Abstract
Many areas of the ocean are nutrient-poor yet support large microbial populations, leading to intense competition for and recycling of nutrients. Organic phosphonates are frequently found in marine waters, but require specialist enzymes for catabolism. Previous studies have shown that the genes that encode these enzymes in marine systems are under Pho regulon control and so are repressed by inorganic phosphate. This has led to the conclusion that phosphonates are recalcitrant in much of the ocean, where phosphorus is not limiting despite the degradative genes being common throughout the marine environment. Here we challenge this paradigm and show, for the first time, that bacteria isolated from marine samples have the ability to mineralise 2-aminoethylphosphonate, the most common biogenic marine aminophosphonate, via substrate-inducible gene regulation rather than via Pho-regulated metabolism. Substrate-inducible, Pho-independent 2-aminoethylphosphonate catabolism therefore represents a previously unrecognised component of the oceanic carbon, nitrogen and phosphorus cycles.
Introduction
Inorganic nitrogen and phosphorus levels in the oceans are often limiting for microbial growth, leading to a heavy reliance on organic nutrient consumption to obtain these elements [1]. One class of organic phosphorus compounds is the phosphonates, molecules with a direct carbon-phosphorus bond: ~25% of high molecular weight dissolved organic phosphorus in the oceans is in phosphonate form [2, 3], while up to 16% of the phosphorus taken up by marine plankton is converted to reduced phosphorus compounds including phosphonates [4]. These studies demonstrate that large quantities of reduced phosphorus compounds—more than the annual pre-anthropogenic riverine input of phosphorus to the oceans—are being continuously and rapidly synthesised globally in the oceans, although the functions of these compounds are currently unknown. Many roles have been proposed for biogenic phosphonates including facilitating bacterial parasitism, altering the permeability of membranes, as a signalling molecule and as a way of increasing the resistance of membranes to phospholipases [5, 6]. In addition, when Van Mooy et al. [4] noted that a Trichodesmium sp. released the majority of newly synthesised reduced phosphorus compounds into the environment they suggested that this may be a way of passing nutrients to epibiontic cells. However, to date there have been no studies which empirically demonstrate any of these functions, and therefore the ecological role of aminophosphonate compounds remains enigmatic. These molecules cannot be catabolised using organic phosphate degrading enzymes, but instead require phosphonate-specific enzyme systems for mineralisation. Previous studies using terrestrial microbes have suggested that these enzymes are, for the most part, under Pho regulon control and are therefore induced by inorganic phosphate starvation [7]. Phosphonates have thus only been considered in marine nutrient cycling models as sources of phosphorus, and even then only in phosphorus-limited waters, despite also containing carbon and frequently nitrogen. As a result these compounds are hypothesised to be recalcitrant in non-phosphate limited waters and the carbon, nitrogen and phosphorus within them removed from biologically cycled nutrient pools via sedimentation to the ocean floor. Consistent with this, phosphonate metabolism has only been previously demonstrated in marine microorganisms under phosphorus starvation conditions [8, 9], and comparatively little consideration has been given to the possibility of organic phosphorus consumption as a carbon or nitrogen source in the presence of inorganic phosphate [10]. However, metagenomic studies have shown that genes for phosphonate metabolism are common throughout the ocean even in regions with relatively high inorganic phosphate concentrations [11, 12]. In 2010, Martinez et al. [11] showed that a phnWX-containing fosmid from a marine metagenomic library enabled an E. coli heterologous host to degrade 2AEP in the presence of inorganic phosphate when the transcriptional regulator was bypassed. This is consistent with previous studies which have shown that it is the repression of gene expression by inorganic phosphate which normally controls phosphonate degradation, rather than inhibition of the enzymes themselves [13]. As a result an alternative regulation system similar to that previously observed in a terrestrial organism [14] could allow the genes already known to be present in the ocean to metabolise phosphonates in an inorganic phosphate-insensitive manner, rather than requiring novel degradation proteins. An NMR study of an anoxic basin found that phosphonates were preferentially metabolised relative to the more labile phosphate esters and, importantly, that this mineralisation occurred in the presence of >0.5 μM soluble reactive phosphorus, although no investigation into the organisms or specific phosphonate compounds involved was performed [15]. The Pho regulon is generally considered to be inactive at low-micromolar concentrations of inorganic phosphate [16], suggesting that inorganic phosphate-insensitive phosphonate degradation does occur in marine systems, analogous to that previously observed in a small number of terrestrial systems [14]. In addition, Clark et al. [3] showed that the ratio of phosphonates to phosphate esters throughout the oceanic water column remains relatively stable despite the total quantity of organic phosphorus decreasing with depth, suggesting that phosphonates must be metabolised proportionately to phosphate esters even in deeper waters where inorganic phosphate tends to be more abundant.
The catabolism of aminophosphonates in non-phosphate-limited waters could be an important coping mechanism for marine microbes in waters of varying nutrient limitation, and provide an alternative nutrient niche for organisms which harbour such inorganic phosphate-insensitive phosphonate catabolism pathways. Large areas of the ocean surface are primarily nitrogen limited, and while phosphorus is expected to be the ultimate limiting nutrient in the oceans, nitrogen is more likely to be the proximate limiting nutrient [1, 17]. Long-term studies in the North Pacific Ocean suggest that oligotrophic environments shift back-and-forth between nitrogen and phosphorus limitation [17, 18]. The induction of phosphonate-degrading enzymes by conditions other than inorganic phosphate starvation would allow the use of aminophosphonates to supply microbial phosphorus, nitrogen and carbon requirements.
This metabolism would also provide a previously unrecognised route for the return of elements to actively cycled nutrient pools rather than loss via sedimentation. With oceanic primary productivity often constrained by nutrient supply [18], the return of aminophosphonate derived carbon, nitrogen and phosphorus to more bioavailable forms via phosphate-insensitive catabolism would extend the contribution of microbial turnover to nutrient cycling beyond that previously described for Pho-regulated metabolism. Such phosphate-insensitive aminophosphonate degradation would also be expected to provide a competitive advantage to microbes in nutrient depleted environments. Given that phosphonate degradation models were developed using terrestrial microbes, we set out to examine the hypothesis that currently known phosphonate degradation systems exist in the marine environment under phosphate-insensitive regulation, allowing marine microbes to consume phosphonates as a nitrogen source under high phosphate conditions which would previously have been expected to preclude this.
Materials and methods
Reagents
1-hydroxy-2-aminoethylphosphonate was kindly provided by F. Hammerschmidt and K. Pallitsch, University of Vienna, Austria. Synthetic phosphonoacetaldehyde was kindly provided by the late H.B.F Dixon, Department of Biochemistry, Cambridge University, UK. Unless stated otherwise all other reagents were from Sigma Aldrich Co. (Poole, Dorset, UK).
Collection of oceanic surface water samples from around the island of Ireland
Environmental surface water samples (≤ 50 cm) were collected from coastal water bodies around the northern shores of Ireland and used to inoculate enrichment cultures. The sampling sites were: Portmuck Beach, Islandmagee, County Antrim; Mulroy Bay, County Donegal; Strangford Lough, County Down; and Kilkeel Harbour, County Down. Isolates from each water body were designated IMG, MRB, SGF or KKW respectively according to their place of origin. All samples were collected in sterile 50 or 15 mL centrifuge tubes (Sarstedt, Nümbrecht, Germany) or 30 mL sterile Universal tubes (Medline Industries Ltd, Cheshire, UK) and stored immediately at 4 °C until they arrived at the lab, where they were inoculated into enrichment cultures without delay.
Inoculation of water samples into medium to enrich for and isolate 2AEP degrading bacteria
To promote the growth of microbes from the seawater samples which could use 2AEP as a nitrogen source enrichment cultures were performed in minimal marine medium. This consisted of: 42.26 mM MgSO4·7H2O, 166.83 mM NaCl, 8.54 mM CaCl2·2H2O, 5.69 mM KCl and 50 mM HEPES sodium salt adjusted to pH 7.4. Ferric ammonium citrate at a final concentration of 25 μM and 1 mL per litre each of trace elements [19] and vitamin solution [20] were added after autoclaving.
Standard nutrients for cultures consisted of 5 mM sodium succinate, 5 mM sodium acetate and 5 mM glucose as carbon sources, with 2 mM NH4Cl and 1 mM KH2PO4 as nitrogen and phosphorus sources, respectively.
Enrichment cultures were carried out in 30 mL sterile universal tubes using 5 mL volumes of minimal marine medium and synthetic, 99%-pure 2 mM 2-aminoethylphosphonate as the sole nitrogen source. In total 25 μL of the water samples were inoculated into separate minimal marine medium volumes and incubated at 28 °C with 100 revolutions per minute (RPM) shaking in a Sanyo ORBI-SAFE incubator (Sanyo Europe Ltd., Watford, UK). After 7 serial transfers morphologically different colonies were isolated from YTSSA plates (per litre 2 g yeast extract, 1.25 g vegetable peptone, 20 g Sigma Sea Salts, 1.5% w/v agar) and 7 isolates which appeared to grow using 2-aminoethylphosphonate as the sole nitrogen source were selected for further study.
Growth studies of isolates with 2AEP as the sole nitrogen source
The ability of each isolate to use 2AEP as the sole nitrogen source was confirmed via growth studies. Colonies of each isolate were inoculated into 15 mL of YTSS medium (as per YTSSA but without agar) in a sterile 50 mL centrifuge tube and grown overnight at 28 °C with 100 RPM shaking. The cells were centrifuged at 10,000 × g for 15 min and washed twice in nutrient-free minimal marine medium and used to make triplicate 5 mL cultures with and without 2 mM 2-aminoethylphosphonate as the sole nitrogen source in sterile 30 mL Universal tubes at a starting O.D. 650 nm of ~0.05, and incubated at 28 °C and 100 RPM. Optical density measurements at 650 nm were taken at T0 and regular intervals using 200 μL samples with a Tecan GENios plate reader (Tecan Group Ltd., Männedorf, Switzerland), before being centrifuged and pellets and supernatants frozen separately. The phosphate concentration in the culture supernatants was measured using BIOMOL Green (Enzo Life Sciences, Exeter, UK) while the pellets from T0 and the end of the exponential growth period were used for protein quantification using Bradford reagent, both according to the manufacturer’s instructions.
Preparation of cell-free extracts from 2AEP-degrading bacteria
Cell-free extracts of each isolate were prepared for use in subsequent enzyme assays. A 250 mL culture of each isolate in YTSS was grown overnight at 28 °C and 100 RPM. The cells were centrifuged at 10,000 × g for 10 min, washed twice in nutrient-free minimal marine medium, re-suspended in 250 mL minimal marine medium with 2 mM 2-aminoethylphosphonate as the sole nitrogen source and incubated at 28 °C and 100 RPM for 16 h. The cells were then centrifuged and the pellet frozen at −80 °C. After a minimum of 2 h the pellet was defrosted on ice for 15 min, re-suspended in 3 mL 50 mM MOPS buffer (pH 7.5) and sonicated in an MSE Soniprep 150 Plus sonicator (MSE UK Ltd., London, UK) for 10 bursts of 20 s at 16.5 μm amplitude on ice with 20 s rests in between. The suspension was centrifuged at 18,000 × g for 30 min at 4 °C and the supernatant passed through a 0.22 μm syringe filter (EMD Millipore Corp., MA, USA). The extract was supplemented with 20% w/v glycerol, the protein concentration quantified using Bradford reagent and then stored at −20 °C.
Enzyme assays to quantify phosphonate degrading activities in cell-free extracts
The PhnA, X and Z-like activities in cell-free extracts prepared from each isolate were examined via enzyme assays. Assay components for “PhnA”-type assays were 50 mM MOPS buffer (pH 7.5), 1 mM ZnSO4 and 10 mM phosphonoacetic acid. “PhnX”-type assays contained 50 mM Tris buffer (pH 8.5), 5 mM MgCl2 and 10 mM phosphonoacetaldehyde. 1-hydroxy-2-aminoethylphosphonate-degrading activity was determined using a modified version of the method described by McSorley et al. [21] and contained 50 mM MOPS buffer (pH 7.5), 2 mM α-ketoglutarate, 0.2 mM sodium ascorbate, 10 mM 1-hydroxy-2-aminoethylphosphonate, 0.1 mM Fe(NH4)2(SO4)2 and 0.1 mM CaCl2. Although McSorley et al. do not state that PhnZ has a requirement for calcium ions they observed that PhnZ appeared to bind half as much calcium as iron, and so the CaCl2 was included as a precaution.
In total 250 μL of reaction mix was prepared for each assay replicate. Triplicate samples were prepared for each assay, as well as triplicate extract-free and substrate-free controls. Addition of isolate protein extract (final concentration 0.5 mg protein mL−1) was always performed last and the tube briefly vortexed. A 125 μL T0 sample was immediately removed and, as PhnA, X and Z are all metalloenzymes, the reaction in this sample was stopped by mixing with 25 μL 100 mM EDTA, vortexing and freezing. The remaining volume was incubated for 16 h in a Sanyo ORBI-SAFE incubator at 28 °C and 100 RPM. At the end of the incubation period tubes were stopped as per the T0 samples.
Phosphate release was quantified using BIOMOL Green. Acetic acid quantification was carried out using an Acetic Acid Kit (Acetate Kinase Manual Format; Megazyme International Ireland, Co. Wicklow, Ireland) via the manufacturer’s recommended protocol for a microplate assay.
Determination of the nutrient conditions which induce the phosphonoacetic acid-cleaving protein in cell-free extracts of a 2AEP-degrading isolate by zymography
A 500 mL YTSS culture of isolate IMG22 was incubated at 28 °C and 150 RPM overnight. The cells were centrifuged at 10,000 × g for 10 min and washed twice in nutrient-free minimal marine medium before being resuspended in 6 × 50 mL volumes of minimal marine medium with no carbon, no nitrogen, no phosphorus, with 2 mM 2-aminoethylphosphonate as the sole nitrogen source, with all standard nutrients (replete) or with all nutrients and 2 mM 2-aminoethylphosphonate. Cultures were incubated and processed, as described above. Protein extracts were concentrated in Amicon Ultra centrifugal filters (Merck Chemicals Ltd., East Midlands, UK; 3 kDa nominal molecular weight limit) as per the manufacturers recommendations until a volume of <0.5 mL remained, at which point all extracts were made up to 0.5 mL using 50 mM pH 7.5 MOPS buffer with 20% w/v glycerol. The extracts were run on duplicate Novex WedgeWell 8–16% Tris-Glycine Mini 1 mm native-PAGE gels in a Novex X Cell 2 mini electrophoresis system (Life Technologies Ltd., Paisley, UK) using the protocol for native PAGE gels described by the manufacturer. Lane 1 contained Novex NativeMark unstained protein standards (Life Technologies Ltd., Paisley, UK), and the protein extracts were loaded into the other lanes in equal (200 μg) quantities. The gels were run at 125 V for 80 min and then removed from the cassettes as recommended by the manufacturer. Coomassie Brilliant Blue staining and destaining were carried out on one gel, as described by the manufacturer.
The second gel was used to assay phosphonoacetic acid cleaving activity via zymography. The gel was covered in 50 mM pH 7.5 MOPS buffer, 1 mM ZnSO4 and 10 mM phosphonoacetic acid and incubated at 30 °C for 2 h. The PhnA buffer was removed, the gel briefly rinsed in water and then covered with Fiske and Subbarow reagent (800 mg Fiske and Subbarow reducer in 5 mL water combined with 15 mL 21.5 mM ammonium molybdate tetrahydrate in 2.5 M H2SO4) to stain for phosphate release. After incubation at room temperature for 20 min the gel was briefly washed in water and then imaged.
Identification of the 2AEP degrading isolates by 16S rDNA sequencing
The sequence of the gene encoding 16S rRNA in each isolate was obtained using the 63f and 1387r universal primers described by Marchesi et al. [22]. PCR mixtures (25 μL) consisted of 1× Sigma PCR-buffer without MgCl2, 2.5 mM MgCl2, 3.5 pmol 63f primer, 7 pmol 1387r primer, 200 μM dATP, dCTP, dGTP and dTTP each (Life Technologies Ltd., Paisley, UK), 0.25 units of Taq DNA polymerase and a 1 μL tip of colony from a growing YTSSA plate of the isolate. The PCR was run on an Eppendorf Mastercycler Pro S PCR machine (Eppendorf UK Limited, Stevenage, UK) programmed for an initial 95 °C step for 4 min, and then following the protocol described by Marchesi et al. [22].
The PCR products were purified using a Wizard SV Gel and PCR Clean-Up System (Promega UK, Southampton, England) according to the manufacturer’s instructions and Sanger sequenced by GATC Biotech AG (Cologne, Germany) before being compared to the NCBI non-redundant nucleotide database excluding uncultured/environmental sample sequences [23].
Statistics
In all quantitative figures the data are the mean of three biological replicates and the error bars are presented as the Standard Error of the Mean. p-values were calculated using a Student’s t-test to a significance level of 95%.
Results
Inorganic phosphate starvation is not required for growth using 2-aminoethylphosphonate as a nitrogen source
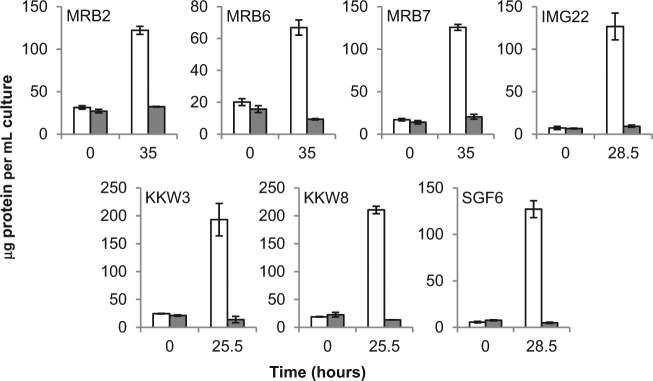
Although analytical limitations have hindered attempts to identify precisely which phosphonates are present in ocean water [24] many studies have identified 2-aminoethylphosphonate (2AEP) in common marine organisms [25–29] and it is believed to be the most abundant phosphonate in the oceans [11]. To investigate the possibility of phosphate-insensitive 2AEP mineralisation, selective enrichment cultures of Atlantic marine surface water samples taken from around the island of Ireland were set up with 2 mM 2AEP provided as the sole nitrogen source. Seven bacterial isolates were obtained which showed significantly higher optical densities (p ≤ 0.001 at the start of stationary phase, Supplementary Figure 1) and protein concentrations (p ≤ 0.002 in all cases, Fig. 1) when grown with 2 mM 2AEP as the sole nitrogen source than in non 2AEP-augmented controls, suggesting 2AEP catabolism despite the presence of 1 mM KH2PO4. In all seven isolates the metabolism of 2AEP was accompanied by the concomitant release of up to 86% of the aminophosphonate-derived phosphorus as inorganic phosphate (Supplementary Figure 1). No significant inorganic phosphate release was observed in control cultures in the absence of 2AEP.
Fig. 1.
Protein concentrations of isolate cultures with (white bars) and without (grey bars) 2 mM 2AEP as the sole nitrogen source at the start of growth and at the approximate beginning of stationary phase. Y axis shows protein concentration in μg per mL culture, X axis show time in hours, isolate identifiers are shown in the top left of each subfigure. Data are averages of biological triplicates; error bars show standard error of the mean
The phylogenetic identity of each isolate was determined by sequencing the gene for 16 S rRNA and comparing it to the NCBI non-redundant database (Supplementary Table 1). Three were closely related to the genus Falsirhodobacter, and the remaining four were members of the genera Rhodobacter, Sphingorhabdus, Terasakiella or Stappia. All of these isolates were therefore α-Proteobacteria, consistent with published data showing that marine α-Proteobacteria are a major source of phosphonate catabolism genes, and in particular of the phnA gene, in the ocean [12, 30].
The concentration of inorganic phosphate supplied in these experiments (1 mM) was more than two orders of magnitude greater than the peak concentration of soluble reactive phosphorus frequently found throughout the oceans (~3 μM: [31, 32]). The point at which microorganisms become phosphorus-starved is unclear: while Escherichia coli begins to express phosphorus starvation genes at ~4 μM inorganic phosphate [16], studies of phytoplankton communities in the Red Sea have shown that 0.1 μM soluble reactive phosphorus induces only low levels of phosphorus starvation-inducible protein production [33]. This is presumably due to communities from oligotrophic environments being more adapted to survival in low nutrient conditions than terrestrial or lab-adapted enteric bacteria such as E. coli.
Only the products of the PhnA enzyme are detected in cell-free extract assays
A number of microbial phosphonate degradation pathways, three of which specifically catabolise 2AEP, are known to exist (Supplementary Figure 2). Cell-free assays of the phosphonate-cleavage enzymes involved, termed PhnA, PhnX and PhnZ, have been performed in previous studies and act on the terminal substrates of different degradative pathways: phosphonoacetic acid as part of the PhnWYA pathway, phosphonoacetaldehyde as part of the PhnWX pathway, and 1-hydroxy-2-aminoethylphosphonic acid as part of the PhnYZ pathway (Supplementary Figure 2: [14, 21, 34, 35]). While the non-specific C-P lyase pathway has also been shown to break down 2AEP, in-vitro activity is lost when the cells are lysed [36], as in this cell-extract based analysis.
Cell-free extracts of each of the 7 isolates grown on 2AEP as a nitrogen source were prepared and phosphonate bond cleaving activity was assayed by measuring the amount of inorganic phosphate and organic product liberated from the terminal intermediate of each pathway. Only incubation of cell extracts with phosphonoacetic acid (10 mM) resulted in the release of organophosphonate derived inorganic phosphate (Fig. 2). Phosphonate cleavage was confirmed by the detection of equimolar concentrations of acetic acid (p ≥ 0.063 in all cases), the organic product of phosphonoacetic acid mineralisation (Fig. 3). The inorganic phosphate insensitive mineralisation of 2AEP in these isolates is therefore consistent with the phosphonoacetate hydrolase (PhnA) pathway, where 2AEP is sequentially converted to phosphonoacetaldehyde, phosphonoacetate and then acetic acid and inorganic phosphate by the PhnW, Y and A enzymes respectively (Supplementary Figure 2: [34]). The phnA gene has previously been shown to be the most abundant phosphonate cleavage gene in marine metagenomes, with up to 11.2% of expected bacterial genome equivalents containing a copy [12]. While a single instance of a phosphate-insensitive PhnA enzyme responsible for the direct catabolism of phosphonoacetate has been reported [34], no previous studies have shown the existence of a complete PhnWYA pathway for 2AEP degradation under phosphate-insensitive regulation.
Fig. 2.
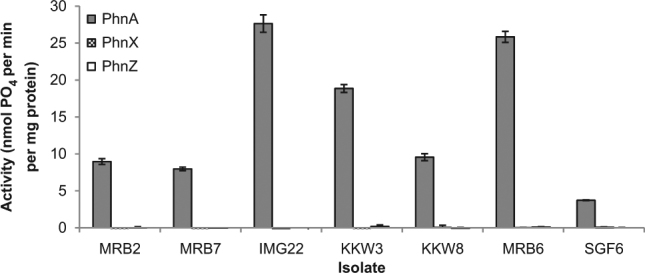
Inorganic phosphate producing activities (in nmol released per minute per mg protein) of isolate protein extracts when given substrates for PhnA (phosphonoacetic acid, grey bars), PhnX (phosphonoacetaldehyde, chequered bars) or PhnZ (1-hydroxy-2-aminoethylphosphonic acid, white bars) enzymes. Data are averages of biological triplicates; error bars show standard error of the mean
Fig. 3.
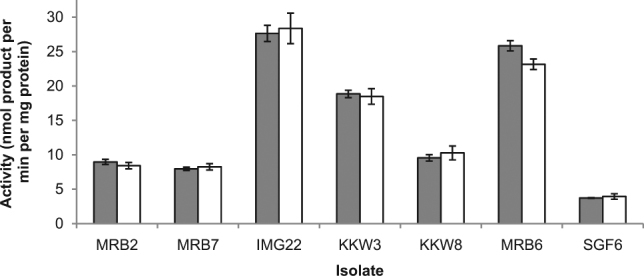
Inorganic phosphate and acetic acid producing activities (in nmol released per minute per mg protein) of isolate protein extracts. Inorganic phosphate is shown in the solid grey bars, and acetic acid in white bars. Data are averages of biological triplicates; error bars show standard error of the mean
The phosphonoacetic acid-cleaving enzyme is induced by the presence of 2AEP, not by starvation
To investigate the induction of the phosphonoacetic acid-degrading activity, isolate IMG22—an isolate closely related to the genus Falsirhodobacter—was selected for further study as it gave the highest activity levels in the initial experiments. A single pre-culture of IMG22 was split between 6 subcultures and each exposed to a different nutrient regime: one culture was supplied with all macronutrients (5 mM glucose, succinate and acetate each as carbon sources, 2 mM NH4Cl as a nitrogen source and 1 mM KH2PO4 as a phosphorus source); three cultures lacked any carbon or nitrogen or phosphorus sources, respectively, but were otherwise replete; one was replete but with the 2 mM NH4Cl replaced by 2 mM 2AEP; and one culture was replete and also contained 2 mM 2AEP as an additional nitrogen source. After 16 h of incubation proteins were extracted and a PhnA zymogram was prepared and stained for inorganic phosphate release [37] (Fig. 4).
Fig. 4.
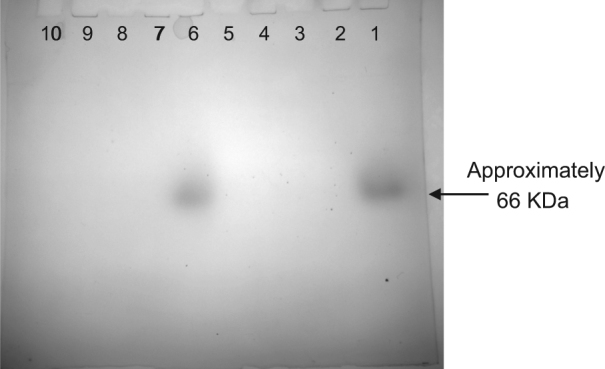
PhnA Zymogram of protein extracts from IMG22 cells exposed to different nutrient conditions. The dark spots in lanes 1 and 6 indicate phosphate release. Lane 1: cells in replete medium +2 mM 2AEP, lane 2: cells in replete medium, lane 4: -phosphorus, lane 6: 2 mM 2AEP as sole nitrogen source, lane 7: -nitrogen, lane 9: -carbon, lane 10: Novex NativeMark protein standards, lanes 3, 5 and 8: no sample
Only protein extracts from cells grown in the presence of 2AEP showed a zone of phosphate release in the zymogram after incubation in the presence of 10 mM phosphonoacetic acid, indicating phosphonoacetic acid cleavage and thus PhnA-like activity, regardless of the other nutrient conditions those cells were exposed to. No activity was observed in phosphorus, carbon or nitrogen starved samples (Fig. 4). Phosphonoacetate cleavage activity correlated with the position of a 66 kDa protein standard (Supplementary Figure 3), consistent with the size of previously characterised PhnA proteins [37].
Discussion
The synthesis of 2AEP and distribution of its catabolic genes appears to be almost ubiquitous in the marine environment [38]. This paired with data showing that many areas of the ocean are nitrogen and/or phosphorus limited, would suggest that 2AEP could be used as an organic nitrogen or organic phosphorus source by microorganisms which harbour aminophosphonate degradative gene clusters. Indeed, Karl [38] notes that common phosphonate degradation genes are present in distantly related bacterial groups, likely the result of horizontal gene transfer driven by strong selective pressures for nutrient acquisition systems. Despite this, no microorganism has been shown to degrade 2AEP via any pathway in inorganic phosphate-replete marine waters even though a single instance of PhnWX-mediated phosphate-insensitive 2AEP catabolism has been described in a terrestrial microbe [14]. If such activity were present in marine organisms this would further extend the potential of aminophosphonates to contribute to the marine carbon, nitrogen and phosphorus cycles.
To our knowledge this study is the first demonstration of growth via inorganic phosphate-insensitive 2AEP degradation by any pathway in aquatic bacteria, and suggests that aminophosphonate compounds are metabolically and biogeochemically active over a wider spectrum of inorganic phosphate concentrations than previously thought. In addition, this is the first demonstration of inorganic phosphate-insensitive 2AEP catabolism by a PhnWYA-like pathway in any organism: the pathway has previously only been demonstrated as a method for obtaining phosphorus from 2AEP under inorganic phosphate starvation conditions [34, 35]. This metabolism appeared to be relatively rapid, with the majority of phosphate release from 2AEP completed within 24 h (Supplementary Figure 1). This is mirrored by the rapid marine phosphonate synthesis demonstrated by Van Mooy et al. [4], and is consistent with their suggestion that phosphorus is quickly cycled between reduced and oxidised forms in the ocean. The exact concentration of 2AEP in bulk ocean water—and the concentration necessary to induce phosphate-insensitive catabolism—is not known, but previous studies have identified many 2AEP-synthesising marine organisms [29] and NMR studies have shown that phosphonates are continuously mineralised throughout the water column [3], in line with the almost ubiquitous distribution of 2AEP catabolic genes in marine systems [12]. Although lab-cultivatable microbes may not necessarily be representative of environmental communities as a whole, these isolates clearly demonstrate that phosphate insensitive aminophosphonate metabolism is present in the oceans.
The ability of some organisms to cycle aminophosphonates regardless of local inorganic phosphate concentrations would accelerate the remineralisation of the carbon, phosphorus and nitrogen contained within them and return these to nutrient pools which are more available to non-phosphonate degrading organisms. Some studies suggest that marine nitrogen limitation is more prevalent than marine phosphorus limitation [1], and so the catabolism of 2AEP as a nitrogen source may be more relevant to microbial function, nutrient turnover and biogeochemical cycling than the catabolism of 2AEP (or other aminophosphonates) as a phosphorus source. Furthermore, aminophosphonate nitrogen supply may explain the previously noted NMR study which showed preferential phosphonate consumption relative to phosphate esters under relatively high-phosphorus conditions [15].
It has been suggested that our understanding of the role of organic phosphorus in aquatic ecosystems is unduly focussed on fuelling phosphorus-limited planktonic growth, something termed the “phosphorus-limited planktonic view” [10]. This study, which demonstrated that some marine bacteria can catabolise the aminophosphonate 2AEP regardless of cellular phosphorus status, would suggest a wider role for phosphonates within the marine nutrient pool than is currently recognised. Our finding of inorganic phosphate-insensitive aminophosphonate catabolism parallels several studies which found relatively high levels of alkaline phosphatase activity in deep waters where marine inorganic phosphate concentrations tend to be highest [39–41]. This growing body of research showing that organic phosphorus catabolism is not always controlled by environmental phosphate concentrations suggests a wider role than that previously ascribed by the “phosphorus limited planktonic view”, one encompassing the supply and recycling of bioavailable carbon and nitrogen in addition to phosphorus to the marine ecosystem.
Electronic supplementary material
Acknowledgements
We thank F. Hammerschmidt and K. Pallitsch, University of Vienna, Austria, and the late H.B.F. Dixon, Cambridge University, UK, for the provision of several phosphonate substrates, and J. Megaw, Queen’s University Belfast, UK, for the provision of a marine water sample. JPC was funded by the Department for Employment and Learning, Northern Ireland, UK.
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
The online version of this article (10.1038/s41396-017-0031-7) contains supplementary material, which is available to authorized users.
References
- 1.Moore CM, Mills MM, Arrigo KR, Berman-Frank I, Bopp L, Boyd PW, et al. Processes and patterns of oceanic nutrient limitation. Nat Geosci. 2013;6:701–10. doi: 10.1038/ngeo1765. [DOI] [Google Scholar]
- 2.Kolowith LC, Ingall ED, Benner R. Composition and cycling of marine organic phosphorus. Limnol Oceanogr. 2001;46:309–20. doi: 10.4319/lo.2001.46.2.0309. [DOI] [Google Scholar]
- 3.Clark LL, Ingall E, Benner R. Marine organic phosphorus cycling; novel insights from nuclear magnetic resonance. Am J Sci. 1999;299:724–37. doi: 10.2475/ajs.299.7-9.724. [DOI] [Google Scholar]
- 4.Van Mooy BAS, Krupke A, Dyhrman ST, Fredricks HF, Frischkorn KR, Ossolinski JE, et al. Major role of planktonic phosphate reduction in the marine phosphorus redox cycle. Science. 2015;348:783–5. doi: 10.1126/science.aaa8181. [DOI] [PubMed] [Google Scholar]
- 5.Mukhamedova KS, Glushenkova AI. Natural phosphonolipids. Chem Nat Compd. 2000;36:329–41. doi: 10.1023/A:1002804409503. [DOI] [Google Scholar]
- 6.Steiner S, Conti SF, Lester RL. Occurrence of phosphonosphingolipids in Bdellovibrio bacteriovorus strain UKi2. J Bacteriol. 1973;116:1199–211. doi: 10.1128/jb.116.3.1199-1211.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 8.Martínez A, Ventouras LA, Wilson ST, Karl DM, DeLong EF. Metatranscriptomic and functional metagenomic analysis of methylphosphonate utilization by marine bacteria. Front Microbiol. 2013;4:340. doi: 10.3389/fmicb.2013.00340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB, et al. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature. 2006;439:68–71. doi: 10.1038/nature04203. [DOI] [PubMed] [Google Scholar]
- 10.Heath RT. Microbial turnover of organic phosphorus in aquatic systems. In: Turner BL, Frossard E, Baldwin D, editors. Organic phosphorus in the environment. Cambridge MA: CABI Publishing; 2005. p. 185–204.
- 11.Martinez A, Tyson GW, DeLong EF. Widespread known and novel phosphonate utilization pathways in marine bacteria revealed by functional screening and metagenomic analyses. Environ Microbiol. 2010;12:222–38. doi: 10.1111/j.1462-2920.2009.02062.x. [DOI] [PubMed] [Google Scholar]
- 12.Villarreal-Chiu JF, Quinn JP, McGrath JW. The genes and enzymes of phosphonate metabolism by bacteria, and their distribution in the marine environment. Front Microbiol. 2012;3:19. doi: 10.3389/fmicb.2012.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGrath JW, Chin JP, Quinn JP. Organophosphonates revealed: new insights into the microbial metabolism of ancient molecules. Nat Rev Microbiol. 2013;11:412–9. doi: 10.1038/nrmicro3011. [DOI] [PubMed] [Google Scholar]
- 14.Ternan NG, Quinn JP. Phosphate starvation-independent 2-aminoethylphosphonic acid biodegradation in a newly isolated strain of Pseudomonas putida, NG2. Syst Appl Microbiol. 1998;21:346–52. doi: 10.1016/S0723-2020(98)80043-X. [DOI] [PubMed] [Google Scholar]
- 15.Benitez-Nelson CR, O’Neill L, Kolowith LC, Pellechia P, Thunell R. Phosphonates and particulate organic phosphorus cycling in an anoxic marine basin. Limnol Oceanogr. 2004;49:1593–604. doi: 10.4319/lo.2004.49.5.1593. [DOI] [Google Scholar]
- 16.Wanner BL. Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int. 1996;49:964–7. doi: 10.1038/ki.1996.136. [DOI] [PubMed] [Google Scholar]
- 17.Tyrrell T. The relative influences of nitrogen and phosphorus on oceanic primary production. Nature. 1999;400:525–31. doi: 10.1038/22941. [DOI] [Google Scholar]
- 18.Karl DM, Björkman KM, Dore JE, Fujieki L, Hebel DV, Houlihan T, et al. Ecological nitrogen-to-phosphorus stoichiometry at station ALOHA. Deep Sea Res Part II Top Stud Oceanogr. 2001;48:1529–66. doi: 10.1016/S0967-0645(00)00152-1. [DOI] [Google Scholar]
- 19.Krieg NR, Holt JG. Enrichment and isolation. In: Gerhardt P editor. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. p. 112–42. .
- 20.Difco Laboratories. Difco manual of dehydrated culture media and reagents for microbiological and clinical laboratory procedures. 9th edn, Detroit: Difco Laboratories; 1953.
- 21.McSorley FR, Wyatt PB, Martinez A, DeLong EF, Hove-Jensen B, Zechel DL. PhnY and PhnZ comprise a new oxidative pathway for enzymatic cleavage of a carbon–phosphorus bond. J Am Chem Soc. 2012;134:8364–7. doi: 10.1021/ja302072f. [DOI] [PubMed] [Google Scholar]
- 22.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, et al. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol. 1998;64:795–9. doi: 10.1128/aem.64.2.795-799.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Young CL, Ingall ED. Marine dissolved organic phosphorus composition: insights from samples recovered using combined electrodialysis/reverse osmosis. Aquat Geochem. 2010;16:563–74. doi: 10.1007/s10498-009-9087-y. [DOI] [Google Scholar]
- 25.Hori T, Arakawa I, Sugita M. Distribution of ceramide 2-aminoethylphosphonate and ceramide aminoethylphosphate (sphingoethanolamine) in some aquatic animals. J Biochem. 1967;62:67–70. doi: 10.1093/oxfordjournals.jbchem.a128637. [DOI] [PubMed] [Google Scholar]
- 26.Kittredge JS, Hughes RR. The Occurrence of α-Amino-β-phosphonopropionic Acid in the Zoanthid, Zoanthus sociatus, and the Ciliate, Tetrahymena pyriformis. Biochemistry. 1964;3:991–6. doi: 10.1021/bi00895a026. [DOI] [PubMed] [Google Scholar]
- 27.Kittredge JS, Roberts E, Simonsen DG. The occurrence of free 2-aminoethylphosphonic acid in the Sea Anemone, Anthopleura elegantissima. Biochemistry. 1962;1:624–8. doi: 10.1021/bi00910a013. [DOI] [PubMed] [Google Scholar]
- 28.Quin LD. The presence of compounds with a carbon-phosphorus bond in some marine invertebrates. Biochemistry. 1965;4:324–30. doi: 10.1021/bi00878a022. [DOI] [Google Scholar]
- 29.Quin LD, Quin GS. Screening for carbon-bound phosphorus in marine animals by high-resolution 31P-NMR spectroscopy: coastal and hydrothermal vent invertebrates. Comp Biochem Physiol B Biochem Mol Biol. 2001;128:173–85. doi: 10.1016/S1096-4959(00)00310-9. [DOI] [PubMed] [Google Scholar]
- 30.Shilova IN, Robidart JC, James Tripp H, Turk-Kubo K, Wawrik B, Post AF, et al. A microarray for assessing transcription from pelagic marine microbial taxa. ISME J. 2014;8:1476–91. doi: 10.1038/ismej.2014.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karl DM, Björkman KM. Dynamics of DOP. In: Hansell DA, Carlson CA, editors. Biogeochemistry of marine dissolved organic matter. Academic Press Inc, London, UK; 2002. p. 249–366.
- 32.Paytan A, McLaughlin K. The oceanic phosphorus cycle. Chem Rev. 2007;107:563–76. doi: 10.1021/cr0503613. [DOI] [PubMed] [Google Scholar]
- 33.Mackey KRM, Labiosa RG, Calhoun M, Street JH, Post AF, Paytan A. Phosphorus availability, phytoplankton community dynamics, and taxon-specific phosphorus status in the Gulf of Aqaba, Red Sea. Limnol Oceanogr. 2007;52:873–85. doi: 10.4319/lo.2007.52.2.0873. [DOI] [Google Scholar]
- 34.Cooley NA, Kulakova AN, Villarreal-Chiu JF, Gilbert JA, McGrath JW, Quinn JP. Phosphonoacetate biosynthesis: in vitro detection of a novel NADP(+)-dependent phosphonoacetaldehyde-oxidizing activity in cell-extracts of a Marine Roseobacter. Microbiology. 2011;80:335–40. doi: 10.1134/S0026261711030076. [DOI] [PubMed] [Google Scholar]
- 35.Borisova SA, Christman HD, Metcalf MEM, Zulkepli NA, Zhang JK, van der Donk WA, et al. Genetic and biochemical characterization of a pathway for the degradation of 2-aminoethylphosphonate in Sinorhizobium meliloti 1021. J Biol Chem. 2011;286:22283–90. doi: 10.1074/jbc.M111.237735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Metcalf WW, Wanner BL. Mutational analysis of an Escherichia coli fourteen-gene operon for phosphonate degradation, using TnphoA’ elements. J Bacteriol. 1993;175:3430–42. doi: 10.1128/jb.175.11.3430-3442.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McGrath JW, Quinn JP. A plate assay for the detection of organophosphonate mineralization by environmental bacteria, and its modification as an activity stain for identification of the carbon-phosphorus bond cleavage enzyme phosphonoacetate hydrolase. Biotechnol Tech. 1995;9:497–502. doi: 10.1007/BF00159565. [DOI] [Google Scholar]
- 38.Karl DM. Microbially mediated transformations of phosphorus in the sea: new views of an old cycle. Annu Rev Mar Sci. 2014;6:279–337. doi: 10.1146/annurev-marine-010213-135046. [DOI] [PubMed] [Google Scholar]
- 39.Hoppe H, Ullrich S. Profiles of ectoenzymes in the Indian Ocean: phenomena of phosphatase activity in the mesopelagic zone. Aquat Microb Ecol. 1999;19:139–48. doi: 10.3354/ame019139. [DOI] [Google Scholar]
- 40.Koike I, Nagata T. High potential activity of extracellular alkaline phosphatase in deep waters of the central Pacific. Deep Sea Res Part II Top Stud Oceanogr. 1997;44:2283–94. doi: 10.1016/S0967-0645(97)00025-8. [DOI] [Google Scholar]
- 41.Baltar F, Arístegui J, Sintes E, van Aken HM, Gasol JM, Herndl GJ. Prokaryotic extracellular enzymatic activity in relation to biomass production and respiration in the meso- and bathypelagic waters of the (sub)tropical Atlantic. Environ Microbiol. 2009;11:1998–2014. doi: 10.1111/j.1462-2920.2009.01922.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.