Abstract
Sex steroid receptors act as ligand activated nuclear transcription factors throughout the body, including the brain. However, post-translational modification of these receptors can direct them to extranuclear sites, including the plasma membrane, where they are able to initiate rapid signaling. Because of the conserved domain structure of these receptors, alternative exon splicing can result in proteins with altered nuclear and extranuclear actions. Although much attention has focused on internal and C-terminal splice variants, both estrogen and androgen receptors undergo N-terminal truncations, as well. These truncated proteins not only influence the transcriptional activity of the full-length receptors, but also associate with caveolin and initiate signaling at the plasma membrane. Such actions may have important physiological consequences in neuronal, endothelial, and cancer signaling and cell survival.
Keywords: Membrane steroid receptor, Splice variant, Estrogen receptor, Androgen Receptor
Introduction
Receptors for androgens, estrogens, and progestins (ARs, ERs, PRs) belong to the steroid receptor superfamily of nuclear transcription factors that share a common structural organization (for reviews see: [1–3]). The defining feature of these receptors is a three domain structure centered around two zinc finger motifs in the DNA binding domain (DBD) that allow interaction with specific promoter elements termed hormone responsive elements (HREs). The carboxy-terminal domain (CTD) contains the ligand binding domain (LBD) and ligand dependent activation function 2 (AF2), the region responsible for recruiting coactivators and interacting with the basal transcriptional machinery to initiate activation of regulated genes. The amino-terminal domain (NTD) is the most variable region, and is primarily associated with the ligand-independent activation function 1 (AF1), that also interacts with AF2 in the ligand-activated state. In addition to direct DNA binding, these receptors can regulate transcription through protein-protein interactions with other transcription factors to modulate their activity.
As nuclear transcription factors, sex steroid receptors (SRs) are primarily localized to the nucleus, but nuclear-cytoplasmic shuttling and post-translational modifications act to localize a portion of SRs to the plasma membrane. The best-described examples of these actions are from cancer and endothelial cells that demonstrate a small proportion of SRs are trafficked to the plasma membrane through palmitoylation of the CTD [4] and interactions with caveolin-1 [5–7]. A role for rapid steroid actions in neurons has been recognized since the 1970’s, but the nature of the receptors underlying the various effects of steroids in the brain continues to evolve [8].
In addition to trafficking of full-length receptors to the membrane and the discovery of structurally unrelated membrane SRs, evidence from studies of ERs and PRs supports a role for N-terminal truncated receptors as mediators of extra-nuclear and membrane actions. Recent evidence from our [9, 10] lab supports the role of membrane-associated NTD truncated androgen receptor in membrane lipid rafts of neurons (Figure 1). In this mini-review, we examine the potential role of SR NTD splice variants in rapid neuronal signaling (Figure 2).
Figure 1.

N27 cells express AR45 protein in membrane lipid rafts. N27 cells were treated with 100 nM testosterone for 24 hours to stabilized the androgen receptor. N27 cells were homogenized and separated into membrane, cytosol, and nuclear fractions. The membrane portion of the cells were further separated into 9 fractions using a sucrose gradient and ultracentrifugation in order to examine lipid rafts. Primary antibodies targeting AR45 (Santa Cruz sc-815/AR-C19 androgen receptor antibody) and lipid raft markers (Cell Signaling 3267 caveolin-1 antibody) were used. AR45 was only observed in caveolin positive lipid raft fractions. N=3 per treatment group.
Figure 2.
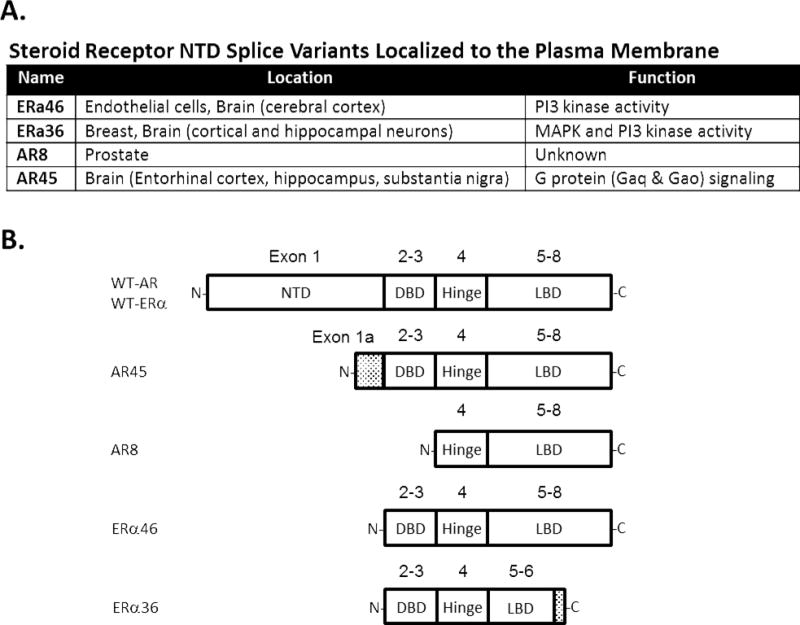
Characterized N-terminal deletions of AR and ER. A: Location and signaling function of N-terminal deleted variants. B: Structure of variants ARs and ERs relative to the wild-type (WT) full-length receptors. Exons are noted above domain schematics. Shaded areas represent unique sequences not present in the WT receptors.
Steroid receptor splice variants
In addition to steroid receptor isoforms encoded by different genes (i.e ERα, ERβ) steroid receptors undergo extensive alternative splicing. [11]. Alternative 5′ untranslated regions (UTRs)/exons appear to be a common characteristic of SRs including glucocorticoid receptors [12, 13], mineralocorticoid receptors [14, 15], ERs [16–18], and ARs [19]. In most cases, these alternative exon sequences do not affect the coding sequence of the SR, but rather tissue specific distribution or expression levels. However, as discussed below, in some cases, alternative splicing of the NTD yields SRs with altered expression, localization, and function. With respect to the functional properties of SR splice variants, those that result in changes to the coding region have been the subject of more study. Numerous splice variants in the coding sequence of SRs have been detected in peripheral tissues, and are abundant in cancer cells. For example, the sensitivity of RT-PCR allows the detection of 18 different ERα splice variants and 17 different PR splice variants in the human endometrium [20, 21] and brain [22]. Depending on the exon(s) spliced out of the coding sequence, these splice variants can result in transcripts that produce either functional or non-functional proteins. The domain structure of SRs and the close overlap of exons with functional domains allow functional splice variants to act as constitutively active, dominant negative, or modulatory factors for full-length receptors [11]. Some of these variants result in internal start codons that produce functional proteins. However, beyond their transcriptional actions, SR variants may also mediate various non-genomic actions depending on their ability to interact with cytoplasmic and membrane proteins through their remaining domains.
Estrogen receptor NTD splice variants
Using RT-PCR, numerous ERα splice variants have been detected in the rat and human brain. In the rat brain, these include deletions of exon 3 [23], exon 4 [24, 25], exons 3/4 [25], and exons 5/6 [25]. Alternative promoter expression can also result in an NTD truncation of exon 1 in ERα [26]. This deletion produces a 46 kDa protein (ERα46) from a start codon early in exon 2, and is also found in mice and humans [26]. In transfected cells overexpressing ERα46, this variant appears predominantly nuclear and can stimulate transcription in response to estrogen, but inhibits the activity of the full-length ERα66 [26]. However, in human endothelial cells ERα46 localizes to the plasma membrane, where it can mediate the estrogen-induced stimulation of endothelial nitric oxide synthase (eNOS) through interactions with PI3 kinase [27, 28]. Similarly, ERα46 interacts with PI3K p85a subunit in rat cerebral cortex and this interaction declines with age [29]. ERα46 and additional isoforms are also localized to rat cerebral endothelial cells [30], but it is unclear whether it is localized to neurons.
A second major NTD truncated ERα, first noted in endometrium [27] and cloned from human breast tumors [31], is ERα36. Like ERα46, ERα36 lacks exon 1 [27, 31–33]. However, it also skips exons 6 and 7 of the full-length ERα and has a unique 27 amino acid CTD [33]. Thus, the resulting protein lacks both the AF1 and much of the AF2/LBD. In the breast, ERα36 is predominantly localized to the plasma membrane where it mediates estrogen activation of MAPK and PI3/Akt signaling [34, 35]. Further evidence from transfection experiments supports a role for ERα36 in the mobilization of calcium in breast cancer cells [36]. In addition to its role at the membrane, there is also evidence that ERα36 can inhibit transcriptional activation by the full-length ERα66 [34]. Although initially found in peripheral tissues peripheral tissues, ERα36 has also been localized to neurons in the cortex and hippocampus of rats where it is mostly extra-nuclear [37]. In both the rat and human brain ERα36 is associated with caveolin-1 in cortical and hippocampal neurons, confirming membrane localization [37]. Ovariectomy and cerebral ischemia both appear to reduce ERα36 levels in the rat hippocampus, and actions of a selective agonist for ERα36 suggest that it may be involved in neuroprotection afforded by estrogen and tamoxifen [37, 38]. In human neuroblastoma cells, knockdown of ERα36 attenuates estrogen-induced activation of MAPK and Akt and neuroprotection against H2O2 toxicity, further supporting a rapid signaling and protective role for this splice variant [39]. In addition to neurons, ERα36 is also found in glioblastoma cells where it may be involved in tamoxifen resistance [40].
A third NTD splice variant, termed MB-1, has also been observed in the human brain [41]. MB-1 lacks 168 nucleotides in the middle of exon 1, significantly reducing transcriptional activity [42]. Antibodies raised against MB-1 demonstrate that it present in astrocytes, endothelial cells, and neurons in several areas of the human brain including the hypothalamus, hippocampus, and amygdala and appears primarily cytoplasmic [43]. A role for MB-1 in rapid estrogen signaling has not been examined.
In rodents, truncated estrogen receptor products (TERPs) lacking exons 1–4 and having unique untranslated N-terminal region were first identified in the rat pituitary [44]. In the mouse TERPs transcripts are present in several tissue, including the brain [45]. An internal start codon in exon 5 leads to the production of a protein containing most of the LBD. TERPs have been localized to both the nucleus [46] and extra-nuclear compartments [45]. TERPs show some basal transcriptional activity and can inhibit transcriptional actions of both ERα and ERβ [45–47]. However, rapid effects of these truncated receptors have not been examined.
Several splice variants of the ERβ are also present in the human and rodent brain. However, in the human all of the variants identified are truncations of the CTD [48] and result in dominant negative transcriptional effects on full length ERs [49]. Similarly, several ERβ splice variants have been detected in the rat brain, including both insertions and deletions [50]. However, like the human, none of the variants identified thus far include truncations of the NTD, and all except ERβ1Δ4 appear nuclear [50, 51].
The membrane-dependent actions of estrogens are not solely dependent on the classical ERs, as other ER-binding proteins, namely the G protein-coupled ER, can mediate rapid effects on kinase signaling [52, 53].
Progesterone receptor NTD splice variants
Alternate translational start sites in the PR gene lead to the expression of two functional proteins, PR-A and PR-B. PR-B has an additional 165 amino acids at the N- terminus and an additional activation domain, AF-3 [54]. Both isoforms are expressed in neurons in addition to a large number of splice variants [55, 56]. As such, PR-A can be considered an NTD partial deletion. However, rapid extranuclear actions of progesterone, including activation of MAPK, appear to be mediated by PR-B, rather than PR-A [57, 58] even though both isoforms have an Src-interacting domain that is able to bind Src and activate MAPK [59]. Despite clear evidence for rapid membrane actions mediated through PR-A and PR-B [60], the role of N-terminal splice variants is less clear. In contrast, most CTD variants act as dominant negative regulators of the nuclear effects of PR-A and PR-B [61]. Three additional NTD deleted PRs have been identified in peripheral tissues including one lacking exon 1 (PR-C), and two lacking exons 1–3 (PR-S and PR-T) [55, 56]. PR-S and PR-T transcripts contain different intronic 5′UTRs, but would likely produce the same AF1/AF3/DBD deleted protein from an internal start codon in exon 4 [55]. An additional N-terminal truncated PR with a potential signal peptide, PR-M, was also cloned from a human aortic cDNA library [62]. However, careful analysis by Samalecos and Gellersen suggest that none of the proposed NTD truncated variants produced functional proteins [63]. In ultrastructural studies extra-nuclear PRs are found in neurons and glial cells in the rat and mouse brain [64, 65], but the nature of these PRs is not known since the antibodies used in these studies would detect PR-A, PR- B, and NTD truncated variants. Evidence for several additional progesterone-activated membrane receptors, suggests that the rapid actions of progestins may not depend on membrane trafficking of the classical PRs [66]. Instead, additional progesterone-binding proteins (mPRs) originally identified in fish have been localized in humans and several other vertebrates [67]. Among these 7-transmembrane proteins is mPRβ, which appears to be enriched in the human brain [67] and at least three mPRs that are present in the rodent spinal cord, including in glia and neurons [68, 69], and the PR membrane component, PGRMC1, that is widely expressed in the brain [70].
Androgen receptor NTD splice variants
Increasing evidence strongly supports AR involvement in rapid non-genomic signaling, such as membrane-associated AR (mAR) activation of different signaling cascades in peripheral cell types. Common signaling cascades initiated by mAR include intracellular calcium release via the phospholipase C (PLC) pathway and G-protein coupled receptor signaling [71–77]. Similarly, non-genomic AR action has been observed in the brain. Studies have shown that a neuronal mAR can also activate several signaling pathways involved in intracellular calcium release, oxidative stress, cell survival, inflammation, and even pathways associated with addiction [10, 78–84]. Interestingly, the classical antagonist for the AR (flutamide) does not alter membrane-associated AR’s non-genomic effects, regardless of cell type [10, 80, 83, 85]. Therefore, the membrane-associated AR may have a different structure or confirmation than the classical AR.
Splice variants have been proposed as candidates for the mAR. Since the mAR is responsive to androgenic agonists, the AR splice variant must contain a ligand binding domain. Only two AR splice variants, AR8 and AR45, meet this qualification. These two AR splice variants have truncated N-terminal domains [86–89]. AR8 has only been observed in prostate cell lines, and has no transcriptional activity due to lacking a DNA binding domain. AR8 function is unknown but AR8 is proposed to be associated with non-genomic signaling, since it is localized to the plasma membrane via palmitoylation [89, 90]. AR45 splice variant seems to have cell specific localization. In peripheral cells, AR45 can dimerize with full length AR and act as a negative regulator via competitive inhibition of the ARE [88]. Interestingly, AR45 is the only reported splice variant in the brain. In contrast to peripheral cells, we show that neuronal AR45 localizes to caveolin positive lipid rafts in the plasma membrane (Figure 1) and interacts with G-protein coupled receptor signaling that can initiate intracellular calcium release [9]. Furthermore, studies using cell-impermeable androgens in a neuronal cell line containing membrane-associated AR45 found rapid non- genomic signaling that was unaffected by classical AR antagonists [9, 10, 80]. Therefore, it is plausible that the neuronal mAR could be AR45.
Ligands of NTD deleted SR splice variants
In addition to the cognate ligands shown to activate membrane-initiated signaling at membrane SRs, membrane effects of androgens and estrogens have been explored with cognate ligands coupled to large membrane impermeable molecules, such as bovine serum albumin (BSA). These conjugates have helped to distinguish membrane actions from those associated with cytoplasmic or nuclear actions. Thus, membrane ERs are activated by BSA-estradiol and membrane ARs are activated by BSA-testosterone. Currently, no antagonist has been found to block membrane associated AR, regardless of cell type [10, 80, 83, 85, 91–95] or ER [96]. In contrast to AR, the ER antagonist ICI 182,780 has no effect on eNOS activity in ERα46 transfected cells, but can inhibit estrogen and BSA-estrogen activated membrane ERα46 [27]. Unlike ERα46, the estrogen receptor antagonists tamoxifen and ICI 182,780 can both act as agonists for ERα36 to stimulate kinase signaling in breast cancer [32, 35] and neurons [38] despite the fact that part of the LBD is missing.
Most of the SR antagonists are targeted to the ligand binding domain located in the CTD of the receptor [97] indicating that the ligand binding domain may not be accessible to the SR antagonist. Interestingly, prior studies have shown that the plasma membrane can alter agonist and antagonist kinetics of receptors due to surrounding protein-protein interactions [98, 99]. For example, a study using the same full length ER cDNA in several cell lines with diverse cellular localization observed different ER agonist and antagonist responses that was dependent on where the ER was localized within the cell [100]. Similarly, inhibition of AR45, AR splice variant missing NTD, by antagonists is dependent on cellular localization. The AR antagonist, flutamide, via binding to the LBD can block nuclear AR45 action in HEK293 and CHO cells [101]. However, flutamide was unable to inhibit plasma membrane AR45 action in a neuronal cell line [10, 80].
Since it possible that the LBD of SRs located within the plasma membrane is not accessible to antagonists, other binding sites of the SR need to explored. Targeting the NTD domain of the SR for antagonists is a current area of research. However, NTD targeted SR antagonists are not appropriate for SRs that lack the NTD. Little progress has been made at targeting SR antagonists outside of the ligand binding domain and the NTD, due to the close similarity of the DBD regions between all the SRs [102]. Recently investigators have discovered a unique residue sequence in the AR DBD that can be targeted [97, 103–105], which has been shown to inhibit full length AR and an AR splice variant missing only the ligand binding domain. DBD targeted AR antagonists have not been examined on the function of splice variants missing the NTD or in splice variants localized to the plasma membrane.
Conclusion
Depending on cellular localization, N-terminal truncated SRs can have diverse effects ranging from transcription to modulating rapid cell signaling. Indeed, these understudied truncated proteins may have important physiological consequences that can influence cell survival. Further, these proteins could be potential therapeutic targets for disorders that no longer respond to conventional treatments that target full length SRs, such as cancer.
Acknowledgments
This study was funded by NIH National Institute of Neurological Disorders and Stroke (R01 NS088514) to RLC and NIH National Institute of Aging (R03 AG049255) to DAS and RLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Bibliography
- 1.Evans RM. The nuclear receptor superfamily: a rosetta stone for physiology. Mol Endocrinol. 2005;19:1429–38. doi: 10.1210/me.2005-0046. [DOI] [PubMed] [Google Scholar]
- 2.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–72. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helsen C, Claessens F. Looking at nuclear receptors from a new angle. Mol Cell Endocrinol. 2014;382:97–106. doi: 10.1016/j.mce.2013.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Meitzen J, Luoma JI, Boulware MI, Hedges VL, Peterson BM, Tuomela K, et al. Palmitoylation of estrogen receptors is essential for neuronal membrane signaling. Endocrinology. 2013;154:4293–304. doi: 10.1210/en.2013-1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levin ER. Extranuclear steroid receptors are essential for steroid hormone actions. Annu Rev Med. 2015;66:271–80. doi: 10.1146/annurev-med-050913-021703. [DOI] [PubMed] [Google Scholar]
- 6.Levin ER, Hammes SR. Nuclear receptors outside the nucleus: extranuclear signalling by steroid receptors. Nat Rev Mol Cell Biol. 2016;17:783–97. doi: 10.1038/nrm.2016.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arnal JF, Lenfant F, Metivier R, Flouriot G, Henrion D, Adlanmerini M, et al. Membrane and Nuclear Estrogen Receptor Alpha Actions: From Tissue Specificity to Medical Implications. Physiol Rev. 2017;97:1045–87. doi: 10.1152/physrev.00024.2016. [DOI] [PubMed] [Google Scholar]
- 8.Rudolph LM, Cornil CA, Mittelman-Smith MA, Rainville JR, Remage-Healey L, Sinchak K, et al. Actions of Steroids: New Neurotransmitters. J Neurosci. 2016;36:11449–58. doi: 10.1523/JNEUROSCI.2473-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garza-Contreras J, Duong P, Snyder BD, Schreihofer DA, Cunningham RL. Presence of Androgen Receptor Variant in Neuronal Lipid Rafts. eNeuro. 2017:4. doi: 10.1523/ENEURO.0109-17.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmes S, Abbassi B, Su C, Singh M, Cunningham RL. Oxidative stress defines the neuroprotective or neurotoxic properties of androgens in immortalized female rat dopaminergic neuronal cells. Endocrinology. 2013;154:4281–92. doi: 10.1210/en.2013-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hirata S, Shoda T, Kato J, Hoshi K. Isoform/variant mRNAs for sex steroid hormone receptors in humans. Trends Endocrinol Metab. 2003;14:124–9. doi: 10.1016/s1043-2760(03)00028-6. [DOI] [PubMed] [Google Scholar]
- 12.McCormick JA, Lyons V, Jacobson MD, Noble J, Diorio J, Nyirenda M, et al. 5′-heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol Endocrinol. 2000;14:506–17. doi: 10.1210/mend.14.4.0438. [DOI] [PubMed] [Google Scholar]
- 13.Reyer H, Ponsuksili S, Wimmers K, Murani E. Transcript variants of the porcine glucocorticoid receptor gene (NR3C1) Gen Comp Endocrinol. 2013;189:127–33. doi: 10.1016/j.ygcen.2013.04.038. [DOI] [PubMed] [Google Scholar]
- 14.Kwak SP, Patel PD, Thompson RC, Akil H, Watson SJ. 5′-Heterogeneity of the mineralocorticoid receptor messenger ribonucleic acid: differential expression and regulation of splice variants within the rat hippocampus. Endocrinology. 1993;133:2344–50. doi: 10.1210/endo.133.5.8404687. [DOI] [PubMed] [Google Scholar]
- 15.Castren M, Damm K. A functional promoter directing expression of a novel type of rat mineralocorticoid receptor mRNA in brain. J Neuroendocrinol. 1993;5:461–6. doi: 10.1111/j.1365-2826.1993.tb00508.x. [DOI] [PubMed] [Google Scholar]
- 16.Hirata S, Shoda T, Kato J, Hoshi K. The multiple untranslated first exons system of the human estrogen receptor beta (ER beta) gene. The Journal of Steroid Biochemistry and Molecular Biology. 2001;78:33–40. doi: 10.1016/s0960-0760(01)00071-1. [DOI] [PubMed] [Google Scholar]
- 17.Kos M, Denger S, Reid G, Gannon F. Upstream open reading frames regulate the translation of the multiple mRNA variants of the estrogen receptor alpha. J Biol Chem. 2002;277:37131–8. doi: 10.1074/jbc.M206325200. [DOI] [PubMed] [Google Scholar]
- 18.Kobayashi M, Ishii H, Sakuma Y. Identification of novel splicing events and post-transcriptional regulation of human estrogen receptor alpha F isoforms. Mol Cell Endocrinol. 2011;333:55–61. doi: 10.1016/j.mce.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Dehm SM, Tindall DJ. Alternatively spliced androgen receptor variants. Endocr Relat Cancer. 2011;18:R183–96. doi: 10.1530/ERC-11-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marshburn PB, Zhang J, Bahrani-Mostafavi Z, Matthews ML, White J, Hurst BS. Variant progesterone receptor mRNAs are co-expressed with the wild-type progesterone receptor mRNA in human endometrium during all phases of the menstrual cycle. Mol Hum Reprod. 2005;11:809–15. doi: 10.1093/molehr/gah244. [DOI] [PubMed] [Google Scholar]
- 21.Skrzypczak M, Merx I, Schuler-Toprak S, Weber F, Inwald EC, Ortmann O, et al. Molecular profiling of estrogen receptor alpha and progesterone receptor transcript variants in endometrial cancer. Steroids. 2015;104:122–8. doi: 10.1016/j.steroids.2015.09.004. [DOI] [PubMed] [Google Scholar]
- 22.Weickert CS, Miranda-Angulo AL, Wong J, Perlman WR, Ward SE, Radhakrishna V, et al. Variants in the estrogen receptor alpha gene and its mRNA contribute to risk for schizophrenia. Hum Mol Genet. 2008;17:2293–309. doi: 10.1093/hmg/ddn130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqualini C, Guivarc’h D, Boxberg YV, Nothias F, Vincent J-D, Vernier P. Stage- and region-specific expression of estrogen receptor alpha isoforms during ontogeny of the pituitary gland. Endocrinology. 1999;140:2781–9. doi: 10.1210/endo.140.6.6752. [DOI] [PubMed] [Google Scholar]
- 24.Skipper JK, Young LJ, Bergeron JM, Tetzlaff MT, Osnorn CT, Crews D. Identification of an isoform of the estrogen receptor messenger RNA lacking exon four and present in the brain. Procedings of the National Academy of Sciences, USA. 1993;90:7172–5. doi: 10.1073/pnas.90.15.7172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friend KE, Resnick EM, Ang LW, Shupnik MA. Specific modulation of estrogen receptor mRNA isoforms in rat pituitary throughout the estrous cycle and in response to steroid hormones. Molecular and Cellular Endocrinology. 1997;131:147–55. doi: 10.1016/s0303-7207(97)00098-1. [DOI] [PubMed] [Google Scholar]
- 26.Hattori Y, Ishii H, Morita A, Sakuma Y, Ozawa H. Characterization of the fundamental properties of the N-terminal truncation (Delta exon 1) variant of estrogen receptor alpha in the rat. Gene. 2015;571:117–25. doi: 10.1016/j.gene.2015.06.086. [DOI] [PubMed] [Google Scholar]
- 27.Li L, Haynes MP, Bender JR. Plasma membrane localization and function of the estrogen receptor alpha variant (ER46) in human endothelial cells. Proc Natl Acad Sci U S A. 2003;100:4807–12. doi: 10.1073/pnas.0831079100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haynes MP, Li L, Sinha D, Russell KS, Hisamoto K, Baron R, et al. Src kinase mediates phosphatidylinositol 3-kinase/Akt-dependent rapid endothelial nitric-oxide synthase activation by estrogen. J Biol Chem. 2003;278:2118–23. doi: 10.1074/jbc.M210828200. [DOI] [PubMed] [Google Scholar]
- 29.Moran J, Garrido P, Cabello E, Alonso A, Gonzalez C. Effects of estradiol and genistein on the insulin signaling pathway in the cerebral cortex of aged female rats. Exp Gerontol. 2014;58:104–12. doi: 10.1016/j.exger.2014.07.018. [DOI] [PubMed] [Google Scholar]
- 30.Stirone C, Duckles SP, Krause DN. Multiple forms of estrogen receptor-alpha in cerebral blood vessels: regulation by estrogen. Am J Physiol Endocrinol Metab. 2003;284:E184–92. doi: 10.1152/ajpendo.00165.2002. [DOI] [PubMed] [Google Scholar]
- 31.Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. Identification, cloning, and expression of human estrogen receptor-alpha36, a novel variant of human estrogen receptor-alpha66. Biochem Biophys Res Commun. 2005;336:1023–7. doi: 10.1016/j.bbrc.2005.08.226. [DOI] [PubMed] [Google Scholar]
- 32.Shi L, Dong B, Li Z, Lu Y, Ouyang T, Li J, et al. Expression of ER-{alpha}36, a novel variant of estrogen receptor {alpha}, and resistance to tamoxifen treatment in breast cancer. J Clin Oncol. 2009;27:3423–9. doi: 10.1200/JCO.2008.17.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rao J, Jiang X, Wang Y, Chen B. Advances in the understanding of the structure and function of ER-alpha36,a novel variant of human estrogen receptor-alpha. J Steroid Biochem Mol Biol. 2011;127:231–7. doi: 10.1016/j.jsbmb.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z, Zhang X, Shen P, Loggie BW, Chang Y, Deuel TF. A variant of estrogen receptor-{alpha}, hER-{alpha}36: transduction of estrogen- and antiestrogen-dependent membrane-initiated mitogenic signaling. Proc Natl Acad Sci U S A. 2006;103:9063–8. doi: 10.1073/pnas.0603339103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SL, Yan LY, Zhang XT, Yuan J, Li M, Qiao J, et al. ER-alpha36, a variant of ER-alpha, promotes tamoxifen agonist action in endometrial cancer cells via the MAPK/ERK and PI3K/Akt pathways. PLoS One. 2010;5:e9013. doi: 10.1371/journal.pone.0009013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kang L, Zhang X, Xie Y, Tu Y, Wang D, Liu Z, et al. Involvement of estrogen receptor variant ER-alpha36, not GPR30, in nongenomic estrogen signaling. Mol Endocrinol. 2010;24:709–21. doi: 10.1210/me.2009-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Fang C, Zou P, Ma YN, Han DN, Ji ZH, et al. [Diverse expression of ER-alpha36, a novel variant of ER-alpha, in hippocampus and cortex of neonatal and adult rats] Sheng Li Xue Bao. 2013;65:263–8. [PubMed] [Google Scholar]
- 38.Zou W, Fang C, Ji X, Liang X, Liu Y, Han C, et al. Estrogen Receptor (ER)-alpha36 Is Involved in Estrogen- and Tamoxifen-Induced Neuroprotective Effects in Ischemic Stroke Models. PLoS One. 2015;10:e0140660. doi: 10.1371/journal.pone.0140660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Han S, Zhao B, Pan X, Song Z, Liu J, Gong Y, et al. Estrogen receptor variant ER-alpha36 is involved in estrogen neuroprotection against oxidative toxicity. Neuroscience. 2015;310:224–41. doi: 10.1016/j.neuroscience.2015.09.024. [DOI] [PubMed] [Google Scholar]
- 40.Liu Y, Huang L, Guan X, Li H, Zhang QQ, Han C, et al. ER-alpha36, a novel variant of ERalpha, is involved in the regulation of Tamoxifen-sensitivity of glioblastoma cells. Steroids. 2016;111:127–33. doi: 10.1016/j.steroids.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 41.Ishunina TA, Swaab DF, Fischer DF. Estrogen receptor-alpha splice variants in the medial mamillary nucleus of Alzheimer’s disease patients: identification of a novel MB1 isoform. J Clin Endocrinol Metab. 2005;90:3757–65. doi: 10.1210/jc.2004-1858. [DOI] [PubMed] [Google Scholar]
- 42.Ishunina TA, Sluiter AA, Swaab DF, Verwer RW. Transcriptional activity of human brain estrogen receptor-alpha splice variants: evidence for cell type-specific regulation. Brain Res. 2013;1500:1–9. doi: 10.1016/j.brainres.2012.12.050. [DOI] [PubMed] [Google Scholar]
- 43.Ishunina TA, Swaab DF. Age-dependent ERalpha MB1 splice variant expression in discrete areas of the human brain. Neurobiol Aging. 2008;29:1177–89. doi: 10.1016/j.neurobiolaging.2007.02.018. [DOI] [PubMed] [Google Scholar]
- 44.Friend KE, Ang LW, Shupnik MA. Estrogen regulates the expression of several different estrogen receptor mRNA isoforms in rat pituitary. Proceedings of the National Academy of Science. 1995;92:4367–71. doi: 10.1073/pnas.92.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ishii H, Shoda Y, Yomogida K, Hamada T, Sakuma Y. Identification of C-terminally and N-terminally truncated estrogen receptor alpha variants in the mouse. J Steroid Biochem Mol Biol. 2011;124:38–46. doi: 10.1016/j.jsbmb.2011.01.003. [DOI] [PubMed] [Google Scholar]
- 46.Resnick EM, Schreihofer DA, Periasamy A, Shupnik MA. Truncated estrogen receptor product-1 suppresses estrogen receptor transactivation by dimerization with estrogen receptors alpha and beta. Journal of Biological Chemistry. 2000;275:7158–66. doi: 10.1074/jbc.275.10.7158. [DOI] [PubMed] [Google Scholar]
- 47.Schreihofer DA, Resnick EM, Soh AY, Shupnik MA. Transcriptional regulation by a naturally occurring truncated rat estrogen receptor (ER), truncated ER product-1 (TERP-1) Molecular Endocrinology. 1999;13:320–9. doi: 10.1210/mend.13.2.0236. [DOI] [PubMed] [Google Scholar]
- 48.Moore JT, McKee DD, Slentz-Kesler K, Moore LB, Jones SA, Horne EL, et al. Cloning and characterization of human estrogen receptor beta isoforms. Biochemical & Biophysical Research Communications. 1998;247:75–8. doi: 10.1006/bbrc.1998.8738. [DOI] [PubMed] [Google Scholar]
- 49.Mott NN, Pak TR. Characterisation of human oestrogen receptor beta (ERbeta) splice variants in neuronal cells. J Neuroendocrinol. 2012;24:1311–21. doi: 10.1111/j.1365-2826.2012.02337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Price RH, Jr, Lorenzon N, Handa RJ. Differential expression of estrogen receptor beta splice variants in rat brain: identification and characterization of a novel variant missing exon 4. Molecular Brain Research. 2000;80:260–8. doi: 10.1016/s0169-328x(00)00135-2. [DOI] [PubMed] [Google Scholar]
- 51.Chung WC, Pak TR, Suzuki S, Pouliot WA, Andersen ME, Handa RJ. Detection and localization of an estrogen receptor beta splice variant protein (ERbeta2) in the adult female rat forebrain and midbrain regions. J Comp Neurol. 2007;505:249–67. doi: 10.1002/cne.21490. [DOI] [PubMed] [Google Scholar]
- 52.Filardo EJ. Epidermal growth factor receptor (EGFR) transactivation by estrogen via the G-protein-coupled receptor, GPR30: a novel signaling pathway with potential significance for breast cancer. J Steroid Biochem Mol Biol. 2002;80:231–8. doi: 10.1016/s0960-0760(01)00190-x. [DOI] [PubMed] [Google Scholar]
- 53.Revankar CM, Cimino DF, Sklar LA, Arterburn JB, Prossnitz ER. A transmembrane intracellular estrogen receptor mediates rapid cell signaling. Science. 2005;307:1625–30. doi: 10.1126/science.1106943. [DOI] [PubMed] [Google Scholar]
- 54.Jacobsen BM, Horwitz KB. Progesterone receptors, their isoforms and progesterone regulated transcription. Mol Cell Endocrinol. 2012;357:18–29. doi: 10.1016/j.mce.2011.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Brinton RD, Thompson RF, Foy MR, Baudry M, Wang J, Finch CE, et al. Progesterone receptors: form and function in brain. Front Neuroendocrinol. 2008;29:313–39. doi: 10.1016/j.yfrne.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cork DM, Lennard TW, Tyson-Capper AJ. Alternative splicing and the progesterone receptor in breast cancer. Breast Cancer Res. 2008;10:207. doi: 10.1186/bcr2097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Faivre EJ, Lange CA. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol Cell Biol. 2007;27:466–80. doi: 10.1128/MCB.01539-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boonyaratanakornkit V, Bi Y, Rudd M, Edwards DP. The role and mechanism of progesterone receptor activation of extra-nuclear signaling pathways in regulating gene transcription and cell cycle progression. Steroids. 2008;73:922–8. doi: 10.1016/j.steroids.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 59.Edwards DP, Wardell SE, Boonyaratanakornkit V. Progesterone receptor interacting coregulatory proteins and cross talk with cell signaling pathways. J Steroid Biochem Mol Biol. 2002;83:173–86. doi: 10.1016/s0960-0760(02)00265-0. [DOI] [PubMed] [Google Scholar]
- 60.Thomas P, Pang Y. Protective actions of progesterone in the cardiovascular system: potential role of membrane progesterone receptors (mPRs) in mediating rapid effects. Steroids. 2013;78:583–8. doi: 10.1016/j.steroids.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 61.Richer JK, Lange CA, Wierman AM, Brooks KM, Tung L, Takimoto GS, et al. Progesterone receptor variants found in breast cells repress transcription by wild-type receptors. Breast Cancer Res Treat. 1998;48:231–41. doi: 10.1023/a:1005941117247. [DOI] [PubMed] [Google Scholar]
- 62.Saner KJ, Welter BH, Zhang F, Hansen E, Dupont B, Wei Y, et al. Cloning and expression of a novel, truncated, progesterone receptor. Mol Cell Endocrinol. 2003;200:155–63. doi: 10.1016/s0303-7207(02)00380-5. [DOI] [PubMed] [Google Scholar]
- 63.Samalecos A, Gellersen B. Systematic expression analysis and antibody screening do not support the existence of naturally occurring progesterone receptor (PR)-C, PR-M, or other truncated PR isoforms. Endocrinology. 2008;149:5872–87. doi: 10.1210/en.2008-0602. [DOI] [PubMed] [Google Scholar]
- 64.Waters EM, Torres-Reveron A, McEwen BS, Milner TA. Ultrastructural localization of extranuclear progestin receptors in the rat hippocampal formation. J Comp Neurol. 2008;511:34–46. doi: 10.1002/cne.21826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitterling KL, Spencer JL, Dziedzic N, Shenoy S, McCarthy K, Waters EM, et al. Cellular and subcellular localization of estrogen and progestin receptor immunoreactivities in the mouse hippocampus. J Comp Neurol. 2010;518:2729–43. doi: 10.1002/cne.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zuo L, Li W, You S. Progesterone reverses the mesenchymal phenotypes of basal phenotype breast cancer cells via a membrane progesterone receptor mediated pathway. Breast Cancer Res. 2010;12:R34. doi: 10.1186/bcr2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y, Bond J, Thomas P. Identification, classification, and partial characterization of genes in humans and other vertebrates homologous to a fish membrane progestin receptor. Proc Natl Acad Sci U S A. 2003;100:2237–42. doi: 10.1073/pnas.0436133100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Labombarda F, Meffre D, Delespierre B, Krivokapic-Blondiaux S, Chastre A, Thomas P, et al. Membrane progesterone receptors localization in the mouse spinal cord. Neuroscience. 2010;166:94–106. doi: 10.1016/j.neuroscience.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 69.Castelnovo LF, Magnaghi V, Thomas P. Expression of membrane progesterone receptors (mPRs) in rat peripheral glial cell membranes and their potential role in the modulation of cell migration and protein expression. Steroids. 2017 doi: 10.1016/j.steroids.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 70.Thomas P. Characteristics of membrane progestin receptor alpha (mPRalpha) and progesterone membrane receptor component 1 (PGMRC1) and their roles in mediating rapid progestin actions. Front Neuroendocrinol. 2008;29:292–312. doi: 10.1016/j.yfrne.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorczynska E, Handelsman DJ. Androgens rapidly increase the cytosolic calcium concentration in Sertoli cells. Endocrinology. 1995;136:2052–9. doi: 10.1210/endo.136.5.7720654. [DOI] [PubMed] [Google Scholar]
- 72.Machelon V, Nome F, Tesarik J. Nongenomic effects of androstenedione on human granulosa luteinizing cells. J Clin Endocrinol Metab. 1998;83:263–9. doi: 10.1210/jcem.83.1.4523. [DOI] [PubMed] [Google Scholar]
- 73.Wunderlich F, Benten WP, Lieberherr M, Guo Z, Stamm O, Wrehlke C, et al. Testosterone signaling in T cells and macrophages. Steroids. 2002;67:535–8. doi: 10.1016/s0039-128x(01)00175-1. [DOI] [PubMed] [Google Scholar]
- 74.Sun YH, Gao X, Tang YJ, Xu CL, Wang LH. Androgens induce increases in intracellular calcium via a G protein-coupled receptor in LNCaP prostate cancer cells. J Androl. 2006;27:671–8. doi: 10.2164/jandrol.106.000554. [DOI] [PubMed] [Google Scholar]
- 75.Liu D, Dillon JS. Dehydroepiandrosterone activates endothelial cell nitric-oxide synthase by a specific plasma membrane receptor coupled to Galpha(i2,3) J Biol Chem. 2002;277:21379–88. doi: 10.1074/jbc.M200491200. [DOI] [PubMed] [Google Scholar]
- 76.Fix C, Jordan C, Cano P, Walker WH. Testosterone activates mitogen-activated protein kinase and the cAMP response element binding protein transcription factor in Sertoli cells. Proc Natl Acad Sci U S A. 2004;101:10919–24. doi: 10.1073/pnas.0404278101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Losel RM, Falkenstein E, Feuring M, Schultz A, Tillmann HC, Rossol-Haseroth K, et al. Nongenomic steroid action: controversies, questions, and answers. Physiol Rev. 2003;83:965–1016. doi: 10.1152/physrev.00003.2003. [DOI] [PubMed] [Google Scholar]
- 78.Estrada M, Espinosa A, Muller M, Jaimovich E. Testosterone stimulates intracellular calcium release and mitogen-activated protein kinases via a G protein-coupled receptor in skeletal muscle cells. Endocrinology. 2003;144:3586–97. doi: 10.1210/en.2002-0164. [DOI] [PubMed] [Google Scholar]
- 79.Estrada M, Uhlen P, Ehrlich BE. Ca2+ oscillations induced by testosterone enhance neurite outgrowth. J Cell Sci. 2006;119:733–43. doi: 10.1242/jcs.02775. [DOI] [PubMed] [Google Scholar]
- 80.Holmes S, Singh M, Su C, Cunningham RL. Effects of Oxidative Stress and Testosterone on Pro-Inflammatory Signaling in a Female Rat Dopaminergic Neuronal Cell Line. Endocrinology. 2016;157:2824–35. doi: 10.1210/en.2015-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gatson JW, Singh M. Activation of a membrane-associated androgen receptor promotes cell death in primary cortical astrocytes. Endocrinology. 2007;148:2458–64. doi: 10.1210/en.2006-1443. [DOI] [PubMed] [Google Scholar]
- 82.Berg AH, Rice CD, Rahman MS, Dong J, Thomas P. Identification and characterization of membrane androgen receptors in the ZIP9 zinc transporter subfamily: I. Discovery in female atlantic croaker and evidence ZIP9 mediates testosterone-induced apoptosis of ovarian follicle cells. Endocrinology. 2014;155:4237–49. doi: 10.1210/en.2014-1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li S, Kang L, Zhang Y, Feng B, Du J, Cui H. Detecting the presence of hippocampus membrane androgen receptors in male SAMP8 mice and their induced synaptic plasticity. Mol Cell Endocrinol. 2015;414:82–90. doi: 10.1016/j.mce.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 84.Sato SM, Johansen JA, Jordan CL, Wood RI. Membrane androgen receptors may mediate androgen reinforcement. Psychoneuroendocrinology. 2010;35:1063–73. doi: 10.1016/j.psyneuen.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Benten WP, Lieberherr M, Stamm O, Wrehlke C, Guo Z, Wunderlich F. Testosterone signaling through internalizable surface receptors in androgen receptor-free macrophages. Molecular biology of the cell. 1999;10:3113–23. doi: 10.1091/mbc.10.10.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jenster G, van der Korput HA, van Vroonhoven C, van der Kwast TH, Trapman J, Brinkmann AO. Domains of the human androgen receptor involved in steroid binding, transcriptional activation, and subcellular localization. Mol Endocrinol. 1991;5:1396–404. doi: 10.1210/mend-5-10-1396. [DOI] [PubMed] [Google Scholar]
- 87.Ikonen T, Palvimo JJ, Janne OA. Heterodimerization is mainly responsible for the dominant negative activity of amino-terminally truncated rat androgen receptor forms. FEBS Lett. 1998;430:393–6. doi: 10.1016/s0014-5793(98)00701-7. [DOI] [PubMed] [Google Scholar]
- 88.Ahrens-Fath I, Politz O, Geserick C, Haendler B. Androgen receptor function is modulated by the tissue-specific AR45 variant. FEBS J. 2005;272:74–84. doi: 10.1111/j.1742-4658.2004.04395.x. [DOI] [PubMed] [Google Scholar]
- 89.Yang X, Guo Z, Sun F, Li W, Alfano A, Shimelis H, et al. Novel membrane-associated androgen receptor splice variant potentiates proliferative and survival responses in prostate cancer cells. J Biol Chem. 2011;286:36152–60. doi: 10.1074/jbc.M111.265124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wadosky KM, Koochekpour S. Androgen receptor splice variants and prostate cancer: From bench to bedside. Oncotarget. 2017;8:18550–76. doi: 10.18632/oncotarget.14537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kampa M, Papakonstanti EA, Hatzoglou A, Stathopoulos EN, Stournaras C, Castanas E. The human prostate cancer cell line LNCaP bears functional membrane testosterone receptors that increase PSA secretion and modify actin cytoskeleton. FASEB journal : official publication of the Federation of American Societies for Experimental Biology. 2002;16:1429–31. doi: 10.1096/fj.02-0131fje. [DOI] [PubMed] [Google Scholar]
- 92.Papakonstanti EA, Kampa M, Castanas E, Stournaras C. A rapid, nongenomic, signaling pathway regulates the actin reorganization induced by activation of membrane testosterone receptors. Molecular endocrinology. 2003;17:870–81. doi: 10.1210/me.2002-0253. [DOI] [PubMed] [Google Scholar]
- 93.Hatzoglou A, Kampa M, Kogia C, Charalampopoulos I, Theodoropoulos PA, Anezinis P, et al. Membrane androgen receptor activation induces apoptotic regression of human prostate cancer cells in vitro and in vivo. The Journal of clinical endocrinology and metabolism. 2005;90:893–903. doi: 10.1210/jc.2004-0801. [DOI] [PubMed] [Google Scholar]
- 94.Gatson JW, Kaur P, Singh M. Dihydrotestosterone differentially modulates the mitogen-activated protein kinase and the phosphoinositide 3-kinase/Akt pathways through the nuclear and novel membrane androgen receptor in C6 cells. Endocrinology. 2006;147:2028–34. doi: 10.1210/en.2005-1395. [DOI] [PubMed] [Google Scholar]
- 95.Kampa M, Kogia C, Theodoropoulos PA, Anezinis P, Charalampopoulos I, Papakonstanti EA, et al. Activation of membrane androgen receptors potentiates the antiproliferative effects of paclitaxel on human prostate cancer cells. Molecular cancer therapeutics. 2006;5:1342–51. doi: 10.1158/1535-7163.MCT-05-0527. [DOI] [PubMed] [Google Scholar]
- 96.Kuroki Y, Fukushima K, Kanda Y, Mizuno K, Watanabe Y. Putative membrane-bound estrogen receptors possibly stimulate mitogen-activated protein kinase in the rat hippocampus. Eur J Pharmacol. 2000;400:205–9. doi: 10.1016/s0014-2999(00)00425-8. [DOI] [PubMed] [Google Scholar]
- 97.Lallous N, Dalal K, Cherkasov A, Rennie PS. Targeting alternative sites on the androgen receptor to treat castration-resistant prostate cancer. Int J Mol Sci. 2013;14:12496–519. doi: 10.3390/ijms140612496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wu J, Gorenstein DG. Structure and dynamics of cytochrome c in nonaqueous solvents by 2D NH-exchange NMR spectroscopy. Journal of the American Chemical Society. 1993;115:6843–50. [Google Scholar]
- 99.Powell CE, Watson CS, Gametchu B. Immunoaffinity isolation of native membrane glucocorticoid receptor from S-49++ lymphoma cells: biochemical characterization and interaction with Hsp 70 and Hsp 90. Endocrine. 1999;10:271–80. doi: 10.1007/BF02738626. [DOI] [PubMed] [Google Scholar]
- 100.Yoon K, Pellaroni L, Ramamoorthy K, Gaido K, Safe S. Ligand structure-dependent differences in activation of estrogen receptor alpha in human HepG2 liver and U2 osteogenic cancer cell lines. Mol Cell Endocrinol. 2000;162:211–20. doi: 10.1016/s0303-7207(99)00261-0. [DOI] [PubMed] [Google Scholar]
- 101.Wu ZY, Chen K, Haendler B, McDonald TV, Bian JS. Stimulation of N-terminal truncated isoform of androgen receptor stabilizes human ether-a-go-go-related gene-encoded potassium channel protein via activation of extracellular signal regulated kinase 1/2. Endocrinology. 2008;149:5061–9. doi: 10.1210/en.2007-1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caboni L, Lloyd DG. Beyond the ligand-binding pocket: targeting alternate sites in nuclear receptors. Med Res Rev. 2013;33:1081–118. doi: 10.1002/med.21275. [DOI] [PubMed] [Google Scholar]
- 103.Cherian MT, Wilson EM, Shapiro DJ. A competitive inhibitor that reduces recruitment of androgen receptor to androgen-responsive genes. J Biol Chem. 2012;287:23368–80. doi: 10.1074/jbc.M112.344671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dalal K, Roshan-Moniri M, Sharma A, Li H, Ban F, Hassona MD, et al. Selectively targeting the DNA-binding domain of the androgen receptor as a prospective therapy for prostate cancer. J Biol Chem. 2014;289:26417–29. doi: 10.1074/jbc.M114.553818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Ban F, Dalal K, Leblanc E, Frewin K, Ma D, et al. Discovery of small-molecule inhibitors selectively targeting the DNA-binding domain of the human androgen receptor. Journal of medicinal chemistry. 2014;57:6458–67. doi: 10.1021/jm500802j. [DOI] [PubMed] [Google Scholar]


