Abstract
Many organisms benefit from being pre-adapted to niches shaped by human activity, and have successfully invaded man-made habitats. One such species is the dry rot fungus Serpula lacrymans, which has a wide distribution in buildings in temperate and boreal regions, where it decomposes coniferous construction wood. Comparative genomic analyses and growth experiments using this species and its wild relatives revealed that S. lacrymans evolved a very effective brown rot decay compared to its wild relatives, enabling an extremely rapid decay in buildings under suitable conditions. Adaptations in intracellular transport machineries promoting hyphal growth, and nutrient and water transport may explain why it is has become a successful invader of timber in houses. Further, we demonstrate that S. lacrymans has poor combative ability in our experimental setup, compared to other brown rot fungi. In sheltered indoor conditions, the dry rot fungus may have limited encounters with other wood decay fungi compared to its wild relatives. Overall, our analyses indicate that the dry rot fungus is an ecological specialist with poor combative ability against other fungi.
Introduction
Species worldwide are negatively affected by anthropogenic habitat destruction. However, for those few species originally living in natural habitats that resemble the man-made ecosphere, the opposite is also the case. Animals like the Norwegian rat (Rattus norvegicus) and the German cockroach (Blatella germanica) have extended their distribution dramatically [1, 2]. Likewise, many plant pathogenic fungi have become extremely widespread as monotypic crop cultivation creates large habitats, and the trade and transport of these crops aid their dispersal [3–5]. A similar pattern is seen with the few wood decay fungi that have expanded their realized niche into the human built environment.
Probably the best-known example of a successful fungal invader of the built environment is the dry rot fungus Serpula lacrymans var. lacrymans (subsequently referred to as var. lacrymans), which is distributed in houses in temperate and boreal regions worldwide causing brown rot decay. It spreads with human transport of timber over long distances and colonizes new buildings in its vicinity by air-borne spores [6, 7]. Colonization of construction timber in buildings is characterized by rapid vegetative mycelial growth and formation of thick (up to 2 cm diameter, Fig. 1) mycelial cords that mediate the transport of nutrition and water to new wood substrates [8]. This allows quick growth and re-allocation of resources via the transport of nutrition and water to the new wood substrates [8, 9].
Fig. 1. The dry rot fungus Serpula lacrymans and its habitat.
. Serpula lacrymans is one of the most devastating decomposer of houses in temperate and boreal regions worldwide. The species is known to form thick cords and a rapid decay of coniferous wood. In nature the species decompose large logs in dry mountain forests. Photo credits: Top left photo by H. Kauserud, the other photos by Mycoteam AS.
Comparative genomic approaches have shown that var. lacrymans and other brown rot fungi have a reduced set of plant cell wall hydrolyzing enzymes to decompose wood compared to the ancestral white rot fungi [10–14]. A recent study has suggested that the set of secreted enzymes responsible for decomposition of var. lacrymans is even smaller than that of some other brown rot fungi [15]. The loss of enzymes by brown rot fungi is correlated with a strategy in which the initial attack of the wood is mediated by hydroxyl radicals produced by chelator-mediated Fenton (CMF) chemistry [10, 13, 16]. These initial attacks have been suggested to be controlled by differential gene expression of the fungi [14, 15]. The attacked wood structure is then further depolymerized by oxidizing and hydrolyzing enzymes that target cellulose and hemicellulose elements in the wood, while leaving modified lignin behind.
Var. lacrymans has a scattered natural range in high altitude mountain regions of North-East Asia, thriving in moraine-dominated habitats around the treeline where woody resources are heterogeneously distributed [6]. Human transport of infected wood appears to have facilitated the colonization in the human domain in temperate regions worldwide. It is widespread in buildings in Europe and Japan, and it is also found in buildings in temperate parts of North and South America (Chile), Australia and New Zealand, but with less abundance [7, 17]. The large European house-colonizing population of var. lacrymans has low genetic variation [7, 18], suggesting a severe population bottleneck during the colonization of the European built environment [6, 7]. Serpula lacrymans var. shastensis (subsequently referred to as var. shastensis) is a close relative of var. lacrymans, from high altitude mountain regions in the Cascade mountain range (North America), but has not been reported in the built environment [19, 20]. Although genetically well-separated, the two sub-species are able to form a dikaryotic mycelium when paired in vitro [7, 20, 21]. In the habitat close to the treeline in the Cascades (Fig. 1), var. shastensis colonizes and decays large logs of Abies magnifica [6, 22]. Both varieties of S. lacrymans appear to be ecological specialists, thriving in exposed mountainous habitats with patchy resource distribution.
In contrast to the confined niches of S. lacrymans, its sister species Serpula himantioides has a widespread circumboreal distribution in natural habitats in temperate and boreal regions [23]. As with S. lacrymans, S. himantioides causes brown rot of conifers, but decomposes wood more slowly, as shown on spruce [19], and produces smaller fruit bodies and smaller cords. Serpula himantioides is rarely found in buildings, and when it is, it decomposes wood more slowly than var. lacrymans. Unlike var. lacrymans, indoor colonization by S. himantioides, as with the majority of other wood decay fungi, represent random, and repeated colonizations from nature [6].
It is not evident which characteristics have made var. lacrymans such a successful invader of the built environment compared to its wild relatives. Pinpointing contrasting genomic differences among the lineages is a first step toward detecting the genetic basis of var. lacrymans invasiveness and persistence. In this study we, therefore, set out to reveal which genomic features separate var. lacrymans from its predominantly wild relatives. We analyzed which genes have undergone shifts in selective pressure and then, which gene families have expanded or contracted during divergence between variants or species. This was achieved by sequencing and de novo genome assembly of var. lacrymans and var. shastensis strains and comparing these to the genome of the sister species S. himantioides. Genomic analyses were complemented by two growth experiments investigating differences in decomposition ability and interspecific competition, to provide more direct evidence for how each of these factors may contribute to var. lacrymans’ success in the built environment.
Materials and methods
Strains
Three strains were used for physiological experiments and genome comparisons in this study. The S. himantioides strain (MUCL38935) was cultured from soil in the UK in 1994, the var. shastensis strain (SHA17-1) was collected in California, US on Abies in 2004 and the var. lacrymans (SL200) was collected from a house in Poland in 1953. Since these strains have been maintained in culture for extended periods of time, caution should be used when interpreting the results as the strains may have changed their behavior through these years.
DNA extraction, sequencing, assembly and gene predictions
More details of the DNA extraction, library preparation, sequencing procedure, and gene prediction pipeline can be found in the Supplementary text. DNA of all three strains was extracted by a modified phenol-chloroform protocol available at the JGI webpage (http://jgi.doe.gov/collaborate-with-jgi/pmo-overview/protocols-sample-preparation-information/). All strains were sequenced using Illumina technology. The two Serpula lacrymans strains were sequenced on an Illumina GAII at the SNP&SEQ Technology Platform in Uppsala, Sweden, while S. himantioides was sequenced on an Illumina Hiseq 2000 at the JGI (http://genome.jgi.doe.gov/Serla_varsha1/Serla_varsha1.info.html).
The Velvet de novo assembler [24] was used to assemble reads into contigs for var. lacrymans and var. shastensis. JGI assembled S. himantioides with the AllPathsLG assembler [25]. The CEGMA pipeline was used [26] to estimate completeness of all assemblies (Table 1). Protein coding genes in the three Serpula strains were annotated using MAKER2 version 2.27 [27].
Table 1.
Summary statistics of the genome assembly, annotation and CEGMA analyses of the three sequenced genomes of Serpula lacrymans var. lacrymans, S. lacrymans var. shastensis and Serpula himantioides
| Species | Strain | # of contigs | # of scaffolds | N50 | Genome size (Mpb) | Assembler | CEGMA | # of predicted genes |
|---|---|---|---|---|---|---|---|---|
| var. lacrymans | SL200 | 4534 | 1529 | 59,716 | 37 | Velvet | 97.6% | 11,352 |
| var. shastensis | SHA17-1 | 3839 | 1170 | 92,207 | 38 | Velvet | 97.2% | 10,910 |
| S. himantioides* | MUCL38935 | 5964 | 4893 | 20,000 | 46 | AllPathsL | 89.5% | 12,011§ |
*Sequenced by JGI, § Number of genes predicted by Maker annotation tool; however, the JGI annotation pipeline predicted 13,805 gene models
Functional annotation
Genes were given a preliminary description by BLAST alignment toward UniProt. InterProScan was used for functional annotation and classifications of protein families [28]. Protein sequences of var. lacrymans, var. shastensis and S. himantioides were obtained from the MAKER2 predictions.
OrthoMCL clustering
Homologous proteins of the three Serpula strains were clustered using the software OrthoMCL [29]. This tool clusters homologous proteins across the given species using Markov cluster algorithm to group orthologs and paralogs. In total 34,273 protein sequences from three different Serpula genomes were compared.
CAFÉ analysis
CAFÉ estimates a global birth and death rate of gene families and changes in gene family size across a phylogeny [30]. All orthoMCL clusters were used as gene families. CAFÉ was run using a global birth/death parameter (λ). Rapidly evolving gene families were estimated using the best fit λ (0.002) at a p-value threshold of 0.01. The ultrametric three used for CAFÉ analysis was based on a multi-locus maximum likelihood phylogeny of ten loci from [31] that was made ultrametric in the R package APE [32].
Selection pressure
Clusters of single copy orthologs were chosen to screen for branch specific changes in selection pressure. The clusters were aligned with the multiple sequence alignment program PRANK [33] with the ‘codon’ alignment mode, using the species phylogeny [21] as guide tree. PRANK has been shown to provide the most accurate alignments, with the lowest false-positive rates [34]. The Codeml from the PAML package [35] was used to identify changes in selection regime. For each group of orthologs, a single dN/dS ratio (ω) was estimated for all branches on the tree (H0) and for three instances where each one of the species was allowed to evolve at a separate rate (H1), The best fit model was determined using a likelihood ratio test and p-values were adjusted to control the false discovery rate (FDR) for multiple hypothesis testing using a α < 0.05 [36]. All alignments with a significant shift in selection pressure between species were manually examined to remove questionable alignment regions if present and were then rerun in the above outlined analysis.
Functional enrichment analyses
Functional enrichment analysis was used to characterize the genes present in all the genomes compared to gene families that were inferred to be expanded or contracted by CAFÉ. A Python script was used to perform functional enrichment analysis of PFAM domains using Fisher’s exact test (http://cgrlucb.wikispaces.com/Functional+Enrichment+Analysis).
Annotation of genes of specific functions of interest
To predict the secretome of each species, a bioinformatics pipeline consisting of SignalP 4.1 [37], TargetP 2.0 [38], TMHMM 2.0 [39], PS_scan [40] and WolfPSort v. 0.2 [41], was used, as implemented in Kohler et al. [42]. Besides the annotations generated for the entire proteomes (e.g., CAZymes and PFAM domains), the proteolytic enzymes present in each secretome were also annotated through BLAST searches against the MEROPS database [43]. Carbohydrate-active enzymes were predicted bysearching predicted proteomes with the dbCAN tool [44, 45].
As cytochrome P450 (cytP450) is an important class of enzymes involved in specialized metabolism, the clusters annotated with cytP450 PFAM domains in Interproscan were manually curated. Only those of over 300 residues with both the EXXR and CXG motif were accepted as functional, according to the method of Syed and Mashele [46]. According to cytP450 nomenclature, a similarity of 40% was considered sufficient to classify a predicted protein into a particular family. A similarity of 55% would allow allocation to a sub-family. Those with < 40% similarity to named cytP450s were—with those that had no significant matches in the NCBI or UniProt databases—considered to probably belong to novel cytP450 families.
Data availability
All raw sequence reads, and assembled genomes are available on NCBI at Bioproject PRJNA412961. In addition, the S. himantioides MUCL38935 genome is available at the JGI genome browser (http://genome.jgi.doe.gov/Serla_varsha1/Serla_varsha1). The MAKER2 gene predictions, the OrthoMCL clusters and the alignments used as input to the Codeml analyses have been deposited in the Dryad Digital Repository: doi:10.5061/dryad.28sb6.
Combative ability
Var. lacrymans, which is found predominately, if not exclusively, inside houses in Europe, was hypothesized to show decreased ability to combat for limited resources since it faces few competitors in this environment. An antagonistic experiment was used to test this hypothesis, where the three Serpula strains of interest were confronted with each other and other brown rot decomposer fungi, pairswise, by growing two well-colonized blocks side by side (see Supplementary Text for detailed experimental setup). The three Serpula strains, and the three species Antrodia xantha, Coniophora puteana and Fomitopsis pinicola were used. All combinations were repeated 10 times. After the experiment, three small wood pieces from within the wood block were transferred to three new culture plates. The strains that were re-isolated from the wood piece were identified and reported. A Pearson’s χ 2 Goodness of Fit test was used to test whether one species had significantly outcompeted another.
Wood decay
The specialized house-living var. lacrymans was expected to decompose spruce especially fast as it is mostly found on spruce in houses, where it is known to grow quickly [19]. To compare the decomposition ability of the three Serpula strains and A. xantha, F. pinicola and C. puteana, mass loss of wood was determined after 60 days colonization at 20 °C on the three tree species Pinus sylvestris, Picea abies and Abies lasiocarpa (see Supplementary Text for experimental setup; the three non-Serpula species were not tested on Abies lasiocarpa). The significance of the differences in mass loss among strains and among wood species was tested with ANOVA analyses using R [47].
Results
Genome summary
The gene prediction pipeline identified a total of 11,352 gene models in var. lacrymans, 10,910 gene models in var. shastensis and 12,011 gene models in S. himantioides (Table 1). Annotated genes were clustered into gene families, of which 6695 were shared among all three strains, corresponding to approximately 55 to 61% of annotated genes in each genome. Given the close relationship among the three species, the number of singleton clusters inferred for each species was surprisingly high. Of the predicted genes 18% in var. shastensis, 23% in var. lacrymans and 24% in S. himantioides were unique to each of the three lineages. Further analysis of singleton genes showed that singletons predominately represented cases where orthologs were absent in the other two species, either due to gene loss or absence of the corresponding coding region from the respective assemblies (results not shown).
Analyses of selection
The genome-wide estimates of selection yielded a mean estimate of ω = 0.137 for S. himantioides, ω = 0.179 for var. lacrymans and ω = 0.234 for var. shastensis (gene clusters with ω > 2 were omitted from these estimates).
Shifts in selective pressure on individual genes between species may pinpoint genes whose functions have contributed to adaptation by each species to their respective realized niches. For the analyses of shifts in selective pressure on a gene-by-gene basis, three series of tests were run, each one with a different species as the foreground branch. After correction for multiple testing, 100, 129, and 265 genes with significantly different ω between foreground and background branches in var. lacrymans, var. shastensis and S. himantioides were detected, respectively (Fig. 2). Among the sets of genes, 43% were annotated with PFAM domains while the rest were unannotated. Our functional analyses were only focused on the genes that had PFAM annotations. A full list of significant genes is provided in the Supplementary Material (Supplementary Table 1).
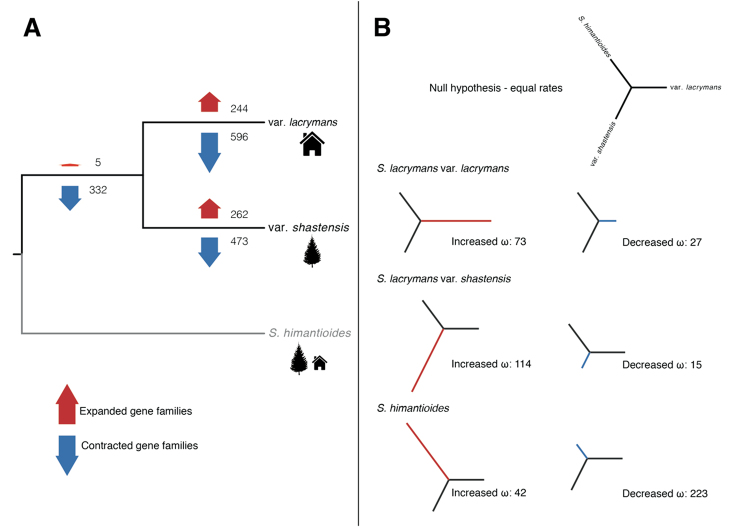
Fig. 2. The comparative genomic differences among the Serpula lacrymans var. lacrymans, Serpula lacrymans var. shastensis and Serpula himantioides.
. a The number of significantly expanded and contracted gene families, based on analyses using a birth-death model of gene family evolution on all gene clusters. The analyses use a rooted ultrametric tree from a 10 loci maximum likelihood analysis, where S. himantioides was the out-group. Thus, only changes in var. shastensis, var. lacrymans and the branch leading to these two, but not the S. himantioides branch were evaluated. b Phylogenetic sketch trees demonstrating the selection analysis. Each tree highlights a branch and the number of genes with significantly increased or decreased ω-values on that branch compared to the expected based on 5866 single gene clusters. The null hypothesis is equal rates on all branches
One of the most pronounced functional signatures detected was the selective shift in many proteins involved in intracellular transport (Table 2) with an elevated ω in S. lacrymans compared to S. himantioides (higher ω in one or both of the S. lacrymans varieties). Several of these proteins identified were involved in the transport of vesicles to the Golgi stack for secretion, (see Supplementary Text for details). In contrast, a protein involved in early endosomal membranes evolved faster in S. himantioides than in S. lacrymans. This, suggested a faster evolution of an endocytic pathway in S. himantioides vs. an exocytic pathway in S. lacrymans.
Table 2.
The gene families that are evolving at a significant different rate (p-value < 0.05 after FDR) among the different Serpula strains and includes a PFAM domain related to intracellular transport
| Cluster.No | Description | PFAM ID | Test | P-value |
|---|---|---|---|---|
| 2435 | Domain_of_unknown_function_(DUF202), SPX_domain | PF02656, PF03105 | Hl, Lh | 0.00899, 0.00044 |
| 1272 | Cofilin/tropomyosin-type_actin-binding_protein, Variant_SH3_domain | PF00241, PF14604 | Hl, Lh | 0.03537, 0.00589 |
| 6654 | RasGEF_N-terminal_motif, RasGEF_domain | PF00618, PF00617 | Hl, Lh | 0.02021, 0.00370 |
| 6080 | SNARE_domain | PF05739 | Hl, Lh | 0.00346, 0.00003 |
| 6147 | RhoGEF_domain | PF00621 | Hl, Lh | 0.00899, 0.00573 |
| 3843 | PX_domain | PF00787 | Ll, Hh | 0.01602, 0.00220 |
| 1940 | Oxysterol-binding_protein | PF01237 | Hs, Lh | 0.01365, 0.00607 |
| 3226 | PH_domain, FHA_domain, Kinesin_motor_domain | PF00169, PF00498, PF00225 | Hs | 0.00279 |
| 6485 | WD_domain, _G-beta_repeat | PF00400 | Lh | 0.00683 |
| 2827 | Sec1_family | PF00995 | Lh | 0.02796 |
| 1406 | FYVE_zinc_finger, TCP-1/cpn60_chaperonin_family | PF01363, PF00118 | Lh | 0.01075 |
H indicates higher omega, L indicates lower omega. l symbolizes Serpula lacrymans var. lacrymans, s symbolizes S. l. var. shastensis and h indicates S. himantioides, thus Hl indicates significant higher omega for var. lacrymans
In addition to the genes related to membrane transport, two regulators of actin polymerization (the guanine nucleotide exchange factors, Rho GEF and Ras GEF) and a gene with a role in actin depolymerization (cofilin) evolved significantly faster in var. lacrymans than in S. himantioides (Table 2).
Expansion and contraction of gene families
All clusters in the data set and a rooted tree were used to infer 244 and 262 gene families that were expanded on the var. lacrymans branch and on the var. shastensis branch, respectively, compared to the rest of the tree (Table 2). Only 5 were expanded on the common branch leading to var. lacrymans and var. shastensis. Compared to the genomic background, CAFÉ inferred 112 and 135 gene families that expanded significantly faster than expected (based on all clusters) in var. lacrymans and var. shastensis, respectively (P-value 0.01). In turn, 596 and 473 gene families were contracted on the var. lacrymans branch and on the var. shastensis branch, respectively, and 332 were contracted on the common branch. Six (var. lacrymans) and four (var. shastensis) gene families showed significantly higher rates of contraction than the genomic background rate.
Functional enrichment of the expanded and contracted gene families demonstrated a change in copy number for gene families related to specialized metabolism amongst all three strains (Supplementary Table 2). In particular, expansions and contractions in a variety of polyketide synthase (PKS) and nonribosomal peptide synthase (NRPS) related PFAM domains were identified (Supplementary Table 2). One NRPS gene family (cluster 0012) was expanded in var. lacrymans, var. shastensis and their common branch. This gene family had nine gene copies in var. shastensis and var. lacrymans, but only one in S. himantioides. The opposite pattern was found for a putative PKS-NRPS hybrid protein gene family of unknown function (cluster 0005), where S. himantioides had ten copies, var. lacrymans eight and var. shastensis six copies. Copy number changes in ATP-binding cassette (ABC) transporters were also detected. These were reduced in var. lacrymans compared to var. shastensis and S. himantioides.
Cytochrome P450s showed expansion in S. lacrymans compared to S. himantioides (Supplementary Table 3). Eighty-nine, 91 and 109 predicted functional cytochrome P450s were identified in S. himantioides, var. shastensis and var. lacrymans respectively. Thus, both var. shastensis and var. lacrymans have experienced expansion of capacity compared to S. himantioides, with an extra five families represented in each. Var. lacrymans and var. shastensis had the same families except that var. shastensis uniquely had one member of CYP5145, and var. lacrymans had one member of CYP6001, a family that was not present in either of the other strains. Thus, in both var. shastensis and var. lacrymans the higher numbers of cytochrome P450 copies were predominantly the result of an increased number of genes from existing families.
Several gene families related to wood decay mechanisms were expanded or contracted (Fig. 2; Supplementary Table 2, Supplementary Text for details). Specifically, the set of CAZymes encoded within the three genomes was very similar, but with a somewhat greater gene complement in S. himantioides (see Supplementary Text for details; Supplementary Table 4). In contrast, an iron reductase with only a CBM1 and a CytB domain was found in S. lacrymans, but not in S. himantioides. (Supplementary Fig. 2).
Evaluating substrate preference
Both S. lacrymans varieties decomposed more of the spruce wood block than S. himantioides, under the conditions tested (50% and 45 vs. 30% mass loss, respectively; Fig. 3). There was no significant difference in the amount of decomposition between var. lacrymans and var. shastensis on spruce or fir (χ 2, p > 0.05). Var. shastensis failed to grow on pine, but it is unknown whether this is due to its inability to decompose pine, or due to other experimental factors, e.g., the experimental setup on moist perlite may not have provided enough minerals. Spruce was more readily degraded by all strains, and this was particularly pronounced for var. lacrymans, which caused a mass loss of 50% of spruce but only 5% of pine wood blocks. See Supplementary Material for the mass loss of the additional species (Supplementary Fig. 1).
Fig. 3. Decomposition rate of Serpula lacrymans var. lacrymans, S. lacrymans var. shastensis and S. himantioides on different wood species.

. Percent mass loss of wood blocks from the three plant species fir (Abies lasiocarpa), pine (Pinus syvestris) and spruce (Picea abies) inoculated by var. lacrymans, var. shastensis and S. himantioides for 60 days. No successful growth was obtained for var. shastensis on pine
Evaluating antagonistic behavior
Serpula himantioides was significantly more combative than var. lacrymans and var. shastensis, as well as the three other brown rot species under the conditions tested (Table 3). Serpula himantioides was present in 79% of the re-isolations from the confrontations against the other species (i.e., as 50% would be a deadlock, S. himantioides took the substrate of the other species in 29% of the cases). The two S. lacrymans varieties were less able to exclude the other species compared to S. himantioides in this experiment, (var. shastensis was found in 40% and var. lacrymans in 41% of the cultures following confrontations, i.e., both lost their substrate in about 10% of the cases). When var. lacrymans and var. shastensis were confronted with C. puteana and A. xantha, the outcomes were close to 50% (i.e., a deadlock), but both S. lacrymans varieties were excluded by S. himantioides and F. pinicola (Table 3).
Table 3.
Results from combat experiments with Serpula lacrymans var. lacrymans, S. lacrymans var. shastensis, S. himantioides and three other fungal species
| var. lacrymans | var. shastensis | S. himantioides | C. puteana | A. xantha | |
|---|---|---|---|---|---|
| var. shastensis | 0.450 (20) | ||||
| S. himantioides | 0.689 (45)* | 0.685 (27) | |||
| C. puteana | 0.430 (43) | 0.500 (34) | 0.155 (45)** | ||
| A. xanta | 0.400 (45) | 0.355 (38) | 0.136 (44)** | 0.154 (39)** | |
| F. pinicola | 0.978 (46)** | 0.889 (45)** | 0.156 (48)** | 0.931 (29)** | 0.292 (48)** |
The proportion of plates with mycelia from the species named in the column after the confrontation test with the species in the row, i.e., read horizontally, higher than 0.5 wins the confrontation with the vertical strain Number of plates (n) used in parenthesis. *indicates significant different (*p < 0.05, **p < 0.005) from expected (E = n/2) by a Person's χ 2 Goodness of fit test, df = 1
Discussion
In this study we aimed to identify which features have made the dry rot fungus Serpula lacrymans var. lacrymans the most successful invasive wood decay fungus in the built environment by comparing its characteristics to its less invasive relatives. Since the successful establishment of an invasive species typically depends on a range of factors, we investigated the contribution of physiological factors (decomposition and combative ability), as well as underpinning genomic features. We detected numerous genomic signatures that may be linked to var. lacrymans invasiveness, including changes in selection pressure and evolution in gene families involved in hyphal growth, transportation, defense and decomposition of wood. Our experimental data suggest that S. lacrymans has poor antagonistic abilities toward other brown rot fungi, but that it has high wood-decomposition ability compared to its largely non-invasive relative S. himantioides. This suggests that S. lacrymans is an ecological specialist while S. himantioides is more of an ecological generalist.
One of our main findings is the differences in genome-wide selection pressure, evaluated by changes in rates of non-synonymous to synonymous substitutions (ω). The ω-values suggested that on average the genes of S. himantioides experienced stronger purifying selection than those of var. lacrymans, and especially those of var. shastensis. However, even if ω-values can detect genes under selection, systematically increased ω at the genome-wide level, can also be the result of demographic history [48]. In organisms with small effective population sizes, selection is less effective in removing deleterious mutations which can lead to elevated genome-wide ω-values. Correspondingly, we suggest that var. lacrymans and var. shastensis, which have higher average ω overall in the genome, may have lower effective population sizes compared to S. himantioides. The differences in effective population size is expected as S. himantioides is distributed worldwide [23], while var. shastensis has limited current distribution and var. lacrymans has gone through a domestication process [7].
In more detail, the genomic analyses revealed a selective shift in genes with functions involved in intracellular transport, growth and reorganization of the cell. Our data suggest that evolutionary changes to these processes may underlie the increase capacity of transportation and growth in var. lacrymans which in turn is likely to be a key factor for its success in the built environment. Buildings are a dry habitat, where the water resources are the most limiting factor. Var. lacrymans can produce the thickest mycelial cords described in the fungal kingdom, up to 2 cm in diameter [8]. In comparison, var. shastensis and S. himantioides produce smaller cords, and S. himantioides has a slower growth rate [19]. The corded network permits the translocation of intracellular resources, e.g., amino acids and water through vacuolar and vesicle trafficking to ensure complete exploitation of large woody substrates [49].
Proteins associated with endomembrane system functioning and hyphal growth had different selection pressure between S. lacrymans and S. himantioides, indicating that changes in resource translocation are important in the adaptation to the different niches. Hyphal growth is dependent on both transport and fusion of secretory vesicles to the plasma membrane and on actin cytoskeleton organization and polarization. Indeed, actin is important for polarized growth and also represents the mechanism for the transport of secretory vesicles that contain materials for the synthesis of new cell wall and membranes in the growing tip [50]. We hypothesize that these genes play a role in the development and maintenance of the mycelial cords, possibly through mediating the re-grouping and re-allocation of resources.
To become a successful colonizer of wood, a fungus has to compete for resources with other decay species. However, the confrontation experiments, where the fungi were growing in fir blocks on moist perlite, revealed that var. lacrymans and var. shastensis have poor combative abilities compared to other wood decay fungi, at least in this nutrient poor setup (Table 3). Species inhabiting more extreme environmental habitats may reduce their antagonistic abilities, following the universal adaptive strategy theory [51]. Thus S. lacrymans inhabiting the dry treeline and built environments may have lost the capacity for broad antagonistic responses. This may also explain why var. lacrymans usually does not spread from colonized buildings into the natural environment, though a few exceptions have been noted in the Czech Republic [52]. In less stressful climates in the boreal and temperate zones, where S. himantioides is typically found, interspecific antagonistic interactions may be more important. Hence, under these conditions, it may have been more advantageous to evolve strong combative ability. This is supported by the increased numbers of PFAM domains possibly related to defense in S. himantioides compared to S. lacrymans, e.g. PKS and ABC transporters. PKS are large synthases particularly involved in the biosynthesis of specialized metabolites with many diverse functions. The gene families are known to expand and contract rapidly in response to adaptation to nutritional and environmental factors, pathogens or interactions with other organisms [53]. ABC transporters are often involved in the efflux of small metabolites [54, 55]. Furthermore, similar expansions of PKS and ABC transporters have been observed in the mycoparasites Clonostachys rosea and Trichoderma virens, and were suggested to be the reason for their extreme combative ability, by producing and transporting toxic compounds from the cells [54]. Serpula himantoides is known to produce antifungal substances, himanimides, that could increase its antagonistic ability [56]. It is unknown if var. lacrymans can produce these substances. More genomic analyses and experiments using different conditions are needed to pinpoint the exact function of the larger number of PKS and ABC transporters in S. himantioides, and whether any of these expansions are related to the previously detected himanimides.
Our growth experiments on wood substrates confirm earlier findings that var. lacrymans is a highly effective decomposer of coniferous wood [19]. In natural environments, S. lacrymans typically occupies large logs of Abies or Tsuga (Fig. 1) and has developed a unique capacity for rapid decay during a short season of favorable growth conditions. Resource availability and utilization of nutrients involve a diverse chemistry for saprotrophic fungi. The varying levels of extractives, such as terpenoids and other phenolic compounds, and the recalcitrant nature of the carbohydrates of wood imply that specialization and adaptation to these conditions are essential to utilize this niche. Our findings suggest that S. lacrymans is a more successful decomposer of spruce and fir than pine, and is more specialized for these specific substrates than S. himantioides. A more narrow substrate range was also suggested in a recent study of var. lacrymans and Gloeophyllum trabeum, where they found gene expression of a wider CAZyme complement in G. trabeum than in S. lacrymans [15]. Furthermore, the speed and efficiency with which S. lacrymans decomposes spruce, compared to S. himantioides, could be related to a more efficient CMF chemistry. The iron reductase (with a CBM1 domain and a cytochrome B domain) found in var. lacrymans and var. shastensis, but not S. himantioides has previously been suggested to have an electron transfer function [57]. Thus, it can target reduced iron directly to the cellulose substrate for efficient CMF. In previous analyses of S. lacrymans, this iron reductase was specifically pinpointed as important in the early oxidative degradation steps of the CMF chemistry [10]. This could contribute to more efficient utilization of carbohydrates from its habitat.
The content of inhibitory extractives is greater in pine wood than in spruce wood [58], which makes pine a less favorable food source for fungi. Differential gene expression analyses of a white rot fungus (Phlebiopsis gigantea) grown on wood where extractives were removed showed several genes potentially related to the processing of extractives [59]. These differentially expressed genes encoded glutathione-S transferase, ABC transporters, lipases, cytochrome P450s and aldehyde dehydrogenase. We found accelerated evolution in S. lacrymans for aldehyde dehydrogenase, an ABC transporter, and cytochrome P450s, and loss of copies of glutathione-S transferase and ABC transporters. The ability to process a diversity of extractives found in wood and secrete their breakdown products may, therefore, also play an important role in substrate specialization and hence adaptation of S. lacrymans to a different habitat. Furthermore, the loss of laccases and the increase of cytochrome P450s in the branch leading to S. lacrymans could be related to both community interactions and the processing of toxic phenolic derivatives produced during the decomposition of lignocellulose. Brown rot fungi do not utilize lignin, however, they depolymerize lignin to gain access to the cellulose and hemicellulose. Thus, as part of adapting to a specialized niche S. lacrymans may have lost genes important for exploitation of some woody substrates in nature, but rather specialized for a more streamlined decomposition of specific substrates. Cytochrome P450s have been suggested to easily duplicate, and to be important in the colonization of new environments and in the breakdown of novel compounds [60]. Moreover, it has been suggested that the large gene repertoire of cytochrome P450s evolved in Phanerochaete chrysosporium increased its resource availability [61], thus the expansion of cytochrome P450s could be related to an expansion of biochemical capacity in var. lacrymans as it invades timber wood. Timber wood is similar to the wood encountered naturally by primary decay species, containing more plant-derived compounds than partially degraded wood that is often available in the forest.
The chemistry of defense and foraging is a recurring issue in our data set. However, without in-depth functional analysis, it is unclear whether the product moved by a particular ABC transporter or metabolized by a cytochrome P450 gene is of importance to the species’ competitive ability and the decomposition of different substrates. Thus, further analyses of the increased set of cytochrome P450s in S. lacrymans, and the increased set of PKS and ABC transporters in S. himantioides, can pinpoint in which functions these gene expansions are involved.
Our results indicate that the devastating dry rot fungus is an ecological specialist that has developed highly effective brown rot decay and effective systems for transportation and growth. Common traits identified between genetically related var. lacrymans and var. shastensis when compared with the sister taxon S. himantioides suggest that var. lacrymans was pre-adapted to the built environment and that the requirements of the mountainous, dry, treeline habitat and the patchy nutrient environment of a house, including a blend of wood and mineral materials, share similar features important for S. lacrymans. This enabled var. lacrymans to opportunistically exploit the built environment when given the opportunity by human activity. Particularly, the evolution of the thick cords and rapid growth may be linked to its natural substrates, to maximize resource translocation and effectively decay the enormous logs. The lower combative ability, suggested from both physiological and genomic data and the narrower enzymatic assortment of our selected strains might explain why var. lacrymans rarely has been able to move from its new building niche back into temperate and boreal woodlands. As var. shastensis is very similar to var. lacrymans both in genetic and physiologic features, we conclude it has the potential to invade buildings, but has not done so because its native range has not been widely exploited by humans and so has not been transferred to the built environment.
Electronic supplementary material
Acknowledgements
We thank Sarah Watkinson for fronting the JGI genome, Ella Thoen for technical help, Anikó Várnai for discussions, and Skui Christmas Tree Plantation for the Abies lasiocarpa wood. S.V.B, J.H., H.K., L.T., and I.S. acknowledge the University of Oslo and Norwegian Research Council (project 221840) for funding. N.H. thanks the Swedish University of Agricultural Sciences and FORMAS (project 2010-1354) for funding. D.C.E., S.C.M., and L.B. thank the UK Natural Environment Research Council, (award NE/K011588/1) for support. Sequencing of SL200 and SHA17-1 was performed by the SNP&SEQ Technology Platform in Uppsala. The facility is part of the National Genomics Infrastructure (NGI) Sweden and Science for Life Laboratory. The SNP&SEQ Platform is also supported bu the Swedish Research Council and the Knut and Alice Wallenberg Foundation. The work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231.
Author contributions
I.S., J.H., H.K., D.C.E., N.H., and L.B. conceived and designed the research. L.T, I.S., and J.H. analyzed physiological properties. I.S. and N.H. extracted DNA., K.L., A.A., K.B., and I.V.G. sequenced and analyzed the S. himantioides genome at JGI., S.V.B., M.B.D., J.H., C.P., I.S. and S.C.M. analyzed genomic data. S.V.B., J.H., D.C.E,, H.K. and I.S. wrote the paper and all other authors discussed and modified the paper.
Compliance with ethical standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0006-8) contains supplementary material, which is available to authorized users.
References
- 1.Nentwig W (ed) (2008) Biological Invasions. Ecological Studies. Berlin: Springer-Verlag; 193.
- 2.Robinson R. In: Genetics of the Norway Rat. International Series of Monographs in Pure and Applied Biology Zoology Division. Oxford: Pergamon Press; 1965;24.
- 3.Anderson PK, Cunningham AA, Patel NG, Morales FJ, Epstein PR, Daszak P. Emerging infectious diseases of plants: pathogen pollution, climate change and agrotechnology drivers. Trends Ecol Evol. 2004;19:535–44. doi: 10.1016/j.tree.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 4.Grunwald NJ, Goss EM, Press CM. Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol Plant Pathol. 2008;9:729–40. doi: 10.1111/j.1364-3703.2008.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stukenbrock EH, Bataillon T, Dutheil JY, Hansen TT, Li R, Zala M, et al. The making of a new pathogen: Insights from comparative population genomics of the domesticated wheat pathogen Mycosphaerella graminicola and its wild sister species. Genome Res. 2011;21:2157–66. doi: 10.1101/gr.118851.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kauserud H, Knudsen H, Hogberg N, Skrede I. Evolutionary origin, worldwide dispersal, and population genetics of the dry rot fungus Serpula lacrymans. Fungal Biol Rev. 2012;26:84–93. doi: 10.1016/j.fbr.2012.08.001. [DOI] [Google Scholar]
- 7.Kauserud H, Svegarden IB, Saetre GP, Knudsen H, Stensrud O, Schmidt O, et al. Asian origin and rapid global spread of the destructive dry rot fungus Serpula lacrymans. Mol Ecol. 2007;16:3350–60. doi: 10.1111/j.1365-294X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 8.Jennings DH, Bravery AF (eds) In: Serpula lacrymans: Fundamental Biology and Control Strategies. Hoboken: Wiley-Blackwell; 1991.
- 9.Boddy L, Frankland J, van West P (eds) (2007) In: Ecology of Saprotrophic Basidiomycetes.Academic Press, London
- 10.Eastwood DC, Floudas D, Binder M, Majcherczyk A, Schneider P, Aerts A, et al. The plant cell wall-decomposing machinery underlies the functional diversity of forest fungi. Science. 2011;333:762–5. doi: 10.1126/science.1205411. [DOI] [PubMed] [Google Scholar]
- 11.Floudas D, Held BW, Riley R, Nagy LG, Koehler G, Ransdell AS, et al. Evolution of novel wood decay mechanisms in Agaricales revealed by the genome sequences of Fistulina hepatica and Cylindrobasidium torrendii. Fungal Genet Biol. 2015;76:78–92. doi: 10.1016/j.fgb.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arantes V, Goodell B. Deterioration Prot Sustain Mater. 2014;1158:3–21. doi: 10.1021/bk-2014-1158.ch001. [DOI] [Google Scholar]
- 13.Riley R, Salamov AA, Brown DW, Nagy LG, Floudas D, Held BW, et al. Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc Natl Acad Sci USA. 2014;111:9923–8. doi: 10.1073/pnas.1400592111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang J, Presley GN, Hammel KE, Ryu JS, Menke JR, Figueroa M, et al. Localizing gene regulation reveals a staggered wood decay mechanism for the brown rot fungus Postia placenta. Proc Natl Acad Sci USA. 2016;113:10968–73. doi: 10.1073/pnas.1608454113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Presley GN, Schilling JS. Distinct growth and secretome strategies for two taxonomically divergent brown rot fungi. Appl Environ Microbiol 2017;83: e-pub ahead of print, 10.1128/AEM.02987-16. [DOI] [PMC free article] [PubMed]
- 16.Floudas D, Binder M, Riley R, Barry K, Blanchette RA, Henrissat B, et al. The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science. 2012;336:1715–9. doi: 10.1126/science.1221748. [DOI] [PubMed] [Google Scholar]
- 17.White NA, Dehal PK, Duncan JM, Williams NA, Gartland JS, Palfreyman JW, et al. Molecular analysis of intraspecific variation between building and ‘wild’ isolates of Serpula lacrymans and their relatedness to S. himantioides. Mycol Res. 2001;105:447–52. doi: 10.1017/S0953756201003781. [DOI] [Google Scholar]
- 18.Skrede I, Maurice S, Kauserud H. Molecular characterization of sexual diversity in a population of Serpula lacrymans, a tetrapolar basidiomycete. G3 (Bethesda) 2013;3:145–52. doi: 10.1534/g3.112.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harmsen L. Taxonomic and cultural studies on brown spored species of the genus Merulius. Friesia. 1960;6:233–77. [Google Scholar]
- 20.Palfreyman JW, Gartland JS, Sturrock CJ, Lester D, White NA, Low GA, et al. The relationship between ‘wild’ and ‘building’ isolates of the dry rot fungus Serpula lacrymans. FEMS Microbiol Lett. 2003;228:281–6. doi: 10.1016/S0378-1097(03)00783-3. [DOI] [PubMed] [Google Scholar]
- 21.Skrede I, Engh IB, Binder M, Carlsen T, Kauserud H, Bendiksby M. Evolutionary history of Serpulaceae (Basidiomycota): molecular phylogeny, historical biogeography and evidence for a single transition of nutritional mode. BMC Evol Biol. 2011;11:230. doi: 10.1186/1471-2148-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kauserud H, Hogberg N, Knudsen H, Elborne SA, Schumacher T. Molecular phylogenetics suggest a North American link between the anthropogenic dry rot fungus Serpula lacrymans and its wild relative S. himantioides. Mol Ecol. 2004;13:3137–46. doi: 10.1111/j.1365-294X.2004.02307.x. [DOI] [PubMed] [Google Scholar]
- 23.Carlsen T, Engh IB, Decock C, Rajchenberg M, Kauserud H. Multiple cryptic species with divergent substrate affinities in the Serpula himantioides species complex. Fungal Biol. 2011;115:54–61. doi: 10.1016/j.funbio.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 24.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gnerre S, MacCallum I, Przybylski D, Riberio F, Burton JN, Walker BJ, et al. High-quality draft assemblies of mammalian genomes of massively parallel sequence data. Proc Natl Acad Sci USA. 2011;108:1513–8. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23:1061–7. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 27.Holt C, Yandell M. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics. 2011;12:491. doi: 10.1186/1471-2105-12-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones P, Binns D, Chang HY, Fraser M, Li W, McAnulla C, et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30:1236–40. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L, Stoeckert CJJ, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–89. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–71. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- 31.Balasundaram SV, Engh IB, Skrede I, Kauserud H. How many DNA markers are needed to reveal cryptic fungal species? Fungal Biol. 2015;119:940–5. doi: 10.1016/j.funbio.2015.07.006. [DOI] [PubMed] [Google Scholar]
- 32.Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 33.Loytynoja A. Phylogeny-aware alignment with PRANK. Methods Mol Biol. 2014;1079:155–70. doi: 10.1007/978-1-62703-646-7_10. [DOI] [PubMed] [Google Scholar]
- 34.Fletcher W, Yang Z. The effect of insertions, deletions, and alignment errors on the branch-site test of positive selection. Mol Biol Evol. 2010;27:2257–67. doi: 10.1093/molbev/msq115. [DOI] [PubMed] [Google Scholar]
- 35.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–91. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 36.Benjamini Y, Krieger AM, Yekutieli D. Adaptive linear step-up procedures that control the false discovery rate. Biometrika. 2006;93:491–507. doi: 10.1093/biomet/93.3.491. [DOI] [Google Scholar]
- 37.Petersen TN, Brunak S, Heijne von G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–6. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 38.Emanuelsson O, Brunak S, Heijne von G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–71. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 39.Krogh A, Larsson B, Heijne von G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–80. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 40.Hulo N. The PROSITE database. Nucleic Acids Res. 2006;34:D227–D230. doi: 10.1093/nar/gkj063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Horton P, Park KJ, Obayashi T, Fujita N, Harada H, Adams-Collier CJ, et al. WoLF PSORT: protein localization predictor. Nucleic Acids Res. 2007;35:W585–7. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;42:410–5. [DOI] [PubMed]
- 43.Rawlings ND, Waller M, Barrett AJ, Bateman A. MEROPS: the database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 2014;42:D503–9. doi: 10.1093/nar/gkt953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42:D490–5. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Y, Mao X, Yang J, Chen X, Mao F, Xu Y. dbCAN: a web resource for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 2012;40:W445–51. doi: 10.1093/nar/gks479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Syed K, Mashele SS. Comparative Analysis of P450 Signature Motifs EXXR and CXG in the Large and Diverse Kingdom of Fungi: Identification of Evolutionarily Conserved Amino Acid Patterns Characteristic of P450 Family McCluskey K (ed) PLoS ONE. 2014;9:e95616–14. doi: 10.1371/journal.pone.0095616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 48.Tajima F. The effect of change in population size on DNA polymorphism. Genetics. 1989;123:597–601. doi: 10.1093/genetics/123.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watkinson SC, Bebber D, Darrah P, Fricker M, Tlalka M, Boddy L. The role of wood decay fungi in the carbon an nitrogen dynamics of the forest floor. In: Gadd GM (ed). Fungi in Biochemical Cycles. Cambridge University Press; 2006. p 1–31.
- 50.Berepiki A, Lichius A, Read ND. Actin organization and dynamics in filamentous fungi. Nat Rev Microbiol. 2011;9:876–87. doi: 10.1038/nrmicro2666. [DOI] [PubMed] [Google Scholar]
- 51.Grime JP, Pierce S. The evolutionary strategies that shape ecosystems. Hoboken: Wiley-Blackwell; 2012
- 52.Kotlaba F. (1992) Nalezy drevomorky domaci - Serpula lacrymans v prirode. Ceska Mycologie.
- 53.Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26. doi: 10.1186/1471-2148-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karlsson M, Durling MB, Choi J, Kosawang C, Lackner G, Tzelepis GD, et al. Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol Evol. 2015;7:465–80. doi: 10.1093/gbe/evu292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Klein C, Kuchler K, Valachovic M. ABC proteins in yeast and fungal pathogens. Essays Biochem. 2011;50:101–19. doi: 10.1042/bse0500101. [DOI] [PubMed] [Google Scholar]
- 56.Aqueveque P, Anke T, Sterner O. The himanimides, new bioactive compounds from Serpula himantoides (Fr.) Karst. Z Naturforsch C. 2002;57:257–62. doi: 10.1515/znc-2002-3-410. [DOI] [PubMed] [Google Scholar]
- 57.Yoshida M, Igarashi K, Wada M, Kaneko S, Suzuki N, Matsumura H, et al. Characterization of carbohydrate-binding cytochrome b562 from the white-rot fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 2005;71:4548–55. doi: 10.1128/AEM.71.8.4548-4555.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sjöström E. (1993) Wood chemistry: fundamentals and applications. London: Academic Press.
- 59.Hori C, Ishida T, Igarashi K, Samejima M, Suzuki H, Master E, et al. Analysis of the Phlebiopsis gigantea genome, transcriptome and secretome provides insight into its pioneer colonization strategies of wood. PLoS Genet. 2014;10:e1004759. doi: 10.1371/journal.pgen.1004759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Syed K, Shale K, Pagadala NS, Tuszynski J. Systematic identification and evolutionary analysis of catalytically versatile Cytochrome P450 monooxygenase families enriched in model basidiomycete fungi Yu J-H (ed) PLoS ONE. 2014;9:e86683–18. doi: 10.1371/journal.pone.0086683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Syed K, Yadav JS. P450 monooxygenases (P450ome) of the model white rot fungus Phanerochaete chrysosporium. Crit Rev Microbiol. 2012;38:339–63. doi: 10.3109/1040841X.2012.682050. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw sequence reads, and assembled genomes are available on NCBI at Bioproject PRJNA412961. In addition, the S. himantioides MUCL38935 genome is available at the JGI genome browser (http://genome.jgi.doe.gov/Serla_varsha1/Serla_varsha1). The MAKER2 gene predictions, the OrthoMCL clusters and the alignments used as input to the Codeml analyses have been deposited in the Dryad Digital Repository: doi:10.5061/dryad.28sb6.