Abstract
Forest management practices often severely affect forest ecosystem functioning. Tree removal by clearcutting is one such practice, producing severe impacts due to the total reduction of primary productivity. Here, we assessed changes to fungal community structure and decomposition activity in the soil, roots and rhizosphere of a Picea abies stand for a 2-year period following clearcutting compared to data from before tree harvest. We found that the termination of photosynthate flow through tree roots into soil is associated with profound changes in soil, both in decomposition processes and fungal community composition. The rhizosphere, representing an active compartment of high enzyme activity and high fungal biomass in the living stand, ceases to exist and starts to resemble bulk soil. Decomposing roots appear to separate from bulk soil and develop into hotspots of decomposition and important fungal biomass pools. We found no support for the involvement of ectomycorrhizal fungi in the decomposition of roots, but we found some evidence that root endophytic fungi may have an important role in the early stages of this process. In soil, activity of extracellular enzymes also decreased in the long term following the end of rhizodeposition by tree roots.
Introduction
Forest soils represent globally important organic carbon (C) pools, which act as C sinks during steady-state functioning [1]. Soil C increases through the accumulation of dead wood and litter on the soil surface and root litter in soil, as well as during translocation of current photosynthates below ground by trees. The latter part of C input is considerable, representing between 25 and 63% of gross primary production [2]. In addition to being temporarily immobilized in fungal mycelia [3], allocation of plant C to roots may also stimulate the turnover of soil organic C, initiating the decomposition of complex organic compounds; this is referred to as the ‘priming effect’ [4, 5]. Furthermore, the rhizodeposited C serves as a biomass source for soil fungi and bacteria. In coniferous forest stands, labile C from trees was shown to be the major source of C for fungal communities and microbial respiration [6, 7].
Forests are often subject to disturbances such as wildfires, insect outbreaks or biomass harvest, which have multiple effects on ecosystem functioning [1, 8]. The cessation of photosynthate flow from trees leads to a complex response that affects multiple habitats within soils. In addition to a general decrease in activity, soil microorganisms become increasingly dependent on more recalcitrant C sources [7]. Ectomycorrhizal fungi (EcMF) in particular are significantly affected by the termination of tree photosynthesis, and due to their important roles in C cycling in ecosystems, they likely mediate the disturbance-induced effects on soil. All their primary roles are substantially affected, including the trafficking of plant C through their mycelia into soil, contribution to the formation of soil organic matter (SOM) and direct or indirect effects on SOM decomposition [9].
The estimates of EcM fungal production and standing biomass indicate high contribution to the microbial biomass, especially in temperate and boreal coniferous forests [1]. Total standing EcM fungal biomass in a Picea abies forest of southern Sweden was estimated at 9.6 t ha−1, which represents approximately one-half of total microbial biomass [10, 11]. When trees are removed, EcMF rapidly lose their source of C and react to this situation. Although coniferous forest soils are full of organic C, it is available in the form of complex compounds that must be decomposed. Several recent studies have investigated whether and under which circumstances decomposition and saprotrophy can be triggered in EcMF and assessed its ecological significance [9, 12–14]. These authors hypothesized that saprotrophic C acquisition by EcMF may be an alternative strategy: (1) during periods with low photosynthate supply, (2) during periods of high photosynthate supply, but when a supplementary resource for massive mycelial production is required, or (3) during decomposition of dying tree roots [14]. While recent views consider it unlikely that EcMF can obtain C from SOM [15], the question whether decomposition of dead roots provides C for EcMF remains unresolved. For a limited time, dead roots may represent a resource that allows the EcMF to disperse to another living roots or generate spores that allow them to survive unfavourable conditions. Due to their intimate symbiosis with fine roots, they may potentially benefit from priority in this substrate and utilize the dead root necromass.
In contrast, EcMF in the soil lose competitive potential after disruption of root carbon transport [16]. It has been proposed that saprotrophic fungi and EcMF have overlapping fundamental niches, but that antagonistic EcMF exclude more efficient saprotrophic decomposers from the deeper organic layers [17] and that competition between these guilds might reduce decomposition rates [18–20]. On the other hand, EcMF may 'prime' decomposition by transferring plant C into bulk soil through hyphal exudation, production of organic acids or simply due to biomass turnover [9, 21]. Similarly, EcMF may decompose soil C as a consequence of mining soil for organic nutrient [14]. These effects would be theoretically relieved after tree removal.
According to a recent metastudy, clearcut harvesting decreases microbial biomass by 14–33% and fungal biomass by 20–40%, having a lesser impact than wildfire but a greater impact than insect outbreaks [22]. Negative short-term effects of clearcutting on ectomycorrhizal fungal biomass and diversity have been found [23, 24], and even in young regenerating stands, the ratio of saprotrophic to EcMF remains high [25]. The effects of tree harvesting on soil microbial communities are most rapid in the initial phases, but long-lasting, recognizable effects can persist from 12 up to 50 years after harvest [25, 26].
While the flow of photosynthates into soil ceases after clearcutting, forest stands are subjected to one-time input of root litter. The standing mass of roots is stand-specific, but may be very high. For example, the root systems in a Pinus taeda plantation contained approximately 32% (17.2 Mg ha−1) of the soil organic carbon [27]. Dead roots appear to contain a substantial amount of labile C, as apparent from the observation that approximately 35% of standing root mass was lost during 1 year after clearcutting of a spruce–fir stand and it is also supported by the observation that in the first season after the clearcut, respiration was 16% higher than in the uncut stand [28]. Compared to the fate of the aboveground litter, the decomposition of dead tree roots has so far received less attention [29, 30]. The mass of EcM fungal mycelia was found to be in the same range as that of fine roots [31], and EcM mycelia thus represent another important labile resource [21]. High respiration rates observed after clearcutting may be due to rapid decomposition of these mycelia.
The aim of this work was to monitor the response of the fungal community after clearcutting and to assess the associated consequences for SOM decomposition. A P. abies stand was subjected to clearcutting, after which fungal biomass, community composition and enzyme activity were assessed and compared to data from before clearcutting. We hypothesized that (1) the utilization of complex compounds in soil should be accompanied by an increase in enzyme activity, and (2) tree removal will be followed by shift in fungal community composition towards saprotrophic fungi. We expected that (3) decomposing fine roots would be partly subject to colonization by soil fungi and (4) partly subjected to temporary utilization by EcMF with saprotrophic capability.
Materials and methods
Study area, sample collection and characterization
Samples were collected in the Training Forest Enterprise Masaryk Forest Křtiny of Mendel University in Brno, a managed mixed temperate forest located north of Brno, Czech Republic (latitude 16°45′E, longitude 49°19′N, 475 m a.s.l.) in a 1-ha 80-year-old spruce (P. abies) stand. In June 2013, all trees in the stand were cut. All aboveground biomass of trees including small branches was removed, while the stumps and root systems in soil were left as well as the forest floor, according to the common management practice in Central European production forests (see Supplementary Figure 1). The clearcut area was not planted with new trees until the end of the experiment. Sampling was performed in May 2013 (before clearcutting, month 0), in July, August, October 2013, January, March, June and December 2014 and in June 2015 (months 2, 3, 5, 8, 10, 13, 19 and 25) to cover a 24-month period after clearcutting (see Supplementary Figure 2 for climatic data, obtained from the nearest weather station—latitude 16°41′E, longitude 49°09′N, 241 m a.s.l.).
During the first sampling, 10 trees located >15 m from the stand edge were labelled. Sample collection was performed by taking five soil cores (4.5 cm diameter, 10 cm depth) around each labelled tree/tree stump at evenly spaced points between 1.5 and 2 m from each tree. The soil core material was separated into three compartments: bulk soil until 10 cm depth (not attached to the P. abies roots), P. abies fine roots (<2 mm diameter) and rhizosphere soil (soil attached to P. abies fine roots after their separation from the soil matrix). Samples from five soil cores around a single tree were combined to represent one sample (10 samples per sample type, per sampling time). Soil and rhizosphere soil material was passed through 5-mm sterile mesh. Samples were freeze-dried and stored at −80 °C.
Water content was measured based on the difference between sample fresh mass and mass after freeze-drying, and pH was measured in distilled water (1:10). C and N content was measured using an elemental analyser. Total ergosterol was extracted using 10% KOH in methanol and analysed by high-performance liquid chromatography [32]. Enzyme assays were performed using soil and root homogenates [33].
DNA extraction, sequencing of PCR amplicons and processing of sequence data
Total genomic DNA was extracted from 250 mg of bulk or rhizosphere soil material using a modified Miller method [34]. Root samples were homogenized using a mortar and pestle under liquid nitrogen and DNA was extracted from 150 mg of root tissue using DNeasy Plant Mini Kit (Qiagen, Germany). All DNA extractions were performed in triplicate, and extracts were combined into one sample and polymerase chain reaction (PCR) amplified in triplicate to reduce PCR bias. PCR amplification of the fungal ITS2 region from DNA was performed using barcoded gITS7 and ITS4 primers [35], as previously described [36]. Sequencing was performed using 250-bp paired-end chemistry on an Illumina MiSeq system.
Sequence data were processed using SEED 2.0 [37] as described previously [36]. After joining pair-end reads, ITS2 extraction and chimera removal, sequences were clustered to Operational Taxonomic Units (OTUs) using UPARSE [38] at a 97% similarity level. Consensus sequences were constructed for each OTU, and the closest hits were identified using UNITE [39]; nonfungal sequences were discarded. Sequence data were deposited in MG-RAST (http://metagenomics.anl.gov/, data set number mgs616649). The relative abundance data are based on sequence abundances and should be taken as proxies of taxon relative abundances only with caution [40]. To assign putative ecological functions to the fungal OTUs, fungal taxa were classified into ecophysiological categories following [41].
Statistical analyses
Past 2.17c (http://folk.uio.no/ohammer/past/) and R v. 3.2.3 [42] were used for statistical analyses. In all cases, differences at P < 0.05 were considered statistically significant. Differences in soil variables and enzyme activities were tested using a Mann–Whitney U-test. Prior to subsequent analyses, fungal communities were standardized using Hellinger transformation. Bray–Curtis dissimilarity was used to construct a fungal community dissimilarity matrix. Because of the repeated sampling of the same sampling sites during the experiment, we took into account the effect of spatial distance by reducing the spatial distance matrix of sampling sites into spatial Principal Coordinates of Neighbour Matrices (PCNM). PCNM vectors were subsequently used as a proxy for spatial autocorrelation in permutational multivariate analysis of variance (PERMANOVA) analysis. PERMANOVA using the adonis function of the ‘vegan’ package in R was used to address the relative importance of time, compartments and water content for fungal community composition. Significance of the environmental characteristics was always tested after accounting for spatial autocorrelation. Pairwise PERMANOVA with 99,999 was performed for fungal communities and fungal ecophysiological cathegories between timepoints and between compartments. Bonferroni correction was used to calculate the corrected P value determined by the pairwise PERMANOVA analysis. Hellinger-transformed fungal OTU and fungal genera communities were used for NMDS analyses using the ‘ecodist’ package in R. Confidence ellipses (95% confidence) were calculated using the ordiellipse function in the ‘vegan’ package. OTU richness was calculated for a data set containing 1000 randomly selected sequences from each sample.
Results
Soil properties, enzyme activity and fungal biomass
P. abies fine root mass in the 10 cm layer of soil was 362 ± 36 g m−2 before clearcutting. It decreased after clearcutting to 299 ± 43 g m−2 in month 8 and to 190 ± 22 g m−2 in month 25, representing 83 and 53% of the initial root mass, respectively. Water content in roots remained high throughout the experiment compared to the soil and rhizosphere, with 77–87% compared to the typical 30–41%. Water content in the rhizosphere was significantly higher than soil until month 3, and beyond that was similar. Soil and rhizosphere pH values were approximately 4.2. Neither water content nor pH showed significant change over time. C and N content did not differ between the soil and rhizosphere and were stable at approximately 5.7 and 0.40%, respectively, with a C/N ratio of 15. The values in roots of the living stand were higher, with 41% C, 0.97% N and a C/N ratio of 43. After clearcutting, the composition of P. abies roots remained the same, with a N content of 0.84–1.08% and a C/N ratio between 37 and 51.
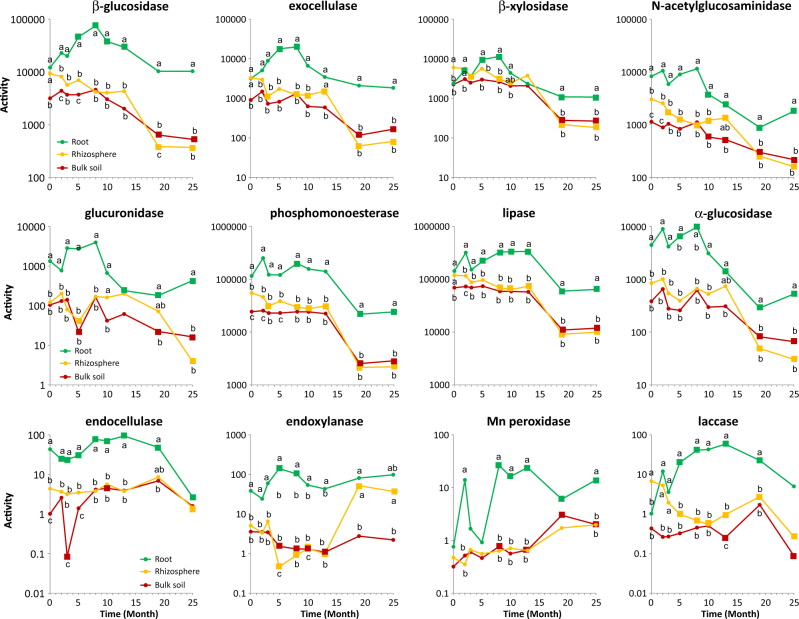
In the P. abies stand, activity of all enzymes was lowest in the bulk soil. Compared to soil, activity of all enzymes except glucuronidase, α-glucosidase, endoxylanase and Mn peroxidase was significantly higher in the rhizosphere. Laccase activity in rhizosphere was 16-fold higher than in bulk soil, while endocellulase, exocellulase, β-glucosidase, β-xylosidase and N-acetylglucosaminidase activity were three-fold to four-fold higher. In the root compartment, the activity of most enzymes was either comparable to or higher than in the rhizosphere (Fig. 1). The activity of endocellulase, endoxylanase and glucuronidase in the root compartment was 8- to 11-fold higher than in the rhizosphere, and α-glucosidase and N-acetylglucosaminidase activity were 5-fold and 3-fold higher, respectively.
Fig. 1.
Activity of extracellular enzymes (µmol min−1 g−1 dry mass) in the soil, rhizosphere and roots of a Picea abies stand before (month 0) and after clearcutting. Values represent means (n = 10) of the enzyme activities. Different letters indicate significant differences in activity between soil, rhizosphere and roots, and squares indicate significantly different abundances compared to the P. abies stand in month 0
After clearcutting, the activity of most enzymes in the soil remained stable for some time, but decreased significantly toward the end of the experiment. Enzyme activity in the rhizosphere decreased faster and reached the same values as in bulk soil at some point during the experiment. In the root compartment, activities of all enzymes except N-acetylglucosaminidase and glucuronidase showed a significant increase in activity (especially laccase, Mn peroxidase, β-glucosidase and exocellulase) for some period after clearcutting but often decreased later (Fig. 1).
Fungal biomass in the P. abies stand significantly differed among the root compartment (113 ± 21 μg ergosterol g−1 soil dry mass), rhizosphere (21 ± 3 μg g−1) and bulk soil (8.8 ± 1.7 μg g−1), and the same pattern was observed 2 months later (Fig. 2). Fungal biomass content in the rhizosphere later decreased and did not significantly differ from bulk soil. Fungal biomass content in the root compartment was always higher than in the rhizosphere and soil, but not significantly higher than initial values. In month 25, the root compartment contained 334 ± 27 μg g−1, and soil contained 8.5 ± 5.0 μg g−1 ergosterol; rhizosphere contained only 6.0 ± 1.4 μg g−1, significantly less than before clearcutting (Fig. 2).
Fig. 2.
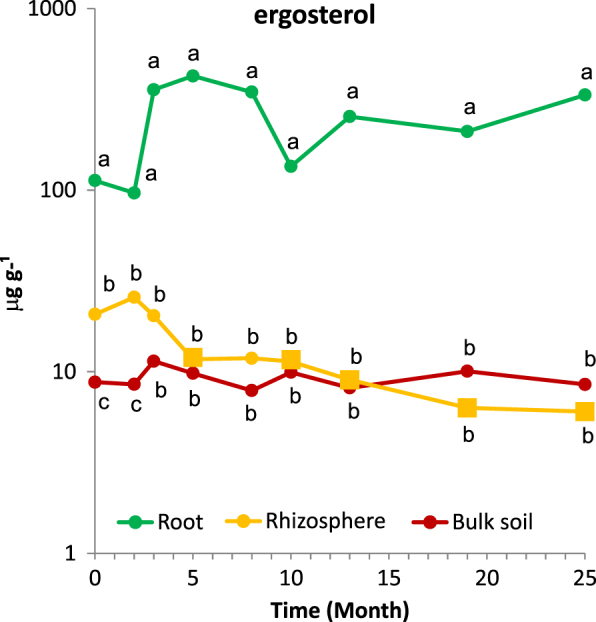
Fungal biomass in the soil, rhizosphere and roots of a Picea abies stand before (month 0) and after clearcutting expressed in μg ergosterol g−1 of dry soil or roots. Values represent means (n = 10) of the ergosterol content. Different letters indicate significant differences in ergosterol content between soil, rhizosphere and roots, and squares indicate significantly different ergosterol content compared to the P. abies stand in month 0
Fungal community in soil and decomposing P. abies roots
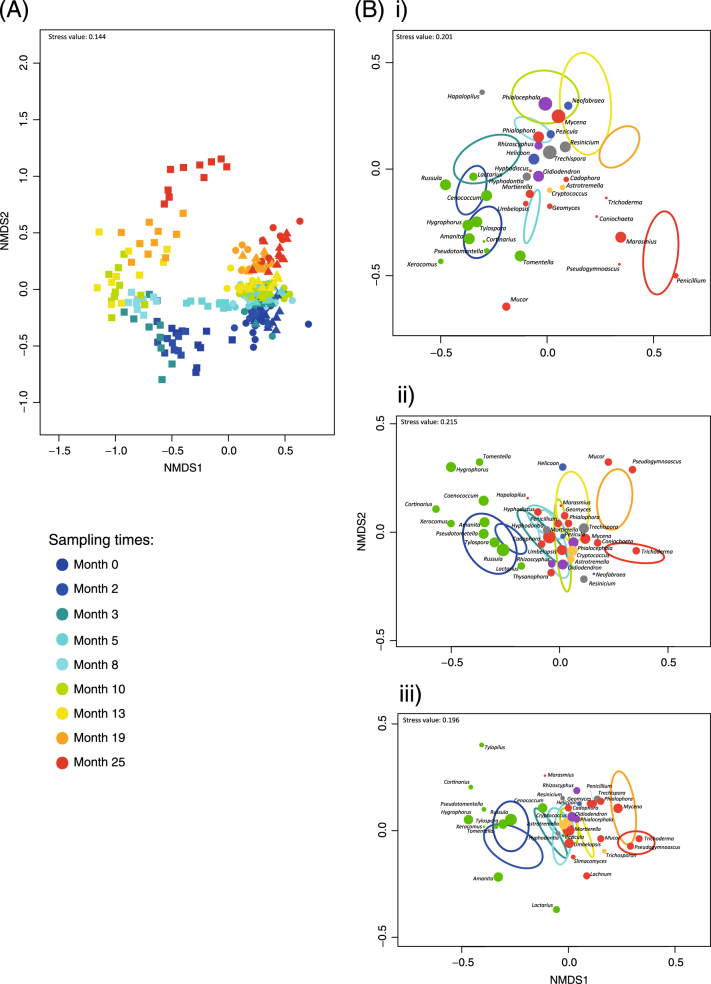
Stepwise selection of factors identified spatial distance (represented by five significant PCNM vectors), compartment (root, rhizosphere and bulk soil) and time after clearcutting as the best explanatory factors for fungal OTU community composition, explaining 7.3, 25.6 and 7.8% of the total variance (based on the Adj R 2 values), respectively. The NMDS ordination revealed greater changes in root-associated communities rather than rhizosphere or soil fungal communities over time after clearcutting (Fig. 3). Time after the clearcutting had a significant effect on fungal community composition, as well as on the composition of fungal ecophysiological categories in all three compartments (Table 1).
Fig. 3.
a Non-metric multidimensional scaling (NMDS) plot of the community composition of fungal genera. Each symbol represents one root (square), rhizosphere (triangle) or soil (circle) sample. Time after clearcutting is indicated by colour. b NMDS plots of the composition of fungal genera communities for three studied compartments: (i) root, (ii) rhizosphere and (iii) soil. Ellipses represent ordination confidence intervals (95%) for different sampling times after clearcutting. The relative abundances of fungal genera are expressed by the size of symbols, and putative fungal ecophysiology is indicated by colours: EcMF—green, endophytic fungi (ENF)—violet, saprotrophs—red, plant pathogens—blue, white-rot—grey and yeast—orange
Table 1.
Factors affecting the composition of fungal communities in three different compartments as revealed by PERMANOVA
| Root | Rhizosphere soil | Bulk soil | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Df | P value | Adj. R 2 | Df | P value | Adj. R 2 | Df | P value | Adj. R 2 | |
| Fungal OTUs | |||||||||
| Spatial distribution | 4 | 0.001 | 0.029 | 5 | 0.001 | 0.089 | 5 | 0.001 | 0.107 |
| Time after clearcutting | 1 | 0.001 | 0.128 | 1 | 0.001 | 0.1 | 1 | 0.001 | 0.103 |
| Water content | NS | NS | NS | 1 | 0.005 | 0.005 | 1 | 0.004 | 0.008 |
| Ecological guild | |||||||||
| Spatial distribution | NS | NS | NS | NS | NS | NS | 1 | 0.006 | 0.021 |
| Time after clearcutting | 1 | 0.001 | 0.232 | 1 | 0.001 | 0.298 | 1 | 0.001 | 0.399 |
| Water content | NS | NS | NS | NS | NS | NS | NS | NS | NS |
Factors significantly affecting composition of the fungal communities on the taxon level (relative sequence abundance of OTUs) and on the ecological guild level (relative sequence abundance of ecological guilds) are in bold. Adjusted R 2 was determined only for statistically significant factors
PERMANOVA permutational multivariate analysis of variance, Df degree of freedom, NS not significant, OTU Operational Taxonomic Unit
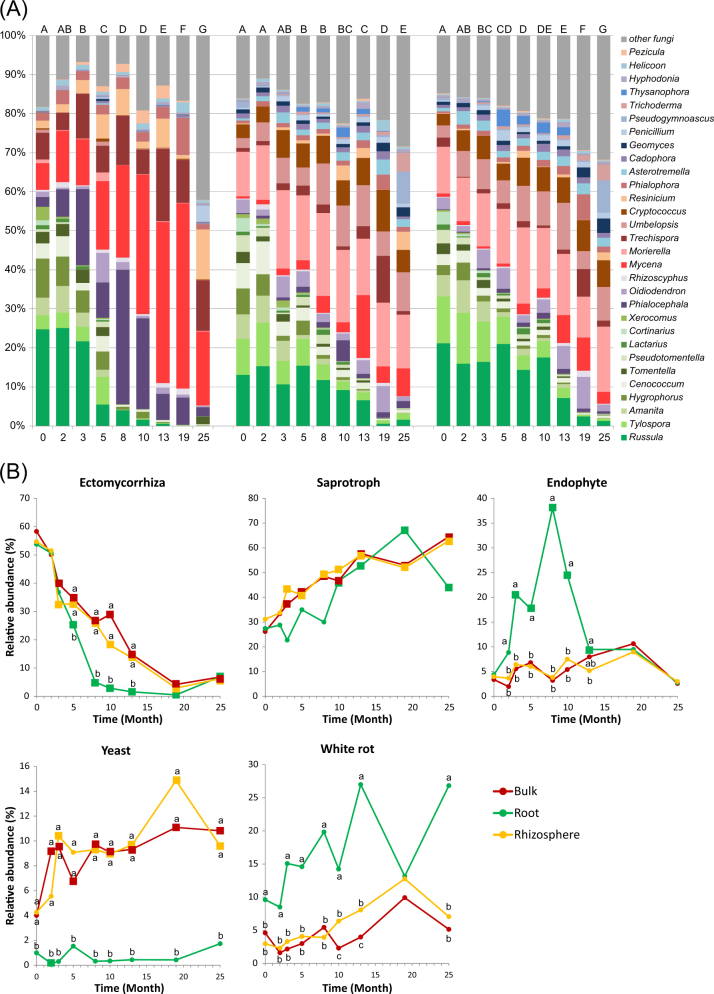
The P. abies stand contained mostly sequences of EcMF (54–59% in all compartments), followed by saprotrophs (30–36%) and root endophytes (3–6%). Bulk soil and rhizosphere also contained 4% yeasts and 3–5% of white-rot fungi, while roots contained more white-rot fungi (10%) and less yeasts (Supplementary Table 1). The first significant shift in fungal community composition and composition of fungal ecophysiological categories was observed 3 or 5 months after clearcutting (Fig. 4). Subsequently, fungal community composition and the proportions of fungal ecophysiological categories changed gradually during succession on decomposing roots. The shift was mostly driven by a decline in the relative abundances of EcM fungal genera (e.g., Russula, Tylospora, Amanita). While the relative abundance of EcMF in the root compartment dropped to only 5% 8 months after clearcutting, it remained higher in rhizosphere (28%) as well as in bulk soil (28%; Fig. 4a). Fungal communities associated with roots were characterized by increased relative abundances of root endophytes (e.g., Phialocephala, Rhizoscyphus) 3 to 13 months after clearcutting (Fig. 3; Fig. 4b). Saprotrophs (e.g., Mycena) dominated in the root compartment 13 months after clearcutting and beyond. Within rhizosphere and bulk soil, the relative abundance of saprotrophs (e.g., Mortierella, Umbelopsis, Pseudogymnoascus) gradually increased (Fig. 3). The proportion of yeasts increased significantly both in the rhizosphere and in bulk soil, reaching values between 10 and 14%. The relative content of white-rot fungi tended to increase, but the increase was not significant due to high variation among samples (Fig. 4b). As a consequence of succession, fungal communities changed profoundly in the 2 years after clearcutting. Fungal OTU richness increased significantly in the rhizosphere and bulk soil, from 100 to 125 OTUs in month 0 to 180–200 OTUs in month 25 (p < 0.0001). Mortierella, Umbelopsis and Pseudogymnoascus had the highest relative abundances in the rhizosphere and bulk soil, while Mycena, Trechispora, Resinicium and Marasmius had the highest relative abundances in the root compartment.
Fig. 4.
Abundances of major fungal genera and functional groups in the soil, rhizosphere and roots of a Picea abies stand before (month 0) and after clearcutting. Values represent means (n = 10) of the relative abundances of ITS2 amplicons expressed as percentages. a Abundance of major fungal genera. Different letters indicate significant differences between timepoints. b Abundance of functional groups. Different letters indicate significant differences in abundance between soil, rhizosphere and root, and square symbols indicate significantly different abundance compared to the P. abies stand on month 0
Community composition of individual functional guilds after removal of spatial effects also significantly changed over time. Saprotrophs and yeasts were the most affected in the rhizosphere, while the EcMF community was the most affected in roots (Table 2). Although the relative abundances of all EcM fungal taxa decreased with time, species did show different levels of persistence (Fig. 3; Supplementary Table 2). While certain EcMF such as Russula torulosa, Russula vesca or Tylopilus felleus disappeared completely, others were still present in the root compartment after 12 months. Other taxa persisted in the soil and rhizosphere even after 18 months. Meliniomyces bicolur in particular exhibited high persistence in soil, even though it disappeared from decomposing roots (Supplementary Table 2).
Table 2.
Factors affecting community composition of the most common fungal guilds in the soil, rhizosphere and roots of a Picea abies stand subjected to clearcutting as revealed by PERMANOVA
| Root | Rhizosphere soil | Bulk soil | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Df | P value | Adj. R 2 | Df | P value | Adj. R2 | Df | P value | Adj. R 2 | |
| Saprotrophic fungi | |||||||||
| Spatial distribution | NS | NS | NS | 4 | 0.001 | 0.068 | 5 | 0.001 | 0.087 |
| Time after clearcutting | 1 | 0.001 | 0.109 | 1 | 0.001 | 0.095 | 1 | 0.001 | 0.081 |
| Water content | NS | NS | NS | 1 | 0.002 | 0.010 | 1 | 0.004 | 0.015 |
| White-rot fungi | |||||||||
| Spatial distribution | 4 | 0.001 | 0.130 | 4 | 0.001 | 0.079 | 4 | 0.001 | 0.105 |
| Time after clearcutting | 1 | 0.004 | 0.030 | 1 | 0.001 | 0.031 | 1 | 0.001 | 0.032 |
| Water content | NS | NS | NS | NS | NS | NS | NS | NS | NS |
| EcM fungi | |||||||||
| Spatial distribution | 3 | 0.001 | 0.057 | 5 | 0.001 | 0.220 | 1 | 0.001 | 0.229 |
| Time after clearcutting | 1 | 0.001 | 0.156 | 1 | 0.001 | 0.070 | 1 | 0.001 | 0.088 |
| Water content | NS | NS | NS | 1 | 0.017 | 0.003 | NS | NS | NS |
| Endophytic fungi | |||||||||
| Spatial distribution | 3 | 0.001 | 0.055 | 4 | 0.001 | 0.121 | 5 | 0.001 | 0.149 |
| Time after clearcutting | 1 | 0.001 | 0.067 | 1 | 0.001 | 0.075 | 1 | 0.001 | 0.066 |
| Water content | NS | NS | NS | 1 | 0.016 | 0.009 | 1 | 0.003 | 0.011 |
| Yeasts | |||||||||
| Spatial distribution | 1 | 0.011 | 0.029 | 2 | 0.002 | 0.027 | 2 | 0.007 | 0.019 |
| Time after clearcutting | 1 | 0.001 | 0.114 | 1 | 0.001 | 0.095 | 1 | 0.001 | 0.080 |
| Water content | NS | NS | NS | 1 | 0.003 | 0.021 | NS | NS | NS |
Factors significantly affecting community composition are in bold. Adjusted R 2 was determined only for statistically significant factors
PERMANOVA permutational multivariate analysis of variance, Df degree of freedom, NS not significant, EcM ectomycorrhizal
Discussion
Response of the P. abies stand to clearcutting
The soil under the P. abies stand was characterized into three distinct compartments: fine roots, rhizosphere and bulk soil. These compartments differed in fungal biomass content and enzyme activity, with both higher in the root compartment over rhizosphere and bulk soil. However, relative proportion of the fine roots and rhizosphere masses were negligible: 0.19 and 0.90%, respectively, or 1.90 and 0.89% if we consider only the organic matter.
Forest clearcutting substantially shifted the composition of available resources from labile, root-derived, recent photosynthates to more recalcitrant forms, represented by senescing tree roots and mycorrhizal fungal mycelia, and caused several changes in the soil habitat. The mass of fine roots decreased to approximately one-half the initial amount 2 years after the disturbance, which is comparable to previous reports on coniferous root decomposition (e.g., [30, 43]). When extrapolated, approximately 5 t ha−1 fine root mass was lost, suggesting that decomposing roots represent an important resource for saprotrophic fungi. While in the uncut stand, 2.8% of fungal biomass was associated with fine roots, 1.9% with rhizosphere and 95.3% with bulk soil, during the peak period of root decomposition, fungal biomass in decomposing roots represented 12.9% of the total. The fungal biomass content in decomposing roots was estimated at approximately 112 mg g−1 in month 5, a value comparable or higher than that of decomposing deadwood or P. abies litter [44, 45] and decomposing roots thus represent a hotspot of decomposition activity. This hotspot seems to be somehow separated from the soil matrix, since both fungal biomass and enzyme activity in rhizosphere declined to values recorded in the bulk soil. Interestingly, although fungal biomass in the bulk soil decreased significantly during the 2 years following clearcutting in the conifer stand [7], as well as in the stand killed by bark beetle infestation [8], we did not observe a significant decrease of fungal biomass; the changes in fungal community composition thus likely included the decrease in certain taxa with a concomitant increase in others.
Previous reports have demonstrated that decomposition may be inhibited by the presence of EcMF [18–20] and the period after clearcutting when EcMF abundance is reduced was hypothesized to be the time of increased C turnover due to relief of this so-called ‘Gadgil effect’ [25]. Contrary to Hypothesis 1, forest clearcutting did not have a positive effect on enzyme activity in rhizosphere or bulk soil, except for Mn peroxidase, whose activity increased in the bulk soil in the late phase of the experiment.
Activity of most enzymes in the rhizosphere significantly decreased soon (3 months) after the clearcutting. This decline suggests that living plant roots stimulate soil enzymatic activity in their rhizosphere. Rhizosphere priming of SOM decomposition was experimentally demonstrated in microcosms with Pinus ponderosa and Populus fremontii [4]. Here, we show that the same can be observed in natural stands where rhizosphere priming may be theoretically counterweighted by the relief of the Gadgil effect and the decomposition of mycelia.
On the contrary, activity of most hydrolytic enzymes significantly decreased in the bulk soil in the late phase of the experiment (20 months after the clearcutting). Decreased enzyme activity in a long-term perspective was previously also observed after reduction of belowground C supply to soil via EcM tree girdling [46] and after tree dieback caused by bark beetle invasion [8]. We hypothesize that the observed long-term decline of hydrolytic enzymes was a response to decrease of a belowground litter production (plant roots and mycorrhizal fungal mycelia). On the contrary, increase of Mn peroxidase activity in the late phase of the experiment may indicate the shift in SOM turnover towards the decomposition of more recalcitrant compounds. Highly significant correlations between enzyme activity and fungal biomass content suggest that fungi might be responsible for a large part of the observed enzyme production in soil. This is in agreement with observations in a mountainous Picea abies forest, where most transcripts of glycoside hydrolases and auxiliary enzymes were of fungal origin [47].
In contrast to soil, clearcutting caused a substantial increase of β-glucosidase, exocellulase, β-xylosidase, endoxylanase and α-glucosidase activities in the root compartment in the early stage of decomposition (5–8 months). The increased activity of these enzymes together with decrease of root mass indicates that early decomposers focus on degradation of less recalcitrant components of plant tissue, such as hemicellulose, pectins and other labile C sources. In later stages (8–13 months), increased endocellulase activity together with continuous high activity of exocellulase and β-glucosidase indicate degradation of more recalcitrant, structural polysaccharides such as cellulose. In the same time period, lignin-degrading enzyme activity (Mn peroxidase and laccase) also increased. These observations suggest that transition from the decomposition of labile forms of C to more recalcitrant forms occurs after approximately 10 months. This sequential degradation of root litter components is energetically favourable and analogous to the decomposition of aboveground litter [48], while the decomposition of bulky deadwood often starts with selective delignification, that is, the removal of the most recalcitrant biopolymer—lignin [48].
Fungal community in the clearcut forest stand
Fungal community composition responded to forest clearcutting in complex ways. While fungal communities in the rhizosphere and soil developed gradually, root fungal communities showed much more dynamics. In all the studied compartments, the proportion of EcMF substantially decreased within 3 months, which corresponds to our Hypothesis 2 and to the previously documented shifts in fungal community composition after belowground carbon disruption induced by clearcutting [25, 49], tree girdling [6, 50], bark beetle outbreaks [8, 51] or defoliation [52]. The decline in the EcMF proportion was faster in the root compartment compared to the soil and rhizosphere compartments. In the rhizosphere, the significant reduction of ergosterol content implies that the relative decrease of EcMF (based on sequence numbers) corresponds to an even more dramatic decline in absolute terms. A similar, although less dramatic, decline in EcMF was observed in the bulk soil. These results may indicate that EcMF in rhizosphere soil are present in forms requiring more host-derived carbon compounds, such as in the form of physiologically active extraradical mycelia. In contrast, the proportion of EcMF forms adapted to unfavourable conditions, such as spores or rhizomorphs, may be higher in bulk soil. The increasing proportion of other fungi in the community during root decomposition must be considered in relation to the decline in EcMF.
Although EcMF persistence differs between taxa, we found no support for the assumption expressed by Talbot et al. [14] that some may decompose dying roots (Hypothesis 4). Along with the fact that EcMF dramatically decrease activity in winter when C is limited [36] and the observation that they often require simple carbon compounds to decompose recalcitrant organic matter [15], it seems that EcMF are rather dependent on C supply from hosts. The only exception might be that Meliniomyces bicolor exhibited high persistence in soil. M. bicolor is an ascomycetous EcM fungus that is closely phylogenetically related to ericoid mycorrhizal fungi [53], which are known to be efficient decomposers of recalcitrant organic compounds [54–56]. Indeed, recent genomic analysis suggests similar enzymatic capabilities of M. bicolor and ericoid mycorrhizal fungi [57], and thus this species may switch to a saprotrophic lifestyle when host C supply terminates.
Between months 3 and 10 post-clearcutting, the proportion of endophytic fungi (ENF) in decaying roots increased. These communities were dominated by Phialocephala fortinii, the well-known dark septate endophytes [58]. Compared to EcMF, ENF generally feature greater enzymatic capabilities for degradation of complex organic compounds [59]. Although the comparative genome analysis showed greater copy numbers of genes encoding enzymes involved in decay of lignin and cellulose in white and brown rot fungi than in root endophytes [55, 59], a colonization priority effect can provide a competitive advantage to the latter. This would complement the recent observations that leaf [60, 61] and wood endophytes [62, 63] are activated after the dieback of host tissues, and contribute significantly to the initial phases of decomposition of dead host tissues [60, 63]. Since the biomass of ENF in living tree organs (leaves, wood and roots) is relatively low and the activity and intensity of utilization of host resources are likely limited [60, 62, 63], we may speculate that the endophytic lifestyle of these fungi actually only represents an adaptation to ensure priority in host tissues for the upcoming decomposition stage in which they rapidly proliferate.
During subsequent decomposition phases (13–25 months), the proportion of ENF sequences in decaying roots declined and was replaced by those of saprotrophic fungi such as Resinicium and Mycena, which are known as white-rot decomposers of recalcitrant organic compounds including lignin and often reported from decomposing deadwood [45]. In a recent study, Smith et al. [64] provided evidence that Mycena spp. may also asymptomatically colonize living coniferous roots, and the presence of Mycena sp. in P. abies roots before clearcutting concurs with the results of that study. It is thus possible that both major fungal groups of putative root decomposers in our study site (based on prevalence in DNA pools from decomposing roots), Phialocephala fortinii and Mycena spp., could have already been present in living roots as asymptomatic endophytes prior to clearcutting. Therefore, contrary to our third hypothesis, we conclude that the composition of fungal communities associated with decomposing roots substantially depends on early colonizers that are capable of association with living plant roots.
Conclusion
This study demonstrates that clearcutting leads to profound changes in soil decomposition processes and fungal community composition. The rhizosphere, as an active compartment of high enzyme activity and high fungal biomass, ceases to exist and starts to resemble bulk soil. Decomposing roots seem to be isolated from the rest of soil and represent a hotspot of decomposition and fungal biomass. In the long term, decomposition in soil decreases, indicating the potential importance of root activity for the decomposition of recalcitrant soil organic matter. While there is no support for the involvement of ectomycorrhizal fungi in the decomposition of roots, we show that root endophytic fungi play an important role in the early stages of this process, indicating the importance of the priority effect.
Electronic supplementary material
Acknowledgements
This work was supported by the Czech Science Foundation (13-06763S and 16-20569S). PK was also supported by Charles University in Prague (NPUI LO1417, MSMT).
Compliance with ethical standards
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0027-3) contains supplementary material, which is available to authorized users.
References
- 1.Baldrian P. Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev. 2017;41:109–30. doi: 10.1093/femsre/fuw040. [DOI] [PubMed] [Google Scholar]
- 2.Litton CM, Raich JW, Ryan MG. Carbon allocation in forest ecosystems. Glob Change Biol. 2007;13:2089–109. doi: 10.1111/j.1365-2486.2007.01420.x. [DOI] [Google Scholar]
- 3.Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, et al. Roots and associated fungi drive long-term carbon sequestration in boreal forest. Science. 2013;339:1615–8. doi: 10.1126/science.1231923. [DOI] [PubMed] [Google Scholar]
- 4.Dijkstra FA, Cheng WX. Interactions between soil and tree roots accelerate long-term soil carbon decomposition. Ecol Lett. 2007;10:1046–53. doi: 10.1111/j.1461-0248.2007.01095.x. [DOI] [PubMed] [Google Scholar]
- 5.Kuzyakov Y. Priming effects: interactions between living and dead organic matter. Soil Biol Biochem. 2010;42:1363–71. doi: 10.1016/j.soilbio.2010.04.003. [DOI] [Google Scholar]
- 6.Högberg P, Nordgren A, Buchmann N, Taylor AFS, Ekblad A, Högberg MN, et al. Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature. 2001;411:789–92. doi: 10.1038/35081058. [DOI] [PubMed] [Google Scholar]
- 7.Churchland C, Grayston SJ, Bengtson P. Spatial variability of soil fungal and bacterial abundance: consequences for carbon turnover along a transition from a forested to clearcut site. Soil Biol Biochem. 2013;63:5–13. doi: 10.1016/j.soilbio.2013.03.015. [DOI] [Google Scholar]
- 8.Štursová M, Šnajdr J, Cajthaml T, Bárta J, Šantrůčková H, Baldrian P. When the forest dies: the response of forest soil fungi to a bark beetle-induced tree dieback. ISME J. 2014;8:1920–31. doi: 10.1038/ismej.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ekblad A, Wallander H, Godbold DL, Cruz C, Johnson D, Baldrian P, et al. The production and turnover of extramatrical mycelium of ectomycorrhizal fungi in forest soils: role in carbon cycling. Plant Soil. 2013;366:1–27. doi: 10.1007/s11104-013-1630-3. [DOI] [Google Scholar]
- 10.Bååth E, Nilsson LO, Göransson H, Wallander H. Can the extent of degradation of soil fungal mycelium during soil incubation be used to estimate ectomycorrhizal biomass in soil? Soil Biol Biochem. 2004;36:2105–9. doi: 10.1016/j.soilbio.2004.06.004. [DOI] [Google Scholar]
- 11.Wallander H, Göransson H, Rosengren U. Production, standing biomass and natural abundance of 15N and 13C in ectomycorrhizal mycelia collected at different soil depths in two forest types. Oecologia. 2004;139:89–97. doi: 10.1007/s00442-003-1477-z. [DOI] [PubMed] [Google Scholar]
- 12.Courty PE, Buéé M, Diedhiou AG, Frey-Klett P, Le Tacon F, Rineau F, et al. The role of ectomycorrhizal communities in forest ecosystem processes: New perspectives and emerging concepts. Soil Biol Biochem. 2010;42:679–98. doi: 10.1016/j.soilbio.2009.12.006. [DOI] [Google Scholar]
- 13.Baldrian P. Ectomycorrhizal fungi and their enzymes in soils: is there enough evidence for their role as facultative soil saprotrophs? Oecologia. 2009;161:657–60. doi: 10.1007/s00442-009-1433-7. [DOI] [PubMed] [Google Scholar]
- 14.Talbot JM, Allison SD, Treseder KK. Decomposers in disguise: mycorrhizal fungi as regulators of soil C dynamics in ecosystems under global change. Funct Ecol. 2008;22:955–63. doi: 10.1111/j.1365-2435.2008.01402.x. [DOI] [Google Scholar]
- 15.Lindahl BD, Tunlid A. Ectomycorrhizal fungi—potential organic matter decomposers, yet not saprotrophs. New Phytol. 2015;205:1443–7. doi: 10.1111/nph.13201. [DOI] [PubMed] [Google Scholar]
- 16.Lindahl BD, de Boer W, Finlay RD. Disruption of root carbon transport into forest humus stimulates fungal opportunists at the expense of mycorrhizal fungi. ISME J. 2010;4:872–81. doi: 10.1038/ismej.2010.19. [DOI] [PubMed] [Google Scholar]
- 17.Bödeker ITM, Lindahl BD, Olson A, Clemmensen KE. Mycorrhizal and saprotrophic fungal guilds compete for the same organic substrates but affect decomposition differently. Funct Ecol. 2016;30:1967–78. doi: 10.1111/1365-2435.12677. [DOI] [Google Scholar]
- 18.Averill C, Hawkes CV. Ectomycorrhizal fungi slow soil carbon cycling. Ecol Lett. 2016;19:937–47. doi: 10.1111/ele.12631. [DOI] [PubMed] [Google Scholar]
- 19.Fernandez CW, Kennedy PG. Revisiting the ‘Gadgil effect’: do interguild fungal interactions control carbon cycling in forest soils? New Phytol. 2016;209:1382–94. doi: 10.1111/nph.13648. [DOI] [PubMed] [Google Scholar]
- 20.Gadgil RL, Gadgil PD. Suppression of litter decomposition by mycorrhizal roots of Pinus radiata. New Zeal J Sci. 1975;5:33–41. [Google Scholar]
- 21.Brabcová V, Nováková M, Davidová A, Baldrian P. Dead fungal mycelium in forest soil represents a decomposition hotspot and a habitat for a specific microbial community. New Phytol. 2016;210:1369–81. doi: 10.1111/nph.13849. [DOI] [PubMed] [Google Scholar]
- 22.Holden SR, Treseder KK. A meta-analysis of soil microbial biomass responses to forest disturbances. Front Microbiol. 2013;4:163. doi: 10.3389/fmicb.2013.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grebenc T, Christensen M, Vilhar U, Cater M, Martin MP, Simoncic P, et al. Response of ectomycorrhizal community structure to gap opening in natural and managed temperate beech-dominated forests. Can J Res. 2009;39:1375–86. doi: 10.1139/X09-072. [DOI] [Google Scholar]
- 24.Jones MD, Durall DM, Cairney JWG. Ectomycorrhizal fungal communities in young forest stands regenerating after clearcut logging. New Phytol. 2003;157:399–422. doi: 10.1046/j.1469-8137.2003.00698.x. [DOI] [PubMed] [Google Scholar]
- 25.Kyaschenko J, Clemmensen KE, Hagenbo A, Karltun E, Lindahl BD. Shift in fungal communities and associated enzyme activities along an age gradient of managed Pinus sylvestris stands. ISME J. 2017;11:863–74. doi: 10.1038/ismej.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cardenas E, Kranabetter JM, Hope G, Maas KR, Hallam S, Mohn WW. Forest harvesting reduces the soil metagenomic potential for biomass decomposition. ISME J. 2015;9:2465–76. doi: 10.1038/ismej.2015.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang GG, Van Lear DH, Hu HF, Kapeluck PR. Accounting carbon storage in decaying root systems of harvested forests. Ambio. 2012;41:284–91. doi: 10.1007/s13280-011-0161-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lytle DE, Cronan CS. Comparative soil CO2 evolution, litter decay, and root dynamics in clearcut and uncut spruce–fir forest. Ecol Manag. 1998;103:121–8. doi: 10.1016/S0378-1127(97)00182-5. [DOI] [Google Scholar]
- 29.Langley AJ, Chapman SK, Hungate BA. Ectomycorrhizal colonization slows root decomposition: the post-mortem fungal legacy. Ecol Lett. 2006;9:955–9. doi: 10.1111/j.1461-0248.2006.00948.x. [DOI] [PubMed] [Google Scholar]
- 30.Li XF, Lange H. A modified soil coring method for measuring fine root production, mortality and decomposition in forests. Soil Biol Biochem. 2015;91:192–9. doi: 10.1016/j.soilbio.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 31.Wallander H, Nilsson LO, Hagerberg D, Bååth E. Estimation of the biomass and seasonal growth of external mycelium of ectomycorrhizal fungi in the field. New Phytol. 2001;151:753–60. doi: 10.1046/j.0028-646x.2001.00199.x. [DOI] [PubMed] [Google Scholar]
- 32.Šnajdr J, Valášková V, Merhautová V, Cajthaml T, Baldrian P. Activity and spatial distribution of lignocellulose-degrading enzymes during forest soil colonization by saprotrophic basidiomycetes. Enzym Microb Technol. 2008;43:186–92. doi: 10.1016/j.enzmictec.2007.11.008. [DOI] [Google Scholar]
- 33.Štursová M, Baldrian P. Effects of soil properties and management on the activity of soil organic matter transforming enzymes and the quantification of soil-bound and free activity. Plant Soil. 2011;338:99–110. doi: 10.1007/s11104-010-0296-3. [DOI] [Google Scholar]
- 34.Sagova-Mareckova M, Cermak L, Novotna J, Plhackova K, Forstova J, Kopecky J. Innovative methods for soil DNA purification tested in soils with widely differing characteristics. Appl Environ Microbiol. 2008;74:2902–7. doi: 10.1128/AEM.02161-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ihrmark K, Bodeker ITM, Cruz-Martinez K, Friberg H, Kubartova A, Schenck J, et al. New primers to amplify the fungal ITS2 region—evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol Ecol. 2012;82:666–77. doi: 10.1111/j.1574-6941.2012.01437.x. [DOI] [PubMed] [Google Scholar]
- 36.Žifčáková L, Větrovský T, Howe A, Baldrian P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ Microbiol. 2016;18:288–301. doi: 10.1111/1462-2920.13026. [DOI] [PubMed] [Google Scholar]
- 37.Větrovský T, Baldrian P. Analysis of soil fungal communities by amplicon pyrosequencing: current approaches to data analysis and the introduction of the pipeline SEED. Biol Fertil Soils. 2013;49:1027–37. doi: 10.1007/s00374-013-0801-y. [DOI] [Google Scholar]
- 38.Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods. 2013;10:996–8. doi: 10.1038/nmeth.2604. [DOI] [PubMed] [Google Scholar]
- 39.Koljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AFS, Bahram M, et al. Towards a unified paradigm for sequence-based identification of fungi. Mol Ecol. 2013;22:5271–7. doi: 10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- 40.Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R, et al. Fungal community analysis by high-throughput sequencing of amplified markers—a user’s guide. New Phytol. 2013;199:288–99. doi: 10.1111/nph.12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R, et al. Global diversity and geography of soil fungi. Science. 2014;346:1256688. doi: 10.1126/science.1256688. [DOI] [PubMed] [Google Scholar]
- 42.R Core Development Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 43.Sariyildiz T. Effects of tree species and topography on fine and small root decomposition rates of three common tree species (Alnus glutinosa, Picea orientalis and Pinus sylvestris) in Turkey. Ecol Manag. 2015;335:71–86. doi: 10.1016/j.foreco.2014.09.030. [DOI] [Google Scholar]
- 44.Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T, et al. Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J. 2012;6:248–58. doi: 10.1038/ismej.2011.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldrian P, Zrustova P, Tlaskal V, Davidova A, Merhautova V, Vrska T. Fungi associated with decomposing deadwood in a natural beech-dominated forest. Fungal Ecol. 2016;23:109–22. doi: 10.1016/j.funeco.2016.07.001. [DOI] [Google Scholar]
- 46.Brzostek ER, Dragoni D, Brown ZA, Phillips RP. Mycorrhizal type determines the magnitude and direction of root-induced changes in decomposition in a temperate forest. New Phytol. 2015;206:1274–82. doi: 10.1111/nph.13303. [DOI] [PubMed] [Google Scholar]
- 47.Žifčáková L, Větrovský T, Lombard V, Henrissat B, Howe A, Baldrian P. Feed in summer, rest in winter: Microbial carbon utilization in forest topsoil. Microbiome. 2017;5:122. doi: 10.1186/s40168-017-0340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Šnajdr J, Cajthaml T, Valášková V, Merhautová V, Petránková M, Spetz P, et al. Transformation of Quercus petraea litter: successive changes in litter chemistry are reflected in differential enzyme activity and changes in the microbial community composition. FEMS Microbiol Ecol. 2011;75:291–303. doi: 10.1111/j.1574-6941.2010.00999.x. [DOI] [PubMed] [Google Scholar]
- 49.Hartmann M, Howes CG, VanInsberghe D, Yu H, Bachar D, Christen R, et al. Significant and persistent impact of timber harvesting on soil microbial communities in Northern coniferous forests. ISME J. 2012;6:2199–218. doi: 10.1038/ismej.2012.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kaiser C, Koranda M, Kitzler B, Fuchslueger L, Schnecker J, Schweiger P, et al. Belowground carbon allocation by trees drives seasonal patterns of extracellular enzyme activities by altering microbial community composition in a beech forest soil. New Phytol. 2010;187:843–58. doi: 10.1111/j.1469-8137.2010.03321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pec GJ, Karst J, Taylor DL, Cigan PW, Erbilgin N, Cooke JEK, et al. Change in soil fungal community structure driven by a decline in ectomycorrhizal fungi following a mountain pine beetle (Dendroctonus ponderosae) outbreak. New Phytol. 2017;213:864–73. doi: 10.1111/nph.14195. [DOI] [PubMed] [Google Scholar]
- 52.Parker TC, Sadowsky J, Dunleavy H, Subke JA, Frey SD, Wookey PA. Slowed biogeochemical cycling in sub-arctic birch forest linked to reduced mycorrhizal growth and community change after a defoliation event. Ecosystems. 2017;20:316–30. doi: 10.1007/s10021-016-0026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hambleton S, Sigler L. Meliniomyces, a new anamorph genus for root-associated fungi with phylogenetic affinities to Rhizoscyphus ericae (Hymenoscyphus ericae), Leotiomycetes. Stud Mycol. 2005;53:1–27. doi: 10.3114/sim.53.1.1. [DOI] [Google Scholar]
- 54.Kerley SJ, Read DJ. The biology of mycorrhiza in the Ericaceae XX. Plant and mycorrhizal necromass as nitrogenous substrates for the ericoid mycorrhizal fungus Hymenscyphus ericae and its host. New Phytol. 1998;139:353–60. doi: 10.1046/j.1469-8137.1998.00189.x. [DOI] [PubMed] [Google Scholar]
- 55.Kohler A, Kuo A, Nagy LG, Morin E, Barry KW, Buscot F, et al. Convergent losses of decay mechanisms and rapid turnover of symbiosis genes in mycorrhizal mutualists. Nat Genet. 2015;47:410–5. doi: 10.1038/ng.3223. [DOI] [PubMed] [Google Scholar]
- 56.Bending GD, Read DJ. Lignin and soluble phenolic degradation by ectomycorrhizal and ericoid mycorrhizal fungi. Mycol Res. 1997;101:1348–54. doi: 10.1017/S0953756297004140. [DOI] [Google Scholar]
- 57.Grelet G, Martino E, Dickie IA, Tajuddin R, Artz R. Ecology of ericoid mycorrhizal fungi. Molecular Mycorrhizal Symbiosis. New York: Wiley; 2016. p. 405-19.
- 58.Grünig CR, Queloz V, Sieber TN, Holdenrieder O. Dark septate endophytes (DSE) of the Phialocephala fortinii s.l.–Acephala applanata species complex in tree roots: classification, population biology, and ecology. Botany. 2008;86:1355–69. doi: 10.1139/B08-108. [DOI] [Google Scholar]
- 59.Schlegel M, Munsterktter M, Guldener U, Bruggmann R, Duo A, Hainaut M, et al. Globally distributed root endophyte Phialocephala subalpina links pathogenic and saprophytic lifestyles. BMC Genom. 2016;17:1015. doi: 10.1186/s12864-016-3369-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Voříšková J, Baldrian P. Fungal community on decomposing leaf litter undergoes rapid successional changes. ISME J. 2013;7:477–86. doi: 10.1038/ismej.2012.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Žifčáková L, Dobiášová P, Kolářová Z, Koukol O, Baldrian P. Enzyme activities of fungi associated with Picea abies needles. Fungal Ecol. 2011;4:427–36. doi: 10.1016/j.funeco.2011.04.002. [DOI] [Google Scholar]
- 62.Parfitt D, Hunt J, Dockrell D, Rogers HJ, Boddy L. Do all trees carry the seeds of their own destruction? PCR reveals numerous wood-decay fungi latently present in sapwood of a wide range of angiosperm trees. Fungal Ecol. 2010;3:338–46. doi: 10.1016/j.funeco.2010.02.001. [DOI] [Google Scholar]
- 63.Song ZW, Kennedy PG, Liew FJ, Schilling JS. Fungal endophytes as priority colonizers initiating wood decomposition. Funct Ecol. 2017;31:407–18. doi: 10.1111/1365-2435.12735. [DOI] [Google Scholar]
- 64.Smith GR, Finlay RD, Stenlid J, Vasaitis R, Menkis A. Growing evidence for facultative biotrophy in saprotrophic fungi: data from microcosm tests with 201 species of wood-decay basidiomycetes. New Phytol. 2017;215:747–55. doi: 10.1111/nph.14551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.