. doi: 10.1126/science.aaa3650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin B, Boumédiene F, Logroscino G, Couratier P, Babron MC, Leutenegger AL, et al. Variation in worldwide incidence of amyotrophic lateral sclerosis: a meta-analysis. Int J Epidemiol. 2017;46:57–74. doi: 10.1093/ije/dyw061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowland LP, Shneider NA. Amyotrophic lateral sclerosis. N Engl J Med. 2001;344:1688–700. doi: 10.1056/NEJM200105313442207. [DOI] [PubMed] [Google Scholar]
- 4.Moujalled D, White AR. Advances in the development of disease-modifying treatments for amyotrophic lateral sclerosis. CNS Drugs. 2016;30:227–43. doi: 10.1007/s40263-016-0317-8. [DOI] [PubMed] [Google Scholar]
- 5.Writing Group, Edaravone (MCI-186) ALS 19 Study Group. Safety and efficacy of edaravone in well defined patients with amyotrophic lateral sclerosis: a randomised, double-blind, placebo-controlled trial. Lancet Neurol. 2017;16:505–12. doi: 10.1016/S1474-4422(17)30115-1. [DOI] [PubMed] [Google Scholar]
- 6.Hardiman O, van den Berg LH. Edaravone: a new treatment for ALS on the horizon? Lancet Neurol. 2017;16:490–1. doi: 10.1016/S1474-4422(17)30163-1. [DOI] [PubMed] [Google Scholar]
- 7.Warren JD, Rohrer JD, Rossor MN. Clinical review. Front Dement BMJ. 2013;347:f4827. doi: 10.1136/bmj.f4827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rabinovici GD, Miller BL. Frontotemporal lobar degeneration: epidemiology, pathophysiology, diagnosis and management. CNS Drugs. 2010;24:375–98. doi: 10.2165/11533100-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hogan DB, Jetté N, Fiest KM, Roberts JI, Pearson D, Smith EE, et al. The prevalence and incidence of frontotemporal dementia: a systematic review. Can J Neurol Sci. 2016;43(Suppl 1):S96–S109. doi: 10.1017/cjn.2016.25. [DOI] [PubMed] [Google Scholar]
- 10.Burrell JR, Kiernan MC, Vucic S, Hodges JR. Motor neuron dysfunction in frontotemporal dementia. Brain. 2011;134:2582–94. doi: 10.1093/brain/awr195. [DOI] [PubMed] [Google Scholar]
- 11.Lomen-Hoerth C, Anderson T, Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–9. doi: 10.1212/WNL.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 12.Ringholz GM, Appel SH, Bradshaw M, Cooke NA, Mosnik DM, Schulz PE. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–90. doi: 10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- 13.Guerreiro R, Brás J, Hardy J. SnapShot: genetics of ALS and FTD. Cell. 2015;160:798.e1. doi: 10.1016/j.cell.2015.01.052. [DOI] [PubMed] [Google Scholar]
- 14.Zhang B, Gaiteri C, Bodea LG, Wang Z, McElwee J, Podtelezhnikov AA, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–20. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, et al. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science. 2014;344:519–23. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–84. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams DB, Windebank AJ. Motor neuron disease (amyotrophic lateral sclerosis) Mayo Clin Proc. 1991;66:54–82. doi: 10.1016/S0025-6196(12)61175-6. [DOI] [PubMed] [Google Scholar]
- 18.Walling AD. Amyotrophic lateral sclerosis: Lou Gehrig’s disease. Am Fam Physician. 1999;59:1489–96. [PubMed] [Google Scholar]
- 19.Gordon PH. Amyotrophic lateral sclerosis: pathophysiology, diagnosis and management. CNS Drugs. 2011;25:1–15. doi: 10.2165/11586000-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 20.Conte A, Lattante S, Zollino M, Marangi G, Luigetti M, Del Grande A, et al. P525L FUS mutation is consistently associated with a severe form of juvenile amyotrophic lateral sclerosis. Neuromuscul Disord. 2012;22:73–75. doi: 10.1016/j.nmd.2011.08.003. [DOI] [PubMed] [Google Scholar]
- 21.Orban P, Devon RS, Hayden MR, Leavitt BR. Chapter 15 juvenile amyotrophic lateral sclerosis. Handb Clin Neurol. 2007;82:301–12. doi: 10.1016/S0072-9752(07)80018-2. [DOI] [PubMed] [Google Scholar]
- 22.Hobson EV, McDermott CJ. Supportive and symptomatic management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2016;12:526–38. doi: 10.1038/nrneurol.2016.111. [DOI] [PubMed] [Google Scholar]
- 23.van Es MA, Hardiman O, Chiò A, Al-Chalabi A, Pasterkamp RJ, Veldink JH et al. Amyotrophic lateral sclerosis. Lancet. 2017. 10.1016/S0140-6736(17)31287-4. [DOI] [PubMed]
- 24.Ellison D, Love S, Chimelli L, Harding B, Lowe J, Vinters H, et al. 3rd ed. Philadelphia: Elsevier – Mosby; 2013. Neuropathology. A Reference Text of CNS Pathology; pp. 308–12.
- 25.Piao YS, Wakabayashi K, Kakita A, Yamada M, Hayashi S, Morita T, et al. Neuropathology with clinical correlations of sporadic amyotrophic lateral sclerosis: 102 autopsy cases examined between 1962 and 2000. Brain Pathol. 2003;13:10–22. doi: 10.1111/j.1750-3639.2003.tb00002.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto K, Murakami N, Kusaka H, Yoshida M, Hashizume Y, Nakazato Y, et al. Ubiquitin-positive intraneuronal inclusions in the extramotor cortices of presenile dementia patients with motor neuron disease. J Neurol. 1992;239:426–30. doi: 10.1007/BF00856806. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto K, Hirai S, Yamazaki T, Sun XY, Nakazato Y. New ubiquitin-positive intraneuronal inclusions in the extra-motor cortices in patients with amyotrophic lateral sclerosis. Neurosci Lett. 1991;129:233–6. doi: 10.1016/0304-3940(91)90469-A. [DOI] [PubMed] [Google Scholar]
- 28.Wightman G, Anderson VE, Martin J, Swash M, Anderton BH, Neary D, et al. Hippocampal and neocortical ubiquitin-immunoreactive inclusions in amyotrophic lateral sclerosis with dementia. Neurosci Lett. 1992;139:269–74. doi: 10.1016/0304-3940(92)90569-S. [DOI] [PubMed] [Google Scholar]
- 29.Kawashima T, Kikuchi H, Takita M, Doh-ura K, Ogomori K, Oda M, et al. Skein-like inclusions in the neostriatum from a case of amyotrophic lateral sclerosis with dementia. Acta Neuropathol. 1998;96:541–5. doi: 10.1007/s004010050932. [DOI] [PubMed] [Google Scholar]
- 30.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–11. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 31.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–3. doi: 10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 32.Liscic RM, Grinberg LT, Zidar J, Gitcho MA, Cairns NJ. ALS and FTLD: two faces of TDP-43 proteinopathy. Eur J Neurol. 2008;15:772–80. doi: 10.1111/j.1468-1331.2008.02195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cairns NJ, Bigio EH, Mackenzie IRA, Neumann M, Lee VMY, Hatanpaa KJ, et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007;114:5–22. doi: 10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Neary D, Snowden JS, Mann DM. Classification and description of frontotemporal dementias. Ann N Y Acad Sci. 2000;920:46–51. doi: 10.1111/j.1749-6632.2000.tb06904.x. [DOI] [PubMed] [Google Scholar]
- 35.Neumann M, Rademakers R, Roeber S, Baker M, Kretzschmar HA, Mackenzie IRA. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–31. doi: 10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–38. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 38.Siddique T, Figlewicz DA, Pericak-Vance MA, Haines JL, Rouleau G, Jeffers AJ, et al. Linkage of a gene causing familial amyotrophic lateral sclerosis to chromosome 21 and evidence of genetic-locus heterogeneity. N Engl J Med. 1991;324:1381–4. doi: 10.1056/NEJM199105163242001. [DOI] [PubMed] [Google Scholar]
- 39.Chiò A, Calvo A, Mazzini L, Cantello R, Mora G, Moglia C, et al. Extensive genetics of ALS: a population-based study in Italy. Neurology. 2012;79:1983–9. doi: 10.1212/WNL.0b013e3182735d36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lattante S, Conte A, Zollino M, Luigetti M, Del Grande A, Marangi G, et al. Contribution of major amyotrophic lateral sclerosis genes to the etiology of sporadic disease. Neurology. 2012;79:66–72. doi: 10.1212/WNL.0b013e31825dceca. [DOI] [PubMed] [Google Scholar]
- 41.Prudencio M, Hart PJ, Borchelt DR, Andersen PM. Variation in aggregation propensities among ALS-associated variants of SOD1: correlation to human disease. Hum Mol Genet. 2009;18:3217–26. doi: 10.1093/hmg/ddp260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mackenzie IRA, Bigio EH, Ince PG, Geser F, Neumann M, Cairns NJ, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–34. doi: 10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 43.Wicks P, Abrahams S, Papps B, Al-Chalabi A, Shaw CE, Leigh PN, et al. SOD1 and cognitive dysfunction in familial amyotrophic lateral sclerosis. J Neurol. 2009;256:234–41. doi: 10.1007/s00415-009-0078-0. [DOI] [PubMed] [Google Scholar]
- 44.Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–72. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kwiatkowski TJ, Bosco DA, Leclerc AL, Tamrazian E, Vanderburg CR, Russ C, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–8. doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 46.Vance C, Rogelj B, Hortobágyi T, De Vos KJ, Nishimura AL, Sreedharan J, et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009;323:1208–11. doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baloh RH. How do the RNA-binding proteins TDP-43 and FUS relate to amyotrophic lateral sclerosis and frontotemporal degeneration, and to each other? Curr Opin Neurol. 2012;25:701–7. doi: 10.1097/WCO.0b013e32835a269b. [DOI] [PubMed] [Google Scholar]
- 48.Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, et al. Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature. 2013;495:467–73. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Johnson JO, Pioro EP, Boehringer A, Chia R, Feit H, Renton AE, et al. Mutations in the Matrin 3 gene cause familial amyotrophic lateral sclerosis. Nat Neurosci. 2014;17:664–6. doi: 10.1038/nn.3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–39. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–77. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 52.Gal J, Kuang L, Barnett KR, Zhu BZ, Shissler SC, Korotkov KV, et al. ALS mutant SOD1 interacts with G3BP1 and affects stress granule dynamics. Acta Neuropathol. 2016;132:563–76. doi: 10.1007/s00401-016-1601-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fecto F, Yan J, Vemula SP, Liu E, Yang Y, Chen W, et al. SQSTM1 mutations in familial and sporadic amyotrophic lateral sclerosis. Arch Neurol. 2011;68:1440–6. doi: 10.1001/archneurol.2011.250. [DOI] [PubMed] [Google Scholar]
- 54.Maruyama H, Morino H, Ito H, Izumi Y, Kato H, Watanabe Y, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–6. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 55.Deng HX, Chen W, Hong ST, Boycott KM, Gorrie GH, Siddique N, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–5. doi: 10.1038/nature10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, et al. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–64. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Freischmidt A, Müller K, Ludolph AC, Weishaupt JH, Andersen PM. Association of mutations in TBK1 with sporadic and familial amyotrophic lateral sclerosis and frontotemporal dementia. JAMA Neurol. 2016;74:110–3. doi: 10.1001/jamaneurol.2016.3712. [DOI] [PubMed] [Google Scholar]
- 58.Freischmidt A, Wieland T, Richter B, Ruf W, Schaeffer V, Müller K, et al. Haploinsufficiency of TBK1 causes familial ALS and fronto-temporal dementia. Nat Neurosci. 2015;18:631–6. doi: 10.1038/nn.4000. [DOI] [PubMed] [Google Scholar]
- 59.Williams KL, Topp S, Yang S, Smith B, Fifita JA, Warraich ST, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee A, Rayner SL, Gwee SSL, De Luca A, Shahheydari H, Sundaramoorthy V, et al. Pathogenic mutation in the ALS/FTD gene, CCNF, causes elevated Lys48-linked ubiquitylation and defective autophagy. Cell Mol Life Sci. 2017;7:11253. doi: 10.1007/s00018-017-2632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–13. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Aulas A, Vande Velde C. Alterations in stress granule dynamics driven by TDP-43 and FUS: a link to pathological inclusions in ALS? Front Cell Neurosci. 2015;9:423. doi: 10.3389/fncel.2015.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chen YZ, Bennett CL, Huynh HM, Blair IP, Puls I, Irobi J, et al. DNA/RNA helicase gene mutations in a form of juvenile amyotrophic lateral sclerosis (ALS4) Am J Hum Genet. 2004;74:1128–35. doi: 10.1086/421054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–75. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, et al. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 2015;163:123–33. doi: 10.1016/j.cell.2015.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, et al. Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol Cell. 2017;65:1044–55.e5. doi: 10.1016/j.molcel.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackenzie IR, Nicholson AM, Sarkar M, Messing J, Purice MD, Pottier C, et al. TIA1 mutations in amyotrophic lateral sclerosis and frontotemporal dementia promote phase separation and alter stress granule dynamics. Neuron. 2017;95:808–16.e9. doi: 10.1016/j.neuron.2017.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Puls I, Jonnakuty C, LaMonte BH, Holzbaur ELF, Tokito M, Mann E, et al. Mutant dynactin in motor neuron disease. Nat Genet. 2003;33:455–6. doi: 10.1038/ng1123. [DOI] [PubMed] [Google Scholar]
- 69.Wu CH, Fallini C, Ticozzi N, Keagle PJ, Sapp PC, Piotrowska K, et al. Mutations in the profilin 1 gene cause familial amyotrophic lateral sclerosis. Nature. 2012;488:499–503. doi: 10.1038/nature11280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Figlewicz DA, Krizus A, Martinoli MG, Meininger V, Dib M, Rouleau GA, et al. Variants of the heavy neurofilament subunit are associated with the development of amyotrophic lateral sclerosis. Hum Mol Genet. 1994;3:1757–61. doi: 10.1093/hmg/3.10.1757. [DOI] [PubMed] [Google Scholar]
- 71.Smith BN, Ticozzi N, Fallini C, Gkazi AS, Topp S, Kenna KP, et al. Exome-wide rare variant analysis identifies TUBA4A mutations associated with familial ALS. Neuron. 2014;84:324–31. doi: 10.1016/j.neuron.2014.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Coppola G, Chinnathambi S, Lee JJ, Dombroski BA, Baker MC, Soto-Ortolaza AI, et al. Evidence for a role of the rare p.A152T variant in MAPT in increasing the risk for FTD-spectrum and Alzheimer’s diseases. Hum Mol Genet. 2012;21:3500–12. doi: 10.1093/hmg/dds161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brenner D, Müller K, Wieland T, Weydt P, Böhm S, Lulé D, et al. NEK1 mutations in familial amyotrophic lateral sclerosis. Brain. 2016;139:e28. doi: 10.1093/brain/aww033. [DOI] [PubMed] [Google Scholar]
- 74.Kenna KP, van Doormaal PTC, Dekker AM, Ticozzi N, Kenna BJ, Diekstra FP, et al. NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48:1037–42. doi: 10.1038/ng.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–56. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–68. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Majounie E, Renton AE, Mok K, Dopper EGP, Waite A, Rollinson S, et al. Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11:323–30. doi: 10.1016/S1474-4422(12)70043-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.van der Zee J, Gijselinck I, Theuns J, Vandenbulcke M, Sieben A, Corsmit E, et al. A pan-European study of the C9orf72 repeat associated with FTLD: geographic prevalence, genomic instability, and intermediate repeats. Hum Mutat. 2013;34:363–73. doi: 10.1002/humu.22244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Gijselinck I, Van Langenhove T, van der Zee J, Sleegers K, Philtjens S, Kleinberger G, et al. A C9orf72 promoter repeat expansion in a Flanders-Belgian cohort with disorders of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum: a gene identification study. Lancet Neurol. 2012;11:54–65. doi: 10.1016/S1474-4422(11)70261-7. [DOI] [PubMed] [Google Scholar]
- 80.Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, et al. The C9orf72GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science. 2013;339:1335–8. doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- 81.Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, et al. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell. 2016;167:774–88.e17. doi: 10.1016/j.cell.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee YB, Chen HJ, Peres JN, Gomez-Deza J, Attig J, Stalekar M, et al. Hexanucleotide repeats in ALS/FTD form length-dependent RNA foci, sequester RNA binding proteins, and are neurotoxic. Cell Rep. 2013;5:1178–86. doi: 10.1016/j.celrep.2013.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Heusler AR, Mistry NA, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–28. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin WL, DeJesus-Hernandez M, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–46. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haeusler AR, Donnelly CJ, Periz G, Simko EAJ, Shaw PG, Kim MS, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, et al. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci. 2016;19:668–77. doi: 10.1038/nn.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lopez-Gonzalez R, Lu Y, Gendron TF, Karydas A, Tran H, Yang D, et al. Poly(GR) in C9ORF72-related ALS/FTD compromises mitochondrial function and increases oxidative stress and DNA damage in iPSC-derived motor neurons. Neuron. 2016;92:383–91. doi: 10.1016/j.neuron.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Sellier C, Campanari ML, Julie Corbier C, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–97. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, et al. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.O’Rourke JG, Bogdanik L, Yáñez A, Lall D, Wolf AJ. Muhammad AKMG et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–9. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Sivadasan R, Hornburg D, Drepper C, Frank N, Jablonka S, Hansel A, et al. C9ORF72 interaction with cofilin modulates actin dynamics in motor neurons. Nat Neurosci. 2016;19:1610–8. doi: 10.1038/nn.4407. [DOI] [PubMed] [Google Scholar]
- 92.Aronica E, Baas F, Iyer A, Asbroek ten AL, Morello G, Cavallaro S. Molecular classification of amyotrophic lateral sclerosis by unsupervised clustering of gene expression in motor cortex. Neurobiol Dis. 2015;74:359–76. doi: 10.1016/j.nbd.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 93.Gjoneska E, Pfenning AR, Mathys H, Quon G, Kundaje A, Tsai LH, et al. Conserved epigenomic signals in mice and humans reveal immune basis of Alzheimer’s disease. Nature. 2015;518:365–9. doi: 10.1038/nature14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Srinivasan K, Friedman BA, Larson JL, Lauffer BE, Goldstein LD, Appling LL, et al. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Chiu IM, Morimoto ETA, Goodarzi H, Liao JT, O’Keeffe S, Phatnani HP, et al. A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 2013;4:385–401. doi: 10.1016/j.celrep.2013.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Krasemann S, Madore C, Cialic R, Baufeld C, Calcagno N, El Fatimy R, et al. The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity. 2017;47:566–81.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boillée S, Yamanaka K, Lobsiger CS, Copeland NG, Jenkins NA, Kassiotis G, et al. Onset and progression in inherited ALS determined by motor neurons and microglia. Science. 2006;312:1389–92. doi: 10.1126/science.1123511. [DOI] [PubMed] [Google Scholar]
- 98.Beers DR, Henkel JS, Xiao Q, Zhao W, Wang J, Yen AA, et al. Wild-type microglia extend survival in PU.1 knockout mice with familial amyotrophic lateral sclerosis. Proc Natl Acad Sci USA. 2006;103:16021–6. doi: 10.1073/pnas.0607423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gerber YN, Sabourin JC, Rabano M, Vivanco MDM, Perrin FE. Early functional deficit and microglial disturbances in a mouse model of amyotrophic lateral sclerosis. PLoS ONE. 2012;7:e36000. doi: 10.1371/journal.pone.0036000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Weydt P, Yuen EC, Ransom BR, Möller T. Increased cytotoxic potential of microglia from ALS-transgenic mice. Glia. 2004;48:179–82. doi: 10.1002/glia.20062. [DOI] [PubMed] [Google Scholar]
- 101.Dibaj P, Steffens H, Zschüntzsch J, Kirchhoff F, Schomburg ED, Neusch C. In vivo imaging reveals rapid morphological reactions of astrocytes towards focal lesions in an ALS mouse model. Neurosci Lett. 2011;497:148–51. doi: 10.1016/j.neulet.2011.04.049. [DOI] [PubMed] [Google Scholar]
- 102.Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165:921–35. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.D’Souza I, Schellenberg GD. Regulation of tau isoform expression and dementia. Biochim Biophys Acta. 2005;1739:104–15. doi: 10.1016/j.bbadis.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 104.Yoshiyama Y, Higuchi M, Zhang B, Huang SM, Iwata N, Saido TC, et al. Synapse loss and microglial activation precede tangles in a P301S tauopathy mouse model. Neuron. 2007;53:337–51. doi: 10.1016/j.neuron.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 105.Asai H, Ikezu S, Tsunoda S, Medalla M, Luebke J, Haydar T, et al. Depletion of microglia and inhibition of exosome synthesis halt tau propagation. Nat Neurosci. 2015;18:1584–93. doi: 10.1038/nn.4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Re DB, Le Verche V, Yu C, Amoroso MW, Politi KA, Phani S, et al. Necroptosis drives motor neuron death in models of both sporadic and familial ALS. Neuron. 2014;81:1001–8. doi: 10.1016/j.neuron.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Nagai M, Re DB, Nagata T, Chalazonitis A, Jessell TM, Wichterle H, et al. Astrocytes expressing ALS-linked mutated SOD1 release factors selectively toxic to motor neurons. Nat Neurosci. 2007;10:615–22. doi: 10.1038/nn1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yamanaka K, Chun SJ, Boillée S, Fujimori-Tonou N, Yamashita H, Gutmann DH, et al. Astrocytes as determinants of disease progression in inherited amyotrophic lateral sclerosis. Nat Neurosci. 2008;11:251–3. doi: 10.1038/nn2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kang SH, Li Y, Fukaya M, Lorenzini I, Cleveland DW, Ostrow LW, et al. Degeneration and impaired regeneration of gray matter oligodendrocytes in amyotrophic lateral sclerosis. Nat Neurosci. 2013;16:571–9. doi: 10.1038/nn.3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.van Rheenen W, Shatunov A, Dekker AM, McLaughlin RL, Diekstra FP, Pulit SL, et al. Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48:1043–8. doi: 10.1038/ng.3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, et al. A survey of human brain transcriptome diversity at the single cell level. Proc Natl Acad Sci USA. 2015;112:7285–90. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, et al. Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature. 2015;518:547–51. doi: 10.1038/nature13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nayak D, Roth TL, McGavern DB. Microglia development and function. Annu Rev Immunol. 2014;32:367–402. doi: 10.1146/annurev-immunol-032713-120240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ueno M, Fujita Y, Tanaka T, Nakamura Y, Kikuta J, Ishii M, et al. Layer V cortical neurons require microglial support for survival during postnatal development. Nat Neurosci. 2013;16:543–51. doi: 10.1038/nn.3358. [DOI] [PubMed] [Google Scholar]
- 115.Meucci O, Fatatis A, Simen AA, Miller RJ. Expression of CX3CR1 chemokine receptors on neurons and their role in neuronal survival. Proc Natl Acad Sci USA. 2000;97:8075–80. doi: 10.1073/pnas.090017497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Limatola C, Ransohoff RM. Modulating neurotoxicity through CX3CL1/CX3CR1 signaling. Front Cell Neurosci. 2014;8:229. doi: 10.3389/fncel.2014.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Brown GC, Neher JJ. Microglial phagocytosis of live neurons. Nat Rev Neurosci. 2014;15:209–16. doi: 10.1038/nrn3710. [DOI] [PubMed] [Google Scholar]
- 118.Paolicelli RC, Bolasco G, Pagani F, Maggi L, Scianni M, Panzanelli P, et al. Synaptic pruning by microglia is necessary for normal brain development. Science. 2011;333:1456–8. doi: 10.1126/science.1202529. [DOI] [PubMed] [Google Scholar]
- 119.Casano AM, Albert M, Peri F. Developmental apoptosis mediates entry and positioning of microglia in the Zebrafish brain. Cell Rep. 2016;16:897–906. doi: 10.1016/j.celrep.2016.06.033. [DOI] [PubMed] [Google Scholar]
- 120.Peri F, Nüsslein-Volhard C. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell. 2008;133:916–27. doi: 10.1016/j.cell.2008.04.037. [DOI] [PubMed] [Google Scholar]
- 121.Neumann H, Kotter MR, Franklin RJM. Debris clearance by microglia: an essential link between degeneration and regeneration. Brain. 2009;132:288–95. doi: 10.1093/brain/awn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Skripuletz T, Hackstette D, Bauer K, Gudi V, Pul R, Voss E, et al. Astrocytes regulate myelin clearance through recruitment of microglia during cuprizone-induced demyelination. Brain. 2013;136:147–67. doi: 10.1093/brain/aws262. [DOI] [PubMed] [Google Scholar]
- 123.Safaiyan S, Kannaiyan N, Snaidero N, Brioschi S, Biber K, Yona S, et al. Age-related myelin degradation burdens the clearance function of microglia during aging. Nat Neurosci. 2016;19:995–8. doi: 10.1038/nn.4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hong S, Beja-Glasser VF, Nfonoyim BM, Frouin A, Li S, Ramakrishnan S, et al. Complement and microglia mediate early synapse loss in Alzheimer mouse models. Science. 2016;352:712–6. doi: 10.1126/science.aad8373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lampron A, Larochelle A, Laflamme N, Préfontaine P, Plante MM, Sánchez MG, et al. Inefficient clearance of myelin debris by microglia impairs remyelinating processes. J Exp Med. 2015;212:481–95. doi: 10.1084/jem.20141656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kawabori M, Kacimi R, Kauppinen T, Calosing C, Kim JY, Hsieh CL, et al. Triggering receptor expressed on myeloid cells 2 (TREM2) deficiency attenuates phagocytic activities of microglia and exacerbates ischemic damage in experimental stroke. J Neurosci. 2015;35:3384–96. doi: 10.1523/JNEUROSCI.2620-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chan G, White CC, Winn PA, Cimpean M, Replogle JM, Glick LR, et al. CD33 modulates TREM2: convergence of Alzheimer loci. Nat Neurosci. 2015;18:1556–8. doi: 10.1038/nn.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Cruchaga C, Kauwe JSK, Harari O, Jin SC, Cai Y, Karch CM, et al. GWAS of cerebrospinal fluid tau levels identifies risk variants for Alzheimer’s disease. Neuron. 2013;78:256–68. doi: 10.1016/j.neuron.2013.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Cady J, Koval ED, Benitez BA, Zaidman C, Jockel-Balsarotti J, Allred P, et al. TREM2 variant p.R47H as a risk factor for sporadic amyotrophic lateral sclerosis. JAMA Neurol. 2014;71:449–53. doi: 10.1001/jamaneurol.2013.6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lill CM, Rengmark A, Pihlstrøm L, Fogh I, Shatunov A, Sleiman PM, et al. The role of TREM2 R47H as a risk factor for Alzheimer’s disease, frontotemporal lobar degeneration, amyotrophic lateral sclerosis, and Parkinson’s disease. Alzheimers Dement. 2015;11:1407–16. doi: 10.1016/j.jalz.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kawamata T, Akiyama H, Yamada T, McGeer PL. Immunologic reactions in amyotrophic lateral sclerosis brain and spinal cord tissue. Am J Pathol. 1992;140:691–707. [PMC free article] [PubMed] [Google Scholar]
- 132.Engelhardt JI, Appel SH. IgG reactivity in the spinal cord and motor cortex in amyotrophic lateral sclerosis. Arch Neurol. 1990;47:1210–6. doi: 10.1001/archneur.1990.00530110068019. [DOI] [PubMed] [Google Scholar]
- 133.Sargsyan SA, Monk PN, Shaw PJ. Microglia as potential contributors to motor neuron injury in amyotrophic lateral sclerosis. Glia. 2005;51:241–53. doi: 10.1002/glia.20210. [DOI] [PubMed] [Google Scholar]
- 134.Alexianu ME, Kozovska M, Appel SH. Immune reactivity in a mouse model of familial ALS correlates with disease progression. Neurology. 2001;57:1282–9. doi: 10.1212/WNL.57.7.1282. [DOI] [PubMed] [Google Scholar]
- 135.Drachman DB, Kuncl RW. Amyotrophic lateral sclerosis: an unconventional autoimmune disease? Ann Neurol. 1989;26:269–74. doi: 10.1002/ana.410260214. [DOI] [PubMed] [Google Scholar]
- 136.Brettschneider J, Libon DJ, Toledo JB, Xie SX, McCluskey L, Elman L, et al. Microglial activation and TDP-43 pathology correlate with executive dysfunction in amyotrophic lateral sclerosis. Acta Neuropathol. 2012;123:395–407. doi: 10.1007/s00401-011-0932-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Zurcher NR, Loggia ML, Lawson R, Chonde DB, Izquierdo-Garcia D, Yasek JE, et al. Increased in vivo glial activation in patients with amyotrophic lateral sclerosis: assessed with [(11)C]-PBR28. Neuroimage Clin. 2015;7:409–14. doi: 10.1016/j.nicl.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Alshikho MJ, Zurcher NR, Loggia ML, Cernasov P, Chonde DB, Izquierdo-Garcia D, et al. Glial activation colocalizes with structural abnormalities in amyotrophic lateral sclerosis. Neurology. 2016;87:2554–61. doi: 10.1212/WNL.0000000000003427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cagnin A, Rossor M, Sampson EL, Mackinnon T, Banati RB. In vivo detection of microglial activation in frontotemporal dementia. Ann Neurol. 2004;56:894–7. doi: 10.1002/ana.20332. [DOI] [PubMed] [Google Scholar]
- 140.Brettschneider J, Toledo JB, Van Deerlin VM, Elman L, McCluskey L. Lee VM-Y et al. Microglial activation correlates with disease progression and upper motor neuron clinical symptoms in amyotrophic lateral sclerosis. PLoS ONE. 2012;7:e39216. doi: 10.1371/journal.pone.0039216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Turner MR, Cagnin A, Turkheimer FE, Miller CCJ, Shaw CE, Brooks DJ, et al. Evidence of widespread cerebral microglial activation in amyotrophic lateral sclerosis: an [11C](R)-PK11195 positron emission tomography study. Neurobiol Dis. 2004;15:601–9. doi: 10.1016/j.nbd.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 142.Butovsky O, Siddiqui S, Gabriely G, Lanser AJ, Dake B, Murugaiyan G, et al. Modulating inflammatory monocytes with a unique microRNA gene signature ameliorates murine ALS. J Clin Invest. 2012;122:3063–87. doi: 10.1172/JCI62636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, et al. C9orf72 BAC Mouse Model With Motor Deficits And Neurodegenerative Features of ALS/FTD. Neuron. 2016;90:521–34. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 144.O’Rourke JG, Bogdanik L, Muhammad AKMG, Gendron TF, Kim KJ, Austin A, et al. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, et al. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron. 2015;88:902–9. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, et al. Neurodegeneration. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science. 2015;348:1151–4. doi: 10.1126/science.aaa9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- 148.Bendotti C, Marino M, Cheroni C, Fontana E, Crippa V, Poletti A, et al. Dysfunction of constitutive and inducible ubiquitin-proteasome system in amyotrophic lateral sclerosis: implication for protein aggregation and immune response. Prog Neurobiol. 2012;97:101–26. doi: 10.1016/j.pneurobio.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 149.Barmada SJ, Serio A, Arjun A, Bilican B, Daub A, Ando DM, et al. Autophagy induction enhances TDP43 turnover and survival in neuronal ALS models. Nat Chem Biol. 2014;10:677–85. doi: 10.1038/nchembio.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–74. doi: 10.1016/0014-5793(93)80398-E. [DOI] [PubMed] [Google Scholar]
- 151.Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- 152.Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000;19:5720–8. doi: 10.1093/emboj/19.21.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–9. doi: 10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 154.Komatsu M, Waguri S, Chiba T, Murata S, Iwata JI, Tanida I, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–4. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 155.Pilli M, Arko-Mensah J, Ponpuak M, Roberts E, Master S, Mandell MA, et al. TBK-1 promotes autophagy-mediated antimicrobial defense by controlling autophagosome maturation. Immunity. 2012;37:223–34. doi: 10.1016/j.immuni.2012.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Matsumoto G, Shimogori T, Hattori N, Nukina N. TBK1 controls autophagosomal engulfment of polyubiquitinated mitochondria through p62/SQSTM1 phosphorylation. Hum Mol Genet. 2015;24:4429–42. doi: 10.1093/hmg/ddv179. [DOI] [PubMed] [Google Scholar]
- 157.Moore AS, Holzbaur ELF. Dynamic recruitment and activation of ALS-associated TBK1 with its target optineurin are required for efficient mitophagy. Proc Natl Acad Sci USA. 2016;113:E3349–58. doi: 10.1073/pnas.1523810113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Richter B, Sliter DA, Herhaus L, Stolz A, Wang C, Beli P, et al. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc Natl Acad Sci USA. 2016;113:4039–44. doi: 10.1073/pnas.1523926113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MMK, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 2004;304:1158–60. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 160.Kitada T, Asakawa S, Hattori N, Matsumine H, Yamamura Y, Minoshima S, et al. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 1998;392:605–8. doi: 10.1038/33416. [DOI] [PubMed] [Google Scholar]
- 161.Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang C, Burman JL, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015;524:309–14. doi: 10.1038/nature14893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, et al. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J Biol Chem. 2007;282:24131–45. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 163.Zhong Z, Umemura A, Sanchez-Lopez E, Liang S, Shalapour S, Wong J, et al. NF-κB restricts inflammasome activation via elimination of damaged mitochondria. Cell. 2016;164:896–910. doi: 10.1016/j.cell.2015.12.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RAK, Levina V, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–95. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–4. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 166.Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–9. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 167.Jian J, Zhao S, Tian QY, Liu H, Zhao Y, Chen WC, et al. Association between progranulin and Gaucher disease. EBioMedicine. 2016;11:127–37. doi: 10.1016/j.ebiom.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Ito Y, Ofengeim D, Najafov A, Das S, Saberi S, Li Y, et al. RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science. 2016;353:603–8. doi: 10.1126/science.aaf6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Chang L, Monteiro MJ. Defective proteasome delivery of polyubiquitinated proteins by ubiquilin-2 proteins containing ALS mutations. PLoS ONE. 2015;10:e0130162. doi: 10.1371/journal.pone.0130162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Rothenberg C, Srinivasan D, Mah L, Kaushik S, Peterhoff CM, Ugolino J, et al. Ubiquilin functions in autophagy and is degraded by chaperone-mediated autophagy. Hum Mol Genet. 2010;19:3219–32. doi: 10.1093/hmg/ddq231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Keren-Shaul H, Spinrad A, Weiner A, Matcovitch-Natan O, Dvir-Szternfeld R, Ulland TK, et al. A unique microglia type associated with restricting development of Alzheimer’s disease. Cell. 2017;169:1276. doi: 10.1016/j.cell.2017.05.018. [DOI] [PubMed] [Google Scholar]