Abstract
Diazotrophs, both Bacteria and Archaea, capable of fixing nitrogen (N2), are present in the tissues and mucous, of corals and can supplement the coral holobiont nitrogen budget with fixed nitrogen (N) in the form of ammonia (NH3). Stylophora pistillata from Heron Island on the Great Barrier Reef collected at 5 and 15 m, and experimentally manipulated in the laboratory, showed that the rates of net photosynthesis, steady state quantum yields of photosystem II (PSII) fluorescence (∆Fv/Fm′) and calcification varied based on irradiance as expected. Rates of N2 fixation were, however, invariant across treatments while the amount of fixed N contributing to Symbiodinium spp. N demand is irradiance dependent. Additionally, both the Symbiodinium and diazotrophic communities are significantly different based on depth, and novel Cluster V nifH gene phylotypes, which are not known to fix nitrogen, were recovered. A functional analysis using PICRUSt also showed that shallow corals were enriched in genes involved in nitrogen metabolism, and N2 fixation specifically. Corals have evolved a number of strategies to derive nitrogen from organic (e.g., heterotrophic feeding) and inorganic sources (e.g., N2 fixation) to maintain critical pathways such as protein synthesis to succeed ecologically in nitrogen-limited habitats.
Introduction
The coral holobiont consists of fungi, viruses, alveolates (dinoflagellates and apicomplexa) and prokaryotes that are associated with the mucous, skeleton and tissues of scleractinian corals (e.g., [1–8]). One of the most commonly reported functions for these symbiotic prokaryotes is nutrient cycling, and in particular nitrogen biogeochemistry [7]. The nitrogen cycle in the oceans is mediated principally by prokaryotic transformations of organic sources of nitrogen (e.g., N2 fixation, nitrification, denitrification) or by inputs of new nitrogen from terrestrial, atmospheric or oceanic sources [9, 10]. In coral reef ecosystems, new nitrogen can be obtained from coastal runoff or atmospheric deposition but the major source of new nitrogen is N2 fixation [11–13]. High rates of N2 fixation have been measured on coral reefs [11, 13] that are mediated by both cyanobacteria and heterotrophic bacteria [10, 12]. Both DON and particulate organic nitrogen (PON) are then released into the benthic environment for use by other microbes and trophic groups such as detrivores [14, 15]. In addition to free-living nitrogen fixers several important members of the reef community have the capability to fix nitrogen through symbiotic associations with nitrogen fixing, or diazotrophic, bacteria [16, 17].
In corals N2 fixation has previously been attributed to prokaryotes known as diazotrophs “associated” with living coral tissue [18]. Diazotrophs, both Bacteria and Archaea, capable of N2 fixation are present in the tissues and mucus of corals and can supplement the coral holobiont nitrogen budget with fixed N [2, 19–22]. These diazotrophs express nitrogenase, a uniquely prokaryotic enzyme, responsible for N2 fixation generally, and in those corals harboring them (e.g., [2, 23–25]). They include both heterotrophic as well as cyanobacterial nitrogen fixers in corals [20, 22], and one of the dominant non-photosynthetic taxa often recovered are rhizobia [20, 22] in the Order Rhizobiales.
The scleractinian coral, Stylophora pistillata, has been the subject of many physiological studies including measurements of N2 fixation. Grover et al. [26] detected virtually no N2 fixation in S. pistillata from the Gulf of Aqaba using tracer gas (15N2) and the “bubble” technique for dissolving the tracer in seawater. One of the important methodological concerns with using the 15N2 tracer approach is adequately dissolving the gas in seawater using the “bubble” compared to “constant enrichment” techniques [27, 28]. Use of the bubble technique significantly underestimates the rate of N2 fixation, and subsequent estimations of new nitrogen introduction into oceanic ecosystems [29, 30]. In regards to the acetylene reduction vs. tracer gas approaches, there are distinct advantages and disadvantages including the fact that the acetylene reduction technique measures gross N2 fixation while the tracer approach measures net N2 fixation into organic material [28, 31].
Here, we use the constant enrichment tracer approach to examine the effects of irradiance on rates of N2 fixation and the contribution of fixed nitrogen products to the N requirements of Symbiodinium spp. [19, 24] in the coral Stylophora pistillata from Heron Island on the Great Barrier Reef. Additionally, on the same samples we quantify the microbiome including Bacteria, Archaea and Symbiodinium spp. diversity, as well as the diazotrophic members of the prokaryotic communities, using a metagenetic approach (i.e., targeted next generation sequencing of 16S rRNA, ITS, and nifH genes). Additional physiological measurements including active fluorescence and rates of photosynthesis and calcification were carried simultaneously on the same samples to provide a quantitative picture of the physiological state of corals fixing nitrogen, who is fixing nitrogen under varying irradiances and the fate of fixed nitrogen in this important and common coral species on the Great Barrier Reef.
Methods
Site characterization and seawater sample processing
All work was conducted during the Austral winter (March 2015) by SCUBA at Tenements Reef northeast of Heron Island Research Station (HIRS), Australia (Lat: S 23° 26′ 33.7535″, Long: E 151° 54′ 54.5983″). Temperature (°C) and irradiance (Ed) of photosynthetically active radiation (PAR: 400–700 nm), reported as μmol quanta m−2 s−1, were taken hourly over several days using HOBO Pendant Temperature/Light loggers at 5 and 15 m depth (n = 3 for each depth). Measurements of irradiance were in lumens m−2 and converted to quanta as described by Piniack and Brown (2008) [61] calibrating against a LiCor cosine-corrected, planar, sensor (LI 192SA).
Seawater samples (n = 5 × 1 l) were collected at 15 m depth and returned to the laboratory where they were each immediately filtered onto GF/F filters. The GF/F filter was placed in a 2 ml cryovial with DNA preservation buffer [32] and stored at −20 °C and DNA extracted (See Supplementary Information). The seawater filtrate was frozen, transported to the University of New Hampshire (UNH) and analyzed for nitrate (NO3, µmol N l−1) and nitrite (NO2, µmol N l−1) using high throughput colorimetric assays on a Smartchem Chemistry Analyzer, and dissolved organic carbon (DOC, µmol C l−1) and dissolved organic nitrogen (DON, µmol N l−1) using high temperature catalytic oxidation and high temperature oxidation with chemiluminescent detection respectively the at the University of New Hampshire Water Quality Analysis Laboratory.
Coral collection, photosynthesis, calcification, and N2 fixation
Branches from individual colonies of Stylophora pistillata were collected at 5 and 15 m depth (n = 7 from each depth). Before collections, and before and after experiments, measurements of ∆Fv/Fm′ quantum yield of PSII fluorescence were measured on each coral using a diving Pulse Amplitude Modulated (PAM, blue actinic version) fluorometer (Walz, Germany) from the side of each colony branch in standard mode at a fixed distance of 1 cm with a universal sample holder [33]. Measurements were made from approximately 0930 to 1130 hours. in a single day and divers wore nitrile gloves to collect branches snipped with bone cutters from individual corals and placed in labeled Whirl-Pak® bags. These samples were returned to HIRS and placed onto plastic holders in running seawater tables at HIRS for <24 h to recover at ~550 µmol quanta m−2 s−1 (~1900–2000 µmol quanta m−2 s−1 ambient under one layer of neutral density cloth) of Ed using a LiCor 192SA cosine-corrected planar sensor.
Branches collected from seven colonies at each depth were incubated reciprocally under two irradiance conditions; low light (~150 μmol quanta m−2 s−1) representative of 15 m depth and high light (~500 μmol quanta m−2 s−1) representative of 5 m depth respectively. The branches from different depths were randomized regardless of irradiance and placed into clear acrylic incubation chambers (height: 10 cm, radius: 4.5 cm) with a water volume of 0.640 l. The incubation chambers were then placed onto submersible stands in a black flow-through plastic bin that served as a water bath with temperature control (~28 °C) and all chambers were continuously mixed using magnetic stir bars (~ 200 rpm). Sterile (0.22 µm) filtered seawater (FSW) was used for all experiments and amended with 15N2 gas (Supplemental Information), and three temperature and oxygen measurements were taken using a calibrated optode electrode (Hach, Inc.) prior to filling incubation chambers. The irradiance exposure was achieved using white (55%) and blue (45%) LEDs (AI Sol White LED Modules, Clear Choice, Los Angeles, USA) mounted above the incubation chambers. The duration of these incubations was 2.0–2.5 h and each run included a blank chamber to correct for any non-coral oxygen fluxes. The rates of net photosynthesis were calculated as described by Strahl et al. [34].
At the completion of each experimental run ∆Fv/Fm′ readings were taken for each coral branch and the oxygen and temperature measurements taken for each chamber including the blank. Seawater (~50 ml) from each chamber, as well as a time zero sample, were collected into glass, acid washed and rinsed, bottles to be used for calcification measurements by the alkalinity anomaly technique (AT, Supplementary Information) which relies on the stoichiometric relationship where two equivalents of alkalinity are removed per mole of carbonate mineral precipitated [35, 36]. All coral tissues were immediately processed and a 1 ml aliquot of homogenized coral tissue was used to quantify chlorophyll concentrations (see Supplementary Information).
Rates of N2 fixation were calculated separately using aliquots of host (2–3 ml) and Symbiodinium spp. (0.5 ml) homogenates. Host tissue was removed from Symbiodinium samples using previously described protocols [62]. These aliquots were placed on baked (450 °C × 24 h) GF/F filters and dried at 60 °C overnight. Dried filters were sent to the Marine Biological Laboratory, Woods Hole, MA for bulk stable isotopic analyses (SIA). See Lesser et al., [23] and Supplementary Information for details of N2 fixation analysis.
For all rate measurements, the coral branches whose tissues were removed were bleached overnight, dried, and wax dipped for surface area measurements [37]. Rates of net photosynthesis, calcification and N2 fixation were normalized to surface area, and expressed as µmol O2 cm−2 h−1, μmol CaCO3 cm−2 h−1 for net photosynthesis and calcification while N2 fixation was expressed as both nmol N cm−2 h−1 and ng N cm−2 h−1. Net photosynthesis was also normalized to chl a for comparison. These experiments were analyzed using a two-factor analysis of variance (ANOVA) with interaction at a significance level of 5%. No unequal variances were detected using the Hartley’s Fmax test while ratios and percentages were a priori arcsine or log transformed for analysis and back transformed for presentation.
DNA extraction, metagenetics, sequencing and bioinformatics
After each experimental run the tissue was removed from individual coral branches and processed immediately for downstream DNA extraction and sequencing. DNA was extracted from filtered seawater (i.e., GF/F filters) and the preserved tissue pellet using the MOBIO PowerSoil® DNA isolation kit (MOBIO Carlsbad, CA). Details of the tissue removal, DNA extraction and downstream analyses, including the functional analysis using PICRUSt, are given in Supplementary Information. Raw 16S, nifH, and Symbiodinium spp. ITS reads were submitted to the NCBI Sequence Read Archive under BioProject accession number PRJNA390752.
Results
Environmental parameters
During the time frame of our collections around Tenements Reef the ambient seawater temperature was 27.9 °C ± 0.6 (SD) at 5 m (>3000 measurements over 3 d) and 27.4 °C ± 1.1 (SD) at 15 m ( > 6000 measurements over 5 d). Irradiance at solar noon (~1200 hours averaged from 1145 to 1215 hours) was 475 µmol quanta m−2 s−1 ± 74 µmol quanta m−2 s−1 (SD) at 5 m and 130 µmol quanta m−2 s−1 ± 32 µmol quanta m−2 s−1 (SD) at 15 m. The concentration of DOC in the seawater (n = 5) at Tenements Reef was 12.6 µmol C l−1 ± 2.7 (SD) while the concentration of DON (n = 5) was 1.1 µmol N l−1 ± 0.64 (SD). The concentration of NOx (NO3− and NO2−) in seawater was 0.71 µmol N l−1 ± 0.5 (SD).
When corals collected from 5 m and 15 m depths were experimentally incubated at 500 µmol quanta m−2 s−1 and 150 µmol quanta m−2 s−1, respectively, as well as the reciprocal irradiance exposure, their rates of photosynthesis and calcification normalized to surface area showed a significant (ANOVA: F3,24 = 15.79, P = 0.0006), and positive, effect of increasing irradiance but no effect for depth of collection or any interaction between the two factors (Fig. 1a). Post hoc multiple comparisons show that corals exposed to higher irradiances are significantly different than corals exposed to lower irradiances regardless of collection depth (Tukeys HSD, P < 0.05). Similarly, when photosynthetic rate is normalized to chl a there is a positive effect of increasing irradiance (ANOVA: F3,24 = 8.53, P = 0.008), but no effect for depth of collection or any interaction between the two factors (Fig. 1b). Corals collected from 15 m and exposed to higher irradiances exhibit a significant depression in photosynthesis when normalized to chl a (Tukeys HSD, P < 0.05). For calcification measurements using the AT approach treatment samples typically decreased by ~85–170 µmol kg−1 SW relative to controls, which was well within the detectible limit of the titrosampler. Rates of calcification also showed a significant (ANOVA: F3,24 = 19.21, P = 0.0002), and positive, effect of only increasing irradiance but no effect for depth of collection or any interaction between the two factors (Fig. 1a). All ∆Fv/Fm′ of PSII fluorescence before both collections and experiments were normal (0.61 ± 0.05, mean ± SD). An analysis of ∆Fv/Fm′ at the end of the experimental exposures revealed a significant (ANOVA: F3,24 = 12.51, P = 0.0017), and negative, effect of increasing irradiance but no effect for depth of collection or any interaction between the two factors (Fig. 2). The 15N2 enrichment (atom% 15Nexcess) for these 2–2.5 h incubations ranged from 0.0038 to 0.0053 which compares well with other studies utilizing 24 h incubations (e.g., [24]). Rates of N2 fixation normalized to surface area expressed on a molar or weight basis showed no significant effect of depth of collection, irradiance or the interaction of these factors (Fig. 3). Looking at the host vs. Symbiodinium spp. compartment, ~95% of the measured fixed N was found in the Symbiodinium spp. compartment, and CZND showed a significant (ANOVA: F3,24 = 13.36, P = 0.011), and positive, effect of increasing irradiance (150 µmol quanta m−2 s−1, 1.16 % ± 0.29 (SE); 500 µmol quanta m−2 s−1, 3.43% ± 0.55 (SD)), but no effect for depth of collection or any interaction between the two factors.
Fig. 1.
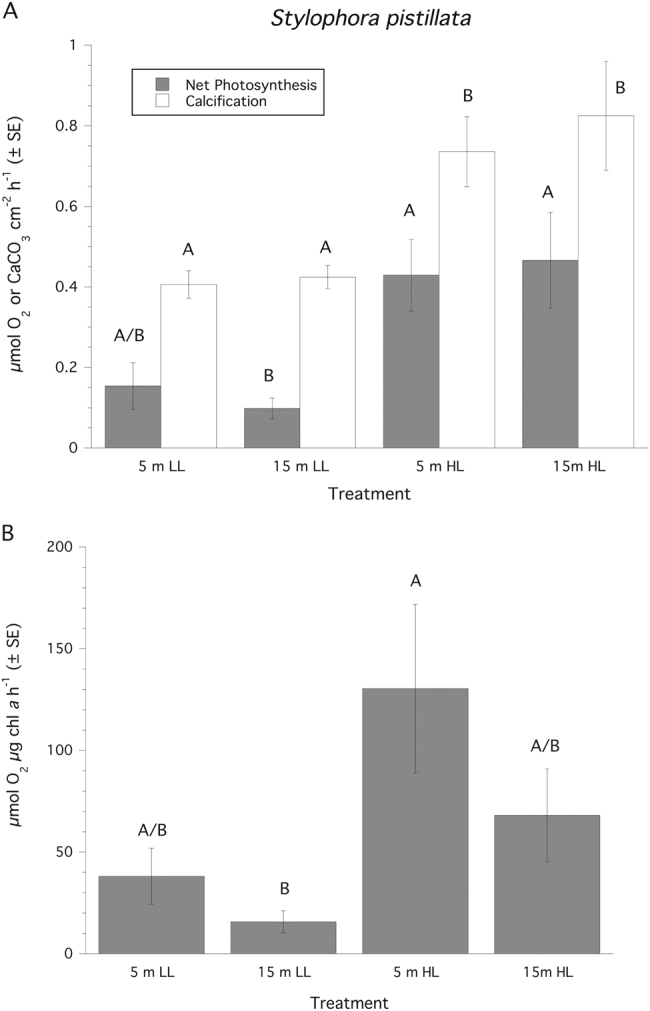
Rates of photosynthesis and calcification normalized to surface area (mean ± SE) for Stylophora pistillata. LL = 150 µmol quanta m−2 s−1, HL = 500 µmol quanta m−2 s−1. Treatment groups (n = 7 samples per group) and for each independent parameter similar superscripts are not significantly different from each other
Fig. 2.
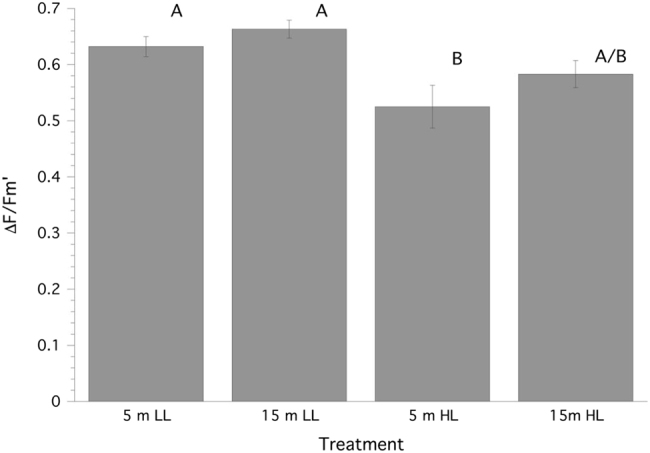
Steady state quantum yield (∆Fv/Fm′) of PSII fluorescence (mean ± SE) for Stylophora pistillata. LL = 150 µmol quanta m−2 s−1, HL = 500 µmol quanta m−2 s−1. Treatment groups (n = 7 samples per group) with similar superscripts are not significantly different from each other
Fig. 3.
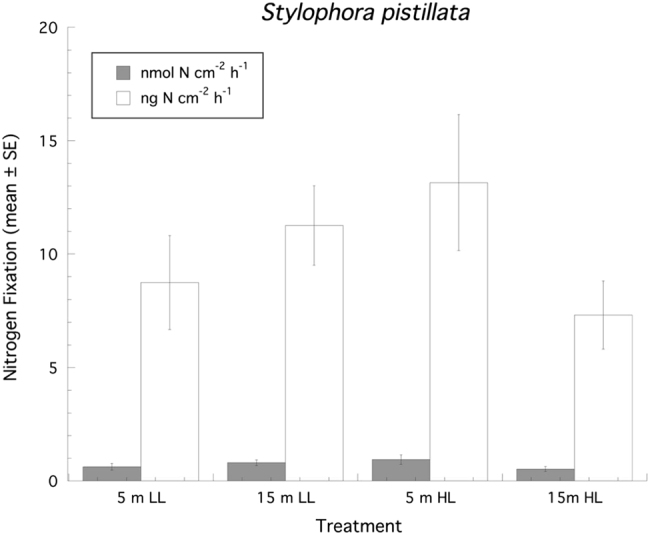
Rates of N2 fixation (n = 7 samples per treatment group) normalized to surface area (mean ± SE) for Stylophora pistillata. LL = 150 µmol quanta m−2 s−1, HL = 500 µmol quanta m−2 s−1
Microbial communities of stylophora pistillata
A total of 1,455,508 reads with an average read length of 253 base pairs were recovered after initial merging and quality trimming, and a total of 1,304,217 reads remained after removing chloroplasts and low diversity OTUs, with an average of 39,522 ± 1,282 reads per sample that ranged from 25,828 to 54,089 total reads per sample. A total of 510,745 OTUs were initially recovered, however 505,557 OTUs had <5 total observations and were removed from the data set, and an additional 169 chloroplast OTUs were removed, leaving a total of 5019 OTUs for subsequent analyses.
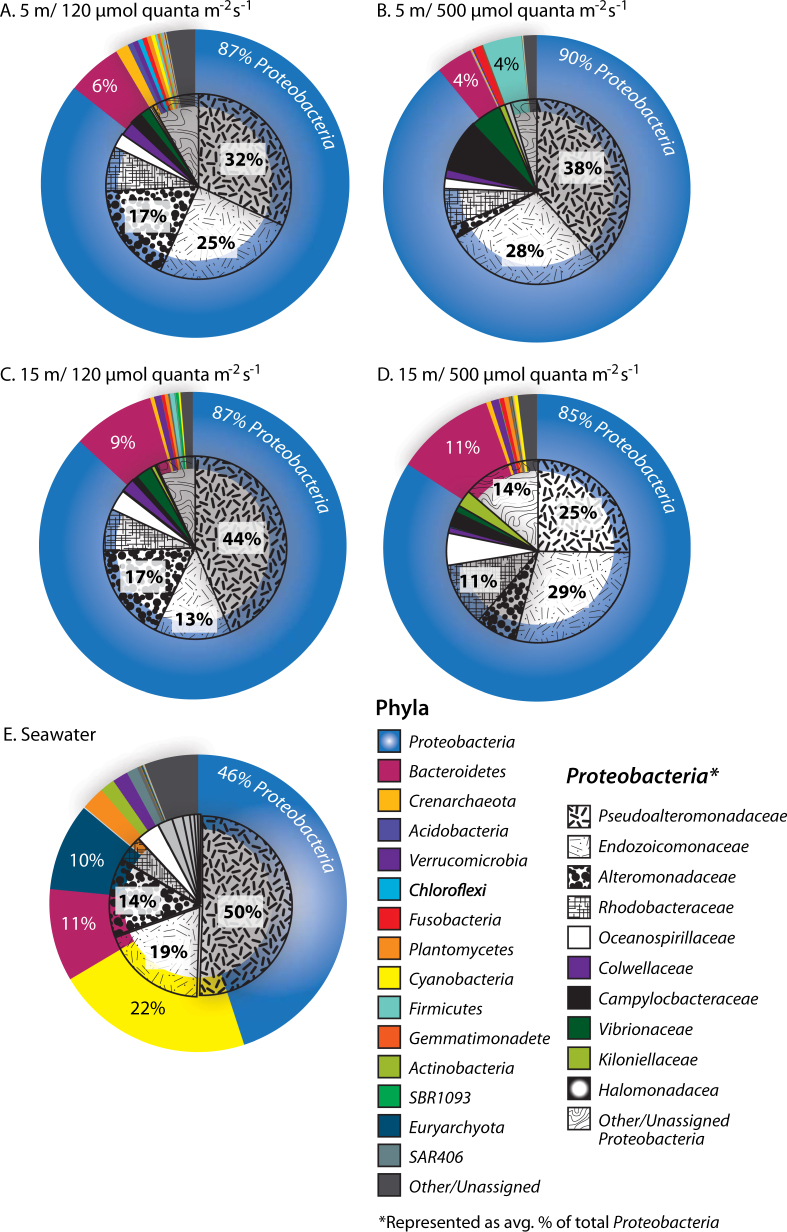
In total, 40 distinct bacterial and archaeal phyla were recovered from Stylophora pistillata experimental samples, which were dominated by OTUs related to members of the Proteobacteria and Bacteroidetes, representing an average of 87% ± 2.0 (SD) and 7.3% ± 3.0, of the total microbiome, respectively (Fig. 4). Within the Proteobacteria (Fig. 4) coral samples were composed of families related to Pseudoalteromonadaceae that averaged 28.5% ± 7.2 (SD) of the total microbiome, as well as Endozoicomonaceae (19.4% ± 5.8), Alteromonadaceae (9.0% ± 6.2), and Rhodobacteraceae (6.7% ± 1.3). Seawater communities (n = 5) were distinct from the coral-associated microbiome (PERMANOVA; F1,31 = 15.6, P < 0.0001, Table S1A), and dominated by OTUs related to members of the Proteobacteria (46%), Cyanobacteria (22%), Bacteroidetes (11%), Euryarchaeota (9.9%), and Planctomycetes (2.6%; Fig. 4).
Fig. 4.
Pie Charts representing average percent contribution of the most dominant prokaryotic phyla (large pies) to the total relative abundance in Stylophora pistillata at: a 5 m, 150 µmol quanta m−2 s−1, b 5 m, 500 µmol quanta m−2 s−1, c 15 m, 150 µmol quanta m−2 s−1, d 15 m, 500 µmol quanta m−2 s−1, e ambient seawater. Additionally, all Proteobacteria families (superimposed small pie charts) are presented across all treatment groups (n = 7 samples per group)
For the 16S derived microbiome, experimental treatments were significantly different from one another (PERMANOVA; F3,24 = 2.4, P < 0.0001; Table S1A). Pairwise comparisons demonstrate that only corals exposed to low irradiances have similar communities while corals exposed to high irradiances, regardless of depth of collection, change their communities significantly. Only 15 one another except corals exposed to the low irradiance (Table S1A). The microbiome of corals exposed to the higher irradiance levels also separate into distinct groups based on sample collection depth (5 vs. 15 m) more readily than corals exposed to low irradiances (Table S1A). Average similarity between groups ranged from ~38% between high irradiance treatments (5 and 15 m) and low irradiance treatments from 5 m to ~45% similarity between low and high irradiance treatments collected from 15 m depth (Table S1A).
Diazotrophic communities using nifH
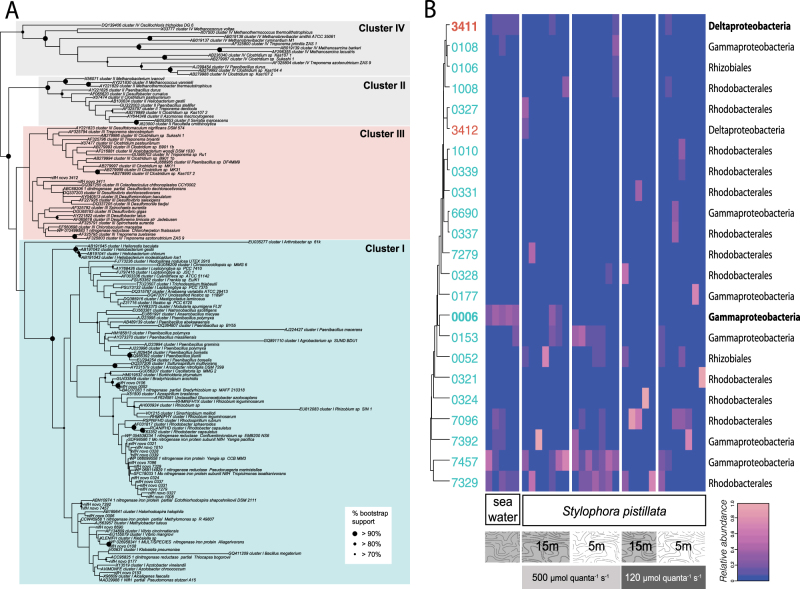
A total of 1,603,178 merged reads were considered during Minimum Entropy Decomposition oligotyping, with an average of 48,581 reads per sample. A total of 219,239 reads (~14%) were assigned to Cluster I phylotypes and 7,035 (~0.4%) were assigned to Cluster III phylotypes. Initial phylotyping by TaxaDiva produced 34 oligotypes that aligned phylogenetically with valid nifH clades and 252 oligotypes that were placed closer to the chlorophyllide reductases Cluster V of nifH phylotypes (Fig. S1) that were more abundant in all coral samples than nifH phylotypes from any other cluster (Fig. S2). Clustering to 95% similarity yielded 23 distinct nifH and 178 non-nifH phylotypes that phylogenetically ally with nifH clusters I–III or cluster V, respectively (Table S2, Fig. 5a, Fig S1). Phylogenetic analysis and nr BLAST searches identified similar taxonomic groups for the de novo nifH phylotypes (Table S3). Notably, of the 23 distinct nifH phylotypes, the majority nested within the Rhodobacterales (12); others nested within Rhizobiales (2), Gammaproteobacteria (7), or Deltaproteobacteria (2). Seawater samples (n = 5) harbored a significantly lower proportion of reads classified as nifH (0.4%) compared to coral samples (12.1%) and were distinct from the coral-associated microbiome (PERMANOVA; F1,31 = 20.14, P < 0.001; Table S1B and Table S3; Fig. S2).
Fig. 5.
nifH diversity recovered from Stylophora pistillata. a Maximum-likelihood topology of nifH sequences, rooted with non-nitrogen-fixing Cluster IV (see Fig. S1 for expanded nifH phylogeny). Novel nifH phylotypes from S. pistillata are denoted as “nifH novo” and boot strap >70% is indicated on applicable nodes. b Relative abundance of nifH phylotypes across S. pistillata samples by treatment. nifH phylotypes are hierarchically clustered by abundance similarity. Newly recovered nifH novo phylotypes are colored to match cluster of origin in b
For nifH phylotypes there was also a significant experimental treatment effect (PERMANOVA; F3,24 = 1.54, P < 0.001; Table S1B). Pairwise comparisons demonstrate that the only treatment having a significant effect on the nifH microbiome was irradiance for both 5 m and 15 m corals (Table S1B). As a result, the average similarity between groups was greatest (~25–47%) between high irradiance treatments for 5 and 15 m corals (Table S1B). When you examine the response of individual phylotypes’ to the experimental variables it revealed that Cluster I/Gammaproteobacteria (nifH _0006) and Cluster III/Deltaproteobacteria types (nifH_3411) were significantly enriched in seawater (31 and 15% compared to 2% each in coral (ANOVA; F1,29 = 109.8, P < 0.0001). During the N2 fixation experiments on S. pistillata, higher irradiances enriched relative abundances of Cluster I/Gammaproteobacteria phylotypes nifH_0153 (1 to 11%; ANOVA; F2,23 = 6.7, P = 0.018) and Cluster I/Gammaproteobacteria nifH_7457 (4 to 24%; ANOVA; F2,23 = 10.9, P = 0.003) but decreased Cluster I/Rhodobacterales nifH_7096 (21–3%; ANOVA; F2,23 = 5.8, P = 0.027).
Symbiodinium communities using ITS2
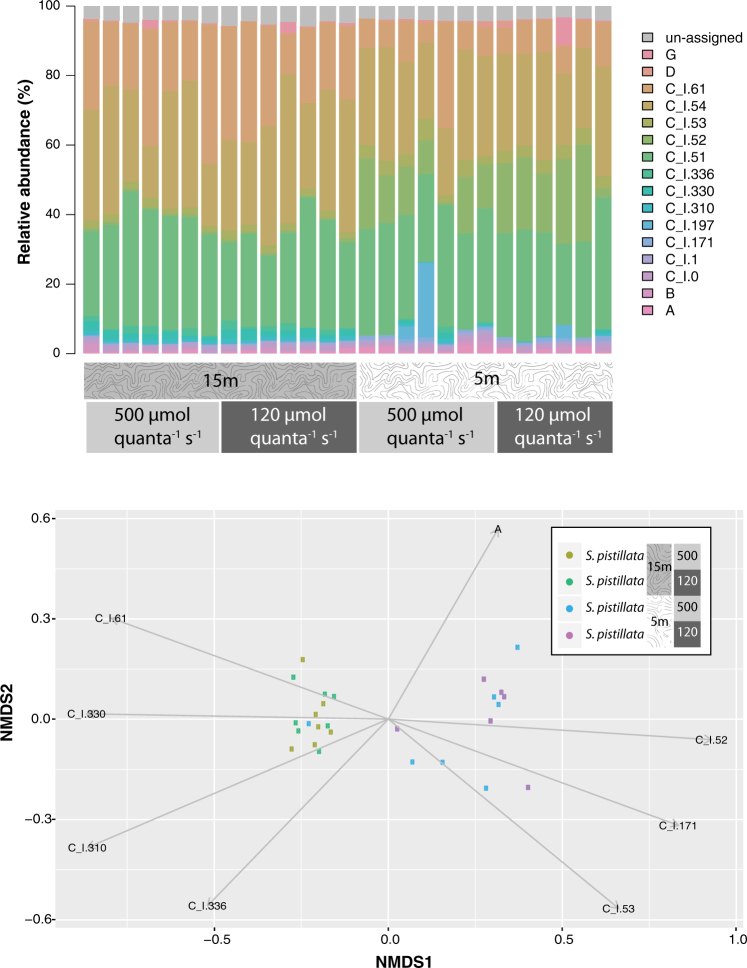
An average of 28,694 reads per sample were considered for SymTyper phylotyping following quality filtering and chimera removal. Read assignment was highly efficient, with 28,470 reads successfully assigned to clades A-I (99.2%, Table S4), with the majority assigned across 39 distinct phylotypes of clade C (Fig. S3) which were condensed to 12 dominant phylotypes (Fig. 6a). PERMANOVA indicated that depth of collection had a significant effect on Symbiodinium diversity (F3,24 = 10.7, P = 0.001), whereas irradiance, and the interaction between depth and irradiance had no effect (F2,24 = 0.74, P = 0.52). Specifically, shallower (5 m) samples were relatively enriched with clade A phylotypes (1% compared to 0.5%, F2,24 = 6.6, corrected P = 0.035), and some clade C phylotypes (C-I.52: 15% compared to 0.7%, F2,24 = 24.2, corrected P = < 0.001; C-I.53: 4% compared to 2%, F2,24 = 20.9, corrected P < 0.0001; C-I.171: 0.9% compared to 0.0%, F2,24 = 49.2, corrected P < 0.001) (Fig. 6b), while deeper samples (15 m) had greater proportions of other C phylotypes (C-I.61: 24% compared to 10%, F2,24 = 14.7, corrected P = 0.003; C-I.310: 1% compared to 0.2%, F2,24 = 19.2, corrected P = 0.006; C-I.330: 2% compared to 0.3%, F2,24 = 20.9, corrected P = 0.005; C-I.336: 1% compared to 0.2%, F2,24 = 8.7, corrected P = 0.02) (Fig. 6b). Relative abundances of phylotypes classified as clades B, D, and G did not differ between depths (Fig. 6a). ANOSIM further supported significant dissimilarity between depths in Symbiodinium spp. communities (R = 0.41, P = 0.001).
Fig. 6.
Symbiodinium spp. diversity recovered from Stylophora pistillata treatment groups (n = 7 samples per group). a. Relative abundances of Symbiodinium phylotypes across S. pistillata coral samples. b. Non-metric MDS plot of Symbiodinium diversity across S. pistillata samples. Vectors are shown for Symbiodinium spp. phylotypes that differed significantly across treatments
Functional predictions using PICRUSt
Linear discriminant analysis following non-parametric significant tests identified 53 KEGG functional categories that discriminated seawater from Stylophora pistillata samples with an absolute LDA score > 2.0 (Fig. S4A). Notably, environmental sensing pathways, transporters, motility, and “pathogenesis” functions were enriched in coral communities. Fewer differences in functional categories distinguished coral communities collected at 5 m from those collected at 15 m (Fig. S4B). Shallower coral communities were enriched for cell motility functions whereas photosynthetic functions are elevated in deeper coral communities. Irradiances during the N2 fixation experiment produced few significant effects (Fig. S4C) on microbiome function: the low irradiance (150 µmol quanta m−2 s−1) exposure was associated with greater carbohydrate metabolism compared to the higher irradiance treatment (500 µmol quanta m−2 s−1). Genes involved in nitrogen metabolism (KEGG pathway 00910) were significantly enriched (Fig. 7) based on depth of collection (ANOVA: F1,24 = 18.6, P = 0.0002) but not by irradiance. There was, however, a significant interaction between depth and irradiance (ANOVA: F1,24 = 11.6, P = 0.002) with post hoc multiple comparisons showing that the corals collected from 5 m and exposed to high irradiances were significantly different than all other treatment groups (Tukey’s HSD, P < 0.05). Several genes involved in N2 fixation (anfG, nifD, nifH, and nifK) were more abundant in coral communities collected from shallower depths (nifK: ANOVA: F2,23 = 5.4, P = 0.029; nifH: ANOVA: F2,23 = 6.1, P = 0.021; nifD: ANOVA: F2,23 = 4.8, P = 0.039; anfG: ANOVA: F2,23 = 1.4, P = 0.24). Predicted abundances of nitrogen-fixing genes did not differ significantly between seawater and coral communities nor was there an independent effect of irradiance or an interaction between depth and irradiance.
Fig. 7.
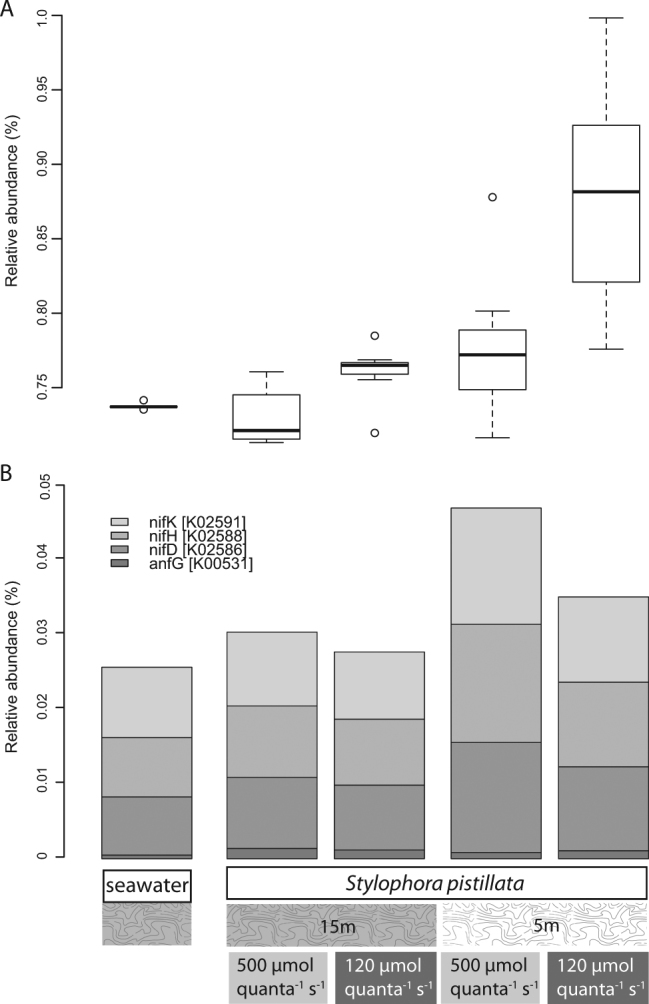
PICRUSt nitrogen metabolism abundance differences. a Relative abundance of OTUs participating in nitrogen metabolism pathways based on PICRUSt prediction. b Relative abundances of PICRUST-predicted OTUs contributing nitrogen-fixing genes (i.e., anfG, nifDHK)
Discussion
Heron Island is an oligotrophic coral cay at the southern end of the Great Barrier Reef. The concentration of DIN (i.e., NOx) is sub-micromolar and DOC/DON is much lower than previously reported for many shallow coral reefs [38]. The rates of photosynthesis and calcification, as well as the quantum yields of PSII fluorescence, for Stylophora pistillata from Heron Island were irradiance dependent while the rates of N2 fixation were not. The absence of any irradiance effect on N2 fixation was unexpected because of the hyperoxia associated with increasing photosynthesis in corals [39–41], and the oxygen sensitivity of nitrogenase [42, 43]. This suggests that while hyperoxia exists in these corals the prokaryotes responsible for N2 fixation in S. pistillata have employed a mechanism (s) to counter the inactivation of nitrogenase by molecular oxygen or, reactive oxygen species (ROS), which corals are known to produce [44]. The strategies to reduce the effects of molecular oxygen or ROS on nitrogenase could include, increased availability of energy in the form of organic carbon, high respiration rates to reduce oxygen pO2 at the site of fixation, reduced rate of access to oxygen to site of fixation, conformational changes in nitrogenase, or high activities of enzymes against ROS [42]. Many of these strategies have been observed in other nitrogen-fixing symbioses such as legumes [45]. Localization studies (i.e., FISH) using nifH probes (sensu [46]) would be useful for identifying potential hypoxic tissue niches where diazotrophs might accumulate.
The rates of N2 fixation in Stylophora pistillata reported here, measured at underwater irradiances equivalent to solar noon, are lower than those reported for Montastraea cavernosa using the acetylene reduction technique during the late evening and early morning where lower oxygen pO2, conditions occur [23]. Given that the isotopic tracer approach measures net uptake [28, 31] it is interesting to note that recent experiments measuring N2 fixation in S. pistillata from a lagoon in New Caledonia, using isotopic tracer techniques, appear to be over twenty times greater than reported here for S. pistillata from Heron Island [24]. Additionally, when compared to S. pistillata from the Red Sea [25] the rates of N2 fixation reported here are orders of magnitude greater when you convert their acetylene reduction measurements to values of nitrogen using conversion ratios [47]. It is also clear that differences between species, and possibly cryptic species (i.e., S. pistillata, [48]), as well as location, depth (=irradiance), time of year and metabolic status may all have important effects on the magnitude of N2 fixation in corals and its contribution to nitrogen budgets [19, 49]. Additionally, differences in the Symbiodiniuum spp. community, the compartment where over 95% of the fixed N is found, may influence rates of N2 fixation through their effect on nitrogen demands. Here, we showed that irradiance had a significant effect on CZND with corals exposed to higher irradiances having almost three time as much of a contribution of N2 fixation to Symbiodiunium spp. N demands. This could reflect the greater needs because of higher productivity at shallower depths reported here, or a function of the differences in Symbiodinium spp. communities where in particular, 5 m corals have a greater abundance of phylotypes in Clade A which are often described as more parasitic then other clades [50].
The prokaryotic communities of Stylophora pistillata has been studied previously [46] but in addition we examined the composition of diazotrophs within the broader prokaryotic community. Both the 16S rRNA and nifH communities recovered showed an effect of the experimental manipulation with irradiances being the most consistent factor affecting those communities. For the recovered 16 S rRNA phylotypes both the 5 m and 15 m corals showed significant shifts to novel 16S communities when exposed to the reciprocal irradiance from their depth of collection. For nifH, only corals from 5 m exposed to high irradiances showed a unique community, consistent with the PICRUSt analysis of nifH genes. When these 5 m corals are exposed to low irradiances there is a shift in their diazotrophic community which is indistinguishable from nifH communities in 15 m corals. The prokaryotic community described here for S. pistillata from Heron Island is more diverse then samples from the shallow waters in the Red Sea (2–5 m), although this could also be due to differences in sequencing chemistry (i.e., Illumina vs. 454 pyrosequencing). Red Sea samples were also dominated by bacteria related to the genus Endozoicomonas (~10–75%) and Burkholderia (~20–70%), some of which are also diazotrophic, while the Heron Island microbiomes were composed of OTUs related to Endozoicomonas (~20%) and OTUs related to the order Burkholderia (~0.5%) across all samples. The prokaryotic community of corals is known to harbor diazotrophic symbionts [2, 20–22], and potentially other prokaryotes that mediate transformations of inorganic nitrogen such as nitrification and denitrification [3, 7]. Although Cyanobacteria are some of the most recognized nitrogen-fixers, they were only a minor constituent of 16S rRNA samples from Heron Island and not reported for Red Sea collections of S. pistillata despite being recovered from other corals on the Great Barrier Reef [20]. An interesting finding of recent studies on diazotrophs in corals is that one of the non-photosynthetic taxa recovered are rhizobia (Alphaproteobacteria, [20, 22]) in the order Rhizobiales. Members of the rhizobia have also been reported from other marine diazotrophic symbioses [51] and in marine waters [52]. The nifH sequences recovered in this study using high throughput sequencing, and predictive functional analyses based on 16 S rRNA (e.g., PICRUSt) also showed low numbers of OTUs related to the Rhizobiales (Bradyrhizobium sp.). While it is intriguing to think that rhizobia may have developed a mutualistic symbiosis with corals [20], it is more likely that plasmid borne nifH genes from this taxon were horizontally transferred into members of the coral microbiome that could then be exploited by the coral host [50, 53]. Rare members of the microbiome (<0.5%) were also related to members of the class Alphaproteobacteria within the Rhodospirillales and Sphingomonadales, orders that often include the nitrogenase encoding genes nifHDK and orthologs of the transcription regulator nifA [54]. A larger proportion of the microbiome was composed of members related to putative nitrogen fixers within the order Rhodobacteriales (~6–9% of the total microbiome based on 16 S rRNA; [54]). Predictive functional results also uncovered genera within the Rhodobacterales (e.g., Yangia, Pseudoruegeria, Confluentimicrobium) with putative nitrogen-fixing capabilities.
Nitrogen-fixing genes from nifH genes Cluster I (Mo-dependent) and III (vanadium-dependent and iron-dependent), as well as Cluster V (nifH homologs involved in pigment biosynthesis; [55]), were recovered from Stylophora pistillata and several varied with irradiance or site of collection as described above. Most Cluster I sequences were placed within the Rhodobacteriales and Rhizobiales, which are both photoheterotrophic, purple non-sulfur bacteria. These bacteria exhibit extreme metabolic flexibility under environmental conditions related to variability in pO2, light availability and carbon source. But most of the nifH sequences recovered were in fact recovered from Cluster V. This Cluster is not known to fix nitrogen and is commonly composed of methanogens and some photosynthetic bacteria involved in photopigment biosynthesis [55, 56]. Despite the fact that this group does not fix nitrogen they are functionally analogous to nitrogen fixers and essential for bacteriochlorophyll biosynthesis in anoxygenic photosynthetic bacteria [55]. To the best of our knowledge the recovery of Cluster V sequences from corals has never been previously reported.
Diazotrophy generally, and specifically in scleractinian corals, had not been widely studied. But renewed interest in the process of N2 fixation in corals [2, 23] has stimulated studies on multiple aspects of N2 fixation in scleractinian corals, and the nitrogen cycle in corals including its effect on primary productivity and its potential interaction with climate change stressors in corals [19, 57, 58, 24, 49]). Diazotrophs have also been recently implicated in the bleaching stress response and have been found to increase in abundance when corals experience thermal stress (Santos et al., 2014; [59, 60, 63]). Some suggest that diazotrophy can cause instability within the holobiont by releasing control over the growth of the nitrogen-limited Symbiodinium, potentially inducing or prolonging bleaching events [57].
Our results demonstrate that bacterial diazotrophs appear to bypass the host and translocate all fixed products to their endosymbiotic Symbiodinium spp., regardless of irradiance or depth of collection. It suggests that there are potentially unique regulatory elements that allow N2 fixation to take place under hyperoxic conditions representative of shallow water/ high irradiance environments that should be the subject of future research.
Electronic supplementary material
Acknowledgments
Thank you to Elizabeth Kintzing and Kerrie Enger for their assistance in the field and laboratory. Thank you to Chris Gaby for assistance with the nifH bioinformatics pipeline. The coral collections were made with the permission of the Great Barrier Reef Marine Park Authority (GBRMPA collecting permit G15/374471.1). The National Science Foundation (OCE 1437054 to MPL) supported this research.
Author contributions
MPL and KMM designed and conducted the collections and experiments. MPL analyzed all physiological and environmental data. KMM conducted all sequencing. SHCN conducted calcification and surface area measurements. KMM and MSP conducted all bioinformatic, statistic, and PICRUSt analyses on sequence data. MPL, KMM, MSP, and SHCN wrote and approved the content of the manuscript.
Compliance with ethical standards
Conflict of interest
The authors declare no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0008-6) contains supplementary material, which is available to authorized users.
References
- 1.Rohwer F, Seguritan V, Azam F, Knowlton N. Diversity and distribution of coral-associated bacteria. Mar Ecol Prog Ser. 2002;243:1–10. doi: 10.3354/meps243001. [DOI] [Google Scholar]
- 2.Lesser MP, Mazel CH, Gorbunov MY, Falkowski PG. Discovery of symbiotic nitrogen-fixing cyanobacteria in corals. Science. 2004;305:997–1000. doi: 10.1126/science.1099128. [DOI] [PubMed] [Google Scholar]
- 3.Wegley L, Edwards R, Rodriguez-Brito B, Liu H, Rohwer F. Metagenomic analysis of the microbial community associated with the coral Porites astreoides. Environ Microbiol. 2007;9:2707–19. doi: 10.1111/j.1462-2920.2007.01383.x. [DOI] [PubMed] [Google Scholar]
- 4.Moore RB, Oborník M, Janouskovec J, Chrudimsky T, Vancová M, Green DH, et al. A photosynthetic alveolate closely relates to apicomplexan parasites. Nature. 2008;451:959–63. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- 5.Ainsworth TD, Vega Thurber R, Gates RD. The future of coral reefs: a microbial perspective. Trends Ecol Evol. 2010;25:233–40. doi: 10.1016/j.tree.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 6.Ainsworth TD, Krause L, Bridge T, Torda G, Raina JB, Zakrzewski M, et al. The coral core microbiome identifies rare bacterial taxa as ubiquitous endosymbionts. ISME J. 2015;9:2261–74. doi: 10.1038/ismej.2015.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fiore CL, Jarett JK, Olson ND, Lesser MP. Nitrogen fixation and nitrogen transformations in marine symbioses. Trends Microbiol. 2010;18:455–63. doi: 10.1016/j.tim.2010.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Bourne DG, Morrow KM, Webster NS. Insights into the coral microbiome: underpinning the health and resilience of reef ecosystems. Ann Rev Microbiol. 2016;70:317–40. doi: 10.1146/annurev-micro-102215-095440. [DOI] [PubMed] [Google Scholar]
- 9.Ward BB, Capone DG, Zehr JP. What’s new in the nitrogen cycle? Oceanography. 2007;20:101–9. doi: 10.5670/oceanog.2007.53. [DOI] [Google Scholar]
- 10.Zehr JP, Kudela RM. Nitrogen cycle of the pen ocean: from genes to ecosystems. Ann Rev Mar Sci. 2011;3:197–25. doi: 10.1146/annurev-marine-120709-142819. [DOI] [PubMed] [Google Scholar]
- 11.Larkum AWD, Kennedy IR, Muller WJ. Nitrogen fixation on a coral reef. Mar Biol. 1988;98:143–45. doi: 10.1007/BF00392669. [DOI] [Google Scholar]
- 12.O’Neil JM, Capone DG. Nitrogen cycling in coral reef environments. In: Capone DG, Bronk DA, Mulholland MR, Carpenter EJ, editors. Nitrogen in The Marine Environment. New York, NY: Academic Press; 2008. pp. 949–89. [Google Scholar]
- 13.Wiebe WJ, Johannes RE, Webb KL. Nitrogen fixation in a reef community. Science. 1975;188:257–59. doi: 10.1126/science.188.4185.257. [DOI] [PubMed] [Google Scholar]
- 14.Wild C, Huettel M, Klueter A, Kremb SG, Rasheed MYM, Jørgensen BB. Coral mucus functions as an energy carrier and particle trap in the reef ecosystem. Nature. 2004;428:66–70. doi: 10.1038/nature02344. [DOI] [PubMed] [Google Scholar]
- 15.Wild C, Woyt H, Huettel M. Influence of coral mucus on nutrient fluxes in carbonate sands. Mar Ecol Prog Ser. 2005;287:87–98. doi: 10.3354/meps287087. [DOI] [Google Scholar]
- 16.Kneip C, Lockhart P, Vob C, Maier UG. Nitrogen fixation in eukaryotes-new models for symbiosis. BMC Evol Biol. 2007;7:55–66. doi: 10.1186/1471-2148-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benavides M, Bednarz VN, Ferrier-Pagès C. Diazotrophs: overlooked key players within coral symbiosis and tropical reef ecosystems? Front Mar Sci. 2017;4:10. doi: 10.3389/fmars.2017.00010. [DOI] [Google Scholar]
- 18.Shashar N, Cohen Y, Loya Y, Star N. Nitrogen fixation (acetylene reduction) in stony corals: evidence for coral-bacterial interactions. Mar Ecol Prog Ser. 1994;111:259–64. doi: 10.3354/meps111259. [DOI] [Google Scholar]
- 19.Cardini U, Bednarz VN, Naumann MS, van Hoytema N, Rix L, Foster RA, et al. Functional significance of dinitrogen fixation in sustaining coral production under oligotrophic conditions. Proc R Soc B. 2015;282:20152257. doi: 10.1098/rspb.2015.2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lema KA, Willis BL, Bourne DG. Corals form characteristic associations with symbiotic nitrogen-fixing bacteria. Appl Environ Microbiol. 2012;78:3136–44. doi: 10.1128/AEM.07800-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olson ND, Ainsworth TD, Gates RD, Takabayashi M. Diazotrophic bacteria associated with Hawaiian Montipora corals: diversity and abundance in correlation with symbiotic dinoflagellates. J Exp Mar Biol Ecol. 2009;371:140–46. doi: 10.1016/j.jembe.2009.01.012. [DOI] [Google Scholar]
- 22.Olson ND, Lesser MP. Diazotrophic diversity in the Caribbean coral. Montastraea Cavern Arch Microbiol. 2013;195:853–59. doi: 10.1007/s00203-013-0937-z. [DOI] [PubMed] [Google Scholar]
- 23.Lesser MP, Falcón LI, Rodríguez-Román A, Enríquez S, Hoegh-Guldberg O, Iglesias-Prieto R. Nitrogen fixation by symbiotic cyanobacteria provides a source of nitrogen for the scleractinian Coral. Montastraea Cavern Mar Ecol Prog Ser. 2007;346:143–52. doi: 10.3354/meps07008. [DOI] [Google Scholar]
- 24.Benavides M, Holbrèque F, Camps M, Lorrain A, Grosso O, Bonnet S. Diazotrophs: a non-negligible source of nitrogen for the tropical coral Stylophora pistillata. J Exp Biol. 2016;219:2608–12. doi: 10.1242/jeb.139451. [DOI] [PubMed] [Google Scholar]
- 25.Pogoreutz C, Rädecker N, Cárdenas A, Gärdes A, Wild C, Voolstra CR. Nitrogen fixation aligns with nifH abundance and expression in two coral trophic groups. Front Microbiol. 2017;8:1187. doi: 10.3389/fmicb.2017.01187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grover R, Ferrier-Pagès C, Maguer JF, Ezzat L, Fine M. Nitrogen fixation in the mucous of Red Sea corals. J Exp Biol. 2014;217:3962–63. doi: 10.1242/jeb.111591. [DOI] [PubMed] [Google Scholar]
- 27.Glibert PM, Bronk DA. Release of dissolved organic nitrogen by marine diazotrophic cyanobacteria, Trichodesmium spp. Appl Environ Microbiol. 1998;60:3996–4000. doi: 10.1128/aem.60.11.3996-4000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilson ST, Böttjer D, Church MJ, Karl DM. Comparative assessment of nitrogen fixation methodologies, conducted in the oligotrophic North Pacific Ocean. Appl Environmen Microbiol. 2012;78:6516–23. doi: 10.1128/AEM.01146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GroBkopf T, Mohr W, Baustian T, Schunck H, Gill D, Kuypers MMM, et al. Doubling of marine dinotrogen-fixation rates based on direct measurements. Nature. 2012;488:361–64. doi: 10.1038/nature11338. [DOI] [PubMed] [Google Scholar]
- 30.Mohr W, GroBkopf T, Wallace WRD, LaRoche J. Methodological underestimation of oceanic nitrogen fixation rates. PLoS ONE. 2010;5:e12583. doi: 10.1371/journal.pone.0012583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McCarthy MD, Bronk DA. Analytical methods for nitrogen chemical characterization and flux rates. In: Capone DG, Bronk DA, Mulholland M, Carpenter EJ, editors. Nitrogen in the Marine Environment. New York, NY: Academic Press; 2008. pp. 1219–76. [Google Scholar]
- 32.Seutin G, White BN, Boag PT. Preservation of avian blood and tissue samples for DNA analyses. Can J Zool. 1991;69:82–90. doi: 10.1139/z91-013. [DOI] [Google Scholar]
- 33.Warner ME, Lesser MP, Ralph PJ. Chlorophyll fluorescence in reef building corals. In: Suggett D, Borowitzka M, Prásil O, editors. Chlorophyll a Fluorescence in Aquatic Sciences: Methods and Applications. Netherlands: Springer; 2010. pp. 209–22. [Google Scholar]
- 34.Strahl J, Stolz I, Uthicke S, Vogel N, Noonan SHC, Fabricius KE. Physiological and ecological performance different in four coral taxa at a volcanic carbon dioxide seep. Comp Biochem Physiol A. 2015;184:179–86. doi: 10.1016/j.cbpa.2015.02.018. [DOI] [PubMed] [Google Scholar]
- 35.Chisholm JRM, Gattusoi JP. Validation of the alkalinity anomaly technique for investigating calcification ad photosynthesis in coral reef communities. Limnol Oceanogr. 1991;36:1232–39. doi: 10.4319/lo.1991.36.6.1232. [DOI] [Google Scholar]
- 36.Gazeau F, Urbini L, Cox TE, Alliouane S, Gattuso JP. Comparison of the alkalinity and calcium anaomaly techniques to estimate rates of net calcification. Mar Ecol Prog Ser. 2015;527:1–12. doi: 10.3354/meps11287. [DOI] [Google Scholar]
- 37.Stimson J, Kinzie RA. The temporal pattern and rate of release of zooxanthellae from the reef coral Pocillopora damicornis (Linnaeus) under nitrogen-enrichment and control conditions. J Exp Mar Biol Ecol. 1991;153:63–74. doi: 10.1016/S0022-0981(05)80006-1. [DOI] [Google Scholar]
- 38.Suzuki Y, Casareto BE. The role of dissolved organic nitrogen (DON) in coral biology and reef ecology. In: Dubinsky Z, editor. Coral Reefs, Ecosystems of the World. Amsterdam: Elsevier; 2011. pp. 207–14. [Google Scholar]
- 39.Dykens JA, Shick JM. Oxygen production by endosymbiotic algae controls superoxide dismutase activity in their animal host. Nature. 1982;297:579–80. doi: 10.1038/297579a0. [DOI] [Google Scholar]
- 40.Kühl M, Cohen Y, Dalsgaard T, Jørgensen BB, Revsbech NP. Microenvironment and photosynthesis of zooxanthellae in scleratinian corals studied with microsensors for O2, pH, and light. Mar Ecol Prog Ser. 1995;117:159–72. doi: 10.3354/meps117159. [DOI] [Google Scholar]
- 41.Shashar N, Cohen Y, Loya Y. Extreme diel fluctuations of oxygen in diffusive boundary layers surrounding stony corals. Bio Bull. 1993;185:455–61. doi: 10.2307/1542485. [DOI] [PubMed] [Google Scholar]
- 42.Fay P. Oxygen relations of N2 fixation in cyanobacteria. Microbiol Rev. 1992;56:340–73. doi: 10.1128/mr.56.2.340-373.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berman-Frank I, Lundgren P, Falkowski P. Nitrogen fixation and photosynthetic oxygen evolution in cyamobacteria. Res Microbiol. 2003;154:157–64. doi: 10.1016/S0923-2508(03)00029-9. [DOI] [PubMed] [Google Scholar]
- 44.Lesser MP. Oxidative stress in marine environments: biochemistry and physiological ecology. Ann Rev Physiol. 2006;68:253–78. doi: 10.1146/annurev.physiol.68.040104.110001. [DOI] [PubMed] [Google Scholar]
- 45.Mus F, Crook MB, Garcia K, Costas AG, Geddes BA, Kouri ED, et al. Symbiotic nitrogen fixation and the challenges to its extension to nonlegumes. Appl Environ Microbiol. 2016;82:3698–10. doi: 10.1128/AEM.01055-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bayer T, Neave MJ, Alsheikh-Hussaine A, Aranda M, Yum LK. The microbiome of the Red Sea coral Stylophora pistillatra is dominated by tissue-associated Endozoicomonas bacteria. Appl Environ Microbiol. 2013;79:4759–62. doi: 10.1128/AEM.00695-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kayanne H, Hirota M, Yamamuro M, Koike I. Nitrogen fixation of filamentous cyanobacteria in a coral reef measured using three different methods. Coral Reefs. 2005;24:197–200. doi: 10.1007/s00338-004-0465-z. [DOI] [Google Scholar]
- 48.Keshavmurthy S, Yang SY, Alamaru A, Chuang YY, Pichon M, Obura D, et al. DNA barcoding reveals the coral “laboratory-rat”, Stylophora pistillata encompasses multiple identities. Sci Rep. 2013;3:1520. doi: 10.1038/srep01520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bednarz VN, Grover R, Maguer JF, Fine M. The assimilation of diazotroph-derived nitrogen by scleractinian corals depends on their metabolic status. mBio. 2017;8:e02058–16. doi: 10.1128/mBio.02058-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lesser MP, Stat M, Gates RD. The endosymbiotic dinoflagellates (Symbiodinium sp.) of corals are parasites and mutualists. Coral Reefs. 2013;32:603–11. doi: 10.1007/s00338-013-1051-z. [DOI] [Google Scholar]
- 51.Summers MM, Katz S, Allen EE, Rouse GW. Association of rhizobia with a marine polychaete. Environ Microbiol Notes. 2013;5:492–98. doi: 10.1111/1758-2229.12043. [DOI] [PubMed] [Google Scholar]
- 52.Le Moal M, Collin H, Biegala IC. Intriguing diversity among diazotrophic picoplankton along a Mediterranean transect: a dominance of rhizobia. Biogeosci. 2011;8:827–40. doi: 10.5194/bg-8-827-2011. [DOI] [Google Scholar]
- 53.Zehr JP, Jenkins BD, Short SM, Steward G. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ Microbiol. 2003;5:539–54. doi: 10.1046/j.1462-2920.2003.00451.x. [DOI] [PubMed] [Google Scholar]
- 54.Tsoy OV, Ravcheev DA, Čuklina J, Gelfand MS. Nitrogen fixation and molecular oxygen: comparative genomic reconstruction of transcription regulation in Alphaproteobacteria. Front Microbiol. 2016;7:1343. doi: 10.3389/fmicb.2016.01343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raymond J, Siefert JL, Staples CR, Blankenship RE. The natural history of nitrogen fixation. Mol Biol Evol. 2004;21:541–54. doi: 10.1093/molbev/msh047. [DOI] [PubMed] [Google Scholar]
- 56.Gaby JC, Buckley DH. A global census of nitrogenase diversity. Environ Microbiol. 2011;13:1790–99. doi: 10.1111/j.1462-2920.2011.02488.x. [DOI] [PubMed] [Google Scholar]
- 57.Rädecker N, Pogoreutz C, Voolstra CR, Wiedenmann J, Wild C. Nitrogen cycling in corals: the key to understanding holobiont functioning? Trends Microbiol. 2015;23:490–97. doi: 10.1016/j.tim.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 58.Cardini U, Bednarz VN, Foster RA, Wild C. Benthic nitrogen fixation in coral reefs and the potential effects of human-induced environmental change. Ecol Evol. 2014;4:1706–27. doi: 10.1002/ece3.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cardini U, van Hoytema N, Bednarz VN, Rix L, Foster RA, Al-Rshaidat MM, et al. Microbial diN2 fixation in coral holobionts exposed to thermal stress and bleaching. Environ Microbiol. 2016;18:2620–33. doi: 10.1111/1462-2920.13385. [DOI] [PubMed] [Google Scholar]
- 60.Lee STM, Davy SK, Tang SL, Kench PS. Mucus sugar content shapes the bacterial community structure in thermally stressed Acropora muricata. Front Microbiol. 2016;7:371. doi: 10.3389/fmicb.2016.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Piniack GA, Brown EK. Growth and mortality of coral transplants (Pocillopora damicornis) along a range of sediment influences in Maui, Hawai'i. Pac Sci. 2008;62:39–55. doi: 10.2984/1534-6188(2008)62[39:GAMOCT]2.0.CO;2. [DOI] [Google Scholar]
- 62.Lesser MP, Shick JM. Effects of irradiance and ultraviolet radiation on photoadaptation in the zooxanthellae of Aiptasia pallida: primary production, photoinhibition and enzymic responses against oxygen toxicity. Mar Biol. 1989;102:243–255. doi: 10.1007/BF00428286. [DOI] [Google Scholar]
- 63.Santos HF, Carmo FL, Duarte G, Dini-Andreote F, Castro CB, Rosado AS, van Elsas JD, Peixoto RS. Climate change affects key nitrogen-fixing bacterial populations on coral reefs. Mar Biol. 2014;8:2272–2279. doi: 10.1038/ismej.2014.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.