Abstract
Although many environments like soils are constantly subjected to invasion by alien microbes, invaders usually fail to succeed, succumbing to the robust diversity often found in nature. So far, only successful invasions have been explored, and it remains unknown to what extent an unsuccessful invasion can impact resident communities. Here we hypothesized that unsuccessful invasions can cause impacts to soil functioning by decreasing the diversity and niche breadth of resident bacterial communities, which could cause shifts to community composition and niche structure—an effect that is likely exacerbated when diversity is compromised. To examine this question, diversity gradients of soil microbial communities were subjected to invasion by the frequent, yet oft-unsuccessful soil invader, Escherichia coli, and evaluated for changes to diversity, bacterial community composition, niche breadth, and niche structure. Contrary to expectations, diversity and niche breadth increased across treatments upon invasion. Community composition and niche structure were also altered, with shifts of niche structure revealing an escape by the resident community away from the invader’s resources. Importantly, the extent of the escape varied in response to the community’s diversity, where less diverse communities experienced larger shifts. Thus, although transient and unsuccessful, the invader competed for resources with resident species and caused tangible impacts that modified both the diversity and functioning of resident communities, which can likely generate a legacy effect that influences future invasion attempts.
Introduction
Biological invasions have the potential to alter the ecological and evolutionary trajectory of the Earth’s ecosystems [1, 2]. Whether such ecosystems are the expanse of corals across the world’s oceans, various grasslands, or the human microbiome, the outcome of an invader’s introduction on the composition and functioning of the resident community is difficult to predict [3, 4]. One common theme that has guided investigations of biological invasions since Elton’s summation of the topic in 1958 is the influence of biological diversity, where he postulated that more diverse ecosystems resist invasion better than less diverse ecosystems [5]. Colloquially known as the ‘diversity-invasion effect’, robust levels of biological diversity contribute to the community’s competitive ability on limiting resources [6] and help create an invasion barrier. Although these theories initially targeted animal and plant communities, their application to understanding microbial invasions has been recently demonstrated [7, 8].
Less is known about the impacts that invaders cause on resident communities, in particular how diversity might buffer any potential impacts that stem from an invasion. Evidence from macroecological studies indicates that robust levels of diversity can buffer against detrimental impacts of invasion, such as decreases of species richness and evenness [9, 10] that can, in turn, alter a community’s composition and functioning [9–12]. Yet, little attention has been paid to the mechanisms that control these impacts. Competition and competitive exclusion are broad mechanisms by which invaders may induce impacts. For instance, the presence of invasive plants, Bromus tectorum and Carpobrotus edulis, induces competition for water and reduces the soil water available for native plants, which likely aids in reducing native biomass and altering community structure [13, 14]. However, revealing mechanisms of impacts at this level of detail or further and determining whether similar competitive mechanisms are invoked across different communities has so far eluded biologists. This is partly because it has been difficult to determine and localize the exact niche preferences of invaders vis-à-vis residents, such as differences in phenology and rooting patterns in the case of plants or carbon utilization patterns in the case of microorganisms. Without the ability to pinpoint what resident species will partake in direct resource competition with the invader and the specific resources for which they strive, predicting and assessing the impacts of invasions on native communities will remain a qualitative affair.
Natural microbial communities frequently experience invasions from alien microbes [7]. Invasions could take the form of fecal shedding into the soil or water; the use of biocontrol and remediation agents; the use of biofertilizers in agriculture; or even probiotics in nutrition. Several studies using in situ, in vitro, and in vivo systems have found evidence of the diversity-invasion effect in microbial communities [15–21]. The mechanisms governing this relationship are, like in plant and animal communities, modulated by the diversity of the resident community and the availability of local resources [7, 16]. There are idiosyncrasies between macro- and microbial invasions as well. The former are often studied when invaders are successful, displaying growth, spreading among the native community, and making up a large portion of community biomass. Microbial invasions, in contrast, are often examined in systems where invaders fail to become autochthonous members of the resident community. For instance, most compost-borne microorganisms added to soil are outcompeted by soil-derived microorganisms [22]. And in situ studies tracking the survival of Escherichia coli and Listeria monocytogenes for extended periods of time (in some cases up to 95 days) showed that invader survival often follows a progressive decline [16, 18, 19]. This begs the question of whether the impacts of invasion can be witnessed in such systems where invasions are ultimately unsuccessful. Given the ubiquity of invasions across a wide variety of ecosystems, a better understanding of what occurs between an invader’s arrival and potential elimination from the community is needed to better comprehend the impacts of unsuccessful invaders on native communities, the mechanisms controlling these impacts, and the ecological implications for future invasion attempts.
The purpose of this study was to assess and quantify the impacts of a microbial invasion by a single model invader, and, in doing so, better understand how this phenomenon affects the diversity and metabolic potential of invaded communities. In accordance with observations of plant systems, we hypothesized that a microbial invasion would decrease microbial community diversity, which would lead to changes in its metabolic potential and create a legacy effect that might influence the outcome of subsequent invasions. These impacts were further hypothesized to scale with the initial diversity of resident communities. We thus constructed a gradient of soil microbial diversity by a removal approach, where natural soils from two different locations were diluted and used to inoculate sterile soil. The resulting microbial communities were subjected to invasion by a non-pathogenic derivative of E. coli O157:H7. E. coli is a model bacterium to study the impact of invasions because although its primary habitat is the vertebrate gut, it must invade secondary habitats like soil or water so that it can get reacquired by a new vertebrate host and perpetuate its lineage [7, 23]. While E. coli has been observed to mount successful invasions and become an autochthonous member of the soil community [24], such occasions are rare and seldom does E. coli breach the barrier of biotic resistance to become a naturalized member of the community [16, 18, 25, 26]. Nonetheless, both experimental and field studies show that it can survive in soil for extended periods time, up to 100 days [25], making it an ideal bacterium to study the impacts of unsuccessful invasions.
In this study, the impacts of invasion were assessed by measuring four community parameters before and after invasion: diversity, community composition, niche breadth, and niche structure. Shifts to any one of these parameters upon invasion would indicate that an invasion attempt could bestow impacts to the resident community, and the consistency of the alterations to these parameters along the diversity gradients could reveal shared mechanisms by which different communities respond to invasion.
Methods
Soil microcosms, experimental set-up, and design
Microbial community inocula were prepared by making an initial 1:2 soil:water mixture, for two source soils: Wildekamp (W) soil and Buinen (B) soil. Both soils were sandy soils (see SI for soil physio-chemical characteristics). We used two different soils and therefore two diversity gradients to increase the variability of community composition.
The initial soil mixtures from each source soil were 1:10 serially diluted in sterile water up to the 10−6 factor, and the soil:water solutions obtained from the 10−1, 10−3, and 10−6 dilutions were inoculated into sterilized W soil (see SI for details on soil preparation and inoculation). After inoculation of the diluted communities, the microcosms were incubated for 79 days in order to establish comparable soil bacterial abundances. We refer to the diversity gradients throughout the text as either W or B gradients and each diversity treatment (dilution level) of a gradient is written as W−1, B−1, W−3, B−3, W−6, and B−6.
After 79 days, E. coli O157:H7 non-pathogenic derivative strain Tn5 luxCDAEB, which has been used in previous studies to examine the diversity-invasibility relationship in soil [16, 18, 26], was introduced into the soil microcosms at a level of 5 × 107 cells per gram soil. This inoculation density is within the norm of a typical invading E. coli population in nature [27] and has been previously used in similar experiments [16, 18, 26], thus providing a robust and comparative model to examine E. coli’s pattern of invasion.
The survival of E. coli was monitored through time, up to 75 days after inoculation. Specifically, the experiment was designed for 6 destructive samplings at days 0, 5, 11, 18, 28, and 75, using three replicates for each treatment at each date. This comprised a total of 126 soil microcosms (i.e., 3 dilution levels × 2 soil gradients × 6 sampling dates × 3 replicates + [3 sterile soil controls × 6 sampling dates]). It is important to note that the microcosms measured at Day 0 were never invaded with E. coli but also measured at Day 28 as controls (referred to as “non-invaded controls”) to examine the effect of incubation time on impacts to niche breadth and structure (further mentioned in the section Quantifying niche breadth and niche structure). Upon sampling, around 2 g of soil from each microcosm was stored at −20 °C for molecular analysis.
Total bacterial enumeration, invader enumeration, and invader population preservation
In order to assure that microcosms had reached an equal number of cells per gram of soil before invasion could take place, total culturable bacteria were checked by dilution plating on Trypticase Soy Agar (TSA) 62 days after they were inoculated with dilution steps. Plates were counted after 4 days of incubation at 28 °C.
Invader survival from samples collected from day 5 onward was tracked from fresh soil via selective dilution plating on TSA supplemented with rifampicin (10 μg/ml) and kanamycin (50 μg/ml), both antibiotics to which this strain of E. coli harbored resistance.
Twenty-eight days after invasion, E. coli colonies growing on the selective medium that were used for counting purposes were re-isolated by streaking on TSA supplemented with the selective antibiotics and incubated at 37 °C. Five colonies from each treatment were isolated and stored at −80 °C in glycerol for further analyses (see Assessing potential invader evolution).
Soil DNA extraction and 16S rRNA gene sequencing: methods to quantify diversity and community composition
Total soil DNA was extracted using 0.5 g of soil with the PowerSoil DNA isolation kit (MOBIO, Carlsbad, California, USA) according to the manufacturer’s instructions, except that 0.25 g of 1 mm glass beads were added in the first step in order to aid in cell lysis. DNA concentration was quantified using PicoGreen dsDNA assay (Invitrogen, Paisley, UK). The DNA retrieved from the 10−1, 10−3, and 10−6 treatments for both gradients collected on Days 0 and 28 was used as template in amplicon sequencing (454 pyrosequencing) reactions targeting the bacterial 16S rRNA gene, using forward primer 16S-515F (5′-TGYCAGCMGCCGCGGTA-3′) and reverse primer 16S-1061R (5′-TCACGRCACGAGCTGACG-3′). For each sample, a unique 10 base pair barcode was used. In the supporting information, we provide detailed information on PCR conditions, sample preparation, sequencing, and subsequent data analyses. Briefly, sequences were filtered using Denoiser (Reeder and Knight 2010), binned into OTUs using UCLUST (Edgar 2010), screened for chimeras using ChimeraSlayer (Haas et al. 2011), aligned using PyNAST (Caporaso et al. 2010), and classified using the Greengenes taxonomy via Ribosomal Database Project classifier (Wang et al. 2007). Samples were rarefied at 2000 sequences (rarefaction curves are provided in Supplementary Fig. S1). The QIIME toolkit was used to estimate diversity (species richness, species evenness, and phylogenetic diversity) and community composition.
Quantifying niche structure and niche breadth
Niche structure (i.e., the pattern and type of resources used) and niche breadth (i.e., the number of resources used) were assessed by measuring the metabolic activity for each microcosm across an array of 71 different (organic) carbon (C) sources using the Biolog GEN III microtiter plate, in accordance with Biolog’s high cell density phenotypic microarray protocol (Biolog, Hayward, California, USA). Briefly, 2 ml of a 1000 times soil dilution in sterile water was mixed with 8 ml of Biolog’s IF-0a inoculation fluid, 0.12 ml of Biolog Dye D, and 1.88 ml of sterile water to inoculate one GEN III plate with 100 µl per well. Assays were done in duplicate and measured every 15 min for 48 h at 23 °C using Biolog’s Omnilog machine. The substrate utilization was measured at day 0 (before invasion) and day 28 (after invasion). Additionally, although samples were destructively taken, we measured non-invaded microcosms from day 0 at day 28, in addition to the planned day 28 sampling, in order to evaluate the effect of incubation to shifts of niche breadth and structure. These microcosms were never inoculated with E. coli and simply called “non-invaded controls.” The data were normalized by the maximum value across all 71 C sources for each sample.
Assessing potential evolution of the invader niche
In order to assess whether any potential metabolic evolution of the invader occurred during its survival in the soil, the niche structure of E. coli colonies that were isolated from the soil microcosms at day 28 were compared to that of the ancestral invader. Specifically, five E. coli colonies from each diversity treatment were pooled at equal cell concentrations (OD600 = 0.04) and inoculated in the GEN III plate using inoculation fluid B, in accordance with the manufacturer’s instructions. Two replicate plates were used, and the duration, frequency, and temperature of the readings were the same as the readings for the soil communities. These protocols have previously been employed to measure either the performance of the soil bacterial community or pure strains [16].
Ordination analyses to assess shifts of niche structure
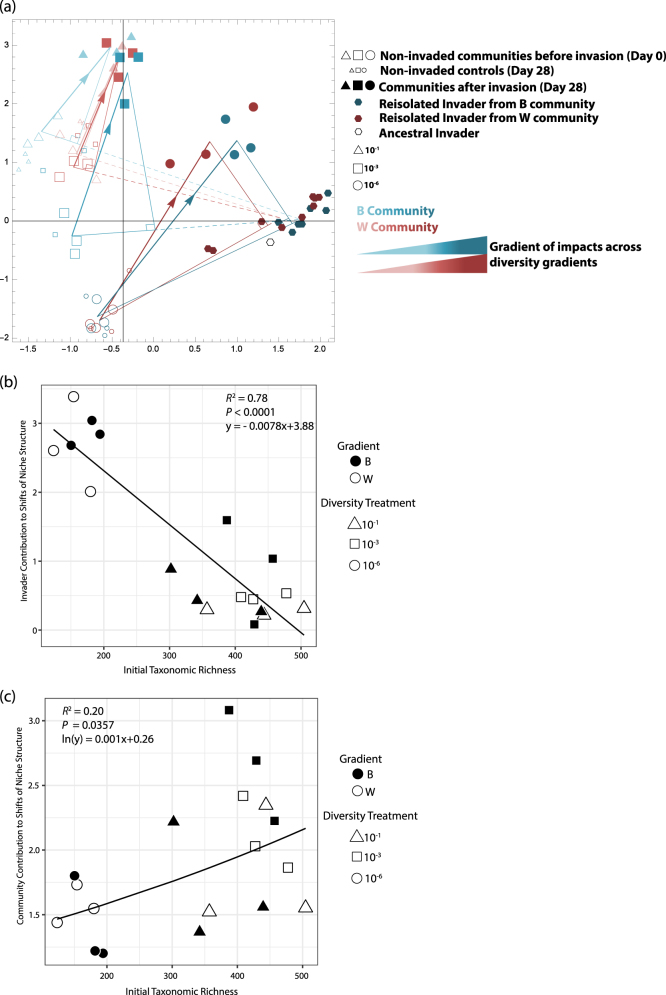
We used a three-step custom ordination analysis (performed in Mathematica 10.1; Wolfram Research Inc., Champaign, Ill., USA. 2015) to visualize the general patterns of shifts to niche structure upon invasion. First, in order to disentangle and quantify the relative contribution of either the invader or resident community to niche shifts, we decomposed the overall change of the invaded community niche, vector µ1 in Fig. 1, into two orthogonal components. The first component, vector µ2, reflects how far the niche of invaded communities—still including the invader population on Day 28—had shifted toward the invader niche along the axis (vector e in Fig. 1) that joins the average niche of the non-invaded community (Blue triangles in Fig. 1) and the average niche of the invader population (green hexagons). (The invader population comprised the metabolic measurements of the ancestral and re-isolated E. coli colonies, described in the section ‘Assessing potential evolution of the invader niche’.) The vector µ2 can thus be interpreted as the direct effect of the invader. The second component, vector µ3 (=µ1−µ2), reflects shifts in niche structure that are orthogonal to the direct invader effect, encompassing changes driven by resident taxa that use different resources than the invader. Therefore, we interpreted this vector as the response and contribution of the resident community to the overall shifts of niche structure.
Fig. 1. Decomposing and quantifying the contribution by the resident community and the invader to shifts of niche structure.
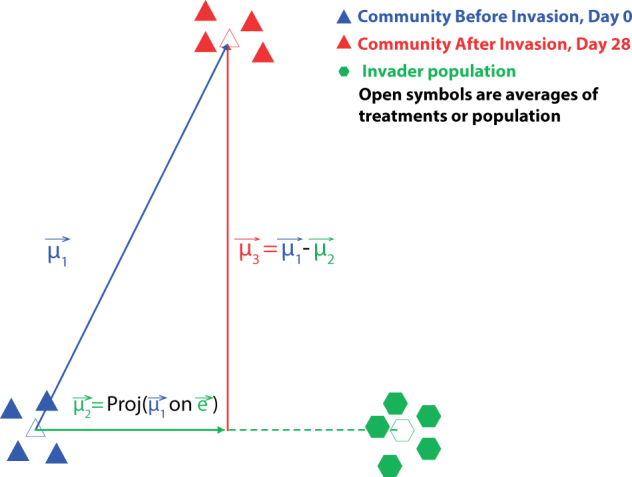
. Vector µ1 is the movement of invaded communities in resource space. Vector µ2 corresponds to the contribution of the E. coli to the invaded community’s niche shift; this vector lies along an axis, vector e, which begins at the mean of communities before invasion and extends to the niche structure of the invader population, which is shown by the dashed green line extending away from vector µ2. Vector µ3 represents the contribution of the resident taxa to the invaded community’s niche shift
Next, the decomposition analysis was complemented by a principal component analysis in order to visualize the variation of niche structure between microcosms of all treatments and the invader. In this way, the original 71-dimensional data set—a single dimension being the performance of a microcosm on one of the 71 carbon sources—was projected onto two principal components (the eigenvectors associated with the two largest eigenvalues of the variance-covariance matrix) that together explained 69.7% of the variation in the niche structure data. The principal component analysis was performed on the complete data set and included measurements from non-invaded communities (Day 0), non-invaded controls (Day 28), invaded communities (Day 28), and both the ancestral and re-isolated E. coli colonies.
Finally, for the purpose of visualization, the 71-dimensional vectors obtained from the separate decomposition analyses (three vectors for each treatment) were projected onto the 2-dimensional principal component space, and the axes were then rotated so as to align the average effect of E. coli across treatments with the horizontal axis (as it appears in Fig. 5).
Fig. 5. Invaded communities are steered away from the invader’s niche.
. (a) is an ordination analysis visualizing the variation of the communities’ niche structures before and after invasion, and it contains vector information that teases apart the effects of the invader and of the resident taxa in regards to shifts of niche structure. Vectors with an arrow head indicate the total shift of communities upon invasion in resource space (equivalent to vector µ1 in Fig. 1). These vectors are decomposed into two orthogonal components, indicated as the legs of a right triangle. The first component of the community shift is due to the impact of the invader (solid portion of vector originating at a non-invaded community and pointing toward E. coli; this is equivalent to vector µ2 in Fig. 1); the second component reflects the response of the resident community (equivalent to vector µ3 in Fig. 1). The invader’s impact vector is continued with a dashed line merely to indicate the entire axis between non-invaded communities and the invader’s niche structure (equivalent to vector e in Fig. 1). The vectors µ2 and µ3 that were obtained separately for each treatment, are orthogonal in the 71-dimensional space of the carbon utilization profile data. Their projections onto the subspace spanned by the first two principal components (which capture 69.7% of the variation in the entire data set) are nearly orthogonal as well, implying that the dominant shifts in niche structure are consistent across treatments. The gradient of color, increasing from the most diverse to least diverse communities and terminating at the invader’s niche structure, indicates that impacts increase as diversity decreases. The least diverse, 10−6 communities are the most impacted and thus have the darkest color. (b) The vector quantifying the impact of the invader to niche shifts or (c) the vector quantifying the response and contribution of resident taxa to niche shifts are regressed with the initial OTU richness of the community
Statistical analyses
Two-way ANOVA analyses with diversity treatment and gradient as factors were used to examine differences of total bacterial abundance, diversity, and niche breadth.Bray–Curtis dissimilarity values were used to assess the extent of shifts to community composition 28 days after invasion. A three-way ANOVA was used to examine differences in E. coli survival stemming from time, diversity treatment, and diversity gradient. Permutation multivariate analysis of variance (PERMANOVA) tests were performed to examine shifts in bacterial community composition. The PERMANOVA was run independently for each soil type (B and W). We used invasion as the main factor and allowed for full permutation of the raw data (total of 105 permutations) with Monte–Carlo tests accounting for type III error (where the fixed effects sum to zero). In order to identify those taxa most affected by shifts due to invasion, we executed a similarity percentages analysis (SIMPER). This analysis identifies those OTUs that most contribute to differences between communities before and after invasion. The SIMPER results were filtered to include only OTUs whose relative abundance had altered (either increased or decreased) a minimum of 1% in relative values upon invasion, in at least two diversity treatments across both gradients. For our purposes, we use the word ‘taxa’ to refer to a group of bacteria assigned to a particular OTU. RELATE, a non-parametric Mantel-type test, was used to examine the relationship between community composition and niche structure, both when (i) considering all 71 screened C sources and all detected taxa, and (ii) only community-exclusive C sources (i.e., sources that cannot be used by the E. coli strain) and those taxa that increased a minimum of 1% in at least two treatments across gradients. This was done by creating two Bray–Curtis dissimilarity matrices of measurements taken at days 0 and 28 for either community composition or niche structure, and these matrices were used in the RELATE test. Bray–Curtis dissimilarity values and the SIMPER tests were obtained and performed using the R package ‘vegan’ [28]. The PERMANOVA and RELATE tests were performed in PRIMER6+ (Clarke and Gorley 2006). All additional analyses were performed using base R functions [29].
Results
Total culturable bacteria and invader survival in soil microcosms
Before the invasion of E. coli, total culturable bacteria were statistically similar between both gradients and all treatments, reaching a mean of 8.0 log CFU/g soil that indicated dilution inocula similarly colonized the sterile soil matrices (Supplementary Fig. S2).
The invasion dynamics of E. coli along the B and W gradients were similar (Supplementary Fig. S3), as evidenced by a lack of interaction between the factors diversity gradient, time, and diversity treatment (ANOVA, F(12,80) = 0.575, P = 0.856). There were significant effects of time (ANOVA, F(4,80) = 259.98, P < 0.0001), diversity treatment (ANOVA, F(3,80) = 163.63, P < 0.0001), and their interaction (ANOVA, F(12,80) = 16.02, P < 0.0001), which was indicative of the consistent temporal decline of E. coli in all communities, the effect being amplified in the 10−3 and 10−1 treatments of both gradients. Upon and throughout the course of invasion, invader survival was highest in the sterile soil and was next highest in the 10−6 treatment (Tukey’s post hoc, P < 0.05). Survival was significantly lower in the 10−1 and 10−3 treatments (Tukey’s post hoc, P < 0.05), which had statistically similar levels of E. coli survival (Tukey’s post hoc, P > 0.05).
Invasion‘s effect on diversity within the community
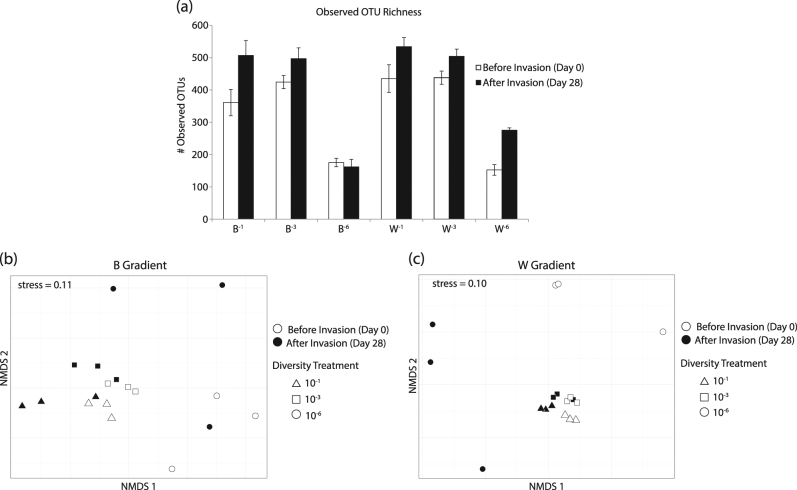
Contrary to our hypothesis, diversity did not decrease after invasion (Fig. 2a, Table 1). Using a two-way ANOVA analysis to test for differences between time (before and after invasion), diversity treatment, and their interaction in each gradient, the observed OTU richness significantly increased after invasion (Table 1). Moreover, OTU richness decreased upon the 10−6 dilution in both diversity gradients, confirming the effectiveness of the dilution-to-extinction approach. The lack of significance between the factors time and diversity treatment indicated that the increase of richness upon invasion was not associated to a particular diversity treatment; rather, the increase was a global effect across each gradient. A similar pattern was observed for Faith’s phylogenetic diversity (Supplementary Fig. S4a, Supplementary Table S1). Phylogenetic diversity increased upon invasion among all treatments in both gradients, and was significantly different between diversity treatments, decreasing as dilution increased. Evenness was not significantly altered by invasion, although evenness tended to increase following invasion (Supplementary Fig. S4b, Supplementary Table S1). Rank abundance curves were generated for each treatment from day 0 and day 28 and fit with a lognormal model, whose fitted parameters and shape indicated that across treatments evenness was either slightly higher or the same after invasion (Supplementary Fig. S5).
Fig. 2. OTU richness increases while community composition shifts upon invasion.
. (a) Observed OTU richness measured via 16S rRNA gene sequencing before (day 0) and after (day 28) invasion. Vertical bars represent the standard error of the mean (n = 3). Results of statistical tests are provided in Table 1. Comparison of community composition in the (b) B gradient and (c) W gradient before and after invasion across all treatments, as measured by examining their 16S rRNA gene structure with non-metric multidimensional scaling
Table 1.
Two-way ANOVA tests examining the effects of the factors time, diversity treatment, and their interaction on species richness
| B Gradient | ||||
|---|---|---|---|---|
| Source of variation | df | MS | F | P |
| OTU richness | ||||
| Time | 1 | 21,054 | 6.987 | 0.0214 |
| Diversity Treatment | 2 | 156,467 | 51.927 | <0.0001 |
| Day × Diversity Treatment | 2 | 9517 | 3.159 | 0.0791 |
| Error | 12 | 3013 | ||
| W Gradient | ||||
|---|---|---|---|---|
| Source of variation | df | MS | F | P |
| OTU richness | ||||
| Time | 1 | 41,530 | 21.618 | <0.001 |
| Diversity Treatment | 2 | 139,602 | 72.669 | <0.0001 |
| Day × Diversity Treatment | 2 | 1206 | 0.628 | 0.55039 |
| Error | 12 | 1921 | ||
Shifts in bacterial community composition and changes in the relative abundance of common bacterial taxa upon invasion
Upon invasion, changes in diversity were associated with significant shifts in bacterial community composition in both gradients (Fig. 2b, c; B gradient: PERMANOVAbefore vs. after, P = 0.01; W gradient: PERMANOVAbefore vs. after, P = 0.01). The Bray–Curtis percent dissimilarity between communities before and after invasion, which quantified the magnitude of shifts to community composition, indicated that the B−6 and W−6 communities after invasion were 70.2 and 73.9% dissimilar, respectively, to their counterparts before invasion (Supplementary Fig. S6). This was higher than the range of 31.3–47.3% observed for the 10−1 and 10−3 communities across both gradients after invasion.
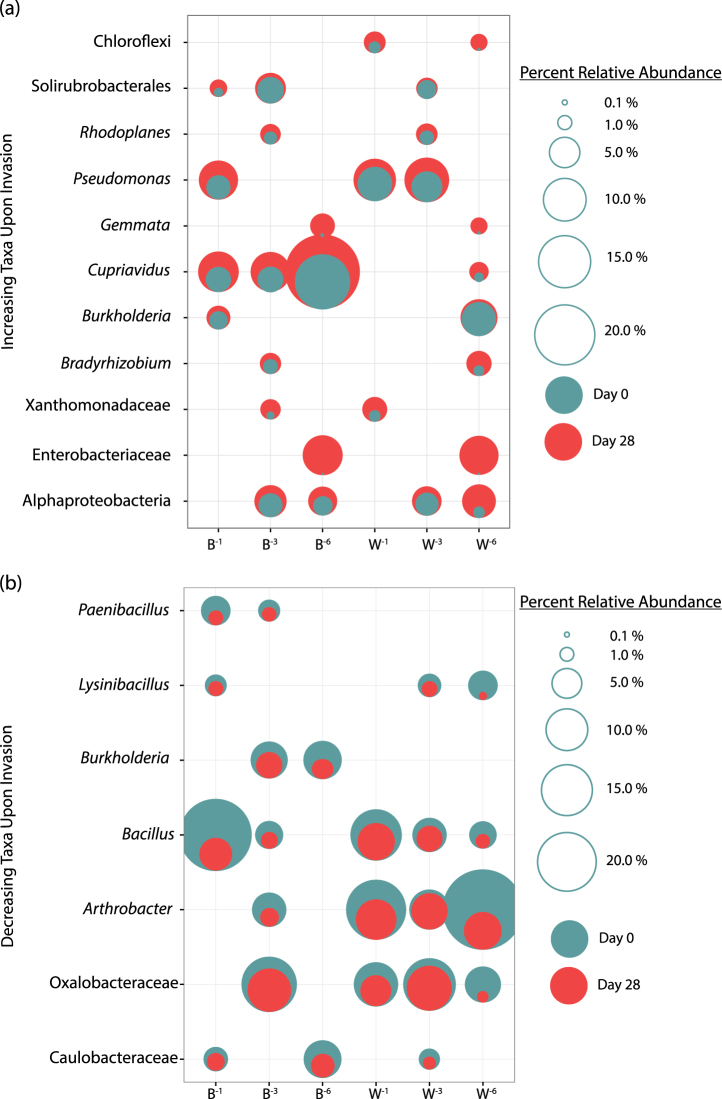
In order to determine the set of OTUs that drove the compositional shifts upon invasion, the SIMPER analysis identified eleven major taxa whose relative abundance consistently increased more than 1% upon invasion in at least two treatments of both gradients (Fig. 3a). OTUs related to Cupriavidus sp. and Alphaproteobacteria increased an average of 4.3 and 3.2-fold, respectively, in four treatments; whereas OTUs affiliated with Pseudomonas and Solirubrobacterales increased an average of 2.2 and 2.3-fold, respectively, in three treatments. The largest increases were related to OTUs classified as Gemmata and Chloroflexi species that increased an average of 129.8 and 107.0-fold, respectively, upon invasion in two of the six treatments. OTUs related to Xanthomonadaceae, Rhodoplanes sp., Burkholderia sp., Enterobacteriaceae, Bradyrhizobium sp., also displayed increases in two treatments, which ranged from 1.5 to 7.9-fold. There were also seven major taxa whose relative abundance consistently decreased a minimum of 1% in relative values in at least two diversity treatments across both gradients upon invasion (Fig. 3b). Among these taxa, an OTU assigned to Bacillus sp. decreased an average of 3.2-fold upon invasion in five of the six treatments. OTUs affiliated to the Oxalobacteraceae and to Arthrobacter sp. declined an average of 4.4 and 3.0-fold in four of six treatments upon invasion, respectively. OTUs assigned to Lysinibacillus sp. and Caulobacteraceae declined 7.8 and 2.6, respectively, in three treatments, and Paenibacillus sp. and Burkholderia sp. declined 3.5 and 2.7-fold, respectively, in two treatments.
Fig. 3. Consistent increases and decreases of bacterial taxa abundances upon invasion.
. (a) Increasing and (b) decreasing bacterial taxa abundances are indicated by overlaid bubble plots depicting a taxa’s relative percent abundance at day 0 (in green) or day 28 (in red). Bubbles are scaled by area to illustrate percent abundance. Each taxa name corresponds to an OTU and its closest related genus, order, or family
Increase in niche breadth of the community upon invasion
When examining the total array of 71 C sources, a two-way ANOVA analysis examining the factors time, diversity treatment, or their interaction indicated that communities after invasion had a significantly higher niche breadth than communities before invasion or the non-invaded controls (Fig. 4a; B gradient: ANOVA, F(2,18) = 56.08, P < 0.0001, Tukey’s post hoc, P < 0.05; W gradient ANOVA, F(2,18) = 76.08, P < 0.0001, Tukey’s post hoc, P < 0.05). A significant interaction between the factors time and diversity treatment further indicated that the effects of invasion were amplified in the 10−6 communities (B gradient: ANOVA, F(2,18) = 5.85, P < 0.01; W gradient ANOVA, F(2,18) = 16.79, P < 0.0001). These effects were not due to the mere presence of the invader, as the same patterns remained significant when the analysis was performed without C sources used by the ancestral invader (Fig. 4b). Across these 35 C sources, there were significant effects of time in both gradients (B gradient: ANOVA, F(2,18) = 56.69, P < 0.0001; W gradient, ANOVA, F(2,18) = 148.7, P < 0.0001). Although there was no significant interaction between time and diversity treatment in the B gradient (ANOVA, F(4,18) = 2.07, P = 0.13), there was a significant interaction of these factors in the W gradient (ANOVA, F(4,18) = 15.0, P < 0.0001).
Fig. 4. Community niche breadth increases upon invasion.
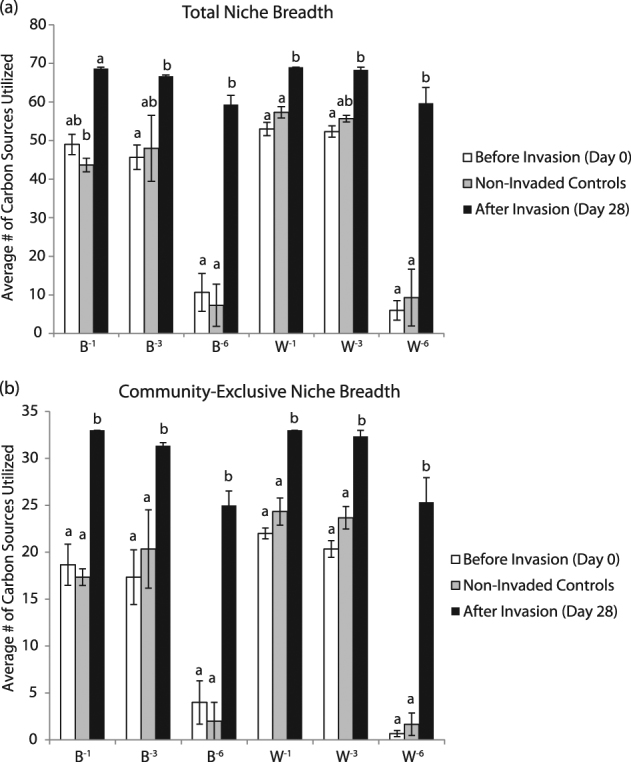
. (a) Total community niche breadth and (b) community-exclusive niche breadth before invasion, after invasion, and in the non-invaded controls. Bars represent standard error of the mean (n = 3), and letters depict significant differences among diversity treatments
Niche structure shifts upon invasion
The invasion of E. coli caused niche structure to shift in all communities (Fig. 5a), and the trajectory of these shifts were a function of both the direct impact of the invader and a response that was solely attributed to the native taxa. In a principal component analysis the non-invaded controls grouped similarly to the communities before invasion, indicating that incubation time throughout the experiment had no influence on the niche shifts. Potential shifts in niche structure caused by E. coli’s potential evolution during the experiment were also ruled out, as the metabolic activity of both the ancestral E. coli strain used for invasion at day 0 and E. coli cells re-isolated from all microcosms at day 28 grouped together, indicating the metabolic capabilities of E. coli remained conserved throughout the experiment.
When the response of each treatment was decomposed into vectors that quantified either the influence of the resident community or invader on niche shifts, the response of each community was to shift aside and away from the invader’s niche. As stated above, the projected vectors thus describe the movement of communities in resource space as a function of either the impact of the invader or response of the resident community: vectors that point toward E. coli quantify the impact of E. coli, and the vectors that point toward the invaded community quantify the contribution and response of the resident community. It was assumed that should an invaded community’s niche only shift toward the invader’s resource use profile (i.e., a shift exclusively corresponding to the vector µ2 in Fig. 1, which are those vectors that point towards the invader population in Fig. 5), this would likely indicate nothing more than a mixing effect between the resident community at day 0 and the invader. Alternatively, deviation from the line passing through the initial community niche and the invader niche would indicate a shift of the resident community’s niche structure aside and away from that of the invader, indicative of an escape away from the invader’s niche preferences. This approach of calculating the shifts to each treatment separately and then comparing shifts among communities in a common space allowed us to account for the uniqueness of each treatments’ initial niche structure. Although such an approach could suffer from the bias that each treatment’s trajectory in resource space is toward opposing directions (i.e., different resources), error estimates that quantified the amount of residual variation after accounting for shifts due to E. coli and the response of the resident community were small (Supplementary Table S2). This indicated that defining orthogonal axes based on the community’s movement toward or away from E. coli captured most of the variation in the data and were thus similar and comparable to principal components one and two of the entire data set.
Regression analyses that aimed to determine the relationship between observed taxonomic richness and either the contribution of the invader or resident community to shifts of niche structure were performed using the length of the aforementioned vectors for all replicates in each community. E. coli’s impact, measured as the component of niche shifts toward E. coli (i.e., the norm of vector μ2 in Fig. 1), strongly and significantly decreased as richness increased (Fig. 5b). On the other hand, the community’s response and contribution to shifts (the norm of vector μ3 in Fig. 1) significantly increased as richness increased (Fig. 5c).
Shifts in community composition explain the shifts in niche structure
The observed shifts in community niche structure were strongly and significantly correlated with changes in bacterial community structure upon invasion across both the W and B gradients (Table 2a). Furthermore, there were also significant correlations between only those OTUs that were found to increase a minimum of 1% in at least two treatments upon invasion and the portion of community-exclusive C sources that were newly used upon invasion (Table 2b).
Table 2.
The relationship between bacterial community composition and niche structure
| Rho | Sig. | |
|---|---|---|
| (a) | ||
| B | 0.825 | <0.01 |
| W | 0.715 | <0.01 |
| (b) | ||
| B | 0.638 | <0.01 |
| W | 0.338 | ≤0.05 |
(a) The correlation between the 16S rRNA gene sequences detected and all 71 C sources used before and after invasion; (b) The correlation between those taxa found to increase at least 1% in at least two of six diversity treatments upon invasion (see Fig. 2a) and the community-exclusive carbon sources upon invasion. RELATE tests were used to calculate the strength of the correlation, Rho, using Spearman’s correlation
Discussion
Microbial invasions are usually an anomaly within the conventional invasion framework. In a typical invasion scenario, as studied in plant communities, invaders start from a relatively small population size and, if they succeed and overcome the pressures of biotic resistance and gain access to available resources, expand their population size whilst displacing members of the resident community [30]. In contrast, either natural or anthropogenic driven microbial invasions may start with an initially large population size. For instance, a release of human feces into soil might discharge the remarkable amount of 109 E. coli cells per gram feces as potential invaders [31]. Despite the initial large density, the introduction of a foreign, invading microbe into a new environment or community is often ephemeral and unsuccessful, meaning that the invader does not present positive net-growth nor spread and naturalize itself into the resident community [16, 18, 19, 22, 24]. Nonetheless, given the ubiquity of microbial invasions across disparate systems and their often transient nature, recent work has called into question whether large invading populations can leave a footprint that impacts the resident community, even if their elimination is imminent [7]. In this study, despite the witnessed trends indicative of invader death and defeat, we demonstrated that the invasion of E. coli caused changes in the diversity, composition, niche breadth, and niche structure of invaded communities. Never before has such an effect been examined in microbial communities facing invasions.
The changes to community composition of all tested communities were rooted in the increases and decreases of several common taxa across treatments. Eleven taxa whose initial relative abundance in the community comprised a range of 7.0 to 17.7% were consistently found to increase upon invasion (reaching 14.3 to 45.0%), which suggests that invasion somehow facilitated their population’s expansion. Also, seven taxa that initially comprised a dominant portion of the community, their total relative abundance before invasion ranging between 15.7 to 52.2%, exhibited large declines upon the introduction of E. coli, declining to abundances that ranged from 5.2 to 23.6%. These declines suggest similar niche preferences with E. coli that, in part due to a high initial population size, was competitively superior at the onset of invasion. In striking opposition to part of our original hypothesis, the net result of the shifts in community composition translated to increases in the diversity metrics of observed OTU richness and phylogenetic diversity, while community evenness was slightly but not significantly altered. A positive relationship between diversity and invasion has been observed for plant systems, but these increases are mostly attributed to the integration of the invader into the resident community or the facilitation of more invasions that collectively increase species richness [32]. In our system, however, the enhanced diversity was not driven by species richness or phylogenetic diversity because, besides the invader, there was no input or recruitment from an external pool of species in the controlled experimental system. Rather, slight but non-significant changes observed in community evenness might have caused the increased detection of taxa at similar sequencing depths across microcosms.
The new community compositions after invasion lead to shifts of niche structure and an increased niche breadth across all diversity treatments. The liaison between community composition and niche structure, as supported by their significant and positive relationships in RELATE tests, is likely a product of resource competition between the invader and resident taxa. This competitive scenario led to changes in community structure, which in turn likely altered niche breadth and niche structure. It is important to highlight that changes to niche breadth were not due to the mere addition of the invader or the potential evolution of its metabolic capacity. While the total niche breadth did increase, 43 to 75% of all newly degraded carbon sources were community-exclusive (i.e., carbon sources that were metabolically inaccessible to the invader). Along that line, while the invader’s impact did strongly influence niche structure, especially if diversity was compromised, the community had its own response and contribution to niche shifts, which was to escape away from the invader’s niche, thereby shifting to use resources that were not accessible to E. coli. This effect was stronger as richness of the community increased. In taking the composition and niche data together, a conceptual and potential mechanistic explanation of the observed impacts is revealed (Fig. 6): upon the initial introduction of E. coli, the taxa that rely heavily on C substrates used by the invader are outcompeted. This, in turn, triggers a competitive release where rare or subordinate taxa could better explore or utilize niches not exploited by E. coli (Fig. 6). In effect, it appears that E. coli has rearranged the structure of the soil microbial community by engaging in resource competition for its preferred substrates, thereby altering the community composition and associated niche differentiation pattern of the resident community via competitive release. Such a mechanism is supported by the fact that the emergence of newly used carbon sources was correlated to the abundance of the main taxa that increased upon invasion. A similar phenomenon has also been reported when the invasion of Limnohabitans planktonicus reduced the abundance of dominant Aeromonas sp., Hydrophila sp., and Brevundimonas sp., resulting in increases of the previously rare Arthrobacter agilis and Flavobacterium sp. [33]. Furthermore, the presence of Pseudomonas fluorescens SBW25 in soil has been shown to reduce the abundance of resident bacteria that favor the same resources, an effect that was stronger if P. fluorescens was allowed to previously adapt to the soil [34]. A more detailed identification and examination of the microbial functional traits [35] of taxa whose abundances increase or decrease could shed further light onto the unique species–species interactions that fuel such a competitive release.
Fig. 6. A conceptual model for understanding an invasion’s impacts and the creation of a legacy.
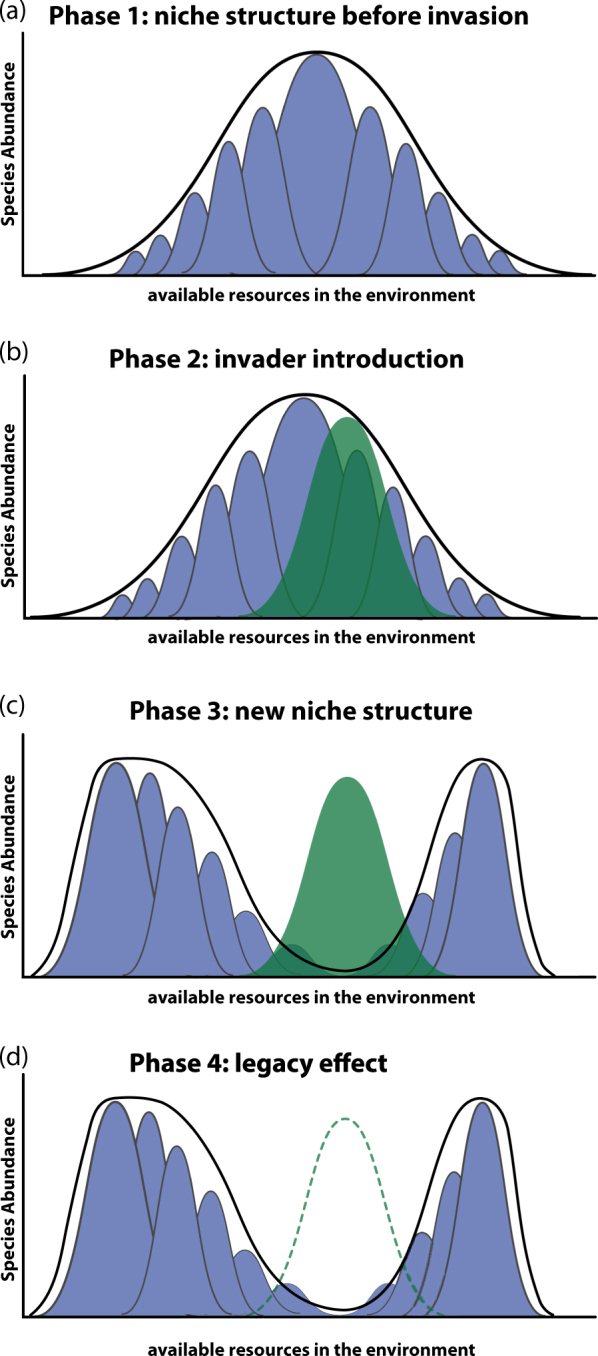
. Consider a community of eleven taxa, each taxa’s population dominant on one of eleven resources, as shown in (a) where the height of each peak represents the abundance of that taxa in the community. The thick black line indicates the entire niche of the native community. (b) When an invader (green peak) is introduced into the community, a zone of competition is created as the invader and resident taxa compete for resources of similar preference. (c) Due to its initial high population size, the invader will manage to outcompete resident taxa in this zone of competition, and alter the niche structure in such a way that residents escape to occupy niches on which the invader has little or no competitive advantage. (d) Even if the invader is eventually eliminated from the community (dashed green line), perhaps due to abiotic, predatory, or antagonistic effects, it will have left a legacy to the community’s niche structure that could influence future invasions of the same or similar invaders
The extent of invasion impacts on soil microbial community across both B and W gradients scaled with community diversity. Just as invasion resistance is weak when microbial diversity is compromised [16, 18], the impacts of invasion were most severe for the less diverse (B−6 and W−6) communities, with larger shifts to community composition and niche structure, as well as larger increases to niche breadth. Thus, the phenomenon of impacts fits squarely within the paradigm of the diversity-invasion effect, where the lower capacity of less diverse communities to exploit available resources decreases their resistance to invasion and, as we show, permits impacts of a greater magnitude. Along these lines, it is important to note that the death of E. coli after 28 days may have influenced some of the alterations to niche breadth and structure. However, considering that the largest impacts of invasion were witnessed in the 10−6 communities, where E. coli survival was at near inoculation abundances, it is likely that this was not an overarching factor in the observed shifts to community niche breadth and structure upon invasion. Furthermore, the death of a transient invader, such as E. coli is a normal phenomenon of many microbial invasions and must be included within the interpretation of the results. In that respect, it is worth noting that the impact of invasion may be dependent on the initial invader density, and although this study did not assess this issue, one can speculate that both the strength of the competition and the invader’s survival time will be greater in response to larger cell densities.
Evoking resource competition, specifically the competition for occupied niches, as the mechanism that drove impacts also implies that E. coli was active and grew in the soil. A myriad of studies have examined E. coli’s survival in soil [18, 26, 36, 37] and, in the absence of a resource pulse or completely sterile environment, only rarely has E. coli growth been detected [24]. Thus, any growth and competition that caused impacts was likely masked behind the cloak of E. coli’s progressive decline and not enough to detect amongst the greater death of the population. However, given that we observed impacts upon invasion, our results indicate that E. coli appears to maintain some semblance of an active lifestyle when invading secondary, external environments like soil. It has recently been shown that different genotypes of E. coli survive longer in soil depending on their niche preferences [38], and across the E. coli species complex there is a large variation in the ratio between stress response and nutrient uptake genes [39]. For the O157:H7 and other forms, future experiments that aim to quantify such genes and their expression in both primary (gut) and external (soil or water) environments could reveal evolved forms that truly exemplify a biphasic lifestyle, capable of growing in both habitats.
In conclusion, our data show that transient and unsuccessful invasions can impact native communities, but the observed impacts have a paradoxical nature. On one hand, invasion-induced increases to diversity and niche breadth may increase invasion resistance of this ecosystem according to the diversity-invasion paradigm. On the other hand, and in accordance with our proposed model of invasion impacts (Fig. 6), the shift of niche structure that steered the community away from resources usable by the invader suggests that a second invasion by the same invader, here E. coli, can last longer or even result in a bona fide successful invasion where the invader crosses the barrier of biotic resistance and is naturalized into the community. It is thus conceivable that the overall invasion resistance of the resident community has increased, except if that invader was E. coli or an invader using similar resources to E. coli. In fact, in practical applications the effectiveness of both biofertilizers [40, 41] and probiotic agents [42] has been shown to hinge on multiple inoculation events and suggests that the legacy of impacts does facilitate future invasion attempts. The work at hand is now to test this hypothesis in order to find support that the mechanisms of resource competition and competitive release provide a window of opportunity for a secondary invasion by a similar invader and, if so, for how long.
Electronic supplementary material
Acknowledgments
We thank Irene Marring, Nadine Guillaumaud, Denis Warshan, and Giovanni Laudanno for their help in the laboratory and with data analyses. Funding was provided by Ecologie Microbienne Lyon (UMR INRA 1418, UMR CNRS 5557) and personal grant to JFS from the University of Groningen. G.S.v.D. was supported by Starting Independent Researcher Grant #309555 of the European Research Council and Vidi Grant 864.11.012 of the Netherlands Organization for Scientific Research (NWO).
Compliance with ethical standards
Conflict of interests
The authors declare that they have no competing interests.
Footnotes
Le Roux, X., Poly, F. and Salles, J.F. Shared senior authorship.
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0003-y) contains supplementary material, which is available to authorized users.
References
- 1.Mack MC, D’Antonio CM. Impacts of biological invasions on disturbance regimes. Trends Ecol Evol. 1998;13:195–8. doi: 10.1016/S0169-5347(97)01286-X. [DOI] [PubMed] [Google Scholar]
- 2.Stuart YE, Campbell TS, Hohenlohe PA, Reynolds RG, Revell LJ, Losos JB. Rapid evolution of a native species following invasion by a congener. Science. 2014;346:463–6. doi: 10.1126/science.1257008. [DOI] [PubMed] [Google Scholar]
- 3.Kumschick S, Gaertner M, Vilà M, Essl F, Jeschke JM, Pyšek P, et al. Ecological impacts of alien species: quantification, scope, caveats, and recommendations. Bioscience. 2014;65:55–63. doi: 10.1093/biosci/biu193. [DOI] [Google Scholar]
- 4.Bradley BA, Blumenthal DM, Wilcove DS, Ziska LH. Predicting plant invasions in an era of global change. Trends Ecol Evol. 2010;25:310–8. doi: 10.1016/j.tree.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 5.Elton C. Ecology of Invasions by Animals and Plants. US: Springer; 1958. [Google Scholar]
- 6.Tilman D. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc Natl Acad Sci USA. 2004;101:10854–61. doi: 10.1073/pnas.0403458101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mallon CA, van Elsas JD, Salles JF. Microbial invasions: the process, patterns, and mechanisms. Trends Microbiol. 2015;23:719. doi: 10.1016/j.tim.2015.07.013. [DOI] [PubMed] [Google Scholar]
- 8.Wei Z, Yang T, Friman V, Xu Y, Shen Q, Jousset A. Trophic network architecture of root-associated bacterial communities determines pathogen invasion and plant health. Nat Commun. 2015;6:8413. doi: 10.1038/ncomms9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flory SL, Clay K. Non-native grass invasion alters native plant composition in experimental communities. Biol Invasions. 2010;12:1285–94. doi: 10.1007/s10530-009-9546-9. [DOI] [Google Scholar]
- 10.Hejda M, Pysek P, Jarosik V. Impact of invasive plants on the species richness, diversity and composition of invaded communities. J Ecol. 2009;97:393–403. doi: 10.1111/j.1365-2745.2009.01480.x. [DOI] [Google Scholar]
- 11.Maron JL, Marler M. Effects of native species diversity and resource additions on invader impact. Am Nat. 2008;172:S18–S33. doi: 10.1086/588303. [DOI] [PubMed] [Google Scholar]
- 12.Zavaleta E, Hulvey K. Realistic species losses disproportionately reduce grassland resistance to biological invaders. Science. 2004;306:1175–7. doi: 10.1126/science.1102643. [DOI] [PubMed] [Google Scholar]
- 13.Melgoza G, Nowak RS, Tausch RJ. Soil water exploitation after fire: competition between Bromus tectorum (cheatgrass) and two native species. Oecologia. 1990;83:7–13. doi: 10.1007/BF00324626. [DOI] [PubMed] [Google Scholar]
- 14.D’Antonio CM, Mahall BE. Root profiles and competition between the invasive, exotic perennial, Carpobrotus edulis, and two native shrub species in California coastal scrub. Am J Bot. 1991;78:885–94. doi: 10.1002/j.1537-2197.1991.tb14492.x. [DOI] [Google Scholar]
- 15.Jousset A, Schulz W, Scheu S, Eisenhauer N. Intraspecific genotypic richness and relatedness predict the invasibility of microbial communities. Isme J. 2011;5:1108–14. doi: 10.1038/ismej.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mallon CA, Poly F, Le Roux X, Marring I, van Elsas JD, Salles JF. Resource pulses can alleviate the biodiversity-invasion relationship in soil microbial communities. Ecology. 2015;96:915. doi: 10.1890/14-1001.1. [DOI] [PubMed] [Google Scholar]
- 17.Stecher B, Chaffron S, Kaeppeli R, Hapfelmeier S, Freedrich S, Weber TC, et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog. 2010;6:e1000711. doi: 10.1371/journal.ppat.1000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Elsas JD, Chiurazzi M, Mallon CA, Elhottova D, Kristufek V, Salles JF. Microbial diversity determines the invasion of soil by a bacterial pathogen. Proc Natl Acad Sci USA. 2012;109:1159–64. doi: 10.1073/pnas.1109326109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vivant A, Garmyn D, Maron P, Nowak V, Piveteau P. Microbial diversity and structure are drivers of the biological barrier effect against listeria monocytogenes in soil. PLoS ONE. 2013;8:e76911. doi: 10.1371/journal.pone.0076991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Roy K, Marzorati M, Negroni A, Thas O, Balloi A, Fava F, et al. Environmental conditions and community evenness determine the outcome of biological invasion. Nat Commun. 2013;4:1383. doi: 10.1038/ncomms2392. [DOI] [PubMed] [Google Scholar]
- 21.Eisenhauer N, Schulz W, Scheu S, Jousset A. Niche dimensionality links biodiversity and invasibility of microbial communities. Funct Ecol. 2013;27:282–8. doi: 10.1111/j.1365-2435.2012.02060.x. [DOI] [Google Scholar]
- 22.Saison C, Degrange V, Oliver R, Millard P, Commeaux C, Montange D, et al. Alteration and resilience of the soil microbial community following compost amendment: effects of compost level and compost‐borne microbial community. Environ Microbiol. 2006;8:247–57. doi: 10.1111/j.1462-2920.2005.00892.x. [DOI] [PubMed] [Google Scholar]
- 23.Savageau MA. Escherichia-Coli habitats, cell-types, and molecular mechanisms of gene-control. Am Nat. 1983;122:732–44. doi: 10.1086/284168. [DOI] [Google Scholar]
- 24.Ishii S, Ksoll WB, Hicks RE, Sadowsky MJ. Presence and growth of naturalized Escherichia coli in temperate soils from lake superior watersheds. Appl Environ Microbiol. 2006;72:612–21. doi: 10.1128/AEM.72.1.612-621.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franz E, Semenov AV, Termorshuizen AJ, De Vos O, Bokhorst JG, Van Bruggen AH. Manure‐amended soil characteristics affecting the survival of E. coli O157: H7 in 36 Dutch soils. Environ Microbiol. 2008;10:313–27. doi: 10.1111/j.1462-2920.2007.01453.x. [DOI] [PubMed] [Google Scholar]
- 26.van Elsas JD, Hill P, Chronakova A, Grekova M, Topalova Y, Elhottova D, et al. Survival of genetically marked Escherichia coli O157: H7 in soil as affected by soil microbial community shifts. Isme J. 2007;1:204–14. doi: 10.1038/ismej.2007.21. [DOI] [PubMed] [Google Scholar]
- 27.Fukushima H, Seki R. High numbers of Shiga toxin-producing Escherichia coli found in bovine faeces collected at slaughter in Japan. FEMS Microbiol Lett. 2004;238:189–97. doi: 10.1016/j.femsle.2004.07.035. [DOI] [PubMed] [Google Scholar]
- 28.Oksanen J, Kindt R, Legendre P, O’Hara B, Stevens MHH, Oksanen MJ, et al. (2007). The vegan package. Community ecology package.
- 29.Team RC. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2014. [Google Scholar]
- 30.Levine J, Adler P, Yelenik S. A meta-analysis of biotic resistance to exotic plant invasions. Ecol Lett. 2004;7:975–89. doi: 10.1111/j.1461-0248.2004.00657.x. [DOI] [Google Scholar]
- 31.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–17. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 32.Theoharides KA, Dukes JS. Plant invasion across space and time: factors affecting nonindigenous species success during four stages of invasion. New Phytol. 2007;176:256–73. doi: 10.1111/j.1469-8137.2007.02207.x. [DOI] [PubMed] [Google Scholar]
- 33.Hornak K, Corno G. Every coin has a back side: invasion by limnohabitans planktonicus promotes the maintenance of species diversity in bacterial communities. Plos ONE. 2012;7:e51576. doi: 10.1371/journal.pone.0051576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gómez P, Paterson S, De Meester L, Liu X, Lenzi L, Sharma M, et al. Local adaptation of a bacterium is as important as its presence in structuring a natural microbial community. Nat Commun. 2016;7. [DOI] [PMC free article] [PubMed]
- 35.Krause S, Le Roux X, Niklaus PA, Van Bodegom PM, Lennon JT, Bertilsson S, et al. Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol. 2014;5:251. doi: 10.3389/fmicb.2014.00251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Semenov AV, Franz E, van Overbeek L, Termorshuizen AJ, van Bruggen AHC. Estimating the stability of Escherichia coli O157: H7 survival in manure-amended soils with different management histories. Environ Microbiol. 2008;10:1450–9. doi: 10.1111/j.1462-2920.2007.01558.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang H, Ibekwe AM, Ma J, Wu L, Lou J, Wu Z, et al. A glimpse of Escherichia coli O157:H7 survival in soils from eastern China. Sci Total Environ. 2014;476:49–56. doi: 10.1016/j.scitotenv.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 38.Ma C, Liu M, Wang H, Chen C, Fan W, Griffiths B, et al. Resource utilization capability of bacteria predicts their invasion potential in soil. Soil Biol Biochem. 2015;81:287–90. doi: 10.1016/j.soilbio.2014.11.025. [DOI] [Google Scholar]
- 39.Ferenci T. Maintaining a healthy SPANC balance through regulatory and mutational adaptation. Mol Microbiol. 2005;57:1–8. doi: 10.1111/j.1365-2958.2005.04649.x. [DOI] [PubMed] [Google Scholar]
- 40.Alves BJ, Boddey RM, Urquiaga S. The success of BNF in soybean in Brazil. Plant Soil. 2003;252:1–9. doi: 10.1023/A:1024191913296. [DOI] [Google Scholar]
- 41.Chanway C, Radley R, Holl F. Inoculation of conifer seed with plant growth promoting Bacillus strains causes increased seedling emergence and biomass. Soil Biol Biochem. 1991;23:575–80. doi: 10.1016/0038-0717(91)90115-Z. [DOI] [Google Scholar]
- 42.Filho-Lima J, Vieira E, Nicoli J. Antagonistic effect of Lactobacillus acidophilus, Saccharomyces boulardii and Escherichia coli combinations against experimental infections with Shigella flexneri and Salmonella enteritidis subsp. typhimurium in gnotobiotic mice. J Appl Microbiol. 2000;88:365–70. doi: 10.1046/j.1365-2672.2000.00973.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.