Abstract
Phytoremediation using willows is thought to be a sustainable alternative to traditional remediation techniques involving excavation, transport, and landfilling. However, the complexity of the interaction between the willow and its associated highly diverse microbial communities makes the optimization of phytoremediation very difficult. Here, we have sequenced the rhizosphere metatranscriptome of four willow species and the plant root metatranscriptome for two willow species growing in petroleum hydrocarbon-contaminated and non-contaminated soils on a former petroleum refinery site. Significant differences in the abundance of transcripts related to different bacterial and fungal taxa were observed between willow species, mostly in contaminated soils. When comparing transcript abundance in contaminated vs. non-contaminated soil for each willow species individually, transcripts for many microbial taxa and functions were significantly more abundant in contaminated rhizosphere soil for Salix eriocephala, S. miyabeana and S. purpurea, in contrast to what was observed in the rhizosphere of S. caprea. This agrees with the previously reported sensitivity of S. caprea to contamination, and the superior tolerance of S. miyabeana and S. purpurea to soil contamination at that site. The root metatranscriptomes of two species were compared and revealed that plants transcripts are mainly influenced by willow species, while microbial transcripts mainly responded to contamination. A comparison of the rhizosphere and root metatranscriptomes in the S. purpurea species revealed a complete reorganization of the linkages between root and rhizosphere pathways when comparing willows growing in contaminated and non-contaminated soils, mainly because of large shifts in the rhizosphere metatranscriptome.
Introduction
Willows (Salix spp.) are particularly interesting for phytoremediation as they grow fast, tolerate high levels of water in soil [1], are extremely diversified with more than 400 species, 200 hybrids [2] and thousands of cultivars, and were shown to harbor significantly more contaminant-degrading bacteria in their rhizosphere as compared to other plant species [3]. However, a better understanding of the response of different willow species and cultivars to contamination stress is necessary to better predict the duration of phytoremediation and refine the management strategies, a sine qua non condition for a wider adoption of this green technology [4, 5]. With this goal in mind, recent studies have started to look at the metatranscriptomic response of willows and their associated microorganisms to contaminant stress. The metatranscriptome of the leaves of willow growing in pots showed that the response to contaminant stress could lead to a cross-tolerance to biotic stresses [6], whereas the root metatranscriptome showed that root-associated fungal endophytes had a greater response to contaminant stress than the willow itself [7]. In addition, the expression of hydrocarbon degradation genes in the root environment was almost exclusively due to bacteria [7]. Similarly, willows growing in pots in contaminated soil induced a large increase in the expression of microbial hydrocarbon degradation genes in their rhizosphere [8], which was subsequently shown to be driven by specific taxonomic groups [9]. Since a large part of the response to contaminant stress and the actual degradation of organic pollutants appears to be due to microorganisms, studying how the microorganisms associated with the roots and the rhizosphere of different cultivars of willows could better inform the choice of an optimal cultivar among the thousands available.
In controlled experiments, plant-associated microbial communities vary with the genotype of their host, even at the subspecies level [10–12]. For willows growing in the field, the effect of the environment (contamination) is often stronger than the effect of cultivar on the microbial communities. For instance, when comparing two willow cultivars, the differences in the fungal and bacterial communities between cultivars were relatively minor compared to the large differences between plant compartments (roots, stems, rhizosphere, and bulk soil) and between differentially contaminated soils [13]. Similarly, contaminant concentration was shown to have an overriding effect on the arbuscular mycorrhizal fungi (AMF) community composition associated with willows, with little effect of cultivar [14]. However, when carefully controlling for environmental variation, it has been shown that North American willows associate more strongly with ectomycorrhizal fungi than Asian or European willows when growing in soil with high levels of hydrocarbon contaminants nearby Montreal, in Canada [15]. At a metal-contaminated site near Quebec city, Canada, the early association of North American willow cultivars with this particular type of fungus resulted in the uptake of larger amounts of zinc [16]. Within the general common responses to contamination, different willow cultivars showed different expression patterns in their leaves, including transcripts annotated as fungal or bacterial [17].
As different willow cultivars show dramatically different capacities to grow under contaminated conditions [18], and that this capacity was shown in a rhizosphere transplantation experiment to be related to the microbial communities [19], it could well be that the association of willow cultivars with different microbial communities results in contrasting phytoremediation ability. However, despite the recent advances detailed above, it is still not known how different willow cultivars and their associated root and soil microbial communities react to contamination stress. We hypothesized that the rhizosphere and root microbial communities of different willow cultivars would show contrasting transcriptomic responses to contaminant presence and that this would be consistent with the phytoremediation potential of the cultivars. Our objective was to analyze the metatranscriptome of the rhizosphere and roots of different willow cultivars growing in the field in contaminated and non-contaminated soils.
Materials and methods
Experimental design
The experimental design has been detailed elsewhere [15, 18]. Briefly, the field experiment was set up on the site of a former petrochemical refinery and consisted of eight non-contaminated blocks and five contaminated blocks. Each block contained 12 plots, 11 of which were planted with 150 cuttings from each of 11 different willow cultivars in June 2011, and one was left unplanted. All willows were coppiced at the end of the first growing season. For the present study, three blocks (C1, C2, and C3) were selected among the contaminated blocks (pH in water: 6.59–7.12, average 6.83) and three blocks (N1, N3, and N7) among the non-contaminated blocks (pH in water: 6.70–7.54, average 7.12). In the contaminated plots, in August 2011, the average polycyclic aromatic hydrocarbon concentrations were 1.0, 0.9, and 14.6 mg kg− 1 of soil for blocks C1, C2, and C3, respectively, whereas the average total hydrocarbon (C10–C50) concentrations were 195.1, 182.3, and 709.2 mg kg− 1 of soil for blocks C1, C2, and C3, respectively. PAHs and hydrocarbon concentrations were below detection limits for the non-contaminated plots. From the 11 cultivars, five willow species were selected for this experiment: Salix caprea hybrid “S365”, S. eriocephala “S25”, S. miyabeana “SX67”, S. nigra “S05”, and S. purpurea “Fish Creek”.
Sampling
Sampling was carried out on 16 August 2012, after two growing seasons. From each plot, three trees were uprooted and the roots were separated from the stem. The roots were shaken to remove loosely adhering soil. Fine roots were then selected and the rhizosphere soil was carefully detached, whereas the roots were rapidly washed in tap water. One root + rhizosphere sample was taken per tree and kept separated for all analyses. Samples were then flash frozen in liquid nitrogen and kept in dry ice for transport to the laboratory, where they were stored at −80 °C until RNA extraction. The whole sampling procedure took a few minutes.
RNA extraction, library preparation, and sequencing
The roots samples of S. purpurea and S. nigra were separately ground in liquid nitrogen with a mortar and pestle and the total RNA was extracted using a CTAB protocol [20] with RNA quantity and quality assessed with a BioAnalyser (Agilent). PolyA containing messenger RNA libraries were constructed using Illumina TruSeq kits with different indexes for each sample. Samples were sequenced on one lane on an Illumina HiSeq 2000 (100 bp paired-end reads) at the McGill University and Genome Quebec Innovation Center.
For each sample from the rhizosphere of S. purpurea, S. eriocephala, S. caprea, and S. miyabeana, 500 mg of frozen soil was separately weighed and extracted using a homemade bead-beating protocol with subsequent phenol–chloroform purification, as previously described [21, 22]. At the phenol addition step, 10 µl of a 307 bp control RNA transcribed from the pSPT18 vector (positions 2867–3104 and 1–70) at a concentration of 0.11 ng µl−1 was added to the frozen soil. RNA extracts were treated with Ambion TURBO DNAse (Life Technologies, Burlington, ON, Canada) and the absence of DNA was confirmed by 16S ribosomal RNA (rRNA) gene universal PCR using primers F1 (positions 11–29) and R2 (positions 519–536) [23]. Libraries were prepared using the Illumina ScriptSeq complete with Ribo-Zero rRNA removal (bacteria) kit following the manufacturer’s instructions. Libraries were multiplexed and sequenced on eight lanes on an Illumina Hiseq 2000 at the McGill University and Genome Quebec Innovation Center.
Bioinformatics analyses
Sequence data were analyzed using an in-house pipeline as previously described in Tremblay et al. [24] and in the Supplementary Methods. Briefly, reads were quality controlled and assembled after which genes were called from the contigs and annotated following the JGI’s guidelines [25]. Quality filtered reads were mapped to assembled contigs to obtain abundance profiles, which were normalized to counts per million (CPM) using EdgeR v3.10.2 [26]. Metatranscriptome read counts and assembly statistics are available in Supplementary Tables S1 and S2, respectively. For the roots, because the samples were not spiked with an internal control during the RNA extraction procedure, the CPM-normalized data sets were used for downstream analyses. For the rhizosphere data sets, the number of sequences related to the spiked-in pSPT18 vector in the filtered data sets was obtained by blasting the sequence of the vector against all the transcripts, which led to the identification of a contig containing the vector, and the number of reads mapping to this contig was then used to normalize the number of transcripts using the method of Moran et al. [27]. One sample (S. eriocephala, non-contaminated, sample GRM_61) for which we did not retrieve the pSPT18 standard was removed from the data set. This pSPT18 normalized data set was used for most downstream analyses of the rhizosphere. The exception was for the root–rhizosphere co-variation analyses where both data sets were normalized to CPM, to maximize comparability.
Statistical analyses
All statistical analyses were carried out in R (v 3.2.3, The R Foundation for Statistical Computing). The effects of contamination and willow species on the general patterns in transcript abundance were visualized using principal coordinate analyses (PCoA) using the “cmdscale” function based on Bray–Curtis dissimilarities obtained from the “vegdist” function of the vegan package (v 2.4–3). The significance of these patterns was tested using multivariate tests of hypothesis based on the Permanova procedure of the “adonis” function of the “vegan” package based on Bray–Curtis dissimilarities. The effect of willow species on the abundance of transcripts related to bacterial and fungal phyla and classes and to KEGG pathways (“ko”) and gene categories involved in various functions was tested using analysis of variance (ANOVA) (“aov” function, for the rhizosphere) or t-tests (functions “t.test”, for roots). These tests were carried out separately for contaminated and non-contaminated soils. The effect of contamination on the abundance of transcripts related to bacterial and fungal phyla and classes and to KEGG pathways (“ko”) and gene categories involved in various functions was tested for each individual willow species using t-tests (function “t.test”). Before running the ANOVAs and t-tests, normal distribution and variance homogeneity of the data were tested using the “shapiro.test” and “bartlett.test” functions, respectively. If the data were not normally distributed or did not show homogeneous variance, they were log transformed before ANOVA and t-tests. If transformation failed, non-parametric Kruskall–Wallis tests (function “kruskall.test”) were used in lieu of ANOVAs and non-parametric Mann–Whitney U-tests (function “wilcox.test”) were used in lieu of t-tests. The function “EBTest” from the “EBSeq” package was used to test all individual transcripts with a target FDR of 0.05 for differences between willow species growing in the same soil or within a single willow species growing in contaminated vs. non-contaminated soil. Co-variation between KEGG pathways from the rhizosphere and willow root metatranscriptomic data sets was calculated based on Spearman r (“cor” function), and the correlation with an absolute r value above 0.8 was kept and imported in Cytoscape (v. 3.4.0) for correlation network visualization.
Data deposition
Data sets are available through NCBI BioProject accession PRJNA390655.
Results
Transcript abundance in the rhizosphere of different willow species
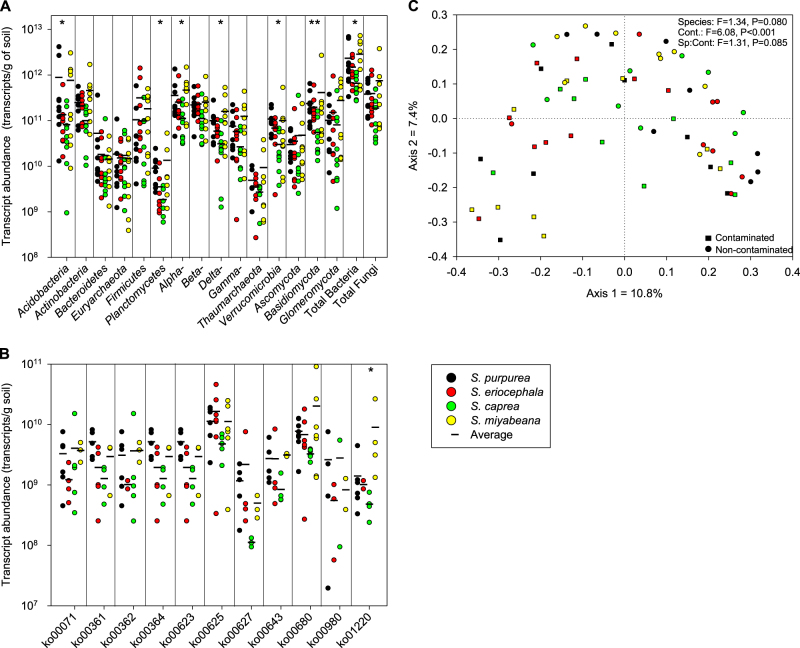
Permanova tests revealed a highly significant effect of soil contamination on the abundance patterns of all transcripts retrieved (F = 6.08, P < 0.001) (Fig. 1c). The effects of species and of the interaction term were nearly significant (F = 1.34, P = 0.080, and F = 1.31, P = 0.085, respectively). When looking separately at samples from contaminated and non-contaminated rhizosphere soils, Permanova tests revealed no significant effects of species on the transcript abundance patterns in contaminated rhizosphere (F = 1.10, P = 0.180). In contrast, the transcript abundance patterns varied significantly per species in the non-contaminated rhizosphere soil (F = 1.30, P = 0.0290).
Fig. 1.
General transcript abundance patterns in the rhizosphere. Normalized transcript abundance summed for a bacterial and fungal phyla and classes and b KEGG pathways (ko’s) involved in hydrocarbon degradation in the rhizosphere of the willow species S. purpurea, S. eriocephala, S. caprea, and S. miyabeana growing in contaminated soils and c principal coordinate analysis of Bray–Curtis dissimilarity calculated from the normalized transcript abundance matrix for the rhizosphere of the willow species S. purpurea, S. eriocephala, S. caprea, and S. miyabeana growing in contaminated and non-contaminated soils. For a and b, dots are values for individual samples and the black bar represents the average. *P < 0.05, **P < 0.01 in ANOVA tests comparing the summed transcript abundance between different species. ko0071: fatty acid degradation; ko00361: chlorocyclohexane and chlorobenzene degradation; ko00362: benzoate degradation; ko00364: fluorobenzoate degradation; ko00623: toluene degradation; ko00625: chloroalkane and chloroalkene degradation; ko00627: aminobenzoate degradation; ko00643: styrene degradation; ko00680: methane metabolism; ko00980: metabolism of xenobiotics by cytochrome P450; ko01220: degradation of aromatic compounds
The transcript abundance of various bacterial phyla/classes (sum of the abundances of all transcripts assigned to these taxa) was significantly different for the different willow species growing in contaminated soil (Fig. 1a). For Acidobacteria, Planctomycetes, Alphaproteobacteria, and total bacteria, transcripts were significantly more abundant in the rhizosphere of S. miyabeana as compared to the rhizosphere of S. caprea, with intermediate values for S. purpurea and S. eriocephala (Fig. 1a). Transcripts affiliated to Deltaproteobacteria were significantly more abundant in the rhizosphere of both S. miyabeana and S. purpurea as compared to S. caprea, with intermediate abundance levels for S. eriocephala (Fig. 1a). The transcripts affiliated to Verrucomicrobia were significantly more abundant in the rhizosphere of S. purpurea as compared to S. caprea, whereas the transcripts linked to Basidiomycota were significantly more abundant in the rhizosphere of S. purpurea, S. eriocephala, and S. miyabeana as compared to S. caprea (Fig. 1a). No significant differences in the summed abundance of transcripts for phyla/classes were observed between the willow species growing in non-contaminated soil (not shown).
We also compared the abundance of all transcripts for each pair of willow genotypes growing in non-contaminated (Supplementary Fig. S1) and contaminated soil (Supplementary Fig. S2) and looked up the assigned taxonomy for the transcripts that showed significant differences between willow species. For the non-contaminated soils, most transcripts that showed significant differences were more abundant in the rhizosphere of S. caprea when compared to S. purpurea (254 vs. 99), S. eriocephala (251 vs. 158), and S. miyabeana (244 vs. 96) (Supplementary Fig. S1). In contrast, most transcripts that showed significant differences were less abundant in the rhizosphere of S. eriocephala as compared to S. purpurea (118 vs. 242) and S. miyabeana (133 vs. 292) (Supplementary Fig. S1). When comparing S. purpurea with S. miyabeana, transcripts that showed significant differences were roughly equally distributed between the two species (171 vs. 165) (Supplementary Fig. S1). Most of the transcripts that showed significant differences in their abundance between willow species could not be assigned taxonomically (1348/2223, 60.6%). The transcripts that showed significant difference in their abundance between willow species and that could be assigned at the domain level were linked to bacteria (589/875, 67.3%), fungi (124/875, 14.2%), and other eukaryotes (135/875, 15.4%). For the contaminated soils, the general trend was that the transcripts showing significant differences were mostly less abundant in S. caprea when compared to S. purpurea (136 vs. 235), S. miyabeana (124 vs. 273), and S. eriocephala (74 vs. 224) (Supplementary Fig. S2). S. miyabeana also showed lower numbers of significant transcripts when compared to S. purpurea (108 vs. 157) and S. eriocephala (102 vs. 263) (Supplementary Fig. S2). When comparing S. purpurea with S. eriocephala, transcripts that showed significant differences were roughly equally distributed between the two species (106 vs. 122) (Supplementary Fig. S2). Here again, most of the transcripts significantly affected by willow species could not be assigned taxonomically (1208/1924, 62.8%). The transcripts that could be assigned at the domain level were linked to bacteria (489/716, 68.3%), fungi (90/716, 12.6%), and other eukaryotes (119/716, 16.6%).
The abundance of transcripts related to KEGG pathways involved in hydrocarbon degradation varied considerably among willow species when growing in contaminated soil, with S. caprea generally having lower abundance (Fig. 1b). However, the only significant difference was KEGG pathway ko01220 “Degradation of aromatic compounds”, where transcripts were significantly more abundant in the rhizosphere of S. miyabeana as compared to S. caprea (Fig. 1b). No significant differences in the abundance of transcripts related to hydrocarbon degradation pathways were observed among species when grown in non-contaminated soils (not shown).
We also looked at the abundance of transcripts related to nutrient uptake, nutrient cycling, and plant–microbe interactions in the rhizosphere, including transcripts annotated as “phosphate regulon transcriptional regulatory protein (phoB)”, “glutamine synthetase (glnA)” or involved in “bacterial chemotaxis”, “sugar (and others) transporters”, “amino acids transport and metabolism”, “nitrogen metabolism” (KEGG pathway ko00910), and “plant-pathogen interactions” (ko04626). For all these transcripts, no significant differences in abundance were observed between willow species both for willows growing in contaminated and non-contaminated soils (not shown). However, there was a nearly significant effect (P < 0.10) of willow species on the abundance of transcripts related to “amino acids transport and metabolism” in contaminated rhizosphere soils (F = 2.47, P = 0.080), with average values of 8.85 × 1010, 7.55 × 1010, 3.38 × 1010, and 1.86 × 1011 transcripts per g of soil for S. purpurea, S. eriocephala, S. caprea, and S. miyabeana, respectively.
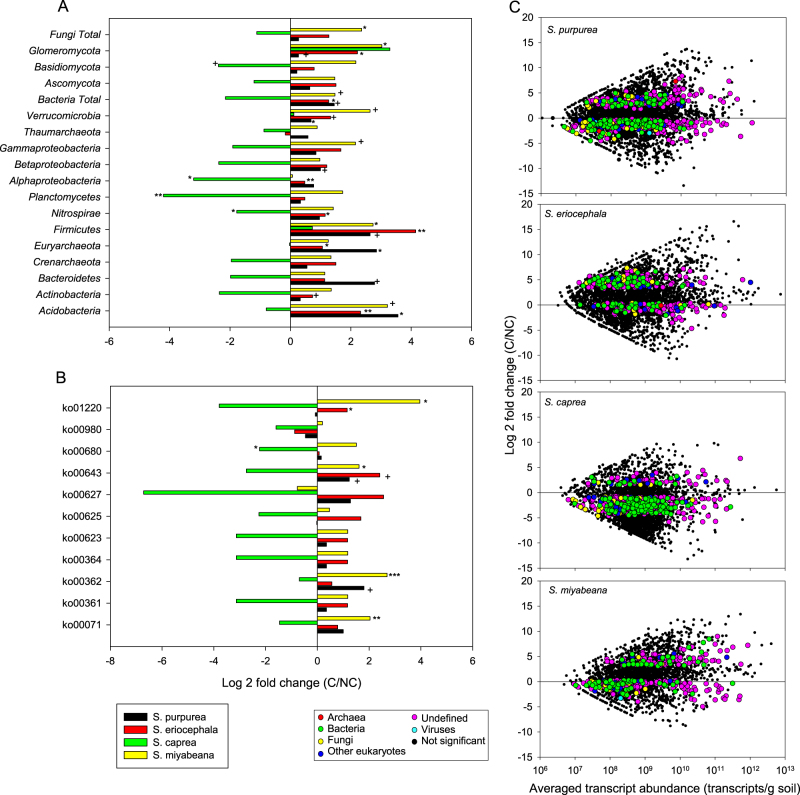
Differences in transcript abundance between contaminated and non-contaminated rhizosphere soil
In addition to looking for differences between willow species, we were also interested in comparing the differences in transcript abundance in the rhizosphere of each willow species when growing in contaminated vs. non-contaminated soil. There were significantly more transcripts (P < 0.05) related to the Acidobacteria, Euryarchaeota, and Verrucomicrobia and nearly significantly more transcripts (P < 0.10) related to the Bacteroidetes, Firmicutes, Betaproteobacteria, total bacteria, and Glomeromycota in the rhizosphere of S. purpurea growing in contaminated soil (Fig. 2a). For S. eriocephala growing in contaminated soil, significantly more transcripts related to the Acidobacteria, Euryarchaeota, Firmicutes, Nitrospirae, Alphaproteobacteria, total bacteria, and Glomeromycota were detected as compared to non-contaminated soil, whereas nearly significantly (P < 0.10) more transcripts related to the Actinobacteria were detected (Fig. 2a). For S. caprea, the trend was very different, with significantly less transcripts related to the Nitrospirae, Planctomycetes, and Alphaproteobacteria and nearly significantly (P < 0.10) less transcripts related to the Basidiomycota when growing in contaminated soil (Fig. 2a). Contamination in the rhizosphere of S. miyabeana resulted in a significant increase in the number of transcripts related to the Firmicutes, Glomeromycota, and total fungi and a nearly significant (P < 0.10) increase in transcripts related to the Acidobacteria, Gammaproteobacteria, Verrucomicrobia, and total bacteria (Fig. 2a).
Fig. 2.
Differences in rhizosphere transcript abundance between contaminated and non-contaminated soils. Fold difference in the normalized transcript abundance summed for a bacterial and fungal phyla and classes, and b KEGG pathways (ko’s) involved in hydrocarbon degradation for the rhizosphere of each individual willow species (S. purpurea, S. eriocephala, S. caprea, and S. miyabeana) growing in contaminated vs. non-contaminated soils. c Fold difference in the normalized transcript abundance for willow species S. purpurea, S. eriocephala, S. caprea, and S. miyabeana growing in contaminated vs. non-contaminated soils. Transcripts that showed significant differences in t-tests are highlighted in color according to their taxonomic affiliation. ko0071: fatty acid degradation; ko00361: chlorocyclohexane and chlorobenzene degradation; ko00362: benzoate degradation; ko00364: fluorobenzoate degradation; ko00623: toluene degradation; ko00625: chloroalkane and chloroalkene degradation; ko00627: aminobenzoate degradation; ko00643: styrene degradation; ko00680: methane metabolism; ko00980: metabolism of xenobiotics by cytochrome P450; ko01220: degradation of aromatic compounds. +P < 0.10, *P < 0.05, **P < 0.01, ***P < 0.001 in t-tests comparing the summed transcript abundance between contaminated and non-contaminated soils
When looking at individual transcripts that showed significant differences in their abundance between contaminated and non-contaminated soils for individual willow species, most of the significant transcripts were more abundant in contaminated soil for S. eriocephala (218/319, 66.3%) and less abundant in contaminated soils for S. purpurea (323/514, 62.8%) and S. caprea (520/594, 87.5%) (Fig. 2c). For S. miyabeana, the number of transcripts significantly affected by contamination was more balanced, with roughly half the transcripts being significantly more abundant and the other half less abundant in contaminated soil (151 vs. 203) (Fig. 2c). Several of the transcripts that showed significant trends could not be classified to a domain (1079/1791, 60.2%), but for the ones that could be classified, bacteria was the most common domain (459/712, 64.5%), followed by fungi (138/712, 19.4%) and other eukaryotes (106/712, 14.8%), whereas Proteobacteria was the most common phylum (284/712, 39.9%) (Fig. 2c).
Transcripts related to KEGG pathways (ko) involved in hydrocarbon degradation were generally more abundant in the rhizosphere of S. purpurea, S. eriocephala, and S. miyabeana growing in contaminated soil and less abundant in the rhizosphere of S. caprea growing in contaminated soil (Fig. 2b). The abundance of transcripts related to KEGG pathway ko01220 “Degradation of aromatic compounds” was significantly higher in the rhizosphere of S. miyabeana and S. eriocephala growing in contaminated soils as compared to the same species growing in non-contaminated soil (Fig. 2b). Similarly, the transcripts related to KEGG pathway ko00643 “Styrene degradation” were significantly more abundant in the rhizosphere of S. miyabeana when growing in contaminated soil, whereas these transcripts were nearly significantly (P < 0.10) more abundant for S. eriocephala and S. purpurea growing in contaminated soil (Fig. 2b). The transcripts of two other pathways were also significantly more abundant in the rhizosphere of S. miyabeana growing in contaminated soils: ko00362 “Benzoate degradation” and ko00071 “Fatty acid degradation” (Fig. 2b). The transcripts associated with the former pathway were also nearly significantly (P < 0.10) more abundant in the rhizosphere of S. purpurea growing in contaminated soil (Fig. 2b). For S. caprea, the abundance of transcripts involved in KEGG pathway ko00680 “Methane metabolism” was significantly lower in contaminated soil (Fig. 2b).
For transcripts related to nutrient uptake, nutrient cycling, and plant–microbe interactions in the rhizosphere, the only significant difference was for the transcripts related to amino-acid transport and metabolism, which were significantly more abundant in the rhizosphere of S. eriocephala growing in contaminated soil as compared to non-contaminated soil. There was also nearly significant (P < 0.10) differences between the abundance of transcripts related to sugar (and other) transporters for S. eriocephala and for transcripts related to amino-acid transport and metabolism for S. miyabeana, with higher abundance in contaminated rhizosphere soil. The general trend observed for the other transcripts in these categories was the same as the ones reported above, with a decreased transcript abundance in contaminated soils for S. caprea and increased transcript abundances in contaminated soils for S. eriocephala, S. purpurea, and S. miyabeana (not shown).
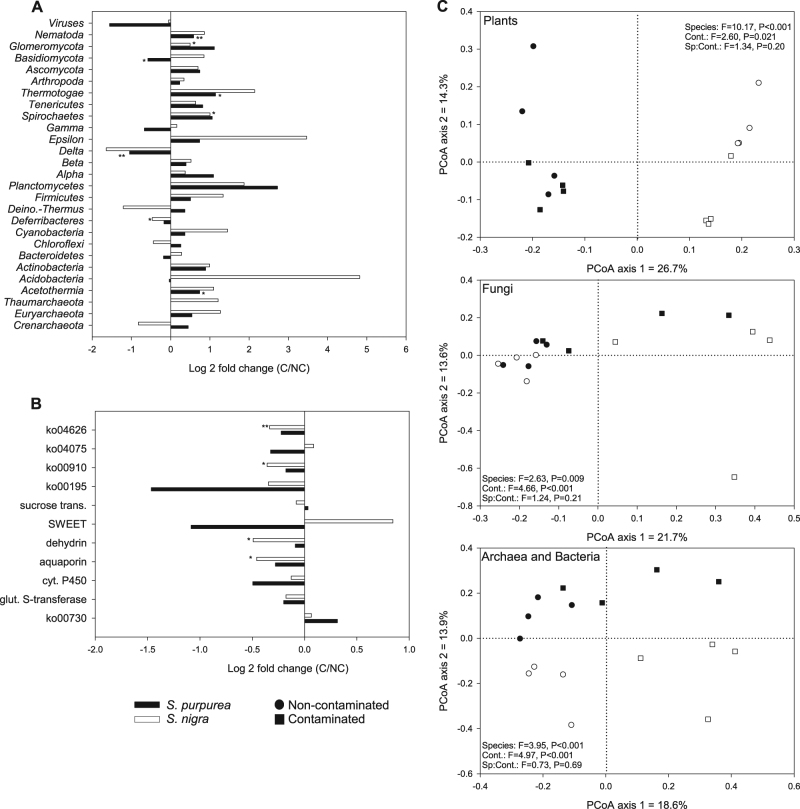
Transcript abundance in the roots of different willow species
The transcript relative abundance patterns in the roots varied according to their taxonomical affiliation (Fig. 3c). For instance, when looking at transcripts annotated as plants (“Streptophyta”), willow species had a much stronger effect on the transcript relative abundance patterns (Permanova F-ratio of 10.17) than soil contamination levels (Permanova F-ratio of 2.60), and the two willow species were clearly separated along the first axis of the PCoA ordination (Fig. 3c). In contrast, for transcripts annotated as fungi and the ones annotated as bacteria and archaea, soil contamination had the strongest effect on the transcript relative abundance patterns (Permanova F-ratios of 4.66 and 4.97, respectively) as compared to the effect of willow species (Permanova F-ratios of 2.63 and 3.95, respectively). Accordingly, the first axes of the PCoA ordinations mostly separated the samples by contamination levels (Fig. 3c), and, for bacteria and archaea, the second axis separated the samples by willow species (Fig. 3c).
Fig. 3.
General transcript relative abundance patterns in the roots and differences in root transcripts relative abundance between contaminated and non-contaminated soils. Fold difference in the normalized transcript relative abundance summed for a bacterial, fungal, and other eukaryotes phyla and classes and b genes, genes categories, and KEGG pathways (ko’s) related to stress response, detoxification, plant hormones, nutrient transport, and communication with microbes for the roots of each individual willow species (S. purpurea and S. nigra) growing in contaminated vs. non-contaminated soils. c Principal coordinate analysis of Bray–Curtis dissimilarity calculated from the normalized transcript relative abundance matrix for the roots of the willow species S. purpurea and S. nigra growing in contaminated and non-contaminated soils. ko04626: plant–pathogen interaction; ko04075: plant hormone signal transduction; ko00910: nitrogen metabolism; ko00195: photosynthesis; ko00730: thiamine metabolism; glut.: glutathione; cyt.: cytochrome; trans.: transporter. *P < 0.05, **P < 0.01, in t-tests comparing the summed transcript relative abundance between contaminated and non-contaminated soils
The summed relative abundance of transcripts for various phyla and classes showed significant differences between the roots of S. purpurea and S. nigra. In roots growing in non-contaminated soil, Thaumarchaeota (t = 5.19, P = 0.014), Chloroflexi (t = 5.28, P = 0.012), Deferribacteres (t = 19.83, P = 0.00028), and Thermotogae (t = 5.23, P = 0.0020) showed significantly higher relative abundance in the roots of S. nigra, whereas transcripts linked to the Epsilonproteobacteria (t = 3.65, P = 0.028), Spirochaetes (t = 3.37, P = 0.018), and Glomeromycota (t = 2.64, P = 0.049) were significantly more abundant in the roots of S. purpurea. For roots growing in contaminated soil, Thaumarchaeota (t = 3.73, P = 0.033), Chloroflexi (t = 10.08, P = 0.00022), and Deferribacteres (t = 8.05, P = 0.0040) showed significantly higher relative abundance in the roots of S. nigra.
The taxonomical affiliation of the individual transcripts that showed significant differences in their relative abundance between the two willow species was determined. In contaminated soil, 7322 transcripts were found to be differentially abundant, of which 1745 (23.8%) were assigned to plants (“Streptophyta”), 43 (0.6%) to bacteria, 1705 (23.3%) to fungi, and 3719 (50.8%) could not be assigned taxonomically. In non-contaminated soil, 7092 transcripts were found to be differentially abundant between the two willow species, of which 2908 (41.0%) were assigned to plants (“Streptophyta”), 12 (0.2%) to bacteria, 380 (5.4%) to fungi, and 3736 (52.7%) could not be assigned taxonomically.
Significant differences in the relative abundance of plant transcript involved in pathways related to stress response, detoxification, plant hormones, nutrient transport, and communication with microbes were observed between willow species when growing both in contaminated and non-contaminated soils. For instance, in non-contaminated soils, the roots of S. nigra showed significantly higher relative abundance transcripts annotated as sucrose transporters (average CPM of 179.9 vs. 79.1, t = 2.90, P = 0.043) and dehydrin (average CPM of 1658.4 vs. 1082.9, t = 3.61, P = 0.023) as compared to the roots of S. purpurea. In contaminated soils, the roots of S. nigra showed significantly higher relative abundance of transcripts annotated as sucrose transporters (average CPM of 170.1 vs. 80.9, t = 3.02, P = 0.024) and significantly lower relative abundance of transcripts related to nitrogen metabolism (KEGG pathway ko00910) and plant–pathogen interactions (KEGG pathway ko04626) (average CPM of 1100.3 vs. 1460.2, t = 5.07, P = 0.0023 and average CPM of 4355.2 vs. 6049.5, t = 3.92, P = 0.011, respectively).
Differences in transcript relative abundance between roots growing in contaminated and non-contaminated soil
Significant differences in the transcript relative abundance of various taxonomic groups were visible when comparing the roots of willows growing in contaminated vs. non-contaminated soils (Fig. 3a). For S. purpurea, roots growing in contaminated soil harbored significantly more Acetothermia, Themotogae, and Nematoda and significantly less Basidiomycota and Deltaproteobacteria as compared to roots growing in non-contaminated soils (Fig. 3a). For S. nigra, roots growing in contaminated soils were significantly enriched in Glomeromycota and Spirochaetes and significantly depleted in Deferribacteres (Fig. 3a).
Individual transcripts that showed significant differences in their relative abundance between the roots of willow growing in contaminated vs. non-contaminated soils were mainly affiliated to fungi. For instance, in the roots of S. nigra, 7725 transcripts showed differential abundance, of which 918 (11.9%) were affiliated to plants (“Streptophyta”), 60 (0.8%) to bacteria, 2816 (36.5%) to fungi, and 3823 (49.5%) could not be classified taxonomically. For S. purpurea, 1249 transcripts showed significant differential patterns in their relative abundance, of which 309 (24.7%) were affiliated to plants (“Streptophyta”), 9 (0.7%) to bacteria, 505 (40.4%) to fungi, and 419 (33.4%) could not be affiliated taxonomically.
For functional gene categories or pathways related to stress response, detoxification, plant hormones, nutrient transport, and communication with microbes, the relative abundance of transcripts in the roots of S. purpurea did not show any significant differences between contaminated and non-contaminated soils (Fig. 3b). However, for S. nigra, transcripts related to aquaporins, dehydrins, nitrogen metabolism (ko00910), and plant–pathogen interactions (ko04626) were significantly less abundant in roots growing in contaminated soils (Fig. 3b).
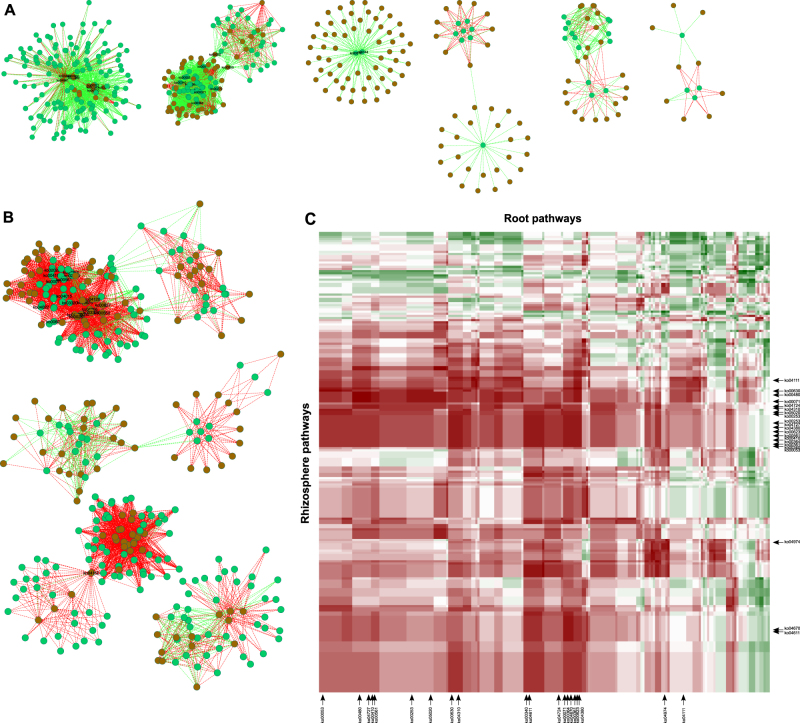
Co-variation of willow root and rhizosphere metatranscriptomes
For S. purpurea, metatranscriptomic data sets from the rhizosphere soil and the associated roots were available from the exact same samples. S. purpurea was selected because it was the only willow species having its genome and transcriptome sequenced at the time and because it was the model plant used by our group in previous greenhouse pot experiments looking at the willow leaf [6], root [7], and rhizosphere [8] (meta-) transcriptome. The correlation networks between the transcripts relative abundance summed at the KEGG pathway level (ko) for the roots and the rhizosphere are shown in Fig. 4. The number of correlations with an absolute Spearman R (R s) value above 0.80 (number of edges) was roughly similar for willows growing in non-contaminated (3071; Fig. 4a) and in contaminated soils (3713; Fig. 4b). The number of nodes (pathways) was also similar for willows growing in non-contaminated (408) and contaminated (350) soils. However, for the non-contaminated environment, the vast majority of the correlations was positive (83.2%, 2554/3071) whereas the majority of the correlations in the contaminated environment was negative (85.6%, 3179/3713). The shifts in correlations between a pathway in the roots and the same pathway in the rhizosphere were further scrutinized (Fig. 4c; Table 1). Twenty root–rhizosphere correlations showed differences of more than 1.5 between their values in non-contaminated and contaminated soils, many of which were related to the metabolism of amino acids (ko00480, ko00340, and ko00410) and to the metabolism and degradation of xenobiotics and lipids (ko00364, k000361, ko00623, ko00561, and ko00071) (Table 1). These shifts in correlation were mainly due to large changes in transcript abundance in the rhizosphere data set between contaminated and non-contaminated soils (Table 1) and accordingly, the pathways clustered more tightly in the rhizosphere data set (Fig. 4c). The pathways that had the highest number of correlations (nodes with the highest number of edges) were different for the non-contaminated and contaminated environments (Fig. 4; Table 2). In non-contaminated soils, the pathways with the highest number of correlations were mostly related to the metabolism of amino acids and to secondary metabolites. In contrast, many of the pathways having the highest number of edges in contaminated soil were related to the degradation of xenobiotic compounds. One should keep in mind that many pathways share genes and that ko’s were often defined in relation to human health, which could explain the presence of ko seemingly unrelated to the plant–soil environment in Tables 1 and 2.
Fig. 4.
Co-variation of willow root and rhizosphere metatranscriptomes. Correlation network showing Spearman correlations between the relative transcript abundance of KEGG pathways (ko’s) in roots (green circles) and in the rhizosphere (brown circles) for the willow S. purpurea growing in a non-contaminated and b contaminated soils. c Heatmap of the change in correlation value between KEGG pathways (ko’s) root and rhizosphere metatranscriptomes when comparing contaminated vs. non-contaminated soil. In a and b, green edges are indicative of positive correlation above 0.8, whereas the red edges are indicative of negative correlations below −0.8. In c, red color indicates large negative values and green color indicates large positive values. In a, b, and c, the pathways highlighted are the ones figuring in Tables 1 and 2
Table 1.
KEGG pathways (ko’s) showing differences of more than 1.5 in their correlation between root and rhizosphere metatranscriptomes when comparing contaminated vs. non-contaminated soils
| |Log 2 fold difference| | |||||
|---|---|---|---|---|---|
| ko | Pathway | Class | |Rs diff.| | Root | Rhizo. |
| ko00480 | Glutathione metabolism | Metabolism of other amino acids | 2.000 | 0.145 | 1.400 |
| ko04670 | Leukocyte transendothelial migration | Immune system | 1.800 | 0.243 | 0.573 |
| ko00364 | Fluorobenzoate degradation | Xenobiotics biodegradation and metabolism | 1.775 | 0.234 | 1.689 |
| ko00340 | Histidine metabolism | Amino-acid metabolism | 1.775 | 0.312 | 3.526 |
| ko00361 | Chlorocyclohexane and chlorobenzene degradation | Xenobiotics biodegradation and metabolism | 1.775 | 0.153 | 1.689 |
| ko00623 | Toluene degradation | Xenobiotics biodegradation and metabolism | 1.775 | 0.184 | 1.689 |
| ko04380 | Osteoclast differentiation | Development | 1.775 | 0.368 | 4.868 |
| ko04724 | Glutamatergic synapse | Nervous system | 1.775 | 0.182 | 0.881 |
| ko04974 | Protein digestion and absorption | Digestive system | 1.632 | 0.851 | 3.379 |
| ko00071 | Fatty acid degradation | Lipid metabolism | 1.600 | 0.172 | 0.304 |
| ko04611 | Platelet activation | Immune system | 1.600 | 0.105 | 0.815 |
| ko00630 | Glyoxylate and dicarboxylate metabolism | Carbohydrate metabolism | 1.600 | 0.242 | 0.493 |
| ko04310 | Wnt signaling pathway | Signal transduction | 1.575 | 0.097 | 3.775 |
| ko00410 | Beta-alanine metabolism | Metabolism of other amino acids | 1.575 | 0.167 | 3.204 |
| ko00561 | Glycerolipid metabolism | Lipid metabolism | 1.575 | 0.152 | 2.731 |
| ko04727 | GABAergic synapse | Nervous system | 1.575 | 0.223 | 1.104 |
| ko00053 | Ascorbate and aldarate metabolism | Carbohydrate metabolism | 1.575 | 0.140 | 3.633 |
| ko04111 | Cell cycle—yeast | Cell growth and death | 1.549 | 0.067 | 0.064 |
| ko00253 | Tetracycline biosynthesis | Metabolism of terpenoids and polyketides | 1.512 | 0.288 | 2.776 |
| ko05020 | Prion diseases | Neurodegenerative diseases | 1.512 | 0.059 | 5.273 |
Table 2.
Top 10 root and rhizosphere nodes with most edges in correlation network analyses for the willow species S. purpurea growing in non-contaminated and contaminated soils
| ko | Pathway | Class | Nb edges |
|---|---|---|---|
| Non-contaminated | |||
| Rhizosphere | |||
| ko00950 | Isoquinoline alkaloid biosynthesis | Biosynthesis of other secondary metabolites | 127 |
| ko00960 | Tropane, piperidine, and pyridine alkaloid biosynthesis | Biosynthesis of other secondary metabolites | 127 |
| ko04961 | Endocrine and other factor-regulated calcium reabsorption | Excretory system | 127 |
| ko03440 | Homologous recombination | Replication and repair | 80 |
| ko00253 | Tetracycline biosynthesis | Metabolism of terpenoids and polyketides | 61 |
| ko05020 | Prion diseases | Neurodegenerative diseases | 61 |
| ko00071 | Fatty acid degradation | Lipid metabolism | 47 |
| ko00250 | Alanine, aspartate, and glutamate metabolism | Amino-acid metabolism | 47 |
| ko00310 | Lysine degradation | Amino-acid metabolism | 47 |
| ko00380 | Tryptophan metabolism | Amino-acid metabolism | 47 |
| Roots | |||
| ko00312 | Beta-lactam resistance | Drug resistance | 60 |
| ko05144 | Malaria | Infectious diseases | 60 |
| ko00052 | Galactose metabolism | Carbohydrate metabolism | 43 |
| ko00071 | Fatty acid degradation | Lipid metabolism | 43 |
| ko00270 | Cysteine and methionine metabolism | Amino-acid metabolism | 43 |
| ko00340 | Histidine metabolism | Amino-acid metabolism | 43 |
| ko00361 | Chlorocyclohexane and chlorobenzene degradation | Xenobiotics biodegradation and metabolism | 43 |
| ko00364 | Fluorobenzoate degradation | Xenobiotics biodegradation and metabolism | 43 |
| ko00380 | Tryptophan metabolism | Amino-acid metabolism | 43 |
| ko00511 | Other glycan degradation | Glycan biosynthesis and metabolism | 43 |
| Contaminated | |||
| Rhizosphere | |||
| ko04152 | AMPK signaling pathway | Signal transduction | 73 |
| ko00281 | Geraniol degradation | Metabolism of terpenoids and polyketides | 62 |
| ko00550 | Peptidoglycan biosynthesis | Glycan biosynthesis and metabolism | 62 |
| ko00625 | Chloroalkane and chloroalkene degradation | Xenobiotics biodegradation and metabolism | 62 |
| ko00627 | Aminobenzoate degradation | Xenobiotics biodegradation and metabolism | 62 |
| ko01220 | Degradation of aromatic compounds | Xenobiotics biodegradation and metabolism | 62 |
| ko02030 | Bacterial chemotaxis | Cell motility | 62 |
| ko03320 | PPAR signaling pathway | Endocrine system | 62 |
| ko04066 | HIF-1 signaling pathway | Signal transduction | 62 |
| ko04120 | Ubiquitin-mediated proteolysis | Folding, sorting, and degradation | 62 |
| Roots | |||
| ko00071 | Fatty acid degradation | Lipid metabolism | 41 |
| ko00364 | Fluorobenzoate degradation | Xenobiotics biodegradation and metabolism | 41 |
| ko00410 | Beta-alanine metabolism | Metabolism of other amino acids | 41 |
| ko00561 | Glycerolipid metabolism | Lipid metabolism | 41 |
| ko00625 | Chloroalkane and chloroalkene degradation | Xenobiotics biodegradation and metabolism | 41 |
| ko02010 | ABC transporters | Membrane transport | 41 |
| ko02020 | Two-component system | Signal transduction | 41 |
| ko03320 | PPAR signaling pathway | Endocrine system | 41 |
| ko04015 | Rap1 signaling pathway | Signal transduction | 41 |
| ko04022 | cGMP-PKG signaling pathway | Signal transduction | 41 |
Discussion
When growing under controlled conditions, with little environmental variation, different genotypes from the same plant species generally associate with distinct microbial communities [10–12]. In contrast, recent studies from our team have shown that the bacterial, fungal, and AMF communities associated with willows growing at the same field site were not significantly affected by cultivar/species in the blocks sampled here (low contamination C1–C3 or non-contaminated N1-N8) [13–15]. These studies have also shown that the microbial community composition is only significantly affected by hydrocarbon contamination at contamination levels much higher (blocks C4 and C5) than the ones found in the contaminated blocks sampled here (blocks C1–C3) (August 2011 PAH averages of 0.9–14.6 vs. 114.4–224.1 mg kg−1 and August 2011 C10–C50 averages of 182–709 vs. 2143–3590 mg kg−1) [15]. Although these previous studies used samples from different trees taken in the same plots, but in August 2011 [14, 15] and November 2012 [13] as compared to August 2012 here, it does suggest that the differences in transcript abundance between willow species and contamination levels observed in the current study are more likely due to a shift in gene expression within similar communities rather than large changes in microbial community composition.
We have shown here a strong overriding effect of soil contamination on transcript abundance patterns in the rhizosphere of willows, as previously reported in a pot study [8]. Similarly, soil contamination level was the most important factor explaining the relative abundance of microbial transcripts in the roots of willows. However, when looking separately at willows growing in highly contaminated and non-contaminated soils in the field, the effect of cultivar can become more apparent [15, 16]. In addition, among a general common response to contamination, different willow cultivars showed different expression patterns in their leaves, including some transcripts annotated as fungal or bacterial [17]. We have further shown here that there are significant differences between the abundance of transcripts related to various microbial taxa in the rhizosphere and in the roots of different field-grown willow species, mostly when grown in contaminated soils. However, this resulted in few significant differences in the abundance of transcripts related to hydrocarbon degradation and to other key rhizosphere functions, such as nutrient cycling and acquisition, chemotaxis, and plant–microbe interactions. The largest and clearest shifts in transcripts abundance between willow species were observed for plant transcripts in the roots. Our study shows that the differences in microbial community composition among closely related genotypes observed elsewhere is also visible at the transcriptomic level, but does not necessarily translate in large shifts in functions. Using DNA-based methods, it was previously reported that gut microbial communities can vary substantially at the taxonomic level, with little changes in the functions [28, 29]
The reported transcriptomic differences between willow roots and the associated rhizosphere microbiota did not lead to significant differences in soil PAH and C10–C50 concentrations in the short term. In fact, in August 2014 (after four growing seasons), analyses of soil PAH in block C3 (the most contaminated block sampled here) showed that the S. caprea plots did contain higher levels of contaminant (PAH average concentrations of 27.0 mg kg−1 for S. caprea vs. 10.3, 2.7, 2.6, and 12.5 mg kg−1 for S. purpurea, S. eriocephala, S. nigra, and S. miyabeana, respectively), mainly in the form of phenanthrene, anthracene, pyrene, and acenaphthylene (unpublished results). Total hydrocarbon concentration in soil (C10–C50) was at its highest in S. purpurea plots (770 mg kg−1), followed by S. caprea (638 mg kg−1), S. miyabeana (551 mg kg−1), S. eriocephala (269 mg kg−1), and S. nigra (253 mg kg−1) (unpublished results). However, high variability in contamination levels made both these trends insignificant, perhaps because soil analyses were done per plot and using bulk soil between willow rows instead of the rhizosphere soil of individual trees.
Not only did the gene expression in the roots and rhizosphere of willows varied according to plant species or soil contamination, but the differences in transcript abundance related to various taxa and pathways in contaminated vs. non-contaminated soils were shown here to also vary according to willow species. Our results are consistent with the reported physiological response of willow cultivars to contamination. Indeed, at the same sampling site used in the current study, S. caprea was previously shown to be particularly sensitive to the presence of soil contaminants, with significant reductions in stem diameter, aerial biomass, leaf chlorophyll concentration, and leaf area [18]. In contrast, S. miyabeana was among the cultivars producing the greatest amount of biomass [18]. The results presented here further suggest that the sensitivity of S. caprea to soil contaminants results in a significant reduction of the transcript abundance of many microbial taxa in its rhizosphere, which leads to a general reduction in the abundance of transcripts related to various pathways. Conversely, the association of the S. caprea species to a suboptimal microbiota [5, 15, 30] could have resulted in its higher sensitivity to pollutant stress. Either way, this could lead to an impaired phytoremediation capacity for S. caprea in the longer term. In contrast, S. miyabeana, S. eriocephala, and S. purpurea generally stimulated the microbial taxa associated with their roots when growing in contaminated soils, leading to increased abundance of transcripts involved in hydrocarbon degradation pathways. Therefore, the increases in plant exudation observed under stressful conditions [31–34] that results in increased microbial biomass [35] and activities [8], appear to be willow species-specific.
S. nigra was shown to increase slightly its leaf size and its number of shoots under contaminated conditions [18], and together with the significant decrease in the transcript relative abundance of two key gene categories related to plant osmotic stresses (dehydrin and aquaporin) in contaminated soils, it suggests a decreased stress levels for S. nigra when growing in contaminated conditions. Interestingly, this was concomitant with a significant increase in the relative abundance of transcripts related to Glomeromycota in the roots of S. nigra growing in contaminated soils. Glomeromycota, the AMF, are well known to improve plant water uptake [36]. Glomeromycota transcripts were also significantly or nearly significantly more abundant in the rhizosphere S. miyabeana, S. purpurea, and S. eriocephala growing in contaminated soils, but not in the rhizosphere of S. caprea. As AMF can help plants to cope with contaminant stresses [37], this difference between the willow species could partly explain the heightened sensitivity of S. caprea to contamination. As for the ectomycorrhizal species (Sphaerosporella brunnea, phylum Ascomycota, class Pezizomycetes) that were previously shown to increase dramatically their abundance in native and naturalized willow species under high contamination conditions at our field site [15], they did not show here any significant differences in their transcript abundance in the rhizosphere and roots, probably because the contamination levels in the blocks sampled here were much lower.
To our knowledge, we are the first to analyze combined plant root and rhizosphere metatranscriptomic data sets. The goal of this approach was to observe if the willow roots and the rhizosphere are co-expressing genes or pathways that could eventually be harnessed for improving phytoremediation efficiency. One striking observation is the almost complete shift in the direction of the correlations (from mostly positive to mostly negative) between root and rhizosphere pathways from non-contaminated to contaminated soil. Our approach also allowed us to highlight some pathways that have a central position in the root–rhizosphere “interactome”. Here again, the contaminated and non-contaminated “interactomes” were completely different, with few pathways being central for both types of soils. Plants are known to secrete, among other things, secondary compounds and amino acids in their rhizosphere [38–40], which are partly responsible for the selective pressure known as the “rhizosphere effect” [41, 42]. It is thus not surprising to find pathways related to these compounds being at the center of the interaction maps when willows are growing in non-contaminated soil. A previous pot study had also showed that the expression of genes involved in pathways related to amino acids increased in the rhizosphere of willows, suggesting indirectly that these compounds are used by the rhizosphere microorganisms [8]. In contaminated soils, pathways that showed the most connections were often related to hydrocarbon degradation. Many plant-derived chemicals (root degradation compounds, secondary metabolites, etc.) stimulate rhizosphere microorganisms to degrade contaminants [43–47], which could explain the central position of these pathways. Interestingly, many of the pathways that showed the largest shifts in correlation were also linked to the degradation of xenobiotics and to the metabolism of amino acids, further highlighting their importance in the root–rhizosphere “interactome”.
Recent work has shown that, inside the roots of willows growing in pots, the main transcriptomic response to organic contaminants is seen in the fungal and bacterial genes, with a much smaller response of the willow itself [7]. Similarly, our results showed that the shifts observed in the “interactome” are mainly due to changes in transcript abundance in the rhizosphere (mainly bacterial transcripts), with little changes in transcript abundance inside willow roots (mainly plant transcripts). Our results also showed that the response of the root metatranscriptome to contaminant is mainly due to fungi, whereas the transcripts affiliated to plants were mostly affected by the willow species. Host–microbiota interactions can be viewed through the lens of the hologenome theory of evolution [48]. One of the implications of this theory is that rapid adaptation of holobionts to stressful conditions is possible through modifications in the microbiota composition, abundance, activities, or gene content [49, 50]. Our results together with recently published work from our group [7] support the notion that the microbial partners of the willow holobiont are the primary responder to environmental stresses. More controlled experiments will, however, be required to fully understand how plants interact with their microbiota at the transcriptomic level, how this interaction is modulated by environmental factors such as soil contamination, and if this fits with the hologenome theory of evolution.
In conclusion, our study further highlights the usefulness and robustness of metatranscriptomic approaches to study plant-associated microbial activities. Although the experiment was carried out in the field with large variations in environmental conditions, the trends observed in the rhizosphere microbial transcript abundance closely mirrored the trends observed in the willow’s physiological response to contaminant. The willow species not only had different abundance of transcripts related to various microbial taxa and functions in their rhizosphere and roots, but these taxa and functions also reacted in a genotype-dependent fashion to the presence of contaminant. The simultaneous analysis of root and rhizosphere metatranscriptomes highlighted that the willow–microbiota “interactome” undergoes a complete overhaul under contamination stress, mainly because of shifts in microbial transcript abundance in the rhizosphere. This analysis has also suggested a central role for several key pathways that could be further scrutinized to optimize the willow–microbiota “interactome” for more efficient and predictable phytoremediation.
Electronic supplementary material
Acknowledgements
This project was funded by the Genome Canada and Genome Québec (2010 Large-Scale Applied Research Project Competition grant 2510). Data analyses were carried out on Compute Canada’s infrastructure through EY resource allocation (2016 Resource Allocation competition). Danielle Ouellette and Julie Marleau are gratefully acknowledged for technical assistance. Pétromont is gratefully acknowledged for allowing us access to their site to carry out our field experiment.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0018-4) contains supplementary material, which is available to authorized users.
References
- 1.Pulford I, Watson C. Phytoremediation of heavy metal-contaminated land by trees—a review. Environ Int. 2003;29:529–40. doi: 10.1016/S0160-4120(02)00152-6. [DOI] [PubMed] [Google Scholar]
- 2.Newsholme C. Willows: the genus Salix. Portland: Timber Press; 2003. p. 256. [Google Scholar]
- 3.Leigh MB, Prouzova P, Mackova M, Macek T, Nagle DP, Fletcher JS. Polychlorinated biphenyl (PCB)-degrading bacteria associated with trees in a PCB-contaminated site. Appl Environ Microbiol. 2006;72:2331–42. doi: 10.1128/AEM.72.4.2331-2342.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El Amrani A, Dumas AS, Wick LY, Yergeau E, Berthomé R. “Omics” insights into PAH degradation toward improved green remediation biotechnologies. Environ Sci Technol. 2015;49:11281–91. doi: 10.1021/acs.est.5b01740. [DOI] [PubMed] [Google Scholar]
- 5.Bell TH, Pitre FE, Joly S, Yergeau E. Increasing phytoremediation efficiency and reliability using novel ‘omics approaches. Trends Biotechnol. 2014;32:271–80. doi: 10.1016/j.tibtech.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 6.Gonzalez E, Brereton NJ, Marleau J, Nissim WG, Labrecque M, Pitre FE, et al. Meta-transcriptomics indicates biotic cross-tolerance in willow trees cultivated on petroleum hydrocarbon contaminated soil. BMC Plant Biol. 2015;15:1. doi: 10.1186/s12870-015-0636-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gonzalez E, Pitre F, Pagé A, Marleau J, Guidi Nissim W, St-Arnaud M et al. Trees, fungi and bacteria: tripartite metatranscriptomics of a root microbiome responding to soil contamination. Microbiome. 2017 (in revision). [DOI] [PMC free article] [PubMed]
- 8.Yergeau E, Sanschagrin S, Maynard C, St-Arnaud M, Greer CW. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J. 2014;8:344–58. doi: 10.1038/ismej.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pagé AP, Yergeau Eacute, Greer CW. Salix purpurea stimulates the expression of specific bacterial xenobiotic degradation genes in a soil contaminated with hydrocarbons. PLoS ONE. 2015;10:e0132062. doi: 10.1371/journal.pone.0132062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488:86–90. doi: 10.1038/nature11237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pérez-Jaramillo JE, Carrión VJ, Bosse M, Ferrão LF, Hollander Md, Garcia AA et al. Linking rhizosphere microbiome composition of wild and domesticated Phaseolus vulgaris to genotypic and root phenotypic traits. ISME J. 2017;11:2244–57. [DOI] [PMC free article] [PubMed]
- 12.Bulgarelli D, Garrido-Oter R, Münch PC, Weiman A, Dröge J, Pan Y, et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe. 2015;17:392–403. doi: 10.1016/j.chom.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tardif S, Yergeau Eacute, Tremblay J, Legendre P, Whyte LG, Greer CW. The willow microbiome is influenced by soil petroleum-hydrocarbon concentration with plant compartment-specific effects. Front Microbiol. 2016;7:1363. doi: 10.3389/fmicb.2016.01363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hassan SED, Bell TH, Stefani FOP, Denis D, Hijri M, St-Arnaud M. Contrasting the community structure of arbuscular mycorrhizal fungi from hydrocarbon-contaminated and uncontaminated soils following willow (Salix spp. L.) planting. PLoS ONE. 2014;9:e102838. doi: 10.1371/journal.pone.0102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bell TH, El-Din Hassan S, Lauron-Moreau A, Al-Otaibi F, Hijri M, Yergeau E, et al. Reduced linkage between bacterial and fungal rhizosphere communities in hydrocarbon-contaminated soils is related to plant phylogeny. ISME J. 2014;8:331–43. doi: 10.1038/ismej.2013.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bell TH, Cloutier-Hurteau B, Al-Otaibi F, Turmel MC, Yergeau E, Courchesne F, et al. Early rhizosphere microbiome composition is related to the growth and Zn uptake of willows introduced to a former landfill. Environ Microbiol. 2015;17:3025–38. doi: 10.1111/1462-2920.12900. [DOI] [PubMed] [Google Scholar]
- 17.Brereton NJB, Gonzalez E, Marleau J, Guidi W, Labrecque M, Joly S, et al. Comparative transcriptomic approaches exploring contamination stress tolerance in Salix sp. reveal the importance for a metaorganismal de novo assembly approach for non-model plants. Plant Physiol. 2016;171:3–24. doi: 10.1104/pp.16.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grenier V, Pitre FE, Nissim WG, Labrecque M. Genotypic differences explain most of the response of willow cultivars to petroleum-contaminated soil. Trees. 2015;29:871–81. doi: 10.1007/s00468-015-1168-5. [DOI] [Google Scholar]
- 19.Yergeau E, Bell TH, Champagne J, Maynard C, Tardif S, Tremblay J, et al. Transplanting soil microbiomes leads to lasting effects on willow growth, but not on the rhizosphere microbiome. Front Microbiol. 2015;6:1436. doi: 10.3389/fmicb.2015.01436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gambino G, Perrone I, Gribaudo I. A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem Anal. 2008;19:520–5. doi: 10.1002/pca.1078. [DOI] [PubMed] [Google Scholar]
- 21.Yergeau E, Bokhorst S, Huiskes AHL, Boschker HTS, Aerts R, Kowalchuk GA. Size and structure of bacterial, fungal and nematode communities along an Antarctic environmental gradient. FEMS Microbiol Ecol. 2007;59:436–51. doi: 10.1111/j.1574-6941.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- 22.Yergeau E, Kowalchuk GA. Responses of Antarctic soil microbial communities and associated functions to temperature and freeze-thaw cycle frequency. Environ Microbiol. 2008;10:2223–35. doi: 10.1111/j.1462-2920.2008.01644.x. [DOI] [PubMed] [Google Scholar]
- 23.Dorsch M, Stackebrandt E. Some modifications in the procedure of direct sequencing of PCR amplified 16S rDNA. J Microbiol Methods. 1992;16:271–9. doi: 10.1016/0167-7012(92)90017-X. [DOI] [Google Scholar]
- 24.Tremblay J, Yergeau E, Fortin N, Cobanli S, Elias M, King TL, et al. Chemical dispersants enhance the activity of oil- and gas condensate-degrading marine bacteria. ISME J. 2017;11:2793–808. [DOI] [PMC free article] [PubMed]
- 25.Huntemann M, Ivanova NN, Mavromatis K, Tripp HJ, Paez-Espino D, Tennessen K, et al. The standard operating procedure of the DOE-JGI Metagenome Annotation Pipeline (MAP v. 4) Stand Genomic Sci. 2016;11:17. doi: 10.1186/s40793-016-0138-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moran MA, Satinsky B, Gifford SM, Luo H, Rivers A, Chan LK, et al. Sizing up metatranscriptomics. ISME J. 2013;7:237–43. doi: 10.1038/ismej.2012.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Human_Microbiome_Project_Consortium. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457:480–4. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Siciliano SD, Germida JJ, Banks K, Greer CW. Changes in microbial community composition and function during a polyaromatic hydrocarbon phytoremediation field trial. Appl Environ Microbiol. 2003;69:483–9. doi: 10.1128/AEM.69.1.483-489.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jones DL, Hodge A, Kuzyakov Y. Plant and mycorrhizal regulation of rhizodeposition. New Phytol. 2004;163:459–80. doi: 10.1111/j.1469-8137.2004.01130.x. [DOI] [PubMed] [Google Scholar]
- 32.Karst J, Gaster J, Wiley E, Landhäusser SM. Stress differentially causes roots of tree seedlings to exude carbon. Tree Physiol. 2017;37:154–64. doi: 10.1093/treephys/tpw090. [DOI] [PubMed] [Google Scholar]
- 33.Naik D, Smith E, Cumming JR. Rhizosphere carbon deposition, oxidative stress and nutritional changes in two poplar species exposed to aluminum. Tree Physiol. 2009;29:423–36. doi: 10.1093/treephys/tpn035. [DOI] [PubMed] [Google Scholar]
- 34.Qin R, Hirano Y, Brunner I. Exudation of organic acid anions from poplar roots after exposure to Al, Cu and Zn. Tree Physiol. 2007;27:313–20. doi: 10.1093/treephys/27.2.313. [DOI] [PubMed] [Google Scholar]
- 35.Esperschutz J, Pritsch K, Gattinger A, Welzl G, Haesler F, Buegger F, et al. Influence of chronic ozone stress on carbon translocation pattern into rhizosphere microbial communities of beech trees (Fagus sylvatica L.) during a growing season. Plant Soil. 2009;323:85–95. doi: 10.1007/s11104-009-0090-2. [DOI] [Google Scholar]
- 36.Jayne B, Quigley M. Influence of arbuscular mycorrhiza on growth and reproductive response of plants under water deficit: a meta-analysis. Mycorrhiza. 2014;24:109–19. doi: 10.1007/s00572-013-0515-x. [DOI] [PubMed] [Google Scholar]
- 37.Glassman SI, Casper BB. Biotic contexts alter metal sequestration and AMF effects on plant growth in soils polluted with heavy metals. Ecology. 2012;93:1550–9. doi: 10.1890/10-2135.1. [DOI] [PubMed] [Google Scholar]
- 38.Badri DV, Weir TL, van der Lelie D, Vivanco JM. Rhizosphere chemical dialogues: plant–microbe interactions. Curr Opin Biotechnol. 2009;20:642–50. doi: 10.1016/j.copbio.2009.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Haichar FeZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, et al. Plant host habitat and root exudates shape soil bacterial community structure. ISME J. 2008;2:1221–30. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 40.Berg G, Zachow C, Lottmann J, Gotz M, Costa R, Smalla K. Impact of plant species and site on rhizosphere-associated fungi antagonistic to Verticillium dahliae Kleb. Appl Environ Microbiol. 2005;71:4203–13. doi: 10.1128/AEM.71.8.4203-4213.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kowalchuk GA, Buma DS, de Boer W, Klinkhamer PGL, van Veen JA. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek. 2002;81:509–20. doi: 10.1023/A:1020565523615. [DOI] [PubMed] [Google Scholar]
- 42.Smalla K, Wieland G, Buchner A, Zock A, Parzy J, Kaiser S, et al. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl Environ Microbiol. 2001;67:4742–51. doi: 10.1128/AEM.67.10.4742-4751.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fletcher JS, Hegde RS. Release of phenols by perennial plant roots and their potential importance in bioremediation. Chemosphere. 1995;31:3009–16. doi: 10.1016/0045-6535(95)00161-Z. [DOI] [Google Scholar]
- 44.Haby PA, Crowley DE. Biodegradation of 3-chlorobenzoate as affected by rhizodeposition and selected carbon substrates. J Environ Qual. 1996;25:304–10. doi: 10.2134/jeq1996.00472425002500020014x. [DOI] [Google Scholar]
- 45.Isidorov V, Jdanova M. Volatile organic compounds from leaves litter. Chemosphere. 2002;48:975–9. doi: 10.1016/S0045-6535(02)00074-7. [DOI] [PubMed] [Google Scholar]
- 46.Miya RK, Firestone MK. Enhanced phenanthrene biodegradation in soil by slender oat root exudates and root debris. J Environ Qual. 2001;30:1911–8. doi: 10.2134/jeq2001.1911. [DOI] [PubMed] [Google Scholar]
- 47.Donnelly PK, Hegde RS, Fletcher JS. Growth of PCB-degrading bacteria on compounds from photosynthetic plants. Chemosphere. 1994;28:981–8. doi: 10.1016/0045-6535(94)90014-0. [DOI] [Google Scholar]
- 48.Zilber-Rosenberg I, Rosenberg E. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev. 2008;32:723–35. doi: 10.1111/j.1574-6976.2008.00123.x. [DOI] [PubMed] [Google Scholar]
- 49.Rosenberg E, Zilber-Rosenberg I. Microbes drive evolution of animals and plants: the hologenome concept. mBio. 2016;7:e01395–15. doi: 10.1128/mBio.01395-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bordenstein SR, Theis KR. Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 2015;13:e1002226. doi: 10.1371/journal.pbio.1002226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.