Abstract
The ecology of antibiotic resistance involves the interplay of a long natural history of antibiotic production in the environment, and the modern selection of resistance in pathogens through human use of these drugs. Important components of the resistome are intrinsic resistance genes of environmental bacteria, evolved and acquired over millennia, and their mobilization, which drives dissemination in pathogens. Understanding the dynamics and evolution of resistance across bacterial taxa is essential to address the current crisis in drug-resistant infections. Here we report the exploration of antibiotic resistance in the Paenibacillaceae prompted by our discovery of an ancient intrinsic resistome in Paenibacillus sp. LC231, recovered from the isolated Lechuguilla cave environment. Using biochemical and gene expression analysis, we have mined the resistome of the second member of the Paenibacillaceae family, Brevibacillus brevis VM4, which produces several antimicrobial secondary metabolites. Using phylogenomics, we show that Paenibacillaceae resistomes are in flux, evolve mostly independent of secondary metabolite biosynthetic diversity, and are characterized by cryptic, redundant, pseudoparalogous, and orthologous genes. We find that in contrast to pathogens, mobile genetic elements are not significantly responsible for resistome remodeling. This offers divergent modes of resistome development in pathogens and environmental bacteria.
Introduction
The current antibiotic crisis, where new antibiotic development is not keeping pace with resistance in pathogens, is driven by mobile resistance genes. The preponderance of evidence implicates non-pathogenic environmental and commensal bacteria as the origin and reservoir of these genes [1–4]. Extensive genotyping of environmental bacteria along with metagenomic studies point to a rich and diverse reservoir of resistance determinants in the environment that includes orthologs of well characterized mobilized genes in addition to novel mechanisms not yet in the clinic [5–12]. Antibiotics have been naturally produced by bacteria for millions of years, which may have caused resistance genes to become widespread in environmental bacteria [1, 13, 14]. The movement of these genes into pathogens is the result of stochastic capture, sometimes before the antibiotic era, but escalated with human use over the past 70 years [13]. While the link between resistance in pathogenic and non-pathogenic bacteria is established, the genetic mechanisms underlying the initial mobilization of resistance elements remains unclear.
The modern use of antibiotics is generally agreed to have selected for mobilizing and disseminating antibiotic resistance genes across almost every type of bacterial pathogen [13, 15]. The direct impact of the non-medical use of antibiotics, such as in agriculture, on the clinical problem of antibiotic resistance, is less certain but is likely significant for disseminating resistance genes in the environment [16–19]. Hotspots for horizontal transfer of resistance genes include wastewater treatment plants, manureing of soils, and other sites where waste products with prior exposure to antibiotics are concentrated and mixed in the environment. Metagenomic studies demonstrate that resistance diversity in the environment is strongly linked to bacterial phylogeny, and is generally not associated with mobile genetic elements [5]. In contrast, phylogenomic investigations have revealed surprising mobility in intrinsic resistomes [9, 12, 20]. Taken together, this suggests that mobilization events are rare, and mobilization of some resistance elements may be linked to the natural expansion and contraction of microbial accessory genomes [21, 22].
We previously identified the edeine biosynthetic cluster from Brevibacillus brevis VM4 and characterized a novel edeine acetyltransferase responsible for self-resistance [23]. While attempting to develop genetic tools for studying edeine biosynthesis, we discovered that B. brevis VM4 was resistant to most antibiotics commonly used as selectable markers. We recently reported an ancient and surprisingly broad drug-resistant genotype in Paenibacillus sp. LC231 [9], which is in the Paenibacillaceae family along with Brevibacillus. We found that resistomes of Paenibacillus sp. LC231 and related strains did not correlate well with species phylogeny, suggesting resistance genes may be mobile in these isolates. In this work, we investigate the intrinsic resistome of B. brevis VM4 using whole-genome sequencing, in vitro enzyme assays, gene expression analysis, and comprehensive phenotype characterization, to reveal cryptic, redundant, and pseudoparalogous genes. Using the experimentally validated B. brevis VM4 and Paenibacillus sp. LC231 resistance genotypes, we expanded resistome predictions to all sequenced Paenibacillaceae and used phylogenomics to study a link between taxonomic, resistome, and secondary metabolite diversity. We ultimately found a complex network of resistance genes weakly clustered by family phylogeny, unlinked to self-produced secondary metabolites, and sometimes associated with mobile genetic elements. Diverse multi-drug resistant genotypes are embedded in all genomes consistent with a long history of antibiotic exposure. This contrasts with the mobilized resistomes of most pathogens where resistance is rapidly acquired and lacks long genomic history.
Results
The Brevibacillus brevis VM4 resistome is diverse and redundant
We found that the edeine producer B. brevis VM4 (ATCC 35690) exhibited an extensive multi-drug resistance phenotype (Supplementary Table 1). Analysis of the draft genome using the Comprehensive Antibiotic Resistance Database [24] (CARD) revealed a complex and redundant resistome. B. brevis VM4 harbors at least 10 predicted resistance genes, including two different resistance determinants for each of chloramphenicol, kanamycin, and rifampin (Supplementary Fig. 1). Predicting the specificity and associated resistance phenotype of efflux pumps that share little sequence identity with related homologs remains difficult [25]. We therefore focused on target protection and antibiotic modification resistance mechanisms. For many antibiotics, the sensitivity profile of B. brevis VM4 correlated well with the resistome genotype. For example, B. brevis VM4 is resistant to penicillins and streptomycin, which correlates with CARD predictions of a TEM-type β-lactamase (bbI) and a streptomycin 6-nucleotidyltransferase (ant(6)-Ic). We also identified two chloramphenicol resistance mechanisms; a chloramphenicol acetyltransferase (catV) and a Cfr-like ribosome methyltransferase (clbB) that is identical (100% at the amino acid level) to an experimentally verified ortholog, ClbB, from B. brevis NBRC 100599 [26]. All of these genes conferred resistance to their respective antibiotics when heterologously expressed in E. coli (Table 1, Supplementary Tables 2, 3, and 4). We also identified genes predicted to encode two 2-deoxystreptamine (2-DOS) aminoglycoside modifying enzymes, consistent with resistance to kanamycin and other 2-DOS aminoglycosides (Table 1). Unexpectedly, we also identified a macrolide kinase and two rifampin phosphotransferases despite phenotypic sensitivity to most macrolide and rifamycin antibiotics.
Table 1.
Heterologous expression in E. coli of resistance genes identified in this study
| Resistance determinant | Name | Antibiotic class | Antibiotic | MIC | Empty vector MIC |
|---|---|---|---|---|---|
| Kanamycin 4′ nucleotidyltransferase | ant(4′)−Ic | Aminoglycoside | Kanamycin | > 256 | 8 |
| Neomycin | > 256 | 16 | |||
| Tobramycin | > 256 | 4 | |||
| Kanamycin 6′ acetyltransferase | aac(6 ’ )−35 | Aminoglycoside | Kanamycin | > 256 | 8 |
| Streptomycin 6 nucleotidyltransferase | ant(6)−Ic | Aminoglycoside | Streptomycin | > 256 | 8 |
| Rifamycin kinase | rphC | Rifamycin | Rifampin | 512 | 4 |
| Rifamycin kinase | rphD | Rifamycin | Rifampin | 256 | 4 |
| TEM β-lactamase | bbI | β-lactam | Ampicillin | > 256 | 16 |
| Piperacillin | > 256 | < 0.5 | |||
| β-lactam/β-lactamase inhibitor | Piperacillin/Tazobactam | 8 | 0.5 | ||
| Cephalosporin | Cephalexin | 4–8 | 4 | ||
| Macrolide kinase | mphJ | Macrolide | Erythromycin | 0.5 | 64 |
| Clarithromycin | 0.5 | 32 | |||
| Azithromycin | 0.063 | 8 | |||
| Telithromycin | 0.125 | > 16 | |||
| Spiramycin | 2 | 4 | |||
| Tylosin | 2 | 512 | |||
| Josamycin | 2–4 | 2 | |||
| Pikromycin | 2 | 4 | |||
| Chloramphenicol acetyltransferase | catV | Phenicol | Chloramphenicol | 16 | 4 |
| Cfr-like ribosomal methyltransferase | clbB | Lincosamide | Clindamycin | 12.8 | 1.6 |
| Lincomycin | 256 | 128 | |||
| Phenicol | Chloramphenicol | 6.4 | 1.6 |
B. brevis VM4 harbors two functional 2-DOS aminoglycoside inactivating enzymes
Genetic redundancy is a characteristic of biological systems [27], and aminoglycoside-producing bacteria commonly express multiple resistance mechanisms for self-protection [28]. B. brevis VM4 harbors two different genes predicted to inactivate 2-DOS aminoglycosides such as kanamycin; an acetyltransferase (aac(6′)-35) that is 54% identical to aac(6′)-34 from Paenibacillus sp. LC231, and a nucleotidyltransferase (ant(4′)-Ic) that is 71% identical to ant(4′)-Ia from Bacillus clausii [29]. B. brevis VM4 lacks genes expected for aminoglycoside biosynthesis. Both aac(6′)-35 and ant(4′)-Ic confer kanamycin resistance when expressed in E. coli (Table 1). Steady-state kinetic analysis of purified enzymes confirmed the predicted activities (Supplementary Table 4).
Using quantitative reverse-transcription PCR (qRT-PCR), we demonstrated that both ant(4’′)-Ic and aac(6’′)-35 are constitutively expressed, but that ant(4′)-Ic is the predominantly expressed resistance determinant (Supplementary Fig. 2). These results suggest that ant(4′)-Ic is the dominant gene responsible for kanamycin resistance in B. brevis VM4. We then determined phenotypic sensitivity to a panel of aminoglycosides where only one of the two enzymes should confer resistance. B. brevis VM4 is resistant to paromomycin and lividomycin, which are both aminoglycosides lacking a 6′-NH2 site for modification by AAC(6′) but have a 4′-OH for ANT(4′) modification (Supplementary Table 1). Conversely, B. brevis VM4 is much more sensitive to sisomicin and netilmicin; both are aminoglycosides with a 6′ amino group but lack the 4′ hydroxyl required for ANT(4′) modification. The resistance phenotype of B. brevis VM4 is consistent with weak aac(6′)-35 expression, and that ant(4′)-Ic is the dominant gene governing 2-DOS aminoglycoside resistance.
B. brevis VM4 is sensitive to macrolide antibiotics despite harboring a macrolide kinase
B. brevis VM4 is sensitive to most macrolides such as erythromycin, yet it harbors a predicted macrolide kinase (mphJ). The most closely related ortholog to MphJ in CARD is MphI from Paenibacillus sp. LC231 (51% identity), which has narrow substrate specificity. MphI is restricted to 16-membered macrolides (tylosin, spiramycin) and 14-membered ketolides (pikromycin, telithromycin), and does not confer resistance to cladinose containing macrolides, including erythromycin [9]. We heterologously expressed mphJ in E. coli and demonstrated that it confers resistance to a wide range of macrolide antibiotics, including erythromycin (14-membered ring), tylosin (16-membered ring), and telithromycin (ketolide), provides some decrease in sensitivity to spiramycin (16-membered ring) and pikromycin (ketolide), and no resistance to josamycin (16-membered ring) (Table 1). Purified MphJ modifies macrolide antibiotics in a GTP-dependent fashion with efficiency comparable to other characterized orthologs (Supplementary Table 5). MphJ is a functional macrolide kinase, and therefore, B. brevis VM4 has the genetic potential for macrolide resistance, though remained sensitive to these antibiotics.
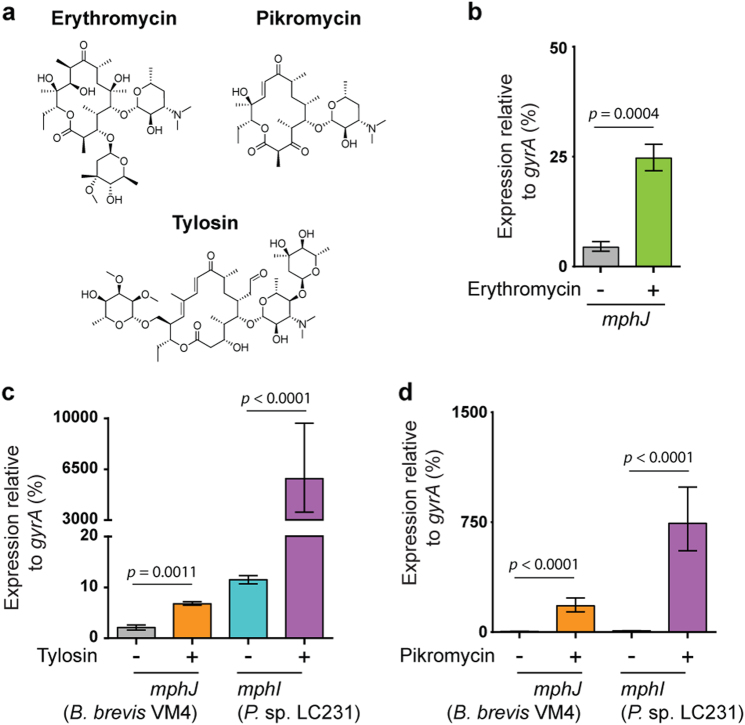
We next investigated mphJ expression using qRT-PCR to determine if the macrolide-sensitive phenotype is a result of lack of gene expression. Constitutive mphJ expression is low but increases almost six-fold when B. brevis VM4 is challenged with ¼ MIC erythromycin (Fig. 1a, b). However, phenotypic sensitivity to erythromycin (MIC) remained unchanged at 0.25 µg mL−1 after preincubation with ¼ MIC erythromycin, indicating that this increase in mphJ expression is not sufficient to confer erythromycin resistance. We then investigated whether mphJ expression is insufficient to confer phenotypic resistance. MphI is actively expressed in Paenibacillus sp. LC231 and serves as a proper calibration for correlating gene expression with macrolide sensitivity phenotypes [9]. We compared mphI and mphJ expression levels with and without tylosin, which is a natural macrolide with a 16-membered ring, a mycarose at 4′ of mycaminose, and a mycinose at position 14 (Fig. 1a). In Paenibacillus sp. LC231, mphI expression is constitutively low but increases > 500-fold when challenged with ¼ MIC tylosin (Fig. 1c). In contrast, mphJ expression increases only three-fold when B. brevis VM4 is challenged with ¼ MIC tylosin. The inducible expression of macrolide resistance genes is known to be dependent on substrate structure [30–32], particularly the presence or absence of sugars on the C3 position, and ring size. Therefore, we also compared mphI and mphJ expression in response to pikromycin, a natural ketolide similar to erythromycin but without a cladinose on the C3 position (Fig. 1a). When challenged with pikromycin, mphJ expression in B. brevis VM4 increases almost 40-fold at ¼ MIC, while mphI expression in Paenibacillus sp. LC231 increases 86-fold at ≤ 0.063 × MIC (Fig. 1a, d). Our findings are consistent with an mphJ activating mechanism that is unable to respond to erythromycin or tylosin but is responsive to pikromycin. mphJ confers limited pikromycin resistance when expressed in E. coli, but high resistance to tylosin and erythromycin. Therefore, the regulation of macrolide resistance in B. brevis VM4 is de-coupled from the in vitro substrate specificity of MphJ, and likely reflects the natural history of macrolide resistance in this organism, where resistance to pikromycin may be the ecological driver.
Fig. 1.
The regulatory system for mphJ expression is dysfunctional in B. brevis VM4. a Chemical structures of the macrolides erythromycin, pikromycin, and tylosin. b Despite phenotypic sensitivity to macrolides, B. brevis VM4 harbors mphJ, whose expression is normally low, and increases in response to ¼ MIC erythromycin. c Comparative expression analysis of mphI in Paenibacillus sp. LC231 and mphJ in B. brevis VM4 in response to ¼ MIC tylosin. d Comparative expression of mphI and mphJ in response to pikromycin. Error bars represent one standard deviation. P-values were determined using the unpaired Students t-test
B. brevis VM4 has two rifampin phosphotransferase pseudoparalogs
Rifampin phosphotransferases (Rph) transfer a phosphate to the C21 position of rifamycins and are widespread in environmental and some pathogenic bacteria, including Listeria monocytogenes [11, 33]. This enzyme family is structurally related to phosphoenolpyruvate (PEP) synthases but has a unique domain architecture [33]. Combinations of different rifampin resistance elements in a single strain are known, but not paralogs of Rphs [10, 11, 34]. CARD analysis of the draft B. brevis VM4 genome predicted two rifampin phosphotransferases that are 78% identical at the amino acid level. We verified that both predicted rphs (rphC and rphD) confer rifampin resistance when heterologously expressed in E. coli (Table 1). Steady-state kinetic analysis of RphC and RphD revealed that both are catalysts for rifampin phosphorylation, and have a catalytic constant similar to other Rphs [9, 11, 33] (Supplementary Table 5). Rifampin directly interacts with RpoB by forming four essential hydrogen bonds. Therefore, we probed the possibility that one Rph acquired the ability to modify a position other than C21 on the antibiotic by performing an in vitro enzyme assay with both RphC and RphD in a single reaction and monitoring for diphosphorylation of the antibiotic. The inactivated rifampin product of a combined reaction is monophosphorylated (Rif-PO4 2− expected mass; 902.3715 Da, combined reaction product; 901.56 m/z in negative-mode), which is identical to the reaction product of each enzyme individually ((RphC product; 901.56 m/z, RphD product; 901.56 m/z), Fig. 2a). Within the B. brevis clade, only B. brevis VM4 and B. brevis GZDF3.1 harbor two Rphs. RphC is conserved across the B. brevis clade, and therefore it is likely that RphD was recently acquired. If RphD arose by duplication of RphC, then they would be expected to cluster together in phylogenetic analysis. However, RphD forms an individual clade unrelated to RphC, and accordingly, these genes should be considered pseudoparalogs (Fig. 2b) [35]. rphD and the immediate upstream region share 95% identity with the region adjacent to an rph in P. lautus FSL F4-0100, which is consistent with horizontal transfer from a Paenibacillus donor (Supplementary Fig. 3a). Genetic context analysis of this locus among B. brevis VM4 relatives reveals that rphD was acquired by the ancestor of B. brevis GZDF3.1 and B. brevis VM4 (Supplementary Fig. 3b).
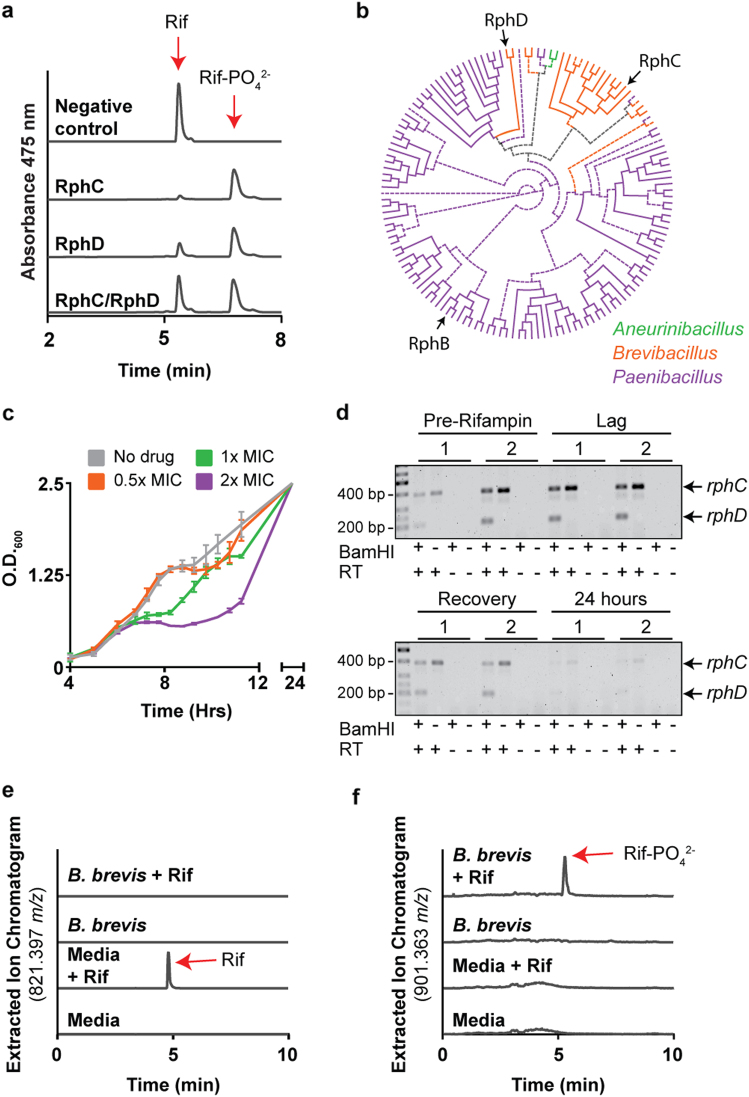
Fig. 2.
B. brevis VM4 has a rifampin sensitive phenotype despite two functional rifampin phosphotransferase pseudoparalogs. a RphC and RphD monophosphorylate rifampin at the same position. b A phylogeny of Paenibacillaceae Rphs demonstrates that RphD is a horizontally acquired resistance element and did not arise from a gene duplication event of RphC. The unrooted Rph phylogeny is displayed as a circular cladogram and with a midpoint root. Branches with low support (bootstrap value < 70) are displayed with dotted lines. RphB, RphC, and RphD are indicated with arrows. c B. brevis VM4 overcomes 1x and 2x MIC rifampin when challenged in mid-exponential phase. Rifampin was added at OD600 of 0.5. d rphC and rphD are constitutively expressed. RNA was isolated from cultures before and after a rifampin challenge and rph genes were amplified using degenerate primers. Expression of each gene was determined after BamHI digestion, which specifically digests RphD. Samples treated identically but void of reverse transcriptase (RT) were used as negative controls. e and f HPLC-high-resolution mass spectrometry analysis of B. brevis VM4 cultured with 1x MIC rifampin showing the degradation of rifampin e and the presence of rifampin phosphate (f)
Paradoxically, despite the presence of two rph elements, B. brevis VM4 is very sensitive to rifampin (MIC 0.063 µg ml−1). In Actinobacteria, rifampin phosphotransferase expression is induced by the presence of rifampin and is regulated by an upstream rifampin-associated element (RAE) [11]. We therefore reasoned that rph expression in B. brevis VM4 might also be inducible despite the absence of an upstream RAE, and investigated this hypothesis by challenging B. brevis VM4 with rifampin at mid-exponential phase. B. brevis VM4 could overcome a rifampin challenge of 1x and 2x MIC after a short lag-phase, which may suggest an inducible resistance mechanism [36] (Fig. 2c). We then probed for rphC and rphD expression before and after a rifampin challenge but found that both genes are constitutively expressed, and are therefore not regulated by rifampin (Fig. 2d). A lag-phase is typically characteristic of inducible resistance, but has only been studied for resistance determinants that do not chemically modify the antibiotic. Therefore, rifampin may be inactivated during this lag phase before B. brevis VM4 returns to exponential growth. We monitored the presence of rifampin and rifampin phosphate in conditioned media with high-resolution mass spectrometry after challenging B. brevis VM4 with 1x MIC rifampin and found that the antibiotic was completely degraded by 24 h (Fig. 2e). We also identified rifampin phosphate; however, the extracted ion chromatogram intensity was lower than expected, consistent with downstream degradation of rifampin phosphate (Fig. 2f). To identify the rifampin degradation product, we added 5 µg ml−1 rifampin to a culture of B. brevis VM4 in stationary phase and monitored rifampin modification over time using HPLC. Rifampin was completely degraded over 4 days, and we were unable to detect the appearance of a product with absorbance at 475 nm, which is characteristic of the naphthoquinone chromophore (Supplementary Fig. 4). We were also unable to detect any other known modified products of rifampin using high-resolution mass spectrometry [10]. Some Actinobacteria are known to degrade rifampin into unidentified products [10], and we can now include the Firmicutes phylum among those bacteria with this capacity.
Paenibacillaceae resistomes are not structured by phylogeny
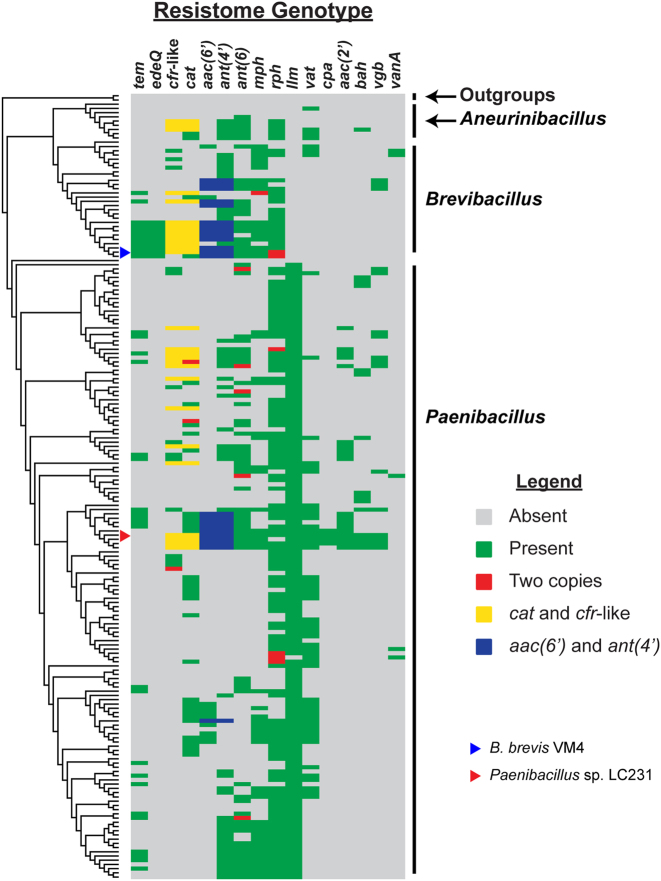
To investigate whether multi-drug resistance genotypes are common in Paenibacillaceae and if their resistomes are vertically inherited, we performed a phylogenomic comparison of resistome genotypes across this family. We first determined the evolutionary relationship between 184 Paenibacillaceae isolates by generating a family tree using 109 well-conserved genes (TIGRFAM “core bacterial protein” GenProp0799) and then comparing the presence or absence of resistance genes in each strain (Fig. 3, Supplementary Fig. 5). In general, multi-drug resistant resistomes are common in Paenibacillaceae. Except for cpa and edeQ, none of the resistance genes are monophyletic, which is indicative of horizontal transfer into discrete clades. The Paenibacillaceae resistomes are therefore not conserved among each related strain as a result of resistance gene gain and loss, which is consistent with environmental selection for optimized resistomes. Redundant chloramphenicol (catV and clbB) and kanamycin (ant(4′)-lc and aac(6′)-35) resistance are common in Paenibacillaceae, especially within Brevibacillus and Aneurinibacillus. However, strains harboring two rph genes are rare; only 6 of 184 Paenibacillaceae strains have two rphs. We also detected two copies of ant(6), cat, clbB, and mph genes in single isolates, but this was also rare.
Fig. 3.
Paenibacillaceae resistomes do not cluster by phylogeny. 15 resistance enzymes and the vanA operon were compared across a family phylogeny of 184 Paenibacillaceae isolates. The tree is presented as a dendrogram. Resistance enzymes were identified in this study, or in the experimental validation of the Paenibacillus sp. LC231 resistome [9]. The functions of each resistance enzymes not identified in this study are; Llm lincosamide resistance methyltransferase; Vat streptogramin A acetyltransferase, Cpa capreomycin acetyltransferase, AAC(2′) kasugamycin acetyltransferase, Bah bacitracin amidohydrolase, Vgb streptogramin B lyase. The complete family phylogeny is found in Supplementary Fig. 5
To investigate whether the resistome genotype of strains was impacted by the isolation source, we compared the generalized habitat (e.g., soil, water, mammalian-associated), if known, with the family phylogeny and resistome comparisons (Supplementary Fig. 6). Resistome genotypes did not appear to depend on the isolation source.
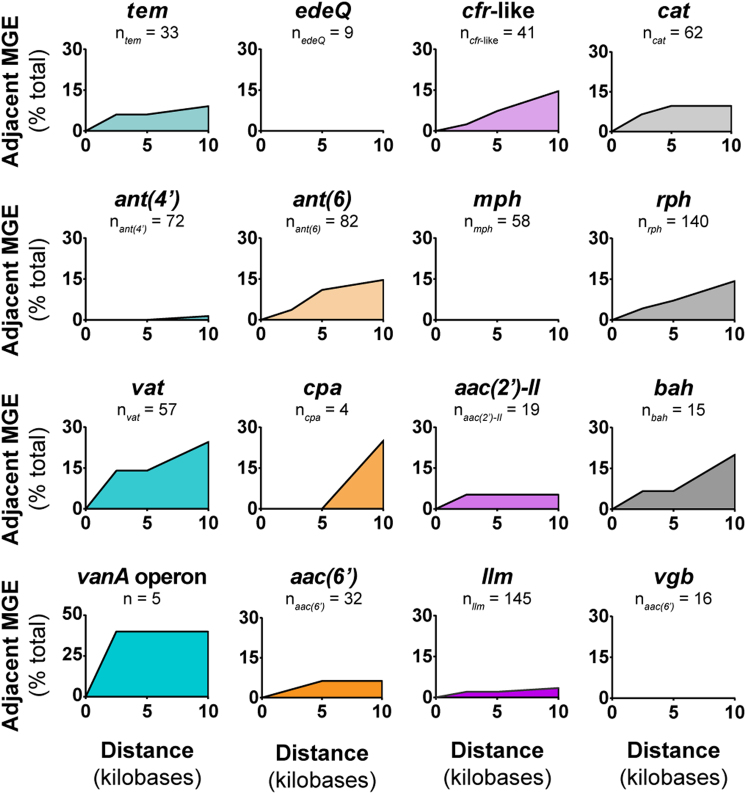
Given that Paenibacillaceae resistomes are variable, we hypothesized that resistance genes could be horizontally transferred on mobile genetic elements (e.g., transposase, resolvase, recombinase). We infer horizontal gene transfer by incongruities between family phylogeny and the presence or absence of resistance genes [37]. Genomic regions that are mobile can be over 100 kb, but we focused on the regions immediately adjacent (< 10 kb) to resistance genes to establish direct genetic linkage and to reduce bias from resistance determinants present near the end of contigs in draft assemblies, similar to previous studies (Supplementary Materials and Methods) [5, 38, 39]. The terms “integrase”, “recombinase”, and “transposase” were the most dominant. Every resistance gene family except for Vgb, Mph, and EdeQ had at least one member within 10 kb of mobile elements, and several had mobile elements within 2.5 kb (Fig. 4). In most cases, resistance genes appear ancestral to genetic association with mobile elements since resistance is more conserved than nearby mobile elements across closely related strains (Supplementary Fig. 7). In general, associated mobile genetic elements do not explain resistance gene acquisition. A caveat is that many of the available genomes are assembled from short-reads, and plasmid detection is difficult. Nevertheless, several Paenibacillus are known to have plasmids [40, 41], but none of the resistance genes identified in this study were identified on known plasmids from completed genomes (Supplementary Fig. 8; see Supplementary Materials and Methods for more details).
Fig. 4.
Paenibacillaceae resistomes are often associated with nearby mobile genetic elements. 2.5 kb, 5 kb, and 10 kb regions upstream and downstream from each resistance gene or operon were searched for annotated mobile genetic elements. The y-axis is the proportion of a resistance genes within that family that has one or more mobile genetic element in a given range
Vancomycin is reserved for serious, multi-drug resistant infections (e.g., Methicillin-Resistant Staphylococcus aureus and enterococci), and the evolution and dissemination of vancomycin resistance in pathogenic bacteria is of significant interest. Vancomycin resistance is the result of a two-component regulatory system (vanRS) and a three-gene operon (vanHAX) that remodels the peptidoglycan drug target by converting d-alanyl-d-alanine to d-alanyl-d-lactate, which is immune to vancomycin binding [42]. Two Paenibacillus strains are known to harbor vancomycin resistance genes that share 79–94% DNA sequence identity with homologs in the vanA operon from vancomycin-resistant enterococci (VRE), leading to the hypothesis that these genes may have originated in the Paenibacillus genus [43]. Paenibacillus sp. LC231 [9] and B. brevis VM4 (this study) are phenotypically sensitive to vancomycin and also to the structurally related antibiotic, teicoplanin (Supplementary Table 1). We were unable to identify a vancomycin resistance operon in either strain. We expanded our search to the Paenibacillaceae family, and identified five vanA operons in total; three in Paenibacillus strains and two in Brevibacillus strains (Fig. 3, Supplementary Fig. 9). Overall, vancomycin resistance operons do not appear widespread in Paenibacillaceae and may have been horizontally transferred into these strains. Furthermore, the two vanA operons identified in Brevibacillus strains are adjacent to mobile genetic elements (transposase, resolvase) (Fig. 4 and Supplementary Figs. 7 and 9). However, the three van operons identified in Paenibacillus strains were not associated with mobile genetic elements, which has been reported previously [43]. The van operon from Paenibacillus macerans was found on a single chromosome of 7.33 Mb, and not on a plasmid, consistent with transfer mediated by an unknown mechanism. Our results support the consensus [44–46] that the vanA operon was mobilized in environmental bacteria before transferring into pathogenic bacteria; however, Paenibacillus strains do not appear to be the origin. The vanF operon has also been identified in Paenibacillus popilliae [47], but it was not found in any of the 184 Paenibacillaceae genomes included in this study.
Resistome genotypes do not correspond with secondary metabolite diversity in Brevibacillus
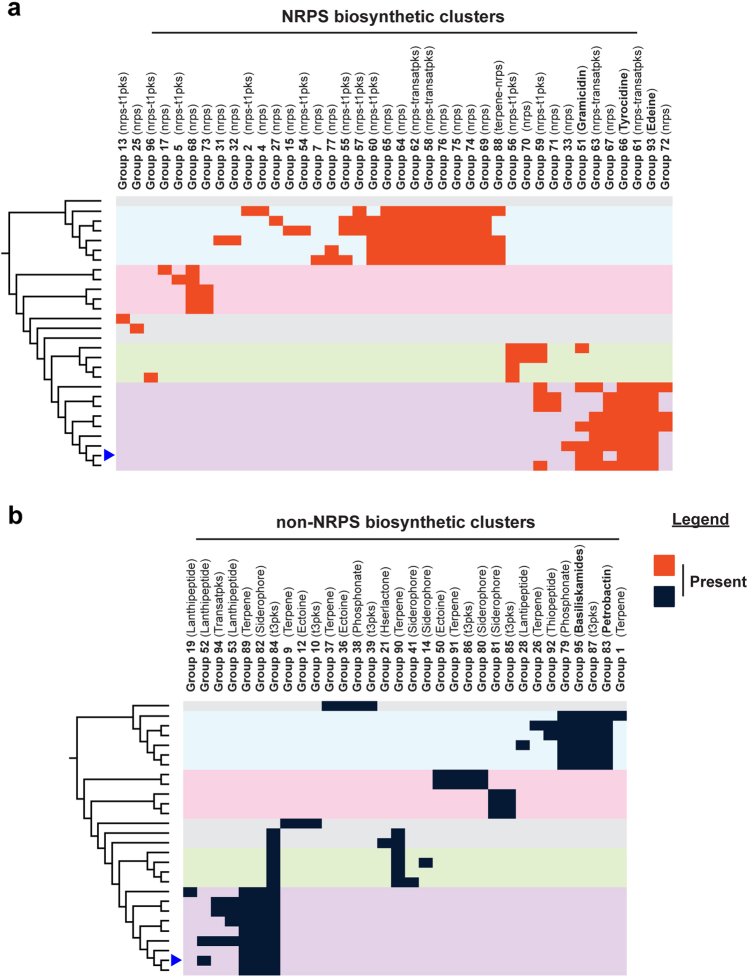
B. brevis VM4 and other Brevibacillus strains are known antibiotic producers, and we therefore investigated a potential link between diversity of secondary metabolism and resistance within this genus. We used antiSMASH [48] to predict 336 biosynthetic clusters in 28 Brevibacillus genomes (Supplementary Dataset 1). Homologous clusters were grouped together using a custom BLASTn-based clustering method, which generated 96 distinct cluster groups. These groups were then mapped onto the Brevibacillus species tree to compare secondary metabolism diversity to species diversity (Fig. 5). In general, biosynthetic clusters distinctly group by phylogeny. Brevibacillus are known producers of several non-ribosomal peptides, and our analysis reveals surprising diversity of non-ribosomal peptide (NRPS) type clusters, most of which are not linked to a known small molecule (Fig. 5a). Each Brevibacillus isolate also has a number of other biosynthetic clusters, including polyketide synthases, terpenes, and siderophores (Fig. 5b). B. brevis VM4 produces gramicidin [49], tyrocidine [50], and edeine [23], and the biosynthetic clusters for these antibiotics are restricted to the B. brevis clade. In Brevibacillus, biosynthetic cluster diversity is therefore a defining feature of species diversity. In contrast, antibiotic resistance is not defined by species diversity and demonstrates that self-produced small molecules do not influence resistance in the Brevibacillus genus.
Fig. 5.
Paenibacillaceae resistomes are not influenced by self-produced secondary metabolites. Paenibacillaceae biosynthetic clusters were annotated using antiSMASH, homologous clusters were grouped together, and the presence or absence of each NRPS-type a and all other cluster types b were mapped onto the family phylogenetic tree, which is presented as a dendrogram of the Brevibacillus phylogeny
The edeine self-resistance mechanism (edeQ) that acetylates edeine is genetically linked within the edeine biosynthetic cluster [23]. edeQ is restricted to strains that have the genetic potential to produce edeine. We further investigated the co-localization of known resistance genes and biosynthetic clusters and uncovered a genetic link between rphC and terpene group 89 (Supplementary Fig. 10). This cluster contains a squalene-hopene synthase, which is a biosynthetic determinant of polycyclic terpenoids such as hopanoids and sporulenes [51]. These compounds are known to play a role in membrane integrity and spore resistance [52–54]. Indeed, several spore germination proteins are immediately adjacent to the squalene-hopene synthase, and rphC, and upstream from rphC is a gene related to known peptidoglycan-glycopolymer ligases [55] (PF03816; LytR-cpsA-psr e-value: 1.7×10−38). RphC is also distantly related to the prodiginine biosynthetic enzymes PigC/RedH (28% identity to PigC), which share a domain architecture with Rphs but catalyze the condensation between a monopyrrole and 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC) by phosphorylating MBC [56, 57]. Therefore, in addition to rifamycin kinase activity, RphC may have a role in secondary metabolite resistance or biosynthesis.
Discussion
The intrinsic resistomes of environmental bacteria are genetically and mechanistically diverse and contain orthologs of resistance genes found in pathogens, indicating that environmental bacteria may be a source of mobile resistance genes in the clinic [2, 5, 6, 9, 13]. In this study, we have expanded the known genetic diversity of antibiotic resistance by eight genes, enabling immediate annotation and detection if they emerge in pathogenic bacteria. In addition, our results point to multi-drug resistant genotypes as being common to members of the Paenibacillaceae family. Further investigations of the intrinsic resistomes of other bacterial families are needed to determine if the Paenibacillaceae family is uniquely enriched for antibiotic resistance genes. Nevertheless, Paenibacillaceae are known to inhabit the human microbiome and other environments where antibiotic use is high [58], and their genomes have GC content similar to some pathogens, which may reduce potential barriers for horizontal transfer because of shared environments and compatible GC content. The honeybee pathogen, Paenibacillus larvae, is known to acquire tetracycline resistance genes on broad-host range plasmids [40]; however, none of the resistance genes identified in this study were associated with known Paenibacillaceae plasmids. Further, Paenibacillus spp. and Brevibacillus spp. are sometimes used as biocontrol agents for promoting plant growth and protection from plant pathogens [58]. Intense antibiotic use in agriculture may select for rapid dissemination of resistance genes from these genera into plant pathogens and other environmental bacteria. Therefore, these organisms should be closely monitored when studying the mobilization of resistance genes in areas where antibiotic use is high.
A significant finding of our study is that resistance phenotype cannot always be inferred from genotype. The resistance phenotype of Paenibacillus sp. LC231 in our previous study mainly correlated with its genotype [9], but this is not true for B. brevis VM4 despite correct predictions of resistance gene function using CARD. We systematically explored the interplay between resistome genotype and phenotype in B. brevis VM4, revealing four cryptic genes (aac(6′)-35, mphJ, rphC, and rphD). These genes confer resistance when expressed in E. coli and are functional in in vitro enzyme assays; however, they are either weakly expressed or are potentially involved in other metabolic processes in B. brevis VM4. These phenomena may impact inference of phenotype from genotype in pathogens [59–61] and is a limitation when studying the resistomes of environmental organisms. The contribution of intrinsic antibiotic resistance genes to the host phenotype is sometimes complex and unclear. For example, in Actinobacteria the expression of Rphs are directly regulated in response to rifampin and involves an upstream palindromic RAE motif [11]; however, in B. brevis VM4 rphC and rphD are constitutively expressed, lack RAE motifs, and appear not to confer a resistant phenotype. RphC is conserved in a hopanoid/sporulene biosynthetic cluster. Therefore, RphC and RphD may not confer rifamycin resistance because of an alternative function in the biosynthesis or resistance of this secondary metabolite. Further elucidation of the biological roles of resistance genes can be accomplished by measuring gene expression, and identifying the presence or absence of regulatory mechanisms, as done in this study. Ultimately, new genetic tools are needed for resistance gene deletion studies in wildtype environmental bacteria.
The impact and risk of environmental antibiotic resistance on medicine remain controversial [3, 59, 62]. The genetic diversity of resistance in pathogens represents only a fraction of the known resistome. This reflects the different ecology of these organisms. In environmental bacteria, exposure to natural antibiotics over millennia has shaped a genetically and mechanistically diverse and rich resistome. In contrast, the resistomes of many pathogens are dominated by resistance genes captured on mobile elements. Gene mobilization from the genomes of environmental bacteria is a primary bottleneck in the movement into the clinic. While previous metagenomic studies have shown that the genetic diversity of resistance genes is directly linked to taxonomic diversity [5], our results demonstrate a broad variation in resistomes between closely related Paenibacillaceae strains. Our previous study uncovered conservation of resistome genotypes in Paenibacillus spp. spatiotemporally separated for over 4 Myr [9]. Taken together, these results suggest that resistomes are reshaped by external factors, likely local chemical ecologies. The apparent movement of antibiotic resistance genes is not unique among non-essential genes; bacterial genomes are usually plastic, and horizontal transfer generates significantly different genetic content between closely related strains [21, 63, 64] (e.g., E. coli [65]). What is unique about resistance genes in a medical context is that they grant immunity to antibiotics, thereby favoring the survival of resistant genotypes. Thus, once associated with mobile elements in a native host, resistance genes rapidly disseminate through bacterial populations with antibiotic selection pressure.
Our results suggest natural movement of resistance genes in Paenibacillaceae spp. over millennia, which does not appear to be driven by obvious mobile elements. However, at least one homolog of all but three of the resistance gene families studied was positioned near genes that could facilitate their movement. If this is also true of other bacterial families, then the transfer of resistance genes from non-pathogens to pathogens may begin with their innocuous co-localization with mobile genetic elements as a result of the natural diversification and rearrangement of bacterial genomes.
Materials and methods
Construction of a Paenibacillaceae family tree
A Paenibacillaceae family tree was generated using 109 well-conserved genes (TIGRFAM “core bacterial protein” GenProp0799). Protein sequences were queried against each Hidden Markov model (HMM) using hmmsearch (HMMER3), and the top sequence from each genome was aligned using hmmalign [66] and concatenated (summarized in Supplementary Dataset 1). Complete and draft genome sequences for all Paenibacillaceae isolates were downloaded from GenBank, and completion was estimated using CheckM [67]. Bacillus cereus ATCC 14579 and Bacillus megaterium NBRC 15308 were used as outgroups. RAxML was used to generate a maximum-likelihood tree using GAMMA model of rate heterogeneity and LG empirical base frequencies (PROTGAMMAAUTO flag) with rapid bootstrap analysis on 100 replicates [68]. All genomes were downloaded from the RefSeq database 19 August, 2016. A comparison of resistome genotypes between habitats was performed by generalizing habitat information found in the “isolation source” qualifier in the respective GenBank files, or in culture collection databases (e.g., ATCC, DSM), if such information was available. The original habitat information was generalized into “not available”, soil, water, endophyte, mammalian-associated, or other. Original and generalized habitats are found in Supplementary Dataset 1.
The rest of the “Materials and methods” section is in the Supplementary information.
Data availability
Nucleotide sequences for Brevibacillus brevis VM4 are deposited into the GenBank WGS database with accession number MXAR00000000. The resistance genes investigated in this study have been deposited into the GenBank Nucleotide database with the accession numbers KY753878 and KY753886. Each resistance determinant sequence was deposited in CARD (https://card.mcmaster.ca/).
Electronic supplementary material
Acknowledgements
We thank Andrew McArthur (McMaster) and members of the CARD team for input into antibiotic resistance sequence analysis and interpretation of phylogenomic correlations. Computer server support provided by the laboratory of Dr. Andrew McArthur and the McMaster Service Lab and Repository (MSLR). We also thank Christine King (McMaster) for genome sequencing, Peter Spanogiannopoulos (UCSF) for early discussion on the rifampin phosphotransferases, and Julie Perry (University of Toronto) for early discussion on gene expression experiments. This research was funded by a Canadian Institutes of Health Research grant (FRN-148463) and by a Canada Research Chair in Antibiotic Biochemistry (to GDW).
Author contributions
ACP, ELW, and GDW designed research. ACP, ELW, KK, and NW performed research. ACP and GDW analyzed data. ACP performed the following experiments; antibiotic susceptibility testing, phylogenetics/phylogenomics, gene expression, growth curves, rifampin degradation studies, and in vitro enzyme assays except for MphJ. ELW performed the following experiments; cloning resistance genes, antibiotic susceptibility testing, and initial studies on gene expression. KK performed HPLC and high resolution mass spectrometry analysis of rifampin degradation. NW assembled the B. brevis VM4 genome and contributed to planning phylogenetics/phylogenomics experiments. ACP and GDW wrote the manuscript with input from all authors.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0017-5) contains supplementary material, which is available to authorized users.
References
- 1.D’costa VM, Mcgrann KM, Hughes DW, Wright GD. Sampling the antibiotic resistome. Science. 2006;311:374–7. doi: 10.1126/science.1120800. [DOI] [PubMed] [Google Scholar]
- 2.Forsberg KJ, Reyes A, Wang B, Selleck EM, Sommer MO, Dantas G. The shared antibiotic resistome of soil bacteria and human pathogens. Science. 2012;337:1107–11. doi: 10.1126/science.1220761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perry JA, Wright GD. The antibiotic resistance “mobilome”: searching for the link between environment and clinic. Front Microbiol. 2013;4:138. doi: 10.3389/fmicb.2013.00138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Salyers AA, Gupta A, Wang Y. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 2004;12:412–6. doi: 10.1016/j.tim.2004.07.004. [DOI] [PubMed] [Google Scholar]
- 5.Forsberg KJ, Patel S, Gibson MK, Lauber CL, Knight R, Fierer N, et al. Bacterial phylogeny structures soil resistomes across habitats. Nature. 2014;509:612–6. doi: 10.1038/nature13377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benveniste R, Davies J. Aminoglycoside antibiotic-inactivating enzymes in actinomycetes similar to those present in clinical isolates of antibiotic-resistant bacteria. Proc Natl Acad Sci USA. 1973;70:2276–80. doi: 10.1073/pnas.70.8.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalan L, Ebert S, Kelly T, Wright GD. Noncanonical vancomycin resistance cluster from Desulfitobacterium hafniense Y51. Antimicrob Agents Chemother. 2009;53:2841–5. doi: 10.1128/AAC.01408-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhullar K, Waglechner N, Pawlowski A, Koteva K, Banks ED, Johnston MD, et al. Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE. 2012;7:e34953. doi: 10.1371/journal.pone.0034953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pawlowski AC, Wang W, Koteva K, Barton HA, Mcarthur AG, Wright GD. A diverse intrinsic antibiotic resistome from a cave bacterium. Nat Commun. 2016;7:13803. doi: 10.1038/ncomms13803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spanogiannopoulos P, Thaker M, Koteva K, Waglechner N, Wright GD. Characterization of a rifampin-inactivating glycosyltransferase from a screen of environmental actinomycetes. Antimicrob Agents Chemother. 2012;56:5061–9. doi: 10.1128/AAC.01166-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Spanogiannopoulos P, Waglechner N, Koteva K, Wright GD. A rifamycin inactivating phosphotransferase family shared by environmental and pathogenic bacteria. Proc Natl Acad Sci USA. 2014;111:7102–7. doi: 10.1073/pnas.1402358111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C, Sui Z, Leclercq SO, Zhang G, Zhao M, Chen W, et al. Functional characterization and phylogenetic analysis of acquired and intrinsic macrolide phosphotransferases in the Bacillus cereus group. Environ Microbiol. 2015;17:1560–73. doi: 10.1111/1462-2920.12578. [DOI] [PubMed] [Google Scholar]
- 13.Davies J, Davies D. Origins and evolution of antibiotic resistance. Microbiol Mol Biol Rev. 2010;74:417–33. doi: 10.1128/MMBR.00016-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright GD. The antibiotic resistome: the nexus of chemical and genetic diversity. Nat Rev Microbiol. 2007;5:175–86. doi: 10.1038/nrmicro1614. [DOI] [PubMed] [Google Scholar]
- 15.Levy SB, Marshall B. Antibacterial resistance worldwide: causes, challenges and responses. Nat Med. 2004;10:S122–9. doi: 10.1038/nm1145. [DOI] [PubMed] [Google Scholar]
- 16.Johnson TA, Stedtfeld RD, Wang Q, Cole JR, Hashsham SA, Looft T, et al. Clusters of antibiotic resistance genes enriched together stay together in swine agriculture. MBio. 2016;7:e02214–15. doi: 10.1128/mBio.02214-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sarmah AK, Meyer MT, Boxall AB. A global perspective on the use, sales, exposure pathways, occurrence, fate and effects of veterinary antibiotics (VAs) in the environment. Chemosphere. 2006;65:725–59. doi: 10.1016/j.chemosphere.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 18.Wang Y, Zhang R, Li J, Wu Z, Yin W, Schwarz S, et al. Comprehensive resistome analysis reveals the prevalence of NDM and MCR-1 in Chinese poultry production. Nat Microbiol. 2017;2:16260. doi: 10.1038/nmicrobiol.2016.260. [DOI] [PubMed] [Google Scholar]
- 19.Witte W. Medical consequences of antibiotic use in agriculture. Science. 1998;279:996–7. doi: 10.1126/science.279.5353.996. [DOI] [PubMed] [Google Scholar]
- 20.Zhang G, Leclercq SO, Tian J, Wang C, Yahara K, Ai G, et al. A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3″)-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 2017;13:e1006602. doi: 10.1371/journal.pgen.1006602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mcinerney JO, Mcnally A, O’connell MJ. Why prokaryotes have pangenomes. Nat Microbiol. 2017;2:17040. doi: 10.1038/nmicrobiol.2017.40. [DOI] [PubMed] [Google Scholar]
- 22.Puigbo P, Lobkovsky AE, Kristensen DM, Wolf YI, Koonin EV. Genomes in turmoil: quantification of genome dynamics in prokaryote supergenomes. BMC Biol. 2014;12:66. doi: 10.1186/s12915-014-0066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Westman EL, Yan M, Waglechner N, Koteva K, Wright GD. Self resistance to the atypical cationic antimicrobial peptide edeine of Brevibacillus brevis Vm4 by the N-acetyltransferase EdeQ. Chem Biol. 2013;20:983–90. doi: 10.1016/j.chembiol.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, Tsang KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 2017;45:D566–d573. doi: 10.1093/nar/gkw1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genee HJ, Bali AP, Petersen SD, Siedler S, Bonde MT, Gronenberg LS, et al. Functional mining of transporters using synthetic selections. Nat Chem Biol. 2016;12:1015–22. doi: 10.1038/nchembio.2189. [DOI] [PubMed] [Google Scholar]
- 26.Hansen LH, Planellas MH, Long KS, Vester B. The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob Agents Chemother. 2012;56:3563–7. doi: 10.1128/AAC.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kitano H. Biological robustness. Nat Rev Genet. 2004;5:826–37. doi: 10.1038/nrg1471. [DOI] [PubMed] [Google Scholar]
- 28.Kudo F, Eguchi T. Biosynthetic genes for aminoglycoside antibiotics. J Antibiot. 2009;62:471–81. doi: 10.1038/ja.2009.76. [DOI] [PubMed] [Google Scholar]
- 29.Bozdogan B, Galopin S, Gerbaud G, Courvalin P, Leclercq R. Chromosomal aadD2 encodes an aminoglycoside nucleotidyltransferase in Bacillus clausii. Antimicrob Agents Chemother. 2003;47:1343–6. doi: 10.1128/AAC.47.4.1343-1346.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mohrle V, Stadler M, Eberz G. Biosensor-guided screening for macrolides. Anal Bioanal Chem. 2007;388:1117–25. doi: 10.1007/s00216-007-1300-5. [DOI] [PubMed] [Google Scholar]
- 31.Vazquez-Laslop N, Klepacki D, Mulhearn DC, Ramu H, Krasnykh O, Franzblau S, et al. Role of antibiotic ligand in nascent peptide-dependent ribosome stalling. Proc Natl Acad Sci USA. 2011;108:10496–501. doi: 10.1073/pnas.1103474108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng Y, Yang S, Jia M, Zhao L, Hou C, You X, et al. Comparative study between macrolide regulatory proteins MphR(A) and MphR(E) in ligand identification and DNA binding based on the rapid in vitro detection system. Anal Bioanal Chem. 2016;408:1623–31. doi: 10.1007/s00216-015-9270-5. [DOI] [PubMed] [Google Scholar]
- 33.Stogios PJ, Cox G, Spanogiannopoulos P, Pillon MC, Waglechner N, Skarina T, et al. Rifampin phosphotransferase is an unusual antibiotic resistance kinase. Nat Commun. 2016;7:11343. doi: 10.1038/ncomms11343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoshino Y, Fujii S, Shinonaga H, Arai K, Saito F, Fukai T, et al. Monooxygenation of rifampicin catalyzed by the rox gene product of Nocardia farcinica: structure elucidation, gene identification and role in drug resistance. J Antibiot. 2010;63:23–8. doi: 10.1038/ja.2009.116. [DOI] [PubMed] [Google Scholar]
- 35.Koonin EV. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 2005;39:309–38. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 36.Hyder SL, Streitfeld MM. Inducible and constitutive resistance to macrolide antibiotics and lincomycin in clinically isolated strains of Streptococcus pyogenes. Antimicrob Agents Chemother. 1973;4:327–31. doi: 10.1128/AAC.4.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philippe H, Douady CJ. Horizontal gene transfer and phylogenetics. Curr Opin Microbiol. 2003;6:498–505. doi: 10.1016/j.mib.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 38.Hu Y, Yang X, Li J, Lv N, Liu F, Wu J, et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Appl Environ Microbiol. 2016;82:6672–81. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li LG, Xia Y, Zhang T. Co-occurrence of antibiotic and metal resistance genes revealed in complete genome collection. ISME J. 2017;11:651–62. doi: 10.1038/ismej.2016.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alippi AM, Leon IE, Lopez AC. Tetracycline-resistance encoding plasmids from Paenibacillus larvae, the causal agent of American foulbrood disease, isolated from commercial honeys. Int Microbiol. 2014;17:49–61. doi: 10.2436/20.1501.01.207. [DOI] [PubMed] [Google Scholar]
- 41.Iiyama K, Mon H, Mori K, Mitsudome T, Lee JM, Kusakabe T, et al. Characterization of KfrA proteins encoded by a plasmid of Paenibacillus popilliae ATCC 14706(T) Meta Gene. 2015;4:29–44. doi: 10.1016/j.mgene.2015.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Courvalin P. Vancomycin resistance in gram-positive cocci. Clin Infect Dis. 2006;42 Suppl 1:S25–34. doi: 10.1086/491711. [DOI] [PubMed] [Google Scholar]
- 43.Guardabassi L, Perichon B, Van Heijenoort J, Blanot D, Courvalin P. Glycopeptide resistance vanA operons in Paenibacillus strains isolated from soil. Antimicrob Agents Chemother. 2005;49:4227–33. doi: 10.1128/AAC.49.10.4227-4233.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guardabassi L, Agerso Y. Genes homologous to glycopeptide resistance vanA are widespread in soil microbial communities. FEMS Microbiol Lett. 2006;259:221–5. doi: 10.1111/j.1574-6968.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- 45.Guardabassi L, Dalsgaard A. Occurrence, structure, and mobility of Tn1546-like elements in environmental isolates of vancomycin-resistant enterococci. Appl Environ Microbiol. 2004;70:984–90. doi: 10.1128/AEM.70.2.984-990.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marshall CG, Broadhead G, Leskiw BK, Wright GD. D-Ala-D-Ala ligases from glycopeptide antibiotic-producing organisms are highly homologous to the enterococcal vancomycin-resistance ligases VanA and VanB. Proc Natl Acad Sci USA. 1997;94:6480–3. doi: 10.1073/pnas.94.12.6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fraimow H, Knob C, Herrero IA, Patel R. Putative VanRS-like two-component regulatory system associated with the inducible glycopeptide resistance cluster of Paenibacillus popilliae. Antimicrob Agents Chemother. 2005;49:2625–33. doi: 10.1128/AAC.49.7.2625-2633.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, et al. antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015;43:W237–43. doi: 10.1093/nar/gkv437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kessler N, Schuhmann H, Morneweg S, Linne U, Marahiel MA. The linear pentadecapeptide gramicidin is assembled by four multimodular nonribosomal peptide synthetases that comprise 16 modules with 56 catalytic domains. J Biol Chem. 2004;279:7413–9. doi: 10.1074/jbc.M309658200. [DOI] [PubMed] [Google Scholar]
- 50.Mootz HD, Marahiel MA. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol. 1997;179:6843–50. doi: 10.1128/jb.179.21.6843-6850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siedenburg G, Jendrossek D. Squalene-hopene cyclases. Appl Environ Microbiol. 2011;77:3905–15. doi: 10.1128/AEM.00300-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ricci JN, Morton R, Kulkarni G, Summers ML, Newman DK. Hopanoids play a role in stress tolerance and nutrient storage in the cyanobacterium Nostoc punctiforme. Geobiology. 2017;15:173–83. doi: 10.1111/gbi.12204. [DOI] [PubMed] [Google Scholar]
- 53.Bosak T, Losick RM, Pearson A. A polycyclic terpenoid that alleviates oxidative stress. Proc Natl Acad Sci USA. 2008;105:6725–9. doi: 10.1073/pnas.0800199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saenz JP, Grosser D, Bradley AS, Lagny TJ, Lavrynenko O, Broda M, et al. Hopanoids as functional analogues of cholesterol in bacterial membranes. Proc Natl Acad Sci USA. 2015;112:11971–6. doi: 10.1073/pnas.1515607112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer K, Matano LM, Qiao Y, Kahne D, Walker S. In vitro reconstitution demonstrates the cell wall ligase activity of LCP proteins. Nat Chem Biol. 2017;13:396–401. doi: 10.1038/nchembio.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chawrai SR, Williamson NR, Mahendiran T, Salmond GPC, Leeper FJ. Characterisation of PigC and HapC, the prodigiosin synthetases from Serratia sp. and Hahella chejuensis with potential for biocatalytic production of anticancer agents. Chem Sci. 2012;3:447–54. doi: 10.1039/C1SC00588J. [DOI] [Google Scholar]
- 57.Chawrai SR, Williamson NR, Salmond GP, Leeper FJ (2008). Chemoenzymatic synthesis of prodigiosin analogues—exploring the substrate specificity of PigC. Chem Commun. 2008;16:1862–4. [DOI] [PubMed]
- 58.Grady EN, Macdonald J, Liu L, Richman A, Yuan ZC. Current knowledge and perspectives of Paenibacillus: a review. Microb Cell Fact. 2016;15:203. doi: 10.1186/s12934-016-0603-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Berendonk TU, Manaia CM, Merlin C, Fatta-Kassinos D, Cytryn E, Walsh F, et al. Tackling antibiotic resistance: the environmental framework. Nat Rev Microbiol. 2015;13:310–7. doi: 10.1038/nrmicro3439. [DOI] [PubMed] [Google Scholar]
- 60.Piddock LJ. Assess drug-resistance phenotypes, not just genotypes. Nat Microbiol. 2016;1:16120. doi: 10.1038/nmicrobiol.2016.120. [DOI] [PubMed] [Google Scholar]
- 61.Thaker MN, Kalan L, Waglechner N, Eshaghi A, Patel SN, Poutanen S, et al. Vancomycin-variable enterococci can give rise to constitutive resistance during antibiotic therapy. Antimicrob Agents Chemother. 2015;59:1405–10. doi: 10.1128/AAC.04490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Martinez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nat Rev Microbiol. 2015;13:116–23. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 63.Oren Y, Smith MB, Johns NI, Kaplan Zeevi M, Biran D, Ron EZ, et al. Transfer of noncoding DNA drives regulatory rewiring in bacteria. Proc Natl Acad Sci USA. 2014;111:16112–7. doi: 10.1073/pnas.1413272111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sela I, Wolf YI, Koonin EV. Theory of prokaryotic genome evolution. Proc Natl Acad Sci USA. 2016;113:11399–407. doi: 10.1073/pnas.1614083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perna NT, Plunkett G, 3rd, Burland V, Mau B, Glasner JD, Rose DJ, et al. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature. 2001;409:529–33. doi: 10.1038/35054089. [DOI] [PubMed] [Google Scholar]
- 66.Eddy SR. Accelerated profile HMM searches. PLoS Comput Biol. 2011;7:e1002195 [DOI] [PMC free article] [PubMed]
- 67.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–55. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Nucleotide sequences for Brevibacillus brevis VM4 are deposited into the GenBank WGS database with accession number MXAR00000000. The resistance genes investigated in this study have been deposited into the GenBank Nucleotide database with the accession numbers KY753878 and KY753886. Each resistance determinant sequence was deposited in CARD (https://card.mcmaster.ca/).