Abstract
Background
A sudden increase in Salmonella Thompson (S. Thompson) cases distributed throughout three border regions in the province of Quebec in November 2016 triggered a provincial investigation to identify a common source of contamination and to put the appropriate control measures into place.
Objective
To report on the outbreak and to describe the use of genomic sequencing to identify the salmonella serotype responsible.
Methods
A descriptive survey of all reported cases of Salmonella serogroup C1 that had occurred between October 1, 2016 and February 15, 2017 was conducted. A case definition was developed. Pulsed field gel electrophoresis supplemented by analyses of genome sequences using the single nucleotide variant phylogenomics method were used to demarcate and manage the outbreak.
Results
Eighteen cases of S. Thompson were identified through whole genome sequencing. The onset dates of symptoms for the 16 cases that presented enteric symptoms were November 21—December 2, 2016. Two cases that presented with atypical symptoms were not reported until February 2017. Among the 18 cases, 16 had eaten or probably eaten chicken shawarma at the same restaurant chain and nine of these cases ate it at the same restaurant. In total, five restaurants from this chain, spread throughout three border regions of Quebec, were identified.
Conclusion
Outbreaks associated with chicken shawarma have been identified in the past. Efforts must be made to ensure that the owners of this type of restaurant know the contamination risk associated with this type of cooking and take the necessary steps to reduce this risk. The use of the genome sequencing method was very useful in defining the outbreak.
Introduction
Salmonella Thompson (S. Thompson) is a salmonella serotype belonging to serogroup C1, which occurs sporadically year-round. Since 2012, an average of 60 to 70 cases are reported annually in the province of Quebec, corresponding to between three and six cases a month, according to data from the mandatory reportable disease system (Maladies à Déclaration Obligatoire, MADO); however, in November 2016 alone, 12 cases of S. Thompson were reported to the Quebec public health agencies (Direction de la Santé Publique, DSPublique).
Details of outbreaks associated with S. Thompson have been published previously (1), (2), (3), (4). One outbreak which occurred in 2012 involved 1,149 confirmed cases in the Netherlands, which was attributed to the consumption of smoked salmon (1). Other studies have identified various vehicles: bread, possibly contaminated by a food handler (2); fresh cilantro (3); and rucola grown in Italy (4). In Canada, two national outbreaks have been investigated. The first outbreak occurred in 2012 with 105 cases, 29 of which were in Quebec, and the source was not identified. The second outbreak occurred in 2014 with 59 confirmed cases, 16 of which were in Quebec, and the most likely source of contamination was chicken (unpublished data).
On December 2, 2016, DSPublique of Montréal notified the Bureau de surveillance et de vigie (BSV) of the Ministère de la Santé et des Services sociaux (MSSS) of a time-place cluster of five cases of Salmonella serogroup C1 detected in a group of persons aged 13 to 19 years. The suspected source of contamination was a fast food restaurant chain serving chicken shawarma (shish taouk). The serotyping of the initial cases enabled detection of the Thompson serotype. On December 15, 2016, after cases appeared in other regions of Quebec surrounding Montréal, the BSV launched and coordinated a provincial investigation. The objective of the investigation was to identify the source of the outbreak and adopt the appropriate control measures.
Methods
Case reporting
In Quebec, salmonellosis is a mandatory reportable disease (MADO). Infections detected by hospital laboratories are reported to regional DSPublique agencies. Isolates are then sent to the Laboratoire de santé publique du Québec (LSPQ) for detailed characterization. Outbreaks and clusters are surveyed by regional DSPublique agencies.
Outbreak detection
The Montréal DSPublique conducts a daily watch of MADOs in its territory. SaTScanTM (version 9.4.2) statistical analysis software is used to detect time and time-place clusters. Clusters of enteric diseases are surveyed based on certain criteria, including the number of cases, the density of the cluster, demographic factors with an unusual distribution and the specificity of the pathogen.
A time-place cluster (spatio-temporal permutation method) of eight cases of salmonellosis was detected on December 2, 2016. Among these cases, five were serogroup C1, one was serogroup D and two are pending identification of the serogroup. Excluding the serogroup D case, the seven remaining salmonellosis cases were considered part of a potential cluster, which included four youths aged 13 to 19 years, who were prioritized for surveying as a common event was suspected. After identifying a restaurant chain as a probable common source for the outbreak, the survey was expanded to other age groups. Subsequent surveys reinforced the suspicion of a common source and the presence of an outbreak.
Epidemiological surveys
The regional survey forms were used prior to launching the provincial survey. The BSV, which coordinates the investigation of provincial outbreaks, asked the DSPublique to survey all cases of Salmonella serogroup C1 using a hypothesis-generating food survey form, prior to obtaining the serotype in order to reduce survey time.
The information gathered on the survey forms was demographic, clinical and dietary food (consumption in the home or at restaurants in the three days prior to the onset of symptoms, etc.). Once completed, the questionnaires were numbered and sent to the BSV and the Ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec (MAPAQ). Data analysis was descriptive in nature. Data were compiled and analyzed using the EXCEL (Microsoft Office 2010) program. The surveys were conducted between December 15, 2016 and February 15, 2017.
Laboratory analyses
Salmonella serogroup C1 strains from the regional laboratories were serotyped at LSPQ. Pulsed field gel electrophoresis (PFGE) was performed at the LSPQ on several S. Thompson isolates received in November and December 2016.
In addition, as S. Thompson behaves in a very clonal manner, whole genome sequencing was used at the LSPQ on S. Thompson isolates with sampling dates between September 22, 2016 and February 3, 2017. A phylogenetic tree, constructed using the maximum likelihood method with the single nucleotide variant phylogenomics (SNVPhyl) pipeline, served to determine the level of proximity of isolates, based on position and the number of robust single nucleotide polymorphisms (SNP) of genomes, allowing identification of the strains that caused the outbreak.
A case definition was developed: a case was confirmed for a resident of, or a visitor to, Quebec who had a S. Thompson infection, whose onset of symptoms or sampling date was on or after October 1, 2016 and whose whole genome sequence was identical or similar (one nucleotide variation). This was designated ST7.
Food safety investigation
The MAPAQ proceeded to conduct a food safety investigation at the targeted establishments in collaboration with its representatives: the City of Montréal’s food inspection division (Division de l’inspection des aliments, DIA) and the Canadian Food Inspection Agency.
Follow up interviews conducted with regard to cases and interventions (in-person and by telephone) took place at the restaurants where the cases had been exposed, from the Montréal, Lanaudière and Montérégie regions, as well as at the central kitchen that supplies these restaurants. At the central kitchen, the Canadian Food Inspection Agency acted as an intermediary of the MAPAQ for the survey.
During interventions at each of the restaurants, an evaluation of critical control points was conducted to determine, among other things, whether the cooking method used for the preparation of chicken shawarma allowed a safe cooking temperature to be reached and to check cross contamination risks, storage temperature and the source of the food. During interventions at the central kitchen, a check was made of, among other things, the source of the targeted foods and the method of preparation of marinated chicken, and the restaurants that had received the batches of the chicken in question were identified. Food samples were taken at some of the chain’s restaurants and at the central kitchen. These samples came from different batches than those consumed by the cases, as the batches distributed during the outbreak period were no longer available.
Results
Descriptive epidemiology
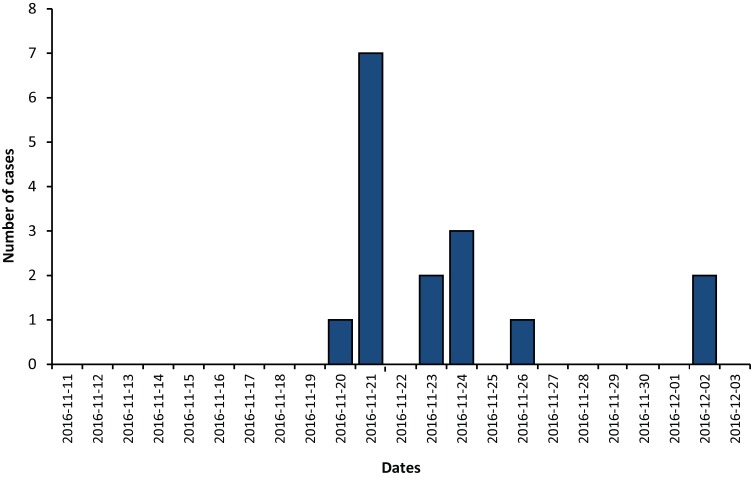
In total, 18 cases were associated with the outbreak, all corresponding to the confirmed case definition. The cases resided in the regions of Montréal (thirteen, all in the same area), Montérégie (three) and Lanaudière (two). Sixteen cases had enteric symptoms (Figure 1). The symptom onset dates were between November 21 and December 2, 2016. Two cases, not reported until February 2017, were found to be associated with the outbreak following laboratory results confirming the presence of the outbreak strain in a hemoculture following osteomyelitis and sampling from an anal abscess.
Figure 1. Epidemic curve according to the date of onset of enteric symptoms, for the outbreak of Salmonella Thompson pulsovar 1 in the Province of Quebec, November-December 2016.
Note: Two cases with atypical symptoms are not represented on the curve because the onset date of symptoms could not be clearly established (n=16). The S. Thompson pulsovar 1 was designated ST7 by whole genome sequencing method
Demographic data were available for all cases. The median age of cases was 25 years and the average was 27.8 years (range: less than one year to 69 years). The male-female ratio was 2:1. Six cases were hospitalized. No deaths were associated with the outbreak.
Food exposure
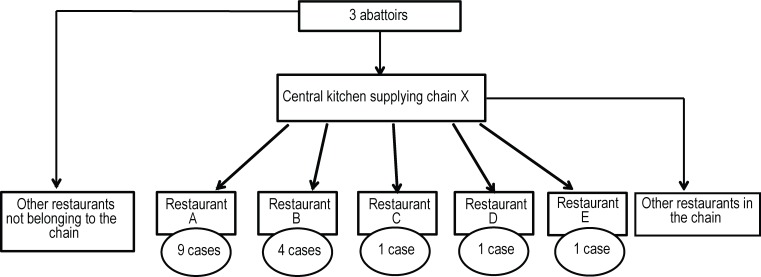
In the survey, 13 out of the 16 cases that presented with enteric symptoms reported having eaten chicken shawarma type food at restaurants associated with the same fast food chain during their exposure period, all within the last two weeks of November 2016. Nine cases ate at the same restaurant. A fourteenth case (out of the 16 cases) reported having consumed chicken shawarma regularly at one of the restaurants of this chain within that period, but was not able to specify the date.
The two cases reported in February 2017 did not present enteric symptoms. It was, therefore, difficult to determine an exact date of the onset of symptoms and establish an exposure period. These two cases reported that they probably consumed shawarma around the end of November at one of the restaurants identified in the outbreak because they ate there regularly.
Sixteen out of 18 cases reported either having eaten shawarma at the same restaurant chain in the three days prior to the onset of their illness or probably having eaten it at the restaurant chain because they ate there regularly. Three restaurants of the chain visited are located in Montréal and two restaurants are in neighbouring regions.
Of the two cases that did not frequent these restaurants, one had eaten chicken at an Asian restaurant and the other had purchased bagged raw turkey at a supermarket.
Laboratory analyses
The S. Thompson isolates analyzed by PFGE that were sampled in November or December 2016 were all pulsovar 1 (Quebec name) and STHXAI.0002/STHBNI.0015 (Canadian name). This pulsovar is common for S. Thompson in Quebec. Indeed, out of 440 strains of S. Thompson typed by EGCP at the LSPQ since 2002, 383 strains were pulsovar 1 (87%).
Genome sequencing was used to better distinguish between strains and to define the outbreak. Among the 25 isolates of S. Thompson sampled between September 22, 2016 and February 4, 2017, that were analyzed at the LSPQ using the SNVPhyl method, 18 had an identical genome sequence (designated ST7), which was the outbreak strain. The sampling dates for the 16 cases with enteric symptoms were between November 22 and December 15, 2016. The two cases with the less common clinical manifestations (osteomyelitis and anal abscess) were sampled on January 29 and February 3, 2017.
The 18 strains of the sequence type designated ST7 had either no nucleotide variation or only one nucleotide variation between them, which constitutes a strong genomic similarity (based on the SNVPhyl method). This cluster of strains is distinct from the other strains of S. Thompson sequenced during the same period, with between three and 771 SNP variations. The strain with 771 SNP is a strain of S. Thompson acquired on a trip, according to data from the public health branch in the region where the case originated.
Food safety investigation
During inspections by Montréal’s DIA and the MAPAQ, shortcomings were observed in the target establishments, including storage temperatures, cleaning and sanitation, and risks of cross contamination.
The restaurant chain in question is supplied by a central kitchen that distributes, among other things, raw marinated chicken to its affiliated restaurants. The central kitchen is supplied by three Quebec abattoirs. The batches of chicken targeted by this survey were slaughtered at these three abattoirs on November 7 and 8, 2016. They would have been marinated on November 10 and 14 at the central kitchen and sent out to the various restaurants in the chain between November 17 and 21, 2016. Several restaurants associated with Chain X, other than those targeted by the surveys and located in the Montréal, Montérégie, Laval and Lanaudière regions, would also have received the batches of chicken involved in the outbreak. According to the food safety surveys, approximately forty restaurants, including Chain X, received the batches of the chicken in question.
DIA inspection of an Asian restaurant, where food was consumed by a person who later exhibited symptoms of infection by S. Thompson belonging to the same strain identified in the outbreak, determined that the chicken supply for the restaurant came from two of the three abattoirs involved (Figure 2).
Figure 2. Distribution of Salmonella Thompson ST7 cases according to the place of consumption, Quebec 2016.
Note: The S. Thompson pulsovar 1 was designated ST7 by whole genome sequencing method
A total of 33 food samples were taken at the restaurants identified and at the central kitchen. No strains of S. Thompson were isolated. One of the samples from the Asian restaurant was found to be positive for S. enteritidis. A summary of the samples taken and the results of microbiological analysis are presented in Table 1.
Table 1. Sampling sites and foods sampled and analysed for Salmonella Thompson outbreak, Province of Quebec, 2016*.
| Sampling site | Foods sampled (# samples) | # Samples | Results of analyses for Salmonella Thompson |
|---|---|---|---|
| Central kitchen (supplier of marinated chicken to affiliated restaurants) | Raw chicken from Abattoir "1" [2x] Raw chicken from Abattoir "2" [2x] Raw marinated chicken [2x] |
6 | Absent |
| Restaurant 1 | Salad RTE mashed potato Cooked rice Cooked chicken Garlic sauce with mayonnaise Hummus |
6 | Absent |
| Restaurant 2 | Salad RTE mashed potato Cooked rice Cooked chicken Garlic sauce with mayonnaise House vinaigrette |
6 | Absent |
| Restaurant 3 | RTE lettuce [3x] RTE tomato Tahini Garlic sauce Hummus RTE turnip Cooked chicken [2x] Raw marinated chicken |
11 | Absent |
| Supplier (Asian restaurant) | Raw chicken | 4 | 3 Absent 1 sample positive for Salmonella enteritidis |
Abbreviation: RTE, ready-to-eat; #, number * The S. Thompson pulsovar 1 was designated ST7 by whole genome sequencing method
Discussion
In November 2016, the sudden increase in the number of reports of S. Thompson and their location in the same area of the Montréal region suggested a common source of contamination. The outbreak was delimited in time and space, all of the cases having occurred in three neighbouring regions. Chicken cooked in the shawarma style (shish taouk) was the common food eaten or probably eaten in the majority of these cases. Chicken shawarma is a specialty food that originates in the Middle East and is prepared by placing marinated meat (chicken, beef or lamb) on a spit to form a cone shape that is roasted in front of a grill. The meat around it is sliced off as needed and served in pita bread or with rice and condiments. This cooking method can lead to insufficient cooking, especially with chicken (5).
The restaurant chain identified in the survey was supplied by a central kitchen, which is itself supplied by three abattoirs in Quebec. It is therefore possible that one batch of chicken from one or more of these three abattoirs could have been contaminated by S. Thompson and distributed over the course of this period throughout this restaurant chain. According to the food safety surveys, about 40 restaurants received the batches of chicken in question; however, only five restaurants were implicated. Shortcomings were observed in the targeted restaurants during the food inspection.
Animals destined for food, including bird species, naturally carry pathogens in their intestinal tract that can contaminate raw meat products during slaughter and processing (6). In Canada, a recent study conducted from December 2012 to December 2013 demonstrated that the national prevalence of salmonella in batches of broiler chickens sampled at the abattoir was as high as 25.6%. Batches from chickens raised in the eastern provinces were more frequently tainted by salmonella. In processed products, which are whole chicken carcasses and parts of carcasses processed in establishments authorized by the federal government, the prevalence of salmonella reached 16.9% (whole chickens) and 29.6% (parts) (6). Similarly, samples of raw chicken products were taken from supermarket chains, butcher shops and independent grocers in 33 large cities in Canada. The prevalence of salmonella was found to be 21% (whole chickens) and 31.6% (parts) (6).
In the United States, limits of acceptability with regard to the percentage of chicken contaminated by salmonella were established by the food inspection agencies in 1996 (7). The percentage of broiler chicken contaminated by salmonella at the abattoir can reach a maximum of 20%. The performance standard is recognized under the Hazard Analysis and Critical Control Points (HACCP) system (8). Between 10 and 19% of abattoirs in the United States have been found to exceed this limit and this percentage is higher for small abattoirs (9), (10), (11), (12).
Several salmonella serotypes can be found in chicken and S. Thompson is part of the group of 12 salmonella serotypes most often found in raw chicken (13), (14).
Although meat-producing chicken is expected to be contaminated by salmonella, adequate cooking should render it inactive. This investigation suggests that the cooking method for making shawarma may represent a risk. Several outbreaks have been associated with this method of preparation (15), (16), (17), (18). With this method of cooking, the raw meat may be in contact with the cooked meat. When a restaurant is busy, it is possible that cooking times may not be adhered to and the meat served may contain parts that are undercooked. To prevent cross contamination or undercooking, restaurants of this type sometimes cook the meat a second time before serving it to customers. Cross contamination may also have occurred in the handling of the raw chicken.
Although the foods sampled did not demonstrate the presence of S. Thompson, the survey strongly suggests a link between the illness occurring and the consumption of chicken shawarma at these restaurants. The time between the onset of symptoms and the reporting of cases to public health authorities is around 10 to 14 days; thus, the chicken sampled at the central kitchen and in the restaurants did not come from the batch delivered and consumed during the cases’ exposure period. This delay is inherent to food outbreak investigations and may explain the negative results.
The isolates were sent to the LSPQ for PFGE to assess their degree of similarity; however, this bacterium shows little diversity and the PFGE pattern 1 is often identified in S. Thompson. Whole genome sequencing was needed to establish the genetic similarity between isolates and allow the outbreak to be delimited. Strains implicated in the outbreak were identical or had only a single nucleotide variation. The other strains of S. Thompson analyzed had three or more nucleotide variations and the epidemiological information available regarding the cases with these exposures was different. Genome sequencing has proven to be effective in several outbreaks (19), (20), (21), (22), (23). The use of the whole genome sequencing technique provides additional powers of discrimination, beyond serotyping and PFGE, to delimit and investigate an outbreak (20), (21), (22), (23), (24), (25). The results of genome sequencing must be interpreted based on available epidemiological information. This report represents one of the first Canadian outbreaks of salmonella to use whole genome sequencing in the case definition.
A survey reports only laboratory-confirmed cases, and it is likely that other people were affected but did not consult a physician or fecal cultures were not obtained. In the mandatory reportable disease system, only a fraction of actual cases are reported, which could explain why only five restaurants in the chain were identified even though the chicken from the same batch was distributed to more than 40 restaurants. Chicken was identified as the probable source of contamination as it is the food that is most likely to have been contaminated with S. Thompson.
In conclusion, we have documented an outbreak of salmonella associated with the consumption of shawarma meals in a series of restaurants. Other outbreaks associated with this type of product have been identified in the past and Health Canada has issued recommendations to prevent future outbreaks of enteric illnesses associated with the preparation of shawarma (5). Additional efforts may assist owners of this type of restaurant to be more aware of the contamination risk associated with this method of cooking and take the necessary steps to reduce the risk. If an outbreak does occur, genome sequencing has proven to be an important tool for defining the outbreak.
Authors’ statement
All of the authors (CG, MF, CD, DR, NS, AU, PAP, VU, SB) participate in enteric disease monitoring. CG, DR and SB prepared the first draft and all of the other authors contributed to the final version by adding comments and suggestions.
Acknowledgements
The authors would like to thank all the individuals in the public health branches who participated in the surveys, as well as the ministère de l’Agriculture, des Pêcheries et de l’Alimentation du Québec staff for their interventions in the various areas involved.
Footnotes
Conflicts of interest: None.
References
- 1.Friesema I, de Jong A, Hofhuis A, Heck M, van den Kerkhof H, de Jonge R et al. Large outbreak of Salmonella Thompson related to smoked salmon in the Netherlands, August to December 2012. Euro Surveill 2014. Oct;19(39):20918. 10.2807/1560-7917.ES2014.19.39.20918 [DOI] [PubMed] [Google Scholar]
- 2.Kimura AC, Palumbo MS, Meyers H, Abbott S, Rodriguez R, Werner SB. A multi-state outbreak of Salmonella serotype Thompson infection from commercially distributed bread contaminated by an ill food handler. Epidemiol Infect 2005. Oct;133(5):823–8. 10.1017/S0950268805004127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JV, Mohle-Boetani J, Reporter R, Abbott S, Farrar J, Brandl M et al. An outbreak of Salmonella serotype Thompson associated with fresh cilantro. J Infect Dis 2001. Mar;183(6):984–7. 10.1086/319254 [DOI] [PubMed] [Google Scholar]
- 4.Nygård K, Lassen J, Vold L, Andersson Y, Fisher I, Löfdahl S et al. Outbreak of Salmonella Thompson infections linked to imported rucola lettuce. Foodborne Pathog Dis 2008. Apr;5(2):165–73. 10.1089/fpd.2007.0053 [DOI] [PubMed] [Google Scholar]
- 5.Canadian Food Inspection Agency. National Microbiological Baseline Study in Broiler Chicken December 2012 – December 2013. http://www.inspection.gc.ca/food/chemical-residues-microbiology/food-safety-testing-reports/2016-08-17/december-2012-december-2013/eng/1471358115567/1471358175297
- 6.Health Canada. Management of the Risks Related to Consumption of Donairs and Similar Products (Gyros, Kebabs, Chawarmas and Shawarmas), 2008. http://www.hc-sc.gc.ca/fn-an/legislation/guide-ld/manage_ris_donair-eng.php
- 7.US Department of Agriculture. Food Safety and Inspection Service, 1996 - Nationwide Broiler Chicken Microbiological baseline data collection program. U.S. Department of Agriculture, Food Safety and Inspection Service, Washington, D.C. http:www.fsis.usda.gov/OPHS/baseline/broiler1.pdf
- 8.U.S. Department of Agriculture, Food Safety and Inspection Service. Pathogen Reduction: hazard analysis and critical control point (HACCP) systems; final rule. Fed Regist 1996;61:38806–989. Available from: http://www.fsis.usda.gov/OA/fr/haccp_rule.htm [Google Scholar]
- 9.Naugle AL, Barlow KE, Eblen DR, Teter V, Umholtz R. U.S. Food Safety and Inspection Service testing for Salmonella in selected raw meat and poultry products in the United States, 1998 through 2003: analysis of set results. J Food Prot 2006. Nov;69(11):2607–14. 10.4315/0362-028X-69.11.2607 [DOI] [PubMed] [Google Scholar]
- 10.Bohaychuk VM, Gensler GE, King RK, Manninen KI, Sorensen O, Wu JT et al. Occurrence of pathogens in raw and ready-to-eat meat and poultry products collected from the retail marketplace in Edmonton, Alberta, Canada. J Food Prot 2006. Sep;69(9):2176–82. 10.4315/0362-028X-69.9.2176 [DOI] [PubMed] [Google Scholar]
- 11.Rose BE, Hill WE, Umholtz R, Ransom GM, James WO. Testing for Salmonella in raw meat and poultry products collected at federally inspected establishments in the United States, 1998 through 2000. J Food Prot 2002. Jun;65(6):937–47. 10.4315/0362-028X-65.6.937 [DOI] [PubMed] [Google Scholar]
- 12.Eblen DR, Barlow KE, Naugle AL. U.S. Food safety and Inspection Service testing for Salmonella in selected raw meat and poultry products in the United States, 1998 through 2003: an establishment-level analysis. J Food Prot 2006. Nov;69(11):2600–6. 10.4315/0362-028X-69.11.2600 [DOI] [PubMed] [Google Scholar]
- 13.Shah DH, Paul NC, Sischo WC, Crespo R, Guard J. Population dynamics and antimicrobial resistance of the most prevalent poultry-associated Salmonella serotypes. Poult Sci 2017. Mar;96(3):687–702. [DOI] [PubMed] [Google Scholar]
- 14.Roy P, Dhillon AS, Lauerman LH, Schaberg DM, Bandli D, Johnson S. Results of salmonella isolation from poultry products, poultry, poultry environment, and other characteristics. Avian Dis 2002. Jan-Mar;46(1):17–24. 10.1637/0005-2086(2002)046[0017:ROSIFP]2.0.CO;2 [DOI] [PubMed] [Google Scholar]
- 15.Honish L, Zazulak I, Mahabeer R, Krywiak K, Leyland R, Hislop N et al. Outbreak of Escherichia coli O157:H7 gastroenteritis associated with consumption of beef donairs, Edmonton, Alberta, May-June 2006. Can Commun Dis Rep 2007. Jan;33(2):14–9. Available from: https://www.canada.ca/en/public-health/services/reports-publications/canada-communicable-disease-report-ccdr/monthly-issue/2007-33/rapid-emergence-methicillin-resistant-staphylococcus-aureus-mrsa-among-children-adolescents-northern-manitoba-2003-2006.html#out [PubMed] [Google Scholar]
- 16.Currie A. Outbreak of E. coli O157:H7 infections in Calgary Health Region, September to October, 2004: Summary report. Calgary: Calgary Health Region, 2005. [Google Scholar]
- 17.Evans MR, Salmon RL, Nehaul L, Mably S, Wafford L, Nolan-Farrell MZ et al. An outbreak of Salmonella typhimurium DT170 associated with kebab meat and yogurt relish. Epidemiol Infect 1999. Jun;122(3):377–83. 10.1017/S0950268899002253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Synnott M, Morse DL, Maguire H, Majid F, Plummer M, Leicester M et al. An outbreak of Salmonella mikawasima associated with doner kebabs. Epidemiol Infect 1993. Dec;111(3):473–81. 10.1017/S0950268800057204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le VT, Diep BA. Selected insights from application of whole-genome sequencing for outbreak investigations. Curr Opin Crit Care 2013. Oct;19(5):432–9. 10.1097/MCC.0b013e3283636b8c [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inns T, Ashton PM, Herrera-Leon S, Lighthill J, Foulkes S, Jombart T et al. Prospective use of whole genome sequencing (WGS) detected a multi-country outbreak of Salmonella Enteritidis. Epidemiol Infect 2017. Jan;145(2):289–98. 10.1017/S0950268816001941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor AJ, Lappi V, Wolfgang WJ, Lapierre P, Palumbo MJ, Medus C et al. Characterization of Foodborne Outbreaks of Salmonella enterica Serovar Enteritidis with Whole-Genome Sequencing Single Nucleotide Polymorphism-Based Analysis for Surveillance and Outbreak Detection. J Clin Microbiol 2015. Oct;53(10):3334–40. 10.1128/JCM.01280-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deng X, Shariat N, Driebe EM, Roe CC, Tolar B, Trees E et al. Comparative analysis of subtyping methods against a whole-genome-sequencing standard for Salmonella enterica serotype Enteritidis. J Clin Microbiol 2015. Jan;53(1):212–8. 10.1128/JCM.02332-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bekal S, Berry C, Reimer AR, Van Domselaar G, Beaudry G, Fournier E et al. Usefulness of High-Quality Core Genome Single-Nucleotide Variant Analysis for Subtyping the Highly Clonal and the Most Prevalent Salmonella enterica Serovar Heidelberg Clone in the Context of Outbreak Investigations. J Clin Microbiol 2016. Feb;54(2):289–95. 10.1128/JCM.02200-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schürch AC, Siezen RJ. Genomic tracing of epidemics and disease outbreaks. Microb Biotechnol 2010. Nov;3(6):628–33. 10.1111/j.1751-7915.2010.00224.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Köser CU, Ellington MJ, Cartwright EJ, Gillespie SH, Brown NM, Farrington M et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathog 2012;8(8):e1002824. 10.1371/journal.ppat.1002824 [DOI] [PMC free article] [PubMed] [Google Scholar]