Abstract
Objective
One source of endogenous reverse transcriptase (eRT) activity in nucleated cells is the Long Interspersed Element-1 (LINE-1/L1), a non-LTR retrotransposon that is implicated in the regulation of gene expression. Nevertheless, the presence and function of eRT activity and LINE-1 in human platelets, an anucleate cell, has not previously been determined.
Approach/Results
We demonstrate that human and murine platelets possess robust eRT activity and identify the source as being LINE-1 ribonucleoprotein-particles (RNPs). Inhibition of eRT in vitro in isolated platelets from healthy individuals or in persons with HIV treated with RT inhibitors enhanced global protein synthesis and platelet activation. If HIV patients were treated with reverse transcriptase inhibitor, we found that platelets from these patients had increased basal activation. We next discovered that eRT activity in platelets controlled the generation of RNA-DNA hybrids, which serve as translational repressors. Inhibition of platelet eRT lifted this RNA-DNA hybrid-induced translational block and was sufficient to increase protein expression of target RNAs identified by RNA-DNA hybrid immunoprecipitation.
Conclusions
Thus, we provide the first evidence that platelets possess L1-encoded eRT activity. We also demonstrate that platelet eRT activity regulates platelet hyperreactivity and thrombosis and controls RNA-DNA hybrid formation, and identify that RNA-DNA hybrids function as a novel translational control mechanism in human platelets.
Keywords: blood platelets, retroelements, protein translation, LINE-1, translational regulation
Subject Code: Basic Science Research
Introduction
Megakaryocytes invest platelets with a complex transcriptome and requisite translational machinery including ribosomes, initiation and termination factors, microRNAs, and messenger RNAs (mRNAs)1, 2. Platelets use a variety of translational and posttranslational mechanisms to finely regulate their protein profile, although only some of these have been identified. For example, a subset of mRNAs are under the translational control of the mTOR pathway3, 4, although this is thought to be mainly mRNAs possessing a 5′ terminal oligopyrimidine tract (5′ TOP)5 or a pyrimidine-rich translational element (PRTE)6. An additional translational regulatory pathway in platelets is cytoplasmic, signal-dependent pre-mRNA splicing7, 8. Nevertheless, only a limited number of pre-mRNAs have been identified in human platelets7–9. Therefore, additional translational control mechanisms likely exist in human platelets. Because of the anucleate nature of platelets, they serve as an ideal model system to study translational regulatory mechanisms.
Due to their ability to insert themselves into various genomic locations, human transposable elements contribute importantly to a highly dynamic genome10. The only functional autonomous non-long terminal repeat retrotransposon in humans is LINE-1 (long interspersed nuclear element-1, L1)11. Almost 17% of the human genome consists of LINE-1 elements12, 13. Active, full-length L1 elements consist of a 5′-UTR with internal promoter activity, two open reading frames (ORFs) separated by a short intergenic region, a 3′-UTR that ends with an AATAAA polyadenylation signal, and a poly(A) tail11, 14, 15. L1 mRNAs are atypical of mammalian mRNAs because they are bicistronic11. The two human ORFs in L1 elements (ORF1 and ORF2p) are in frame, separated by a 63-bp non-coding spacer that contains stop codons11. Translation of ORF1 protein occurs via ribosomal initiation in the 5′-UTR16 while translation of ORF2p occurs by an unconventional termination/reinitiation mechanism17. The COOH-terminal half of the 40-kDa ORF1 protein (p40) is involved in RNA binding and is well conserved across species11, 18–20. ORF2 encodes an ~150-kDa protein with three conserved domains: NH2-terminal endonuclease (EN) domain, central reverse transcriptase (RT) domain, and COOH-terminal zinc finger-like domain11, 21–23. Both L1-encoded proteins preferentially associate with their own encoding RNA (cis preference) to form a ribonucleoprotein (RNP) complex24, 25; a proposed retrotransposition intermediate26, 27. However, L1-encoded reverse transcriptase (RT) is also known to reverse transcribe RNAs encoded by Alu elements28, SVA elements29, and Pol II mRNAs causing processed pseudogene formation (26) in trans. L1 RT can reverse transcribe mRNAs coding for actin, GAPDH, and nucleolin into cDNAs30. In addition to shaping the genome over evolutionary time, L1 retrotransposition events alter gene expression patterns31. The most notable gene altering process involves insertional mutagenesis, which can serve as a functional gene knock-out32. Retrotransposed L1 sequences can also function as alternative promoters that regulate gene transcription33. The insertion of L1 sequences into introns considerably reduces transcriptional elongation of target genes34. L1 retrotransposition events are responsible for diseases including hemophilia, β-thalassemia, and muscular dystrophy35. There is also evidence that endogenous RT activity regulates cellular proliferation, differentiation and gene expression36–41.
Reverse transcription of RNA also generates RNA-DNA hybrid intermediates. The formation of RNA-DNA hybrids has been used in molecular biology strategies as an in vitro technique to identify specifically translated mRNAs42, 43. RNA-DNA hybrids also regulate transcriptional efficiency44, and are required for efficient double-strand break repair45.
Persons with HIV are typically prescribed RT-inhibitor based therapy. The introduction of anti-retroviral therapy (ART) has resulted in improved life expectancy46. However, persons with HIV are at increased risk of thrombotic disorders, including atherothrombosis47–49. The molecular mechanisms underpinning this increased thrombosis risk remain unclear. In addition, whether platelets, which promote thrombosis, are directly affected by RT-inhibitors has not been studied in detail. Accordingly, we sought to determine if human platelets possess RT activity and, if so, its functional effects.
Material and Methods
Material and Methods are available in the online-only Supplement Material.
Results
Endogenous RT activity is present in human platelets
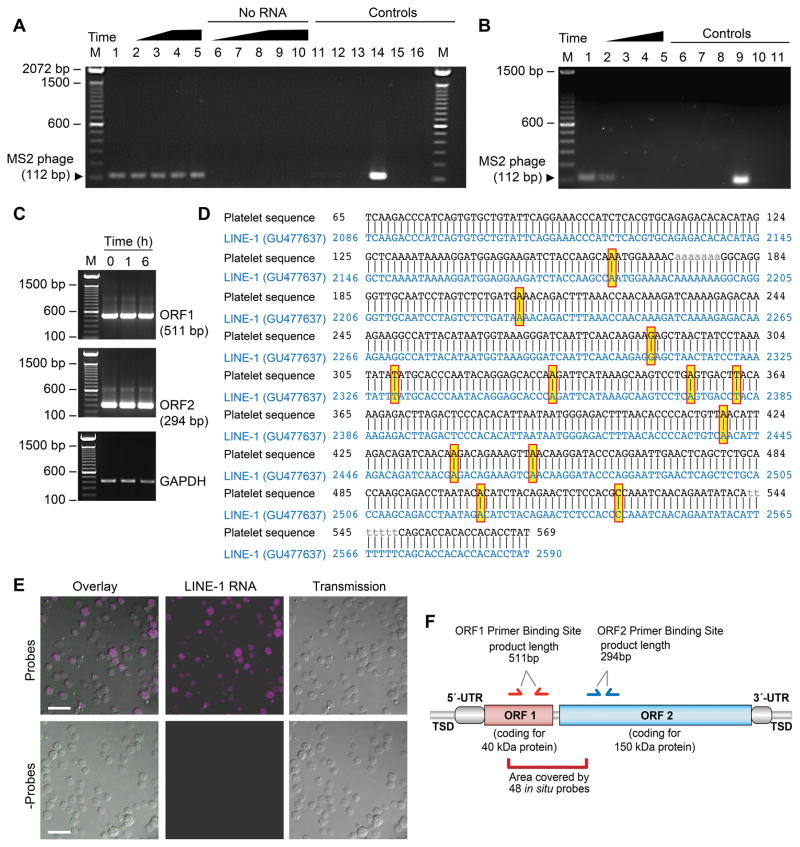
Although in nucleated cells endogenous retrotransposon-encoded RT (eRT) is essential for the mobilization of transposable elements, eRT activity has not previously been examined in human platelets. Therefore, we first determined if human platelets possess eRT activity using a PCR-based in vitro assay37. An exogenous MS2 phage RNA is added to isolated platelet lysates and evidence of reverse transcription is detected by probing for the DNA product of MS2 phage. We found that eRT activity is present in platelets at baseline and is retained over time (Figure 1A). Stimulating platelets with thrombin (t=6 hours) did not visibly alter eRT activity (Figure 1A, lane 5). In parallel experiments, we also confirmed that platelets in vitro transcribe an exogenously added GFP RNA, thus excluding the possibility of contaminating bacteria which are used to produce MS2 phage RNA50 in the PCR reagents (data not shown). Treating platelets with nevirapine, a clinically used RT inhibitor, blocked reverse transcription of exogenous MS2 phage in a dose-dependent manner (Figure 1B). Additional experiments of platelet lysates with 100kDa size-exclusion based spin columns (Supplemental Figure S1), suggested that LINE-1 elements could account for most of the eRT activity (i.e. fully processed HERV-K RT has a MW of 66kDa).
Figure 1. Human platelets possess endogenous RT activity and express LINE-1 ORF1 and ORF2 mRNA.
(A) Platelet lysates were incubated with (lanes 1–5) or without (lanes 6–10) exogenous MS2 phage RNA. Lanes 11–16 represent controls. The following conditions apply: M - marker; lanes 1,6 - freshly-isolated platelets; lanes 2,7 - platelets stored for 2 hours; lanes 3,8 - platelets stored for 4 hours; lanes 4,9 - platelets stored for 6 hours; lanes 5,10 - platelets activated with thrombin (0.1 U/ml) for 6 hours; lane 11 - platelet lysate replaced with the lysis buffer only; lane 12 - omission of the platelet lysate; lane 13 - omission of the MS2 phage RNA; lane 14 - platelet lysate replaced with commercial RT; lane 15 - omission of the reverse MS2 phage primer; lane 16 - negative PCR. This figure is representative of n>10 independent experiments. (B) Platelets were treated with DMSO (Veh, lane 1) or increasing concentrations of nevirapine ([26 μg/ml], [130 μg/ml], and [195 μg/ml], lanes 2–4 respectively) and reverse transcription of exogenous MS2 phage RNA was measured. Controls include: lane 5 - no RNA; lane 6 - platelet lysate replaced with the lysis buffer only; lane 7 - omission of the platelet lysate; lane 8 - omission of the platelet lysate and MS2 phage RNA; lane 9 - platelet lysate replaced with commercial RT; lane 10 - omission of the reverse MS2 phage primer; lane 11 - negative PCR. This figure is representative of 5 independent experiments. (C) Freshly isolated platelets were examined at baseline (t=0 h) or stimulated with thrombin (0.1 U/ml) for 1 or 6 hours. RNA from the platelets was isolated and ORF1 and ORF2 mRNA expression was evaluated by PCR. The bottom panel shows GAPDH as a loading control. This figure is representative of 3 independent experiments. (D) PCR products for LINE-1 were excised and sequenced using Sanger-Sequencing. The resulting sequence was aligned with a reference sequence. Yellow boxes highlight mismatches. (E) Human platelets were fixed in suspension and Stellaris FISH probes specific for LINE-1 (magenta, top panels) were hybridized to detect LINE-1 mRNA in situ. The bottom panels show platelets where the probes were omitted (- Probes) as a control (scale bars = 10 μm). These images are representative of 4 independent experiments. (F) Schematic representation of the structural organization of active L1 retroelements. The boundaries of a retroelement are defined by the presence of tandem site duplications (TSDs). L1 elements are composed of a 5′UTR that contains an internal RNA polymerase II promoter, followed by two open reading frames (ORF1 and ORF2) and a 3′UTR containing a polyadenylation signal (pA). Primer binding sites for ORF1 (red) and ORF2 (blue), and the area of in situ probe binding (dark red) are indicated.
Human platelets express LINE-1 ORF1 and ORF2
Human platelets express mRNA encoding LINE-1 ORF1 and L1 ORF2 (Figures 1C and 1D). Activation of platelets did not visibly alter the expression of LINE-1 ORF1 or ORF2 (Figure 1C). We next performed in situ hybridization experiments using custom designed probes against LINE-1 (Figure 1E). The specificity of the in situ experiments is based on the ability to detect a visible signal only if multiple probes align with their target RNA sequentially (mismatches indicated in Figure 1D are most likely artifacts introduced during the sequencing process). Consistent with our findings by PCR and Sanger-sequencing (Figures 1C and 1D), we found that LINE-1 mRNA is endogenously expressed in situ in the cytoplasm of unstimulated human platelets (Figure 1E, see also Figure 1F demonstrating binding sites of primer pairs and in situ probes). Laser capture microscopy (LCM) of individual human platelets further confirmed LINE-1 ORF1 and ORF2 expression (Supplemental Figure S2). LCM also excludes the possibility that signals detected in the PCR experiments are derived from LINE-1 mRNA or DNA introduced by contaminating nucleated cells.
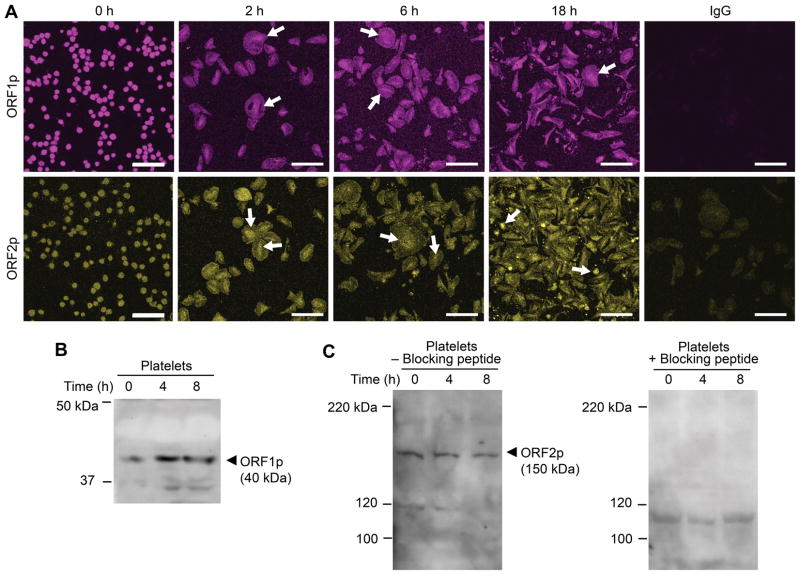
We next investigated the localization of LINE-1 encoded ORF1 (ORF1p) and ORF2 proteins (ORF2p) in human platelets. Both ORF1p and ORF2p are robustly expressed basally in the cytoplasm of unstimulated platelets (Figure 2A). In activated platelets, LINE-1 proteins concentrated and accumulated in the middle of the cell bodies (Figure 2A, arrows), cellular regions rich in mRNAs and ribosomal constituents7, 8. LINE-1 proteins were also detected in megakaryocytes and proplatelet extensions (data not shown). When platelets were allowed to culture overnight (18 hours), ORF2p continued to be robustly expressed and was also identified in the cytoplasm of platelets with extended cell bodies51, 52 (Supplemental Figure S3). Immunoblotting also demonstrated that ORF1p (Figure 2B) and ORF2p (Figure 2C) protein was present in platelets. Specificity of the anti-ORF2p antibody was confirmed by performing a quench experiment using the immunogenic peptide (Figure 2C).
Figure 2. Human platelets express ORF1 and ORF2 protein.
(A) Freshly isolated platelets were fixed in suspension immediately (baseline, 0 h) or adhered to immobilized fibrinogen in the presence of thrombin for 2, 6, or 18 hours. Immunofluorescence staining with an anti-L1 ORF1p (top row, magenta stain), and anti-L1 ORF2p (bottom row, yellow stain) antibody, respectively, demonstrates expression of both L1-encoded proteins ORF1p and ORF2p in platelets, irrespective of presence or absence of thrombin stimulation. White arrows indicate the time-dependent concentration of L1 proteins in central cell areas, rich in mRNA and ribosomes. The isotype specific IgG control is shown on the far right (scale bars = 10 μm). This figure is representative of 3 independent experiments. (B) Immunoblot analysis of platelet lysates using the anti-L1 ORF1p antibody. Freshly isolated platelets were immediately fixed in suspension (t=0 h) or allowed to adhere to immobilized fibrinogen in the presence of thrombin for 4 or 8 hours. The cells were then lysed and proteins separated by SDS-PAGE electrophoresis. (C) Immunoblot analysis of platelet lysates using the anti-L1 ORF2p antibody. Platelets were treated as in B, however the samples were split and processed in parallel in the absence (left panel) or presence (right panel) of an ORF2p quenching peptide. Presence of the blocking peptide prevents the detection of L1 ORF2p with the anti-L1 ORF2p antibody. Panels B and C are representative of 4 and 5 independent experiments, respectively.
Platelets contain ribonucleoprotein particles (RNPs) harboring L1 mRNA, and L1 ORF2p
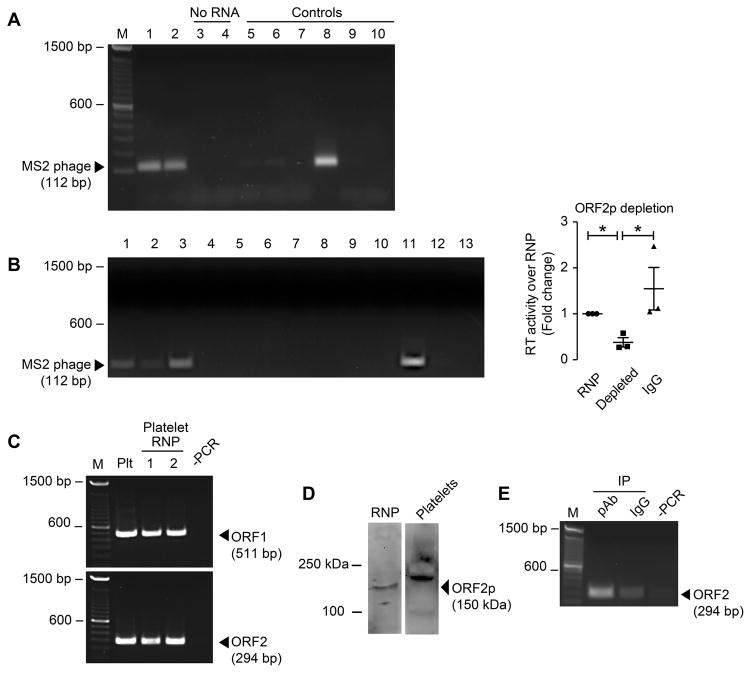
We hypothesized that RNPs in platelets contain functional LINE-1 mRNA and LINE1-ORF2p harboring reverse transcriptase activity. Therefore, to further identify the localization and function of endogenous L1-encoded RT (eRT) in platelets, we focused on the subcellular localization of eRT. Ribonucleoprotein particles (RNPs) are intracellular complexes integral to the regulation of gene expression53. To examine whether the eRT expression and activity we identified (Figures 1–2) is contained within RNPs, we next determined whether RNPs isolated from platelets harbored reverse transcriptase activity. Purified RNPs from platelets were sufficient to reverse transcribe an exogenous MS2 phage RNA in an in vitro RT-activity assay (Figure 3A), indicating that platelet eRT activity is found within RNPs. We next investigated if the eRT activity is ORF2p (which contains the RT domain) dependent. RNP lysates used for RT-activity assays were depleted of LINE-1 ORF2p using the anti-ORF2p antibody characterized in Figure 2. Immunodepletion of LINE-1 ORF2p reduced RT activity by more than 60% (Figure 3B), indicating that the LINE-1 ORF2p RT domain is the major source for platelet eRT. Consistent with the eRT activity associated with RNPs, RNA coding for LINE-1 ORF1 and ORF2 was identified within isolated platelet RNPs (Figure 3C). Furthermore, L1 ORF2p was confirmed in platelet derived RNPs (Figure 3D). To confirm that platelets are capable of forming L1 RNPs, we in vitro translated T7-tagged ORF1p using the plasmid pAD2TE127 as a template. Next, isolated total platelet RNA was incubated with the recombinant T7-tagged ORF1p and ORF1p-bound RNA was subsequently immunoprecipitated using the T7-tag. T7-tagged ORF1p specifically binds to platelet derived LINE-1 mRNA indicating that platelets are capable of forming LINE-1 RNPs (Figure 3E). In addition, the experiments confirmed that the RNPs isolated from platelets (Figure 3A) are composed of LINE-1 encoding mRNAs and encoded proteins. After identifying that the major source of eRT activity in platelets is the L1 ORF2p RT domain in isolated RNPs, we next sought to determine the function of eRT activity in platelets.
Figure 3. Platelet RNPs contain L1 RNA and are a source of eRT activity.
(A) RT assays with platelet-derived RNPs. RNPs were isolated as described in the Methods section and incubated with (lanes 1 and 2) or without (lanes 3 and 4) MS2 phage RNA. Controls include: lane 5 - platelet lysate replaced with the lysis buffer only; lane 6 - omission of the platelet lysate; lane 7 - omission of the platelet lysate and MS2 phage RNA; lane 8 - platelet lysate replaced with commercial RT; lane 9 - omission of the reverse MS2 phage primer; lane 10 - negative PCR. This figure is representative of 5 independent experiments. (B) Depletion of L1 ORF2p from platelet-derived RNPs. RNPs were isolated as described in the Materials and Methods section, and L1 ORF2p was depleted using immunoprecipitation strategies. Non-depleted (lane 1), depleted (lane 2), or IgG-depleted platelet RNPs (lane 3) were incubated with (lanes 1–3) or without (lanes 4–6) MS2 phage RNA. Controls include: lane 7 - platelet lysate without MS2 phage RNA ; lane 8 - RNP replaced with the lysis buffer only; lane 9 - omission of the RNP; lane 10 - omission of the RNP and MS2 phage RNA; lane 11 - platelet lysate replaced with commercial RT; lane 12 - omission of the reverse MS2 phage primer; lane 13 - negative PCR. The dot blot on the right side represents the densitometric analysis of the non-depleted (RNP), depleted, or IgG-depleted platelet RNPs (IgG), indicating a Å60% reduction in RT activity (mean±SEM). This figure is representative of 3 independent experiments. Single asterisk: p<0.05 (C) RT-PCR on RNA isolated from platelet RNPs. RNA from RNPs was isolated and probed for LINE-1 ORF1 (top panel) and ORF2 (bottom panel). RNA in two different RNP samples [1, 2] was analyzed using PCR and specific primer sets. An unfractionated platelet lysate sample (Plt) was analyzed in parallel. This figure is representative of n=5 independent experiments. (D) Immunoblot analysis of platelet-derived RNP (left panel), and platelet lysates (right panel) using the anti-L1 ORF2p antibody. Panels are representative of 3 independent experiments. (E) RT-PCR detecting platelet-derived endogenous L1 RNA/ORF1p interaction. L1 ORF1p was in vitro translated from the expression plasmid pAD2TE1, resulting in a T7 tagged ORF1p. Platelet RNA was isolated and incubated with ORF1p-T7 tag, then immunoprecipitated and analyzed for bound LINE-1 RNA. pAb – IP using anti-T7 antibody, IgG – isotype control, -PCR – control. This figure is representative of 3 independent experiments.
Endogenous RT in platelets alters cytoskeletal dynamics, promotes pro-thrombotic functional responses in vivo, and regulates global protein synthesis
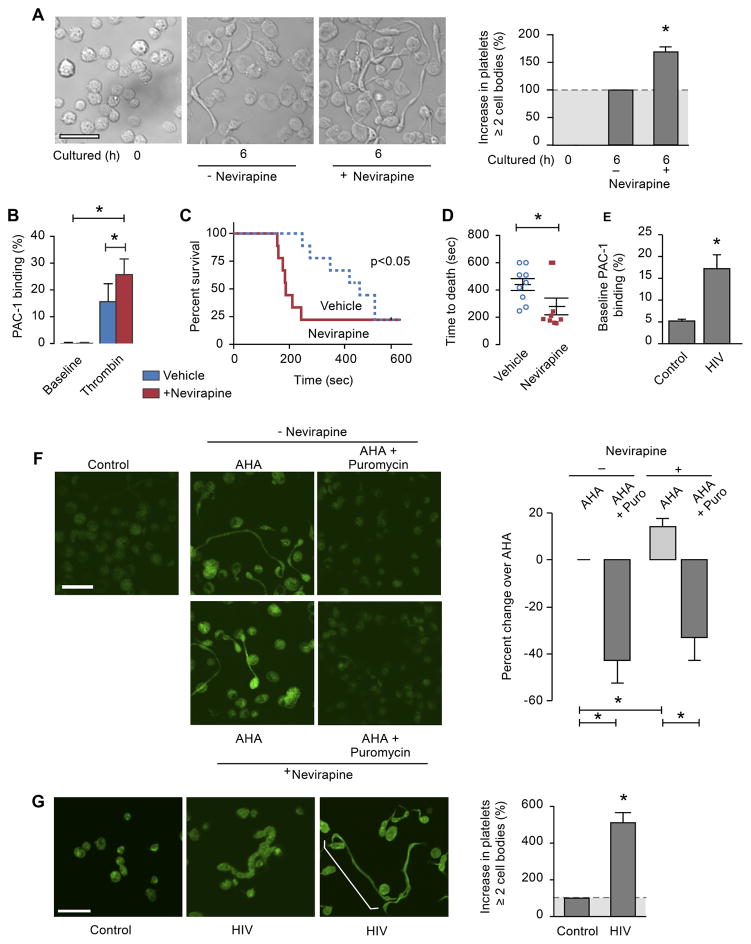
Previous studies by our group and others51, 52 demonstrated that cytoskeletal reorganization can promote the formation of extended platelets with 2 or more cell bodies, a process termed progeny formation that resembles how megakaryocytes form new platelets. In tumor cells, inhibition of eRT activity induces cellular differentiation37, 40, a process that also regulates cytoskeletal events. Therefore, we hypothesized that platelet eRT activity regulates platelet progeny formation; morphological changes in human platelets orchestrated by cytoskeletal dynamics. When platelet eRT activity was inhibited with nevirapine (an RT-inhibitor used to treat persons with HIV), at doses similar to concentrations seen in treated patients, the number of newly-formed cell bodies that extended from cultured platelets significantly increased (Figure 4A).
Figure 4. Inhibition of eRT activity in platelets alters actin cytoskeletal events, promotes pro-thrombotic functional responses, and induces global protein synthesis.
(A) Platelets from healthy individuals were isolated and fixed in suspension (0 hr), or treated with nevirapine (750 μM) or vehicle (control, DMSO) and subsequently incubated for 6 hours. The microscopy panels display representative examples of cultured platelets in the absence (-nevirapine) or presence of nevirapine (scale bar = 10 μm). The bar graph displays the percent increase in the number of platelets with at least two cell bodies in treated versus untreated platelets (mean±SEM; n=8). Single asterisk: p<0.05. (B) Platelets from healthy individuals were pre-treated with nevirapine for 6 hr (750μM, red) or its vehicle control (DMSO, blue). PAC-1 binding to activated integrin αIIbβ3 on the platelet surface was evaluated by flow-cytometry at baseline or after incubation with thrombin (0.1U/mL for 10 min.). The bar graph depicts the mean±SEM. Single asterisk: p<0.05. (C) Wild type mice were treated in vivo with either nevirapine (red) or its vehicle (DMSO, blue) by daily gavage for four days. Pulmonary embolism was induced by retro-orbital injection of collagen/epinephrine as described in the methods. Mortality from respiratory failure was measured to the stopping point of 600 seconds (10 min.). The Kaplan-Meier plot depicts the percent of mice surviving over time. The x-axis shows time in seconds till cessation of breathing (n=8–12 mice/group). (D) Summary blot for the time to death (seconds) for the individual animals from the experiment shown in (C). Single asterisk: p<0.05. (E) Human platelets from healthy individuals or persons with HIV were isolated. PAC-1 binding to activated integrin αIIbβ3 on the platelet surface was evaluated by flow-cytometry without additional, exogenous stimulation (i.e. baseline). The bar graph depicts the mean±SEM. Single asterisk: p<0.05. (F) Analysis of de novo protein synthesis in platelets. Freshly isolated platelets were cultured in the presence of an azido-amino acid analog of alanine (AHA), without (top) or with (bottom) nevirapine. In addition, select samples were pre-incubated with puromycin (AHA + Puromycin) or its vehicle (AHA) for two hours. The platelets were fixed after 6 hours and incorporation of the amino acid analog into protein was visualized (green stain). Unlabeled platelets fixed at baseline (left panel) were used to control for background immunofluorescence (scale bar = 10 μm). The bar graph displays the Cell Profiler analysis of the staining intensity for AHA (green) and is expressed as % change over AHA treated cells (±SEM; n=3). Single asterisk: p<0.05. This figure is representative of n=3 independent experiments. (G) Platelets isolated from healthy individuals (control) and persons with HIV were fixed in suspension. The microscopy panels display representative examples of platelets isolated from the two experimental groups, including representative panels from two persons with HIV (actin stain in green, scale bar = 10 μm). The bar graph displays the percent increase in the number of platelets with at least two cell bodies (as shown by the white bracket in the far right panel) in persons with HIV versus healthy individuals (control) (mean±SEM; n=3). Single asterisk: p<0.05.
Platelet cytoskeletal reorganization and progeny formation can result in platelet activation 51. We next sought to determine if platelet eRT activity regulated functional responses and thrombosis. We first stimulated platelets pre-treated in vitro with nevirapine (or its vehicle control) with thrombin. Inhibition of platelet eRT activity with nevirapine increased activated integrin αIIbβ3 expression on the platelet surface (Figure 4B). To establish whether nevirapine regulated thrombosis in vivo, we first confirmed that similar to human platelets, murine platelets possess eRT activity (Supplemental Figure S4). In a well-established pulmonary thrombosis model54, which is platelet dependent, nevirapine significantly accelerated thrombosis-dependent mortality (Figure 4C and D). To determine the clinical relevance of these findings, we examined activation of integrin αIIbβ3 in platelets isolated from persons with HIV treated with RT inhibitors clinically and matched healthy controls. As shown in Figure 4E, unstimulated platelets from persons with HIV had significantly increased basal activation of integrin αIIbβ3, thus recapitulating our in vitro findings (Figure 4B).
Based on these in vitro and in vivo results and observations51 that platelet cytoskeletal events require protein synthesis, we next determined whether eRT activity regulates de novo protein synthesis. We performed metabolic labeling by incubating platelets with Click-IT AHA (L-Azidohomoalanine51) in the presence or absence of nevirapine. Inhibiting eRT activity significantly increased the incorporation of labeled AHA into platelets (Figure 4F). The specificity of eRT-regulated translation in platelets was demonstrated by using puromycin (a translational inhibitor) as a control (Figure 4F). These data indicate that eRT activity in platelets regulates protein synthetic events.
To establish the relevance of platelet eRT activity in human disease, we examined the activity and function of eRT in platelets isolated from persons with HIV. We confirmed, similar our observations in platelets from healthy subjects, that platelets from persons with HIV (with undetectable viral load) possess eRT activity (data not shown). Next, we incubated platelets isolated from healthy donors or a person with HIV treated with an RT-inhibitor with [35S]methionine and [35S]cysteine. Compared to a healthy control subject, protein synthesis was increased in a person with HIV treated clinically with an RT-inhibitor (Supplemental Figure S5A). Increased protein synthesis was also observed when platelets from a healthy donor were treated with nevirapine ex vivo (Supplemental Figure S5B). Furthermore, platelets from persons with HIV on ART (antiretroviral therapy) demonstrated dramatically increased morphologic changes (Figure 4G) that require cytoskeletal reorganization.
RNA-DNA hybrids are present in human platelets
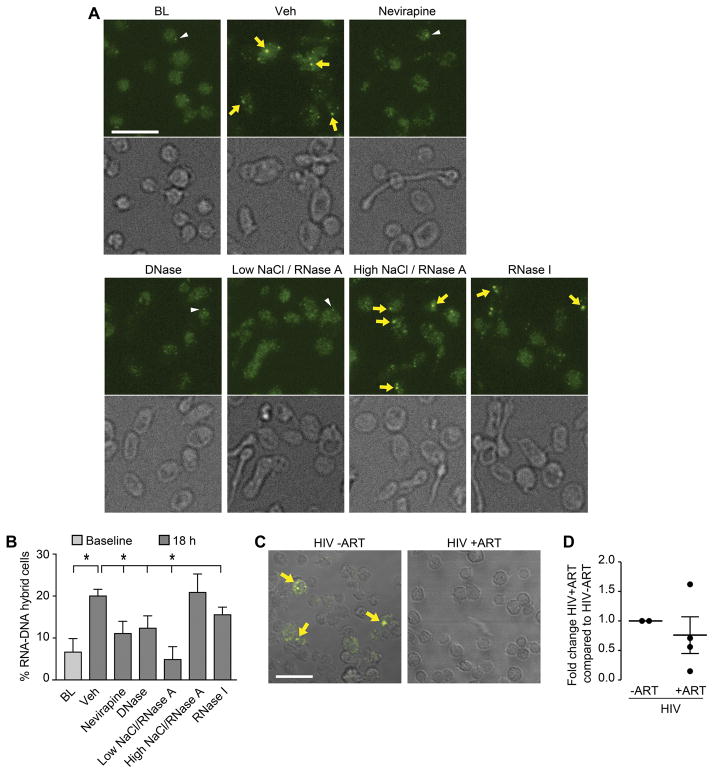
As our findings suggest that platelet eRT activity controls translational events, we hypothesized that one function of eRT in platelets is to generate RNA-DNA hybrids, resulting in a translational block (see graphic abstract). We identified RNA-DNA hybrids in human platelets in situ (Figure 5A and B). We next determined the mechanisms controlling RNA-DNA hybrid formation. In freshly isolated platelets from healthy control subjects, RNA-DNA hybrids were present in 6.6%±3.2% of the cells. When platelets were incubated overnight, the percentage of hybrid-positive cells increased to 20.0%±1.6% (Figure 5, p<0.05). This increase in hybrid formation was RT-dependent, since the treatment of platelets with nevirapine reduced hybrid-positive cells (11.1%±2.9%, Figure 5, p<0.05). To demonstrate the specificity of the antibody-based hybrid detection, we treated platelets with DNase and RNase A in a low concentration NaCl buffer, conditions favoring the digestion of DNA or RNA in RNA-DNA hybrids. As expected, both DNase and RNase A significantly reduced hybrid formation (Figure 5). In contrast, using a high concentration of NaCl in combination with RNase A (conditions under which hybrids are not digested) did not decrease hybrid formation.
Figure 5. RNA-DNA hybrids are present in human platelets and increased in persons with HIV not treated with an RT inhibitor.
(A) Immunofluorescence analysis of platelets stained with an anti-hybrid antibody. Platelets were fixed in suspension (BL), or treated with DMSO (Veh) or nevirapine (750 μM) and subsequently incubated for 18 hr. RNA-cDNA hybrids were detected using an anti-hybrid antibody (yellow arrows and white arrowheads). Post-fixation, platelets were treated with Turbo DNase (digests double strand DNA and DNA in hybrids), RNase A low (NaCl concentration at 70mM, digests single strand RNA, double strand RNA, and RNA in RNA-DNA hybrids), RNase A high (NaCl concentration at 500 mM, digests only single strand RNA and leave hybrids intact), or RNase I (which digests single and double stranded RNA). Scale bar = 10 μm. (B) The bar graph displays the percent of RNA-DNA hybrid positive cells (mean±SEM; n=3). Single asterisk: p<0.05, Veh compared to BL, nevirapine treated, low NaCl/RNase A, and RNase I treated cells. (C) Confocal images of platelets immunostained with an anti-RNA-DNA hybrid antibody (yellow, arrows). Platelets from persons with HIV before the initiation of treatment (HIV -ART) and persons with HIV on treatment (HIV +ART) were isolated and fixed in suspension. Scale bar = 10 μm. (D) The bar graph displays the fold change of RNA-DNA hybrid positive cells in samples from HIV –ART vs. HIV +ART (mean±SEM; n=2–4).
Yoshida and colleagues55 also used exogenous MS2 phage RNA to examine RT activity in washed platelet extracts from patients with myeloproliferative neoplasms. They speculated that RT activity may be found in mitochondria. While not a primary focus of our studies, in additional control experiments, we stained platelets with mitotracker to detect mitochondria, and the anti-RNA-DNA hybrid antibody in parallel, correlating potential intra-mitochondrial transcription events resulting in co-transcriptional RNA-DNA hybrid formation. While some co-staining could be detected in platelet mitochondria, about 50% of the hybrid signals were independent of mitochondrial staining (data not shown). Next, we compared the RNA-DNA hybrid expression levels between platelets isolated from persons with HIV before initiation of ART, and platelets isolated from persons with HIV following treatment with RT-inhibitor based ART. Platelets from persons with HIV had reduced numbers of RNA-DNA hybrids, compared to platelets from persons with HIV not being treated with RT-inhibitor based ART (Figure 5C and D).
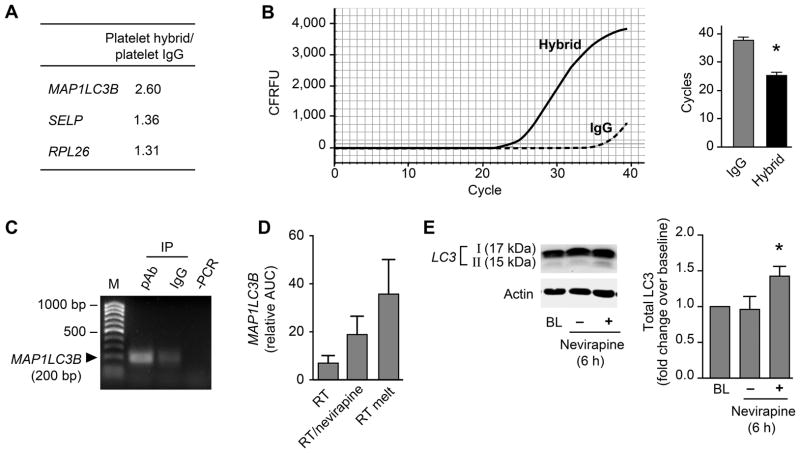
To identify candidates regulated by RNA-DNA hybrids, we immunoprecipitated hybrids from platelet lysates from healthy control subjects. RNA-DNA hybrids were isolated, a cDNA library of these RNAs was created, and sequencing was performed. Sequence analysis of the cDNA library revealed the enrichment of several target RNAs (see Supplemental Table 1). Three of the enriched identified RNAs coded for MAP1LC3B (Microtubule Associated Protein 1 Light Chain 3 Beta), SELP (P-selectin, which regulates platelet activation), and RPL26 (ribosomal protein L26, which regulates gene expression) (Figure 6A). MAP1LC3B mediates autophagy56, 57 and autophagy has been linked to platelet activation58. Accordingly, we focused on validating the presence of MAP1LC3B nucleotide sequences. Enrichment of MAP1LC3B sequences within RNA-DNA hybrids was confirmed in subsequent IP and qPCR experiments (Figure 6B). Thus, MAP1LC3B mRNA is expressed within RNA-DNA hybrids in platelets.
Figure 6. RNA-DNA hybrids serve as translational repressors in human platelets.
(A) Fold enrichment of co-immunoprecipitated RNA-DNA hybrids that were sequenced with targets identified (mRNAs coding for MAP1LC3B, SELP – P-selectin, and RPL26 – ribosomal protein L26) when comparing IgG vs. specific anti-hybrid antibody. (B) MAP1LC3B was verified using qPCR. An example of the real-time tracing is depicted on the left. The bar graph shows the change in cycle number threshold (mean±SEM; n=3, single asterisk: p<0.05). (C) RT-PCR analysis to demonstrate the presence of MAP1LC3B RNA in L1 ORF1p-T7 immunoprecipitates. T7-tagged L1 ORF1p was in vitro translated from the pAD2TE1 expression plasmid. Platelet RNA was isolated and incubated with ORF1p-T7 tag, immunoprecipitated, and analyzed for bound MAP1LC3B RNA. pAb – IP using anti-T7 antibody, IgG – isotype control, -PCR – control for contaminants. This figure is representative of 3 independent experiments. (D) MAP1LC3B was in vitro transcribed, and the mRNA reverse transcribed using an RT-activity assay, however, with shortened extension time (15 min, RT). Select samples were pre-incubated with nevirapine (500μM, 45 min, 37°C, RT+nevirapine), or processed using a 2 min melting step (95°C, RT melt). The mRNA or RNA-DNA hybrids were subsequently introduced into an in vitro translation assay, and resulting MAP1LC3B protein was separated by SDS-PAGE and detected using an anti-myc-DDK tag antibody. The bar graph of the densitometric analysis is shown. (E) Platelets were lysed at baseline (0 hr), or treated with DMSO control or nevirapine (750 μM) and subsequently incubated for 6 hr. Total MAP1LC3B (LC3B) was detected by means of Western blotting. LC3 isoforms I and II were detected and analyzed using ImageJ. A representative Western Blot for LC3 and the corresponding control actin blot are depicted (left). The bar graph (right) displays the fold change of total LC3 of the 6 hr samples over baseline. Single asterisk: p<0.05. This figure is representative of n=4 independent experiments.
LINE-1 ORF1p interacts with MAP1LC3B RNA in trans
To determine if MAP1LC3B mRNA can be a substrate for LINE-1 ORF1p and ORF2p, we in vitro translated T7-tagged ORF1p from the expression plasmid pAD2TE127 as a template (Figure 3E). Platelet RNA was incubated with the recombinant T7-tagged ORF1p and immunoprecipitated using an anti-T7-tag antibody. ORF1p binds to platelet derived MAP1LC3B mRNA (Figure 6C) indicating that this mRNA is associated with LINE-1 RNPs and would therefore be accessible to the RT domain of LINE-1 ORF2p.
RNA-DNA hybrids regulate protein synthetic events in human platelets
We next in vitro transcribed a MAP1LC3B-myc-DDK-tag encoding plasmid, generating an mRNA template for a modified in vitro RT activity assay and allowing us to specifically detect the translated MAP1LC3B fusion protein. Some platelet lysates used in the RT activity assay as RT donor were treated with the RT-inhibitor nevirapine to block hybrid formation. In addition, select samples underwent a melting step, resulting in a separation of RNA-DNA hybrids and releasing the translational block. The cDNA/hybrids were in vitro translated and the resulting proteins were separated by SDS-PAGE using a myc-DDK tag antibody for detection of in vitro translated proteins. The bar graph (Figure 6D) demonstrates that inhibiting hybrid formation (either by nevirapine or by inducing melting of the RNA from the DNA) increased the synthesis of MAP1LC3B protein. As these data suggest that MAP1LC3B synthesis in platelets may be translationally controlled, at least in part, by RNA-DNA hybrids, we next examined MAP1LC3B expression in live platelets pre-treated with nevirapine. Nevirapine significantly increased total MAP1LC3B protein (LC3B I and LC3B II) in platelets (Figure 6E). Similar findings were observed with two other proteins: P-selectin and ribosomal protein L26 (Supplemental Figure S6A and S6B). P-selectin surface translocation to the platelet surface was also increased in persons with HIV treated with RT inhibitors (Supplemental Figure S6C). These data indicate that nevirapine inhibits RNA-DNA hybrid formation and enhances protein synthesis by releasing a translational block.
Discussion
Platelets are anucleate and traditionally viewed as having a fixed molecular signature and associated functional responses. Nevertheless, platelets use an ever increasing, and often unexpected, repertoire of mechanisms to regulate their transcriptional and proteomic signature in health and disease2. Here, we provide the first evidence that human platelets possess LINE-1 derived eRT activity, regulating platelet morphology and protein synthesis, and leading to pro-thrombotic functional responses in vitro and in vivo. We also demonstrate that these findings are concordantly observed in persons with HIV clinically treated with RT inhibitors.
Both LINE-1 ORF1p (RNA binding) and ORF2p (endonuclease and RT domain) are encoded by the bicistronic LINE-1 and expressed in platelets. Concordant with the detected expression of both LINE-1 proteins, we discovered that platelets possess eRT activity (Figure 1). Previous studies have demonstrated the importance of LINE-1 ribonucleoprotein particles27, 30, 59, intracellular complexes that are integral to the regulation of gene expression53 and for retrotransposition in other cell types. Consistent with this, we report that the majority of platelet eRT activity is localized to LINE-1 RNPs (Figure 3). Depleting LINE-1 ORF2p from platelet RNPs demonstrates that LINE-1 RNPs are the major (e.g. ~60% of greater) contributor to platelet eRT activity. Furthermore, we show that exogenously expressed LINE-1 ORF1p containing an epitope tag (T7) at the carboxy terminus binds selectively to its endogenous encoding RNA (cis-preference30) in platelets. These findings indicate that human platelets contain functional LINE-1 RNPs. When platelets were incubated on immobilized fibrinogen and stimulated with thrombin, we observed trafficking of the LINE-1 proteins towards the central areas of platelets (Figure 2), subcellular regions known to be rich in mRNAs and ribosomal constituents7, 8. This shift in localization would allow LINE-1 proteins to interact with endogenous mRNAs that code for other cellular proteins (trans-acting LINE-1), a concept evaluated in previous studies30.
Since LINE-1 ORF2p includes an RT domain, it is worth discussing the functions of RT enzymes. In biochemical in vitro assays, RTs are used to reverse transcribe mRNA into cDNA to enable the subsequent PCR amplification step. This reaction forms mRNA-cDNA hybrids. Here, we demonstrate that platelets dynamically express RNA-DNA hybrids. Hybrid expression was dependent on eRT (ORF2p) and hybrid formation was blocked by nevirapine at doses achieving concentrations relevant to patients treated with this drug.
Based on our findings, we propose a previously unrecognized cellular mechanism. In our model, eRT in platelets generates RNA-DNA hybrids that block translation by partially reverse transcribing mRNA (see graphic abstract). The cellular concept of intentionally stalling translation was used in molecular biology strategies as an in vitro technique to identify specifically translated mRNAs42, 43, but so far has not been identified as an actively used translational control mechanism in primary human cells. In contrast, it is a well-accepted concept that the stability of co-transcriptional RNA-DNA hybrids affects transcriptional efficiency44, and even that stability differs between coding and non-coding regions60. However, this transcriptional mechanism is not based on eRT utilization. Nevertheless, in 2013, Sciamanna et al61 suggested that the regulation of tumor cell biology by LINE-136, 37, 39, 62, an area of intense research, could be induced by the formation of RNA-DNA hybrids. Using CsCl density gradients, they selectively identified Alu and LINE-1 containing RNA-DNA hybrid molecules in cancer but not in normal cells. They concluded that LINE-1 mediated RNA-DNA hybrids would interfere with the formation of dsRNA and therefore, the synthesis of regulatory small RNAs61.
Our findings demonstrate that RNA-DNA hybrid formation regulates synthesis of specific candidate RNAs by the induction of a translational block. This is supported directly by protein de novo synthesis assays, but also indirectly by the formation of platelet progeny, a process strictly dependent on protein synthetic activity51. MAP1LC3B was identified as one of the target mRNAs of trans-acting L1 ORF1p. MAP1LC3B plays an important role in autophagic processes56, 57 and serves as a good autophagy marker63. In addition, this catabolic process maintaining cellular hemostasis, adaptation to starvation, development and pathogen elimination needs to be tightly regulated64. Therefore, the translational control of one of the major players in autophagy by RNA-DNA hybrid formation is an attractive focus of future research. Two other identified hybrids also underscore the importance of this novel regulatory pathway. RPL26 is important for ribosomal hemostasis and regulation of gene expression, while SELP (P-selectin) is one of the major α-granular constituents being translocated to the surface of activated platelets upon stimulation65.
This novel mechanism for translational control has clinical implications. As part of ART (antiretroviral therapy), patients with HIV are typically prescribed RT-inhibitor based therapy. In persons with HIV, the introduction of ART has dramatically improved life expectancy46. As a result, chronic and often accelerated disorders, including atherothrombosis47–49, are replacing opportunistic infections as important causes of morbidity and mortality66. Furthermore, in patients with HIV under ART, and despite undetectable virus levels, a 30-fold increase in mortality when compared to uninfected controls could be demonstrated. The increased mortality was attributed to ongoing innate immune responses and activation of coagulation factors 67–69.
We demonstrated that platelet eRT activity regulates integration activation in vitro and in vivo. Similar findings of platelet integrin activation were observed in persons with HIV prescribed antiretroviral therapies that include RT inhibitors. We postulate that inhibition of platelet eRT activity may contribute to the increased risk of thrombosis observed in persons with HIV70. Furthermore, since we detected LINE-1 expression in other primary human blood cells, including megakaryocytes and T-cells (Supplemental Figure S8), our findings may have implications not only in the megakaryocyte-platelet axis, but also in other cell lineages.
Platelets isolated from persons with HIV were more synthetically active (Supplemental Figure S5) and had altered morphology resembling dumbbells (Figure 4C, as seen in vitro with nevirapine treated platelets), suggesting deregulated translational control mechanisms. Consistent with these observations, hybrids were higher in platelets from patients with HIV not treated with ART than compared to healthy individuals or persons with HIV treated with ART. Our findings suggest that administration of ART to persons with HIV alters platelet protein synthesis by lifting the RNA-DNA hybrid induced translational block; although we cannot exclude the possibility that differences in RNA-DNA hybrids in persons with HIV was also induced by HIV encoded RT. In this context, it is also interesting to note that platelets contain dengue virus DNA when infected with dengue virus, a (+)ssRNA virus (Supplemental Figure S7). Dengue virus DNA is not part of the virus life cycle and these data suggest a reverse transcription event. Furthermore, when eRT was inhibited, dengue DNA could not be detected. One might speculate if inhibiting dengue DNA formation removes a RNA-DNA hybrid induced translational block, leading to increased dengue virus replication.
While our findings confirm and extend prior studies in murine cells and tumor lines37, 39, 71, we recognize that several reports did not confirm that nevirapine blocks LINE-1 RT72. There are several potential explanations for these seemingly contradictory findings. First, we used higher concentrations of nevirapine (750μM), specifically chosen to recapitulate systemic concentrations of nevirapine achieved clinically. Second, we did not use tumor cell lines, but primary human cells (e.g. platelets). Finally, we examined RNA-DNA hybrid formation, rather than retrotransposition events. We also identified that another RT inhibitor, delavirdine, similarly blocked eRT activity (data not shown), indicating that our findings are not confined to nevirapine alone. While size-exclusion experiments (Supplemental Figure S1) did not completely exclude the possibility of eRT activity deriving from proviral insertions of human endogenous retroviruses (HERV), the fully processed HERV-K RT has a molecular weight of 66kDa, making this unlikely.
In conclusion, our data identify a previously unrecognized function for LINE-1 encoded proteins. We propose a model whereby endogenous LINE-1 mediate RNA-DNA hybrid formation controls protein expression for select mRNAs (please see graphical abstract). Because LINE-1 is classically considered to be involved in disease inducing de novo disruption of gene expression by retrotransposition events35, or tumor genesis73, 74, our findings also demonstrate that in human platelets, LINE-1 functions in a newly described constructive and potentially regulated fashion. Finally, our results provide evidence for a new translational regulatory mechanism that is present in platelets and which regulates integrin activation and thrombosis.
Supplementary Material
Highlights.
LINE-1 elements are one source of endogenous RT-activity (eRT).
Presence and function of eRT activity and LINE-1 in human platelets has not previously been determined.
LINE-1 retrotransposon’s RT-activity reverse transcribes select mRNAs, inducing RNA-DNA hybrids.
Inhibition of RT activity regulates platelet activation, thrombosis, and RNA-DNA hybrid formation which is a new mechanism of translational control.
Acknowledgments
We thank J.V. Moran for providing the pAD2TE plasmid. We thank Chris K. Rodesch and Keith R. Carney of the University of Utah Cell Imaging Core for technical assistance. We also thank Diana Lim for preparation of the figures, critical comments, and consultation regarding effective display of the images.
Sources of Funding: HL066277 (ASW) and HL112311 (ASW, GAZ), R37HL044525 (GAZ), AG040631, HL126547, and HL092161 (MTR), and GM103806 (JWR). HS was supported by a Post-Doctoral fellowship (0625098Y), a Beginning-Grant-in-Aid (09BG1A2250381) from the American Heart Association Western States Affiliate, and a Lichtenberg-Professorship from the Volkswagen Foundation. The project described was supported by the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through Grant 8UL1TR000105 (formerly UL1RR025764). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. This material is the result of work supported with resources and the use of facilities at the George E. Wahlen VA Medical Center, Salt Lake City, Utah. The sponsor had no role in the design or preparation of paper. The contents do not represent the views of the U.S. Department of Veterans Affairs or the United States Government.
Nonstandard Abbreviations and Acronyms
- eRT
endogenous reverse transcriptase
- LINE-1
Long Interspersed Element-1
- RNP
ribonucleoprotein-particles
- PRTE
pyrimidine-rich translational element
- UTR
untranslated region
- ORF
open reading frame
- ART
anti-retroviral therapy
- HERV
human endogenous retrovirus
- LCM
laser capture microscopy
- AHA
L-Azidohomoalanine
Footnotes
Disclosures: The authors declare no conflicts or competing financial interests.
References
- 1.Schwertz H, Rowley JW, Tolley ND, Campbell RA, Weyrich AS. Assessing protein synthesis by platelets. Methods Mol Biol. 2012;788:141–153. doi: 10.1007/978-1-61779-307-3_11. [DOI] [PubMed] [Google Scholar]
- 2.Weyrich AS, Schwertz H, Kraiss LW, Zimmerman GA. Protein synthesis by platelets: Historical and new perspectives. J Thromb Haemost. 2009;7:241–246. doi: 10.1111/j.1538-7836.2008.03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weyrich AS, Dixon DA, Pabla R, Elstad MR, McIntyre TM, Prescott SM, Zimmerman GA. Signal-dependent translation of a regulatory protein, bcl-3, in activated human platelets. Proc Natl Acad Sci U S A. 1998;95:5556–5561. doi: 10.1073/pnas.95.10.5556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zimmerman GA, Weyrich AS. Signal-dependent protein synthesis by activated platelets: New pathways to altered phenotype and function. Arterioscler Thromb Vasc Biol. 2008;28:s17–24. doi: 10.1161/ATVBAHA.107.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang H, Hornstein E, Stolovich M, Levy G, Livingstone M, Templeton D, Avruch J, Meyuhas O. Amino acid-induced translation of top mrnas is fully dependent on phosphatidylinositol 3-kinase-mediated signaling, is partially inhibited by rapamycin, and is independent of s6k1 and rps6 phosphorylation. Mol Cell Biol. 2001;21:8671–8683. doi: 10.1128/MCB.21.24.8671-8683.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsieh AC, Liu Y, Edlind MP, et al. The translational landscape of mtor signalling steers cancer initiation and metastasis. Nature. 2012;485:55–61. doi: 10.1038/nature10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Denis MM, Tolley ND, Bunting M, et al. Escaping the nuclear confines: Signal-dependent pre-mrna splicing in anucleate platelets. Cell. 2005;122:379–391. doi: 10.1016/j.cell.2005.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, Tilley RE, Rondina MT, Harris EM, Kraiss LW, Mackman N, Zimmerman GA, Weyrich AS. Signal-dependent splicing of tissue factor pre-mrna modulates the thrombogenecity of human platelets. J Exp Med. 2006;203:2433–2440. doi: 10.1084/jem.20061302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shashkin PN, Brown GT, Ghosh A, Marathe GK, McIntyre TM. Lipopolysaccharide is a direct agonist for platelet rna splicing. J Immunol. 2008;181:3495–3502. doi: 10.4049/jimmunol.181.5.3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. Line-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ostertag EM, Kazazian HH., Jr Biology of mammalian l1 retrotransposons. Annu Rev Genet. 2001;35:501–538. doi: 10.1146/annurev.genet.35.102401.091032. [DOI] [PubMed] [Google Scholar]
- 12.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 13.Penzkofer T, Jager M, Figlerowicz M, Badge R, Mundlos S, Robinson PN, Zemojtel T. L1base 2: More retrotransposition-active line-1s, more mammalian genomes. Nucleic Acids Res. 2017;45:D68–D73. doi: 10.1093/nar/gkw925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loeb DD, Padgett RW, Hardies SC, Shehee WR, Comer MB, Edgell MH, Hutchison CA., 3rd The sequence of a large l1md element reveals a tandemly repeated 5′ end and several features found in retrotransposons. Mol Cell Biol. 1986;6:168–182. doi: 10.1128/mcb.6.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scott AF, Schmeckpeper BJ, Abdelrazik M, Comey CT, O’Hara B, Rossiter JP, Cooley T, Heath P, Smith KD, Margolet L. Origin of the human l1 elements: Proposed progenitor genes deduced from a consensus DNA sequence. Genomics. 1987;1:113–125. doi: 10.1016/0888-7543(87)90003-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McMillan JP, Singer MF. Translation of the human line-1 element, l1hs. Proc Natl Acad Sci U S A. 1993;90:11533–11537. doi: 10.1073/pnas.90.24.11533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alisch RS, Garcia-Perez JL, Muotri AR, Gage FH, Moran JV. Unconventional translation of mammalian line-1 retrotransposons. Genes Dev. 2006;20:210–224. doi: 10.1101/gad.1380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hohjoh H, Singer MF. Sequence-specific single-strand rna binding protein encoded by the human line-1 retrotransposon. Embo J. 1997;16:6034–6043. doi: 10.1093/emboj/16.19.6034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolosha VO, Martin SL. In vitro properties of the first orf protein from mouse line-1 support its role in ribonucleoprotein particle formation during retrotransposition. Proc Natl Acad Sci U S A. 1997;94:10155–10160. doi: 10.1073/pnas.94.19.10155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martin SL, Li J, Weisz JA. Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the orf1 protein of mouse line-1. J Mol Biol. 2000;304:11–20. doi: 10.1006/jmbi.2000.4182. [DOI] [PubMed] [Google Scholar]
- 21.Fanning T, Singer M. The line-1 DNA sequences in four mammalian orders predict proteins that conserve homologies to retrovirus proteins. Nucleic Acids Res. 1987;15:2251–2260. doi: 10.1093/nar/15.5.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human l1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 23.Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. doi: 10.1126/science.1722352. [DOI] [PubMed] [Google Scholar]
- 24.Esnault C, Maestre J, Heidmann T. Human line retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 25.Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV. Human l1 retrotransposition: Cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–1439. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human line-1 protein and rna. Embo J. 1996;15:630–639. [PMC free article] [PubMed] [Google Scholar]
- 27.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for line-1 retrotransposition. Hum Mol Genet. 2005;14:3237–3248. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 28.Dewannieux M, Esnault C, Heidmann T. Line-mediated retrotransposition of marked alu sequences. Nat Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 29.Raiz J, Damert A, Chira S, Held U, Klawitter S, Hamdorf M, Lower J, Stratling WH, Lower R, Schumann GG. The non-autonomous retrotransposon sva is trans-mobilized by the human line-1 protein machinery. Nucleic Acids Res. 2012;40:1666–1683. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kulpa DA, Moran JV. Cis-preferential line-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–660. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 31.Elbarbary RA, Lucas BA, Maquat LE. Retrotransposons as regulators of gene expression. Science. 2016;351:aac7247. doi: 10.1126/science.aac7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen JM, Ferec C, Cooper DN. Line-1 endonuclease-dependent retrotranspositional events causing human genetic disease: Mutation detection bias and multiple mechanisms of target gene disruption. J Biomed Biotechnol. 2006;2006:56182. doi: 10.1155/JBB/2006/56182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matlik K, Redik K, Speek M. L1 antisense promoter drives tissue-specific transcription of human genes. J Biomed Biotechnol. 2006;2006:71753. doi: 10.1155/JBB/2006/71753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the l1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 35.Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landriscina M, Fabiano A, Altamura S, Bagala C, Piscazzi A, Cassano A, Spadafora C, Giorgino F, Barone C, Cignarelli M. Reverse transcriptase inhibitors down-regulate cell proliferation in vitro and in vivo and restore thyrotropin signaling and iodine uptake in human thyroid anaplastic carcinoma. J Clin Endocrinol Metab. 2005;90:5663–5671. doi: 10.1210/jc.2005-0367. [DOI] [PubMed] [Google Scholar]
- 37.Mangiacasale R, Pittoggi C, Sciamanna I, Careddu A, Mattei E, Lorenzini R, Travaglini L, Landriscina M, Barone C, Nervi C, Lavia P, Spadafora C. Exposure of normal and transformed cells to nevirapine, a reverse transcriptase inhibitor, reduces cell growth and promotes differentiation. Oncogene. 2003;22:2750–2761. doi: 10.1038/sj.onc.1206354. [DOI] [PubMed] [Google Scholar]
- 38.Sciamanna I, Barberi L, Martire A, Pittoggi C, Beraldi R, Giordano R, Magnano AR, Hogdson C, Spadafora C. Sperm endogenous reverse transcriptase as mediator of new genetic information. Biochem Biophys Res Commun. 2003;312:1039–1046. doi: 10.1016/j.bbrc.2003.11.024. [DOI] [PubMed] [Google Scholar]
- 39.Sciamanna I, Landriscina M, Pittoggi C, Quirino M, Mearelli C, Beraldi R, Mattei E, Serafino A, Cassano A, Sinibaldi-Vallebona P, Garaci E, Barone C, Spadafora C. Inhibition of endogenous reverse transcriptase antagonizes human tumor growth. Oncogene. 2005;24:3923–3931. doi: 10.1038/sj.onc.1208562. [DOI] [PubMed] [Google Scholar]
- 40.Sinibaldi-Vallebona P, Lavia P, Garaci E, Spadafora C. A role for endogenous reverse transcriptase in tumorigenesis and as a target in differentiating cancer therapy. Genes Chromosomes Cancer. 2006;45:1–10. doi: 10.1002/gcc.20266. [DOI] [PubMed] [Google Scholar]
- 41.Spadafora C. Endogenous reverse transcriptase: A mediator of cell proliferation and differentiation. Cytogenet Genome Res. 2004;105:346–350. doi: 10.1159/000078207. [DOI] [PubMed] [Google Scholar]
- 42.Cooper JA, Moss B. Translation of specific vaccinia virus rnas purified as rna-DNA hybrids on potassium iodide gradients. Nucleic Acids Res. 1979;6:3599–3612. doi: 10.1093/nar/6.11.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dudley K. Hybrid-arrested translation. Methods Mol Biol. 1988;4:39–45. doi: 10.1385/0-89603-127-6:39. [DOI] [PubMed] [Google Scholar]
- 44.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The rna-DNA hybrid maintains the register of transcription by preventing backtracking of rna polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 45.Ohle C, Tesorero R, Schermann G, Dobrev N, Sinning I, Fischer T. Transient rna-DNA hybrids are required for efficient double-strand break repair. Cell. 2016;167:1001–1013. e1007. doi: 10.1016/j.cell.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Mellors JW, Rinaldo CR, Jr, Gupta P, White RM, Todd JA, Kingsley LA. Prognosis in hiv-1 infection predicted by the quantity of virus in plasma. Science. 1996;272:1167–1170. doi: 10.1126/science.272.5265.1167. [DOI] [PubMed] [Google Scholar]
- 47.Massberg S, Brand K, Gruner S, Page S, Muller E, Muller I, Bergmeier W, Richter T, Lorenz M, Konrad I, Nieswandt B, Gawaz M. A critical role of platelet adhesion in the initiation of atherosclerotic lesion formation. J Exp Med. 2002;196:887–896. doi: 10.1084/jem.20012044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mastenbroek TG, van Geffen JP, Heemskerk JW, Cosemans JM. Acute and persistent platelet and coagulant activities in atherothrombosis. J Thromb Haemost. 2015;13(Suppl 1):S272–280. doi: 10.1111/jth.12972. [DOI] [PubMed] [Google Scholar]
- 49.Weyrich AS, Cipollone F, Mezzetti A, Zimmerman GA. Platelets in atherothrombosis: New and evolving roles. Current Pharmaceutical Design. 2007;13:1685–1691. doi: 10.2174/138161207780831374. [DOI] [PubMed] [Google Scholar]
- 50.Kothapalli R, Danyluck GM, Bailey RD, Loughran TP., Jr Problems associated with product enhancement reverse transcriptase assay using bacteriophage ms2 rna as a template. J Virol Methods. 2003;109:203–207. doi: 10.1016/s0166-0934(03)00072-7. [DOI] [PubMed] [Google Scholar]
- 51.Schwertz H, Koster S, Kahr WH, Michetti N, Kraemer BF, Weitz DA, Blaylock RC, Kraiss LW, Greinacher A, Zimmerman GA, Weyrich AS. Anucleate platelets generate progeny. Blood. 2010;115:3801–3809. doi: 10.1182/blood-2009-08-239558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thon JN, Montalvo A, Patel-Hett S, Devine MT, Richardson JL, Ehrlicher A, Larson MK, Hoffmeister K, Hartwig JH, Italiano JE., Jr Cytoskeletal mechanics of proplatelet maturation and platelet release. J Cell Biol. 2010 doi: 10.1083/jcb.201006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mansfield KD, Keene JD. The ribonome: A dominant force in co-ordinating gene expression. Biol Cell. 2009;101:169–181. doi: 10.1042/BC20080055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rowley JW, Chappaz S, Corduan A, et al. Dicer1-mediated mirna processing shapes the mrna profile and function of murine platelets. Blood. 2016;127:1743–1751. doi: 10.1182/blood-2015-07-661371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida S, Yamada H, Sakurai T, Suzuki M, Kojima K. Re-examination by improved reverse transcriptase assay of DNA polymerase in platelets from myelodysplastic disease patients. Biochem Int. 1989;19:1133–1141. [PubMed] [Google Scholar]
- 56.Feng W, Chang C, Luo D, Su H, Yu S, Hua W, Chen Z, Hu H, Liu W. Dissection of autophagy in human platelets. Autophagy. 2014;10:642–651. doi: 10.4161/auto.27832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klionsky DJ, Abdelmohsen K, Abe A, et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition) Autophagy. 2016;12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ouseph MM, Huang Y, Banerjee M, et al. Autophagy is induced upon platelet activation and is essential for hemostasis and thrombosis. Blood. 2015;126:1224–1233. doi: 10.1182/blood-2014-09-598722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, Kopera HC, Athanikar JN, Hasnaoui M, Bucheton A, Moran JV, Gilbert N. Characterization of line-1 ribonucleoprotein particles. PLoS Genet. 2010:6. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kraeva RI, Krastev DB, Roguev A, Ivanova A, Nedelcheva-Veleva MN, Stoynov SS. Stability of mrna/DNA and DNA/DNA duplexes affects mrna transcription. PLoS One. 2007;2:e290. doi: 10.1371/journal.pone.0000290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sciamanna I, Gualtieri A, Cossetti C, Osimo EF, Ferracin M, Macchia G, Arico E, Prosseda G, Vitullo P, Misteli T, Spadafora C. A tumor-promoting mechanism mediated by retrotransposon-encoded reverse transcriptase is active in human transformed cell lines. Oncotarget. 2013;4:2271–2287. doi: 10.18632/oncotarget.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oricchio E, Sciamanna I, Beraldi R, Tolstonog GV, Schumann GG, Spadafora C. Distinct roles for line-1 and herv-k retroelements in cell proliferation, differentiation and tumor progression. Oncogene. 2007 doi: 10.1038/sj.onc.1210214. [DOI] [PubMed] [Google Scholar]
- 63.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. Lc3, gabarap and gate16 localize to autophagosomal membrane depending on form-ii formation. J Cell Sci. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 64.Mizushima N. The pleiotropic role of autophagy: From protein metabolism to bactericide. Cell Death Differ. 2005;12(Suppl 2):1535–1541. doi: 10.1038/sj.cdd.4401728. [DOI] [PubMed] [Google Scholar]
- 65.Smyth SS, McEver RP, Weyrich AS, Morrell CN, Hoffman MR, Arepally GM, French PA, Dauerman HL, Becker RC, Platelet Colloquium P. Platelet functions beyond hemostasis. J Thromb Haemost. 2009;7:1759–1766. doi: 10.1111/j.1538-7836.2009.03586.x. [DOI] [PubMed] [Google Scholar]
- 66.Vachiat A, McCutcheon K, Tsabedze N, Zachariah D, Manga P. Hiv and ischemic heart disease. J Am Coll Cardiol. 2017;69:73–82. doi: 10.1016/j.jacc.2016.09.979. [DOI] [PubMed] [Google Scholar]
- 67.Funderburg NT, Lederman MM. Coagulation and morbidity in treated hiv infection. Thrombosis research. 2014;133(Suppl 1):S21–24. doi: 10.1016/j.thromres.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tenorio AR, Zheng Y, Bosch RJ, Krishnan S, Rodriguez B, Hunt PW, Plants J, Seth A, Wilson CC, Deeks SG, Lederman MM, Landay AL. Soluble markers of inflammation and coagulation but not t-cell activation predict non-aids-defining morbid events during suppressive antiretroviral treatment. The Journal of infectious diseases. 2014;210:1248–1259. doi: 10.1093/infdis/jiu254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Younas M, Psomas C, Reynes J, Corbeau P. Immune activation in the course of hiv-1 infection: Causes, phenotypes and persistence under therapy. HIV medicine. 2016;17:89–105. doi: 10.1111/hiv.12310. [DOI] [PubMed] [Google Scholar]
- 70.Auerbach E, Aboulafia DM. Venous and arterial thromboembolic complications associated with hiv infection and highly active antiretroviral therapy. Semin Thromb Hemost. 2012;38:830–838. doi: 10.1055/s-0032-1328887. [DOI] [PubMed] [Google Scholar]
- 71.Pittoggi C, Sciamanna I, Mattei E, Beraldi R, Lobascio AM, Mai A, Quaglia MG, Lorenzini R, Spadafora C. Role of endogenous reverse transcriptase in murine early embryo development. Mol Reprod Dev. 2003;66:225–236. doi: 10.1002/mrd.10349. [DOI] [PubMed] [Google Scholar]
- 72.Dai L, Huang Q, Boeke JD. Effect of reverse transcriptase inhibitors on line-1 and ty1 reverse transcriptase activities and on line-1 retrotransposition. BMC Biochem. 2011;12:18. doi: 10.1186/1471-2091-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sciamanna I, De Luca C, Spadafora C. The reverse transcriptase encoded by line-1 retrotransposons in the genesis, progression, and therapy of cancer. Front Chem. 2016;4:6. doi: 10.3389/fchem.2016.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiao-Jie L, Hui-Ying X, Qi X, Jiang X, Shi-Jie M. Line-1 in cancer: Multifaceted functions and potential clinical implications. Genet Med. 2016;18:431–439. doi: 10.1038/gim.2015.119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.