Abstract
Endospore formation in the Gram-positive bacterium Bacillus subtilis initiates in response to nutrient depletion and involves a series of morphological changes that result in the creation of a dormant spore. Early in this developmental process, the cell undergoes an asymmetric cell division that produces the larger mother cell and smaller forespore, the latter destined to become the mature spore. The mother cell septal membrane then engulfs the forespore, at which time an essential channel, the so-called feeding-tube apparatus, is thought to cross both membranes to create a direct conduit between the cells. At least nine proteins are required to form this channel including SpoIIQ under forespore control and SpoIIIAA-AH under the mother cell control. Several of these proteins share similarity to components of Type-II, -III and -IV secretion systems as well as the flagellum from Gram-negative bacteria. Here we report the X-ray crystallographic structure of the cytosolic domain of SpoIIIAB to 2.3 Å resolution. This domain adopts a conserved, secretion-system related fold of a six membered anti-parallel helical bundle with a positively charged membrane-interaction face at one end and a small groove at the other end that may serve as a binding site for partner proteins in the assembled apparatus. We analyzed and identified potential interaction interfaces by structure-guided mutagenesis in vivo. Furthermore, we were able to identify a remarkable structural homology to the C-subunit of a bacterial V-ATPase. Collectively, our data provides new insight into the possible roles of SpoIIIAB protein within the secretion-like apparatus essential to bacterial sporulation.
Keywords: Secretion system, SpoIIIAB, Sporulation
Introduction
Bacteria have evolved divergent transport systems to allow the specific selection and passage of small and large molecules across the plasma membrane for various purposes including cell homeostasis, virulence effector protein secretion, cell-cell communication, and pathogenesis (Burkinshaw and Strynadka, 2014; Christie et al., 2014; Costa et al., 2015; Korotkov et al., 2012; Kostakioti et al., 2005). Recently, a unique channel apparatus has been described to function during spore formation (“sporulation”) by the Gram-positive bacterium Bacillus subtilis (reviewed in Crawshaw et al., 2014). Sporulation is an ancient developmental program that allows a starving cell to survive as a dormant cell type, a spore. Early in sporulation, an asymmetric septum divides the rod-shaped bacterium into two cells: a smaller “forespore”, which will become the spore, and a larger “mother cell”, which contributes to the development of the forespore but ultimately dies. At first these two cells lie side-by-side; later the mother cell membrane migrates around the forespore in a phagocytic-like process, resulting in the forespore being engulfed as a double-membraned protoplast within the mother cell cytosol. The forespore then acquires a protective peptidoglycan cortex and protein coat layers, and is finally released as a mature spore into the environment by lysis of the mother cell.
At around the time of engulfment, at least nine proteins assemble into a channel apparatus that spans the two opposing membranes separating the mother cell and forespore (Crawshaw et al., 2014). Eight of these proteins (SpoIIIAA-AH) are encoded in a single operon (spoIIIA) expressed in the mother cell under the control of the compartment-specific alternative sigma factor σE (Guillot and Moran, 2007; Illing and Errington, 1991). A ninth protein, SpoIIQ, is produced in the forespore under the control of σF (Londono-Vallejo et al., 1997). Mutants lacking any one of these channel genes display collapsed forespores that are unable to carry out gene expression directed by the late-acting sigma factor σG (Camp and Losick, 2008; Doan et al., 2009; Kellner et al., 1996; Londono-Vallejo et al., 1997). The block in gene expression is not specific to σG, however, given that the phage T7 RNA polymerase engineered to be expressed in the forespore also requires the channel genes for late activity (Camp and Losick, 2009). Together these data support a model in which the channel transports one or more substrates, yet to be identified, that generally support the physiology of the forespore as well as its capacity to carry out macromolecular synthesis. In one version of this model, the channel has been proposed to act as a “feeding tube” through which small molecule metabolites are transported to the developing spore from the mother cell (Camp and Losick, 2009).
Although many open questions remain regarding the SpoIIIAA-AH•SpoIIQ sporulation channel, recent studies have helped to bring into focus its evolutionary origins and some of its structural features. Intriguingly, the emerging picture is that this mother cell-to-forespore channel represents a novel, hybrid secretion system with proteins that share common elements with those of diverse secretion systems found in Gram-negative bacteria. Homology searches (Söding et al., 2005) reveal that SpoIIIAA resembles AAA+ superfamily ATPases of the type II secretion system (T2SS) and type IV secretion system (T4SS) (Doan et al., 2009; Planet et al., 2001), while SpoIIIAB displays homology with the GspF/PilC family of proteins found in the T2SS and the related Type IV pilus (T4SP) secretion system (Abendroth et al., 2009; Karuppiah et al., 2010; Py et al., 2001). In contrast, SpoIIIAF, SpoIIIAG, and SpoIIIAH harbor domains that are evolutionarily related to the ring-forming PrgK/EscJ protein family of the type III secretion system (T3SS) (Bergeron et al., 2015; Levdikov et al., 2012; Meisner et al., 2012; Worrall et al., 2016; Yip et al., 2005). In the case of SpoIIIAH, structural studies have confirmed that its extracellular domain, which is anchored to the mother cell membrane by an N-terminal transmembrane domain, harbors the PrgK/EscJ “ring building motif” fold that helps to drive multimerization of the ring complex at the base of the T3SS apparatus (Bergeron et al., 2015; Worrall et al., 2016; Yip et al., 2005). This has led to a working model in which two stacked rings comprised of SpoIIIAH and the forespore membrane-anchored protein SpoIIQ, with which SpoIIIAH is known to interact (Blaylock et al., 2004; Doan et al., 2005; Levdikov et al., 2012; Meisner et al., 2012), serve as the “basal platform” upon which other channel components assemble. Recently we have determined the near-atomic resolution cryo-EM structure of SpoIIIAG in the assembled state, showing formation of ring building motif-mediated rings with interfacial packing similar to that observed in the T2SS and T3SS, but with unique stoichiometry and structural features critical to sporulation specific function (Zeytuni et al., 2017). Determining the structure of the remaining individual channel components, as well as how they fit together in a sporulation competent assembly, remains a crucial step toward a complete understanding of this unique channel apparatus.
In this study, we focus upon the essential SpoIIIAB sporulation channel protein. A mutant lacking spoIIIAB, as with other channel mutants, displays collapsed forespores that are unable to support late gene expression (Camp and Losick, 2009, 2008; Doan et al., 2009). Biochemical analysis has shown that SpoIIIAB resides in a complex with four other channel proteins: SpoIIIAD, SpoIIIAE, SpoIIIAF, and SpoIIIAG (Doan et al., 2009). Bioinformatics and structural prediction tools have suggested that SpoIIIAB is similar to regions of the GspF/PilC proteins of the T2SS and evolutionarily-related T4SP (Py et al. 2001; Abendroth et al. 2009; Karuppiah et al. 2010). These polytopic membrane proteins are localized to the inner membrane platform of the T2SS or T4SP although their specific functions remain unknown. Here we report the atomic structure of the cytosolic domain (residues 27-153) of SpoIIIAB protein to 2.3 Å resolution. In good agreement with structure prediction analysis, SpoIIIAB adopts a conserved, secretion-system related fold, comprised of an anti-parallel bundle of six helices. We have investigated potential interaction interfaces by structure-guided mutagenesis in vivo and discuss a surprising similarity to the C-subunit protein from the bacterial V-ATPase complex. Our data and structural analysis provides experimentally validated evidence that the sporulation channel is a hybrid secretion-like machinery.
Results
SpoIIIAB structure
SpoIIIAB is predicted to be a bitopic membrane protein with transmembrane helices at the N- and the C- termini and a soluble domain in between them (residues 27-153). To obtain structural and biochemical information for the soluble domain, recombinant SpoIIIAB27-153 was over-expressed in BL21 Escherichia coli cells and found to be soluble and stable in solution. Crystallization trials using the sitting drop vapor diffusion resulted in the appearance of a single crystal form which diffracted to a resolution of 2.32 Å (see Table 1 for data collection and refinement statistics). Perhaps not unexpectedly, given the relatively low partial sequence identity of SpoIIIAB27-153 to the T2SS/T4P GspF-family members of known structure (28.1% to V. cholerae EpsF, 22.6% to V. cholerae TcpE and 26.4% to T. thermophilus PilC), molecular replacement attempts were unsuccessful. To obtain phase information, SeMet labeled protein was prepared and used for single wavelength anomalous diffraction (SAD) phasing. The collected Se-Met derivative phasing datasets displayed similar resolution to the native crystal form and 8 selenium atoms were found in the crystal unit cell (Table 1). Although both Se-Met derivativized and native crystals were isomorphous with respect to the primitive orthorhombic space group including near identical unit cell dimensions, one of the space group screw axis has altered in unit cell direction a to b (P21221 in the native and P22121 in the Se-Met derivative crystal forms) (Table 1). Accordingly, an initial model was built according to the SeMet derivative experimentally phased electron density map and then was used as an input for molecular replacement against the native dataset. After manual rounds of rebuilding and refinement the collected native data yielded a high-quality structure (Table 1) with close to all the protein sequence clearly resolved.
Table 1. Data collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Protein | Native | SeMet derivative |
|---|---|---|
| SpoIIIAB 27-153 | SpoIIIAB 27-153 | |
| Data collection | Home source | CLS |
| Space group | P21221 | P22121 |
| Cell dimensions | ||
| a, b, c (Å) | 74.54, 82.00, 84.75 | 75.01, 81.97, 83.49 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 2.32 | 2.3 |
| Rsym or Rmerge | 13.1 (60.2) | 7.6 (36.7) |
| I/σI | 26.95 (2.81) | 25.19 (4.26) |
| Completeness (%) | 99.9 (98.1) | 95 (100) |
| Redundancy | 13.1 | 4.8 |
| Wavelength (Å) | 1.541 | 0.978 |
| Refinement | ||
| Resolution (Å) | 2.32 | 2.3 |
| No. reflections | 23068 | 22403 |
| Current Rwork/Rfree | 20.7/25.8 | |
| No. atoms | ||
| Protein | 4020 | |
| Ligand/ion | 40 | |
| Water | 114 | |
| B-factors (Å2) | ||
| Protein | 68.7 | |
| Ligand/ion | 46.1 | |
| Water | 42.9 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.89 |
The asymmetric unit of our crystal contains four monomers that are arranged in an anti-parallel manner with the N- and C- termini of each monomer facing away from those of the adjacent monomer (Fig. S1A). Two distinct crystallographic packing interfaces are formed within the asymmetric unit; the first is the symmetrical interface between chain A/B or C/D and the second is between chains A/C or B/D (Fig. S1A). All four SpoIIIAB27-153 monomers adopt the same overall fold, as the average root mean square deviation (RMSD) among the Cα of the monomers ranges between 0.78 – 1.27 Å (Fig. S1B). Although all four monomers are structurally similar, we found that monomer A better fits to monomer D while monomer B better fits to monomer C (Fig. S1B). Each SpoIIIAB27-153 monomer forms a six-helix bundle, with the antiparallel α-helices arranged in a cylinder, where each helix is tilted relative to the cylinder axis (Fig. 1A). The N- and C-termini are both located in close proximity to each other and contain a number of positively charged residues that supports a likely membrane facing orientation for this region of SpoIIIAB. The core of the cylinder is comprised of hydrophobic side chains, many of which are conserved in homologous SpoIIIAB proteins (Fig. 1B, Fig. S2). The top of the cylinder, facing away from the membrane, presents a small groove lined with hydrophobic side chains of the cylinder core (Fig. 1C).
Figure 1.
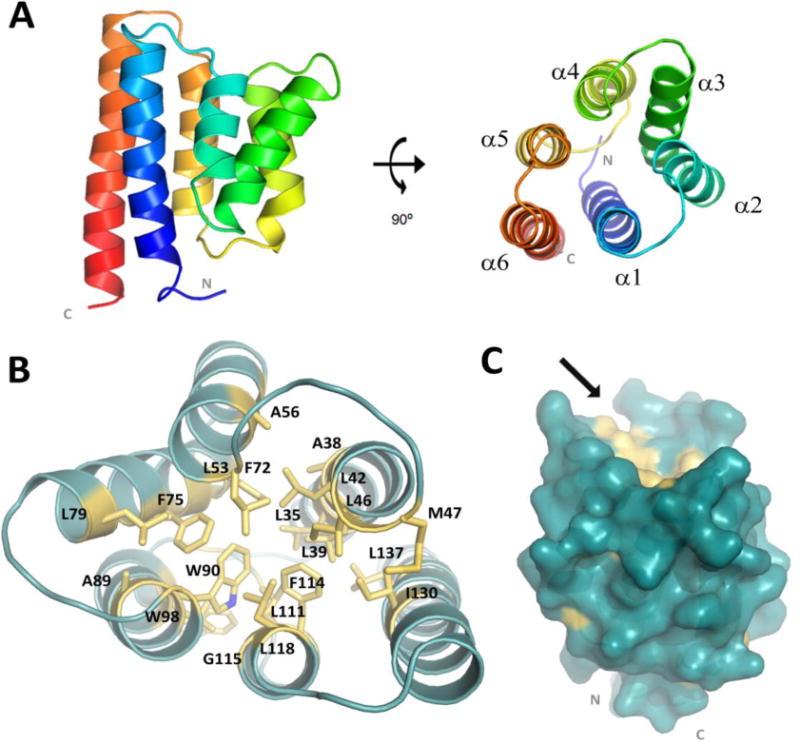
Overall SpoIIIAB27–153 monomeric structure. (A) Representative SpoIIIAB27-153 structure (cartoon drawing) folds as a six-helices bundle with both N and C termini are in close proximity and facing the same direction (membrane). The molecule is shown in two views, related by a 90° rotation. (B) Conserved residues comprising the SpoIIIAB27-153 hydrophobic core presented as yellow sticks. (C) Surface representation of the monomeric SpoIIIAB27-153 in blue whereas the conserved hydrophobic-core related residues are in yellow. Arrow points toward the small groove located at membrane-opposed protein face and lined with hydrophobic side chains associated with the hydrophobic core.
Structure-guided mutagenesis
Multiple sequence alignment (MSA) reveals high similarity between SpoIIIAB orthologs from different species (sequence identity between 36.5% to 95.8%) (Fig. S2) (Rice et al., 2000). To investigate the role of some of these conserved residues we introduced structure-based point mutations and assessed their effect on sporulation in vivo. Mapping the conserved residues on our SpoIIIAB structure revealed that R28 and E107 form a salt bridge interconnecting helix 1 and helix 5 and likely stabilizes their tight conformation around the membrane facing side of the six-helix bundle (Fig. 2A). In order to disrupt this salt bridge, we introduced the mutations R28E or E107A, with each resulting with a ~10,000-fold decrease in spore formation in vivo, the same defect displayed by cells deleted for spoIIIAB (Fig. 2B). Among the numerous strictly conserved hydrophobic residues, L132 is the only one that is surface exposed, located on the external face midway down the ~20 amino acid helix 6 of the bundle (Fig. 2A). Mutating L132 to alanine reduced spore formation in vivo by nearly 100-fold, suggesting this side chain has a significant role in protein-protein interactions essential for assembly or function of the complex and subsequent sporulation (Fig. 2B). To further elucidate the extent and specificity of the L132 containing interface we introduced mutations to other conserved residues in helices 5 and 6. These mutations, which included E109A, E109R, K128A, K128E, E143A and E143R, all had no significant effect on sporulation efficiencies in vivo, implying that the conserved interaction with L132 is not only specific but strictly localized as well (Fig. 2B). According to the MSA (Fig. S2), helix 1 contains a number of conserved charged residues. Mutation of several of these (R33E, E43A and E45R) reduced spore formation in vivo by 100- to 10,000-fold (Fig. 2), revealing another possible protein interaction interface. Although the SpoIIIAB27-153 structure contains six helices interconnected by five turns, conserved residues can only be found in the turns connecting helix 1 to 2 and helix 5 to 6 (Fig. S2). Previous work demonstrated that mutation of Y48A (located at the turn between helix 1 and 2) resulted in severe decrease in sporulation efficiency in vivo (Meisner, 2011). We performed additional mutagenesis of two residues also located at the turn between helix 5 and 6 (G119V or D122A), each also resulting in a notable decrease in sporulation efficiency in vivo (10,000-fold and 30-fold, respectively; Fig. 2B). The positioning of these conserved residues at the turns facing away from the membrane further support that this region has a role in protein-protein interactions with other members of the sporulation ‘feeding-tube’ channel.
Figure 2.
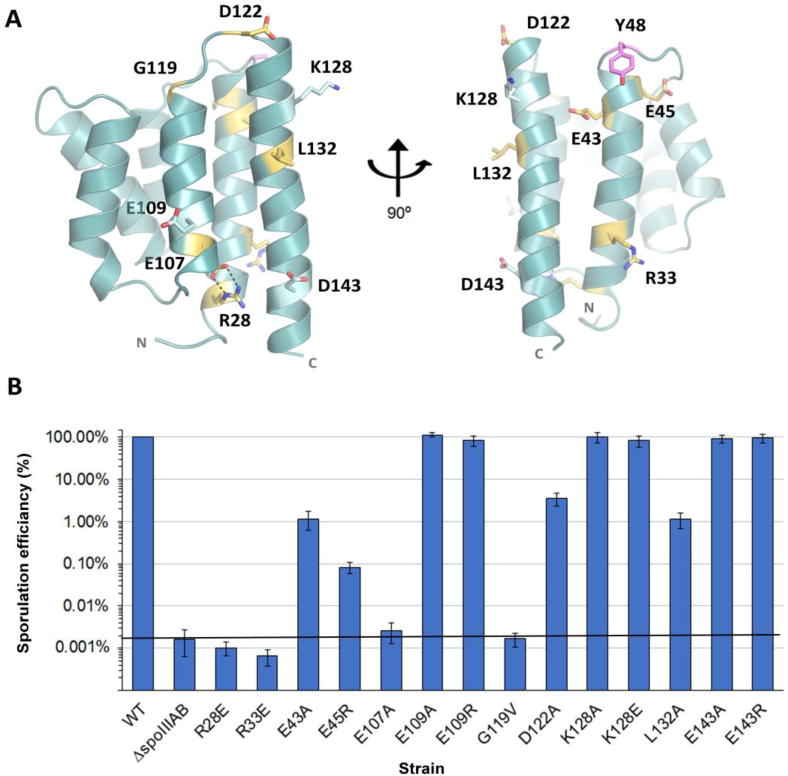
SpoIIIAB Structure-guided mutagenesis. (A) SpoIIIAB27-153 structure in cartoon representation and the mutated conserved residues are presented as sticks and color coded according to their effect on sporulation efficiency in vivo. Yellow-severe effect, light blue- mild/no effect. Magenta – severe effect as was demonstrated by Meisner, 2011. The molecule is shown in two views, related by a 90° rotation. (B) Sporulation efficiency assays of SpoIIIAB variants relative to wild type control cells set to 100%. Data are presented as the mean ± standard deviation.
SpoIIIAB structural homologs
The SpoIIIAB27-153 fold determined here shares significant global structural similarity to homologous proteins from various secretion systems. Structural superposition of the SpoIIIAB27-153 monomer with V. cholerae EpsF (PDB code: 3C1Q, (Abendroth et al., 2009)), V. cholerae TcpE (PDB code: 2WHN, (Kolappan and Craig, 2013)) and T. thermophilus PilC (PDB code: 4HHX, (Karuppiah et al., 2010)) resulted in overall Cα RMSDs of 1.89, 1.77 and 1.97 Å, respectively (on 264, 276 and 192 common atoms respectively; Fig. 3). An interesting variable region in these structural superpositions, however, was observed between helices 4 and 5 and the turn that connects them. Each of the four different structures display increasing length in the analogous region, with TcpE the shortest, followed by PilC and EpsF sharing mid-range lengths and with SpoIIIAB27-153 the longest (Fig. 3). Another unique feature of SpoIIIAB27-153 is a smaller angle between helices 5 and 6 (~31° as measured between the Cα of K105, D122 and Q146 residues) in comparison to that observed for the homologs (~ 43°–47° between analogous Cα locations of EpsF – E131-G144-N173, TcpE - P56-R129-G142 and PilC – D72-G85-D100) leading to a different positioning of helix 6 within the helical bundle. We predict these differing external protrusions and structural differences likely reflect the association of the unique protein binding-partners between the different secretion systems.
Figure 3.
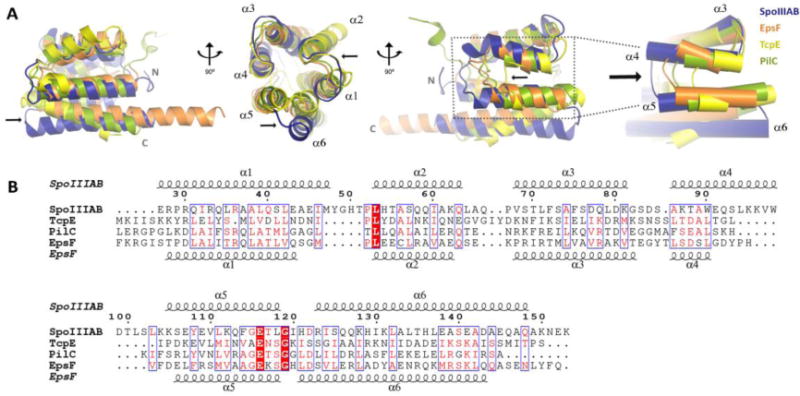
SpoIIIAB shares a similar fold to homologous protein form Type II secretion system and Type IV pilus. (A) Structural overlay of SpoIIIAB27-153 with EpsF56-171, TcpE1-102 and PilC53-168 proteins in blue, orange, yellow and green, respectively (PDB codes: 3C1Q, 2WHN and 4HHX, respectively). Two SpoIIIAB27-153 regions display structural variation and are marked by black arrows; helix 6 angle and the increasing dimensions of helices 4-5 and the loop connecting them. The structures overlay is shown in three views, related by a 90° rotation. (B) Multiple sequence alignment of SpoIIIAB27-153 protein and the structural homologs, residues are numbered according to SpoIIIAB. Top and bottom secondary structures are presented according to SpoIIIAB27-153 and EpsF56-171 crystal structures, respectively.
Notably, different oligomerization states in solution or in crystal have been reported for the structural homologs of SpoIIIAB27-153. While SpoIIIAB27-153 was found to be monomeric in solution and to form a likely non-physiological dimer in its crystal form (as suggested from the observed opposing orientation of the two monomers involved and their predicted transmembrane spanning regions) (Fig. S1, S3), TcpE1–102 was found to be monomeric both in the solution and crystallized forms (Kolappan and Craig, 2013). By contrast, PilC53–168 was reported to form dimers in solution (Karuppiah et al., 2010) and a small-fraction of EpsF56–171 was shown to form dimers in vivo by cross-linking experiments (Abendroth et al., 2009). Both PilC53–168 and EpsF56–171 form dimers in their crystal lattices, which were suggested to be physiologically relevant. Although sharing a significant structural similarity, the observed dimer interfaces of PilC53–168 and EpsF56–171 are entirely distinct from each other (Fig. S3). While the EpsF56–171 dimer is symmetrical with both monomers sharing the same orientation, PilC53–168 forms an asymmetrical dimer with one of the monomers shifted in reference to the other. We note for the latter that such a dimeric interface and orientation does not allow the positioning of the transmembrane helices in the same plane and therefore, as with SpoIIIAB, is physiologically unlikely. Indeed one of the confounding issues with the secretion system components has been the inability to capture their true oligomerization state in the isolated form and in the absence of other components of the system and/or native membrane (PrgK for example displays only dimers in solution, but in the assembled state, as evidenced from near-atomic cryoEM analysis (Worrall et al., 2016), forms clearly 24mer rings with significant interfacial interactions). Although missing a common oligomerization theme, all four protein homologs of SpoIIIAB compared here are members of large tube-shaped protein assemblies that are built from stacked ring-like components that collectively create a complex that can cross membrane(s) and selectively transport substrates through its hollow central channel. Therefore, we propose that SpoIIIAB and all other homologs may, given the appropriate cellular environment and effective concentration in the membrane, form homo-oligomeric structures that can in turn interact with other multimeric protein partners of the assembled sporulation ‘feeding-tube’ channel.
Interestingly, using DALI (Holm and Rosenström, 2010), we were able to detect that the C–subunit of the V-ATPase from T. thermophilus also shares significant structural similarity to SpoIIIAB27-153 (Fig. 4). The V-ATPase Subunit C serves as a “socket” that attaches soluble and cytosolic V1 central stalk subunits onto the L-subunit ring of the membrane-bound V0 domain (Iwata et al., 2004). The overall structure of the C-subunit consists of three domains related by a distorted threefold symmetry that can be superimposed with an RMSD of 2.0 Å and were suggested to be a product of gene triplication, although no obvious sequence similarity was observed. That the resulting pseudo-trimeric protein serves as a docking platform in this system might imply that the basic functional unit of SpoIIIAB and homologs may also have the propensity to trimerize. Previous cross-linking studies demonstrated that the N- and C-termini of the V-ATPase C-subunit are facing the membrane and interact with the L-subunit of the V0 domain, while the opposite face interacts with the cytosolic F-subunit of the V1 domain. Structural alignment revealed best fit between SpoIIIAB27–153 and the first C-subunit domain (residues 78-183) with a RMSD of 3.4 Å over 108 Cα atoms and similar membrane orientation, suggesting by analogy that SpoIIIAB27-153 also faces the cytosol (Fig. 4). Furthermore, the conserved cytosol-facing C-subunit T105 residue that interacts with the F-subunit overlays with Y48 of SpoIIIAB27-153, a conserved sporulation-essential residue (Fig. 4). Overall the observed structural similarity can provide hints for possible roles of SpoIIIAB, oligomerization number and hierarchy in the sporulation channel apparatus. Essentially, by analogy, SpoIIIAB would be well positioned to serve as a structural link between the membrane-bound protein components and other soluble components, such as, the SpoIIIAA ATPase of the feeding tube secretion assembly essential to bacterial sporulation.
Figure 4.
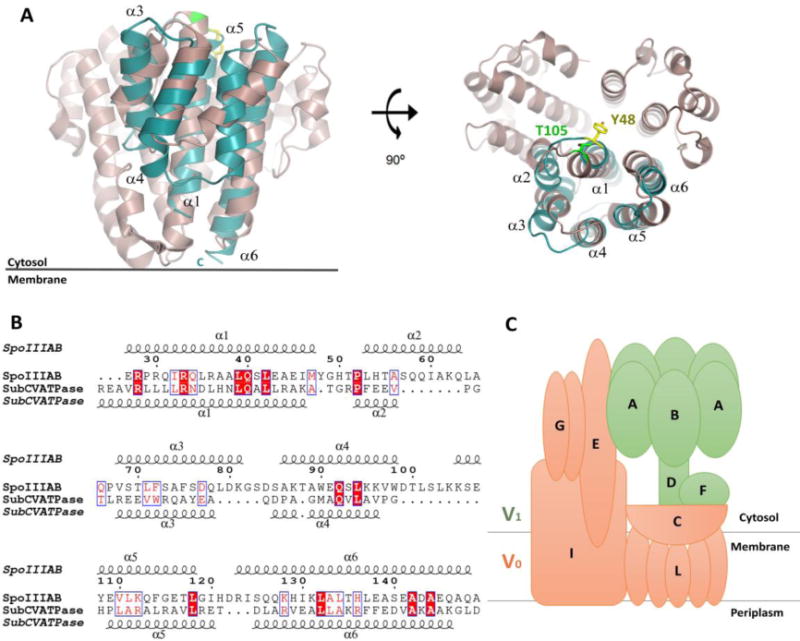
SpoIIIAB and T. thermophilus V-ATPase shares structural similarity. (A) Structural overlay of SpoIIIAB27-153, in blue, and the T. thermophilus V-ATPase C-subunit, in purple, shown in two views, related by a 90° rotation. SpoIIIAB overlaps to the first domain of the V-ATPase (residues 78-183). Specific and functionally conserved residues that are facing the cytosol are presented as sticks, C-subunit T105 in green and SpoIIIAB Y48 in yellow and the numbers of SpoIIIAB helices are numbered. (B) Sequence alignment of SpoIIIAB27-153 and the V-ATPase C-subunit, residues are numbered according to SpoIIIAB. Top and bottom secondary structures are presented according to their crystal structures. (C) Schematic representation of the T. thermophilus V-ATPase subunits composition. The V1 and V0 domains are colored in green and orange, respectively.
Conclusions
The SpoIIIAB atomic structure determined here further demonstrates that the B. subtilis sporulation channel is a hybrid, secretion-like machinery, involving homologous proteins to that of the type II, III, and IV bacterial secretion systems. The striking structural similarity between the SpoIIIAB fold and the V-ATPase C-subunit component reveals new implications for a common role and perhaps, at least in part, shared molecular mechanisms between the bacterial secretion systems and energy producing cellular-machineries. As such, this shared fold can also provide support for a possible role of SpoIIIAB-like proteins as a reversible platform between the soluble and the membrane-bound components of these bacterial nano-machineries. Finally, the SpoIIIAB structure represents another piece of the forespore development puzzle and sets a foundation for further structural and biochemical efforts to elucidate protein hierarchy and interactions in this system.
Methods
Recombinant protein expression and purification
The truncated spoIIIAB gene (UniProt Q01368-1, residues 27-153) was amplified by a polymerase chain reaction (PCR) from Bacillus subtilis strain PY79 genomic DNA. The primers were designed to introduce an NdeI site at the initiation codon, ATG, and a BamHI site after the termination codon. (All primers used in this study are listed in Table S1 of the supplementary material.) The DNA fragments were digested with NdeI and BamHI and cloned into the respective sites of plasmid pET28a (+) to construct pSM16, in which the spoIIAB gene was fused in-frame to express a 6-His tag at the N-terminus of the protein followed by thrombin proteolysis site. (All plasmids used in this study are listed in Table S2 of the supplementary material.)
E. coli BL21 strain cells, harboring the expression plasmid, were cultivated according to an auto-induction protocol (Studier, 2005) in a medium containing kanamycin (50 mg/ml) at 37°C for 8 hours. The cultivation temperature was then shifted from 37° to 22°C for a further 48 hours. To produce selenomethionine-labeled protein, E. coli strain B834 (methionine auxotroph) harboring the expression plasmid was cultivated in 6 L M9 medium (supplemented with 1 mM MgSO4, 0.1 mM CaCl2, 0.01 mM FeCl3, 1 mg thiamine, 1% glucose, 1.2 g each L-lysine, L-threonine and L-phenylalanine, and 0.6 g each L-isoleucine, L-leucine and selenomethionine) with 50 mg/ml kanamycin at 37°C. Overexpression was induced by an addition of 1 mM isopropyl β-D-thiogalactopyranoside (IPTG) after the culture had reached A600 of 0.5. After further cultivation for 18 h at 20°C, cells were harvested by centrifugation at 7,438 g for 8 min at 4°C.
The collected cells were suspended in buffer A (50 mM Tris–HCl, pH 8, 300 mM NaCl and 5 mM imidazole), incubated with DNase I (10 mg/ml) and protease-inhibitor tablets (Roche) at 4°C. The cells were then disrupted by two cycles in a French press pressure cell at 172 MPa. Cell debris was removed by high-speed centrifugation (Beckman, 70Ti rotor, 45k rpm, 45 min, 4°C) and the soluble fraction was applied to a gravity Ni-NTA column (Bio-Rad Econo-Column chromatography column, Thermo Scientific HisPur Ni-NTA resin) pre-equilibrated with buffer A. Bound protein was washed with 100 ml buffer B (20 mM Tris–HCl, pH 8, 1 M NaCl and 5 mM imidazole) and eluted with buffer C (20 mM Tris–HCl, pH 8, 300 mM NaCl, and 500 mM imidazole). To remove the 6-His tag, thrombin was added to the eluted protein and the mixture was dialyzed against buffer D (10 mM Tris–HCl, pH 8 and 300 mM NaCl) for 16 hours at 4°C. The protein was then applied onto a size-exclusion column (Superdex 75 26/60, GE Healthcare Biosciences) pre-equilibrated with buffer D. Finally, purified SpoIIIAB was concentrated to ~11 mg/ml, flash-frozen in liquid nitrogen and stored at −80°C.
Crystallization and structure determination
Crystals were grown using the sitting drop vapor diffusion method at 25°C, using 0.2 µL of protein solution mixed with an equal volume of precipitant. For both the SpoIIIAB native and Se-Met derivative protein, the crystallization precipitant was 0.15 M potassium phosphate and 3.3 M ammonium sulfate. Crystals were harvested without further addition of cryo-protectants and flash-cooled in liquid nitrogen. Native diffraction data collected at the home source using an image plate detector system (MAR 345 mm) (X-Ray Research) and MicroMax-007HF (Rigaku) generator. Data collection was performed at 100 °K and a total of 360 frames (360°) were collected with an oscillation range of 1°. The exposure time was 10 min per image and the crystal-to-detector distance was 220 mm. Se-Met derivative diffraction data was collected at beamline CMCF-BM of the Canadian Light Source in Saskatoon, Saskatchewan. Single-wavelength anomalous dispersion (SAD) data was collected at the peak wavelength of 0.9785 Å. Data collection was performed at 100 K and a total of 240 frames (120°) were collected with an oscillation range of 0.5°. The exposure time was 0.5 sec per image and the crystal-to-detector distance was 280 mm. Data was reduced and scaled using the HKL2000 suite (Otwinowski Z & Minor W, 1997). Phenix AutoSol program (Adams et al., 2010) was used for phasing, and 8 selenium atoms were found in the substructure solution. The model was built using Phenix AutoBuild and Coot (Adams et al., 2010; Emsley et al., 2010) and refined with Refmac5 (Vagin et al., 2004). For the Rfree calculation, 5% of the data was excluded. The obtained SeMet derivative model was then used as a template for molecular replacement in Phaser (42) against the native data (Table 1), The final model was manually edited in Coot (Adams et al., 2010; Emsley et al., 2010) and refined with Refmac5 (Vagin et al., 2004). Structural figures were prepared with PyMOL (DeLano, 2002).
Least-squares overlaps
R.M.S. calculations were performed with SwissPDB viewer (Guex N, Peitsch, 1997) using the domain alternate fit option to align structures and to define the conformational changes between structural homologs.
Electrostatic potential calculations
Electrostatics were calculated with PyMOL (DeLano, 2002), using the Adaptive Poisson-Boltzmann Solver (APBS) plug-in (Baker et al., 2001).
In vivo sporulation assays
spoIIIAB complementation plasmid and mutant construction
The spoIIIAB gene was fused to the promoter region of the spoIIIA operon, including the spoIIIAA ribosome binding site, and the spoIIIA operon terminator. This construct was encoded by two overlapping synthetic gene fragments (gBlocks, Integrated DNA Technologies), which were assembled into EcoRI/BamHI-digested pDG1662 (Guérout-Fleury et al., 1996) using the Gibson Assembly Master Mix (New England Biolabs) to generate the wild-type complementation construct of spoIIIAB, pKF72. (See Table S2 in supplementary material for a full list of plasmids used in this study.) Complementation constructs encoding missense mutants of spoIIIAB were generated by inserting gBlocks into pKF72 vector fragments as follows: To construct the plasmids encoding R28E, R33E, E43A, and E45R, gBlocks were each inserted into EagI/HindIII-double digested pKF72 using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). To construct the plasmids encoding E109R, K128A, K128E, L132A, and E143R, gBlocks were each inserted into HindIII/BamHI-digested pKF72 using NEBuilder HiFi DNA Assembly Master Mix (New England Biolabs). For the remaining spoIIIAB missense mutants E107A, E109A, G119V, D22A and E143A, Q5 Site-Directed Mutagenesis reactions (New England Biolabs) were transformed directly into B. subtilis (see below). The primers used in these reactions with pKF72 as a template were as follows: prKF107/prKF108 (E107A), prKF109/prKF110 (E109A), prKF111/prKF112 (G119V), prKF115/prKF116 (D122A), and prKF117/prKF118 (E143A). (See Table S1 of the supplementary material for the full list of primers used in this study).
Strain construction
All B. subtilis strains were derived from the laboratory strain PY79 (Youngman et al., 1983). The in-frame spoIIIAB deletion mutant, strain AHB1028, was obtained from our laboratory collection. Competent B. subtilis cultures of strain AHB1028 were prepared as previously described (Wilson and Bott, 1968) and transformed with sequenced plasmids or DpnI-treated site-directed mutagenesis reactions. Transformed B. subtilis strains were selected on Luria-Bertani (LB) agar with chloramphenicol (5 μg/mL), and chromosomal integration at the amyE locus was confirmed by a loss of α-amylase activity on LB agar with starch. Recipients of Q5 mutagenesis reactions were subsequently screened by sequencing PCR products of spoIIIAB at the amyE complementation locus. Chromosomal DNA was extracted from the appropriate isolates to transform competent AHB1028 and again select on LB with chloramphenicol. E109A, G119V, D122A and E143A were confirmed for ΔspoIIIAB.
The full names and genotypes of strains used in this study are listed in Table S3 of the supplementary material.
General bacterial methods and sporulation assays
B. subtilis strains were maintained in liquid LB or on solid LB plates with 1.6% agar. E. coli DH5α strains, each with a specific plasmid, were similarly maintained in or on LB with ampicillin (100 μg/ml). Sporulation efficiency assays have been previously described (Nicholson and Setlow, 1990; Schaeffer et al., 1965). Briefly, colonies were inoculated into 2 ml Difco sporulation medium (DSM) and grown for 24 hrs at 37° C. Cultures were then submerged in an 80°C water bath for 20 minutes to select for mature spores. Serial dilutions were plated on DSM agar. Colony-forming units were counted to calculate spores/ml as a percentage of wild-type spores/ml, set to 100%. Presented data are derived from at least three experiments, each in duplicate, and displayed as the mean +/− the standard deviation.
Coordinates
Structures have been submitted to the Protein Data Bank (accession code 6BS9).
Supplementary Material
Acknowledgments
This work was funded by a Banting fellowship to N.Z from the Canadian Institutes of Health Research (CIHR) and operating grants from the National Institutes of Health grant DP2 GM105439 to A.H.C., and operating grants from CIHR the Howard Hughes International Senior Scholar program to N.C.J.S. N.C.J.S. is a Tier I Canada Research Chair in Antibiotic Discovery.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
N.Z. and N.C.J.S. designed the research; N.Z., K.A.F., S.C.M., A.H.C. and N.C.J.S. performed the research; N.Z., K.A.F., A.H.C. and N.C.J.S. analyzed the data and wrote the paper. The authors declare no conflict of interest.
References
- Abendroth J, Mitchell DD, Korotkov KV, Johnson TL, Kreger A, Sandkvist M, Hol WGJ. The three-dimensional structure of the cytoplasmic domains of EpsF from the type 2 secretion system of Vibrio cholerae. J Struct Biol. 2009;166:303–315. doi: 10.1016/j.jsb.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker Na, Sept D, Joseph S, Holst MJ, McCammon Ja. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–41. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergeron JRC, Worrall LJ, De S, Sgourakis NG, Cheung AH, Lameignere E, Okon M, Wasney GA, Baker D, McIntosh LP, Strynadka NCJ. The modular structure of the inner-membrane ring component PrgK facilitates assembly of the type III secretion system basal body. Structure. 2015;23:161–172. doi: 10.1016/j.str.2014.10.021. [DOI] [PubMed] [Google Scholar]
- Blaylock B, Jiang X, Rubio A, Moran CP, Pogliano K. Zipper-like interaction between proteins in adjacent daughter cells mediates protein localization. Genes Dev. 2004;18:2916–2928. doi: 10.1101/gad.1252704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkinshaw BJ, Strynadka NCJ. Assembly and structure of the T3SS. Biochim Biophys Acta - Mol Cell Res. 2014;1843:1648–1693. doi: 10.1016/j.bbamcr.2014.01.035. [DOI] [PubMed] [Google Scholar]
- Camp AH, Losick R. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 2009;23:1014–1024. doi: 10.1101/gad.1781709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp AH, Losick R. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol Microbiol. 2008;69:402–417. doi: 10.1111/j.1365-2958.2008.06289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie PJ, Whitaker N, González-Rivera C. Mechanism and structure of the bacterial type IV secretion systems. Biochim Biophys Acta. 2014;1843:1578– 1591. doi: 10.1016/j.bbamcr.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol. 2015;13:343–359. doi: 10.1038/nrmicro3456. [DOI] [PubMed] [Google Scholar]
- Crawshaw AD, Serrano M, Stanley WA, Henriques AO, Salgado PS. A mother cell-to-forespore channel: Current understanding and future challenges. FEMS Microbiol Lett. 2014;358:129–136. doi: 10.1111/1574-6968.12554. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos: 2002. 0.99. [Google Scholar]
- Doan T, Marquis KA, Rudner DZ. Subcellular localization of a sporulation membrane protein is achieved through a network of interactions along and across the septum. Mol Microbiol. 2005;55:1767–1781. doi: 10.1111/j.1365-2958.2005.04501.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5(7):e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch M. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714– 23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Guillot C, Moran CP. Essential internal promoter in the spoIIIA locus of Bacillus subtilis. J Bacteriol. 2007;189(20):7181–9. doi: 10.1128/JB.00915-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–9. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Errington J. The Spoiiia Operon of Bacillus-Subtilis Defines a New Temporal Class of Mother-Cell-Specific Sporulation Genes under the Control of the Sigma-E Form of Rna-Polymerase. Mol Microbiol. 1991;(8):1927–40. doi: 10.1111/j.1365-2958.1991.tb00816.x. [DOI] [PubMed] [Google Scholar]
- Iwata M, Imamura H, Stambouli E, Ikeda C, Tamakoshi M, Nagata K, Makyio H, Hankamer B, Barber J, Yoshida M, Yokoyama K, Iwata S. Crystal structure of a central stalk subunit C and reversible association/dissociation of vacuole-type ATPase. Proc Natl Acad Sci U S A. 2004;101:59–64. doi: 10.1073/pnas.0305165101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karuppiah V, Hassan D, Saleem M, Derrick JP. Structure and oligomerization of the PilC type IV pilus biogenesis protein from Thermus thermophilus. Proteins Struct Funct Bioinforma. 2010;78:2049–2057. doi: 10.1002/prot.22720. [DOI] [PubMed] [Google Scholar]
- Kellner EM, Decatur A, Moran CP., Jr Two-stage regulation of an anti-sigma factor determines developmental fate during bacterial endospore formation. Mol Microbiol. 1996;21:913–924. doi: 10.1046/j.1365-2958.1996.461408.x. [DOI] [PubMed] [Google Scholar]
- Kolappan S, Craig L. Structure of the cytoplasmic domain of TcpE, the inner membrane core protein required for assembly of the Vibrio cholerae toxin-coregulated pilus. Acta Crystallogr Sect D Biol Crystallogr. 2013;69:513–519. doi: 10.1107/S0907444912050330. [DOI] [PubMed] [Google Scholar]
- Korotkov KV, Sandkvist M, Hol WGJ. The type II secretion system: biogenesis, molecular architecture and mechanism. Nat Rev Microbiol. 2012;10:336–51. doi: 10.1038/nrmicro2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostakioti M, Newman CL, Thanassi DG, Stathopoulos C. MINIREVIEW Mechanisms of Protein Export across the Bacterial Outer Membrane. 2005;187:4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdikov VM, Blagova EV, McFeat A, Fogg MJ, Wilson KS, Wilkinson AJ. Structure of components of an intercellular channel complex in sporulating Bacillus subtilis. Proc Natl Acad Sci U S A. 2012;109:5441–5. doi: 10.1073/pnas.1120087109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Londono-Vallejo JA, Frehel C, Stragier P. spoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol Microbiol. 1997;24:29–39. doi: 10.1046/j.1365-2958.1997.3181680.x. [DOI] [PubMed] [Google Scholar]
- Meisner J. A Junction between Differentiating Bacterial Cells. Emory University; 2011. [Google Scholar]
- Meisner J, Maehigashi T, Andre I, Dunham CM, Moran CP. Structure of the basal components of a bacterial transporter. Proc Natl Acad Sci. 2012;109:5446–5451. doi: 10.1073/pnas.1120113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson W, Setlow P. Sporulation, germination and outgrowth. In: Vander-Horn C, Cutting S, editors. Molecular Biological Methods for Bacillus. John Wiley & Sons; New York: 1990. pp. 391–450. [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol Crystallogr Pt A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Planet PJ, Kachlany SC, DeSalle R, Figurski DH. Phylogeny of genes for secretion NTPases: identification of the widespread tadA subfamily and development of a diagnostic key for gene classification. Proc Natl Acad Sci U S A. 2001;98:2503–2508. doi: 10.1073/pnas.051436598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Py B, Loiseau L, Barras F. An inner membrane platform in the type II secretion machinery of Gram-negative bacteria. EMBO Rep. 2001;2:244–248. doi: 10.1093/embo-reports/kve042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–7. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Vagin AA, Steiner RS, Lebedev AA, Potterton L, McNicholas S, Long F, Murshudov GN. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- Wilson GA, Bott KF. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J Bacteriol. 1968;95:1439–1449. doi: 10.1128/jb.95.4.1439-1449.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worrall LJ, Hong C, Vuckovic M, Deng W, Bergeron JRC, Majewski DD, Huang RK, Spreter T, Finlay BB, Yu Z, Strynadka NCJ. Near-atomic-resolution cryo-EM analysis of the Salmonella T3S injectisome basal body. Nature. 2016;540:597–601. doi: 10.1038/nature20576. [DOI] [PubMed] [Google Scholar]
- Yip CK, Kimbrough TG, Felise HB, Vuckovic M, Thomas Na, Pfuetzner Ra, Frey Ea, Finlay BB, Miller SI, Strynadka NCJ. Structural characterization of the molecular platform for type III secretion system assembly. Nature. 2005;435:702–707. doi: 10.1038/nature03554. [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci U S A. 1983;80:2305–9. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeytuni N, Hong C, Flanagan KA, Worrall LJ, Theiltges KA, Vuckovic M, Huang RK, Massoni SC, Camp AH, Yu Z, Strynadka NC. Near-atomic resolution cryoelectron microscopy structure of the 30-fold homooligomeric SpoIIIAG channel essential to spore formation in Bacillus subtilis. Proc Natl Acad Sci. 2017;114(34):E7073–E7081. doi: 10.1073/pnas.1704310114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


