Abstract
Eukaryotic cells, whether free-living or organismal, rely on metallo-reductases to process environmental ferric iron and cupric copper prior to uptake. In addition, some free-living eukaryotes (e.g. fungi and algae) couple ferri-reduction to ferro-oxidation, a process catalyzed by a small cohort of multi-copper oxidases; in these organisms, the ferric iron product is a ligand for cell iron uptake via a ferric iron permease. In addition to their support of iron uptake in lower eukaryotes, ferroxidases support ferrous iron efflux in Chordata; in this process the release of the ferrous iron from the efflux transporter is catalyzed by its ferroxidation. Last, ferroxidases also catalyze the oxidation of cuprous copper and, as metallo-oxidases, mirror the dual activity of the metallo-reductases. This Perspective examines the teleos of the yin-yang of this redox cycling of iron and copper in their metabolism.
Graphical Abstract
Eukaryotes employ a combination of metallo-reduction, metallo-oxidation and metallo-permeation to get iron where it needs to go without causing mischief.
The Argument
Aqueous iron and copper most commonly cross membranes in their reduced, low-valent oxidation states, Fe2+ and Cu+1, respectively2–7. Although a sizable fraction of microbial eukaryotes (protists) thrive in hypoxic if not anaerobic niches8, 9, fungi, plants and animals are, at the least, facultative aerobes (fungi). These organisms not only contain ATP-generating mitochondria fueled by terminal reduction of dioxygen, but express also the broad spectrum of anti-oxidant defense mechanisms common to aerobic organisms8–11. In general, these mechanisms serve to manage the contributors to what is broadly referred to as “redox stress” or, more specifically, a compromise in the cell’s reduction potential in one or more cell compartments13–15. The neutral pH reduction potentials of the Fe3+/Fe2+ and Cu2+/Cu1+ couples make them good 1e− reductants of O2 (generating the superoxide radical, O2−) and of hydrogen peroxide, H2O2 (generating the hydroxyl radical, OH•) as illustrated in Fig. 117–21. Thus, aqueous Fe2+ and Cu1+ are strong pro-oxidants and managing their abundance would be an essential component of a cell’s redox stress tool-box. This low valent, metal ion-dependent redox biology has been called “a fundamental theme of aerobic life”23. Reasonably, a (facultative) aerobe expressing a metallo-oxidase activity, one that oxidizes these low-valent metal ions by 1-electron without the production of 1e−-reduced dioxygen species, would have selective advantage. The premise of this argument is that multi-copper metallo-oxidases provide this advantage.
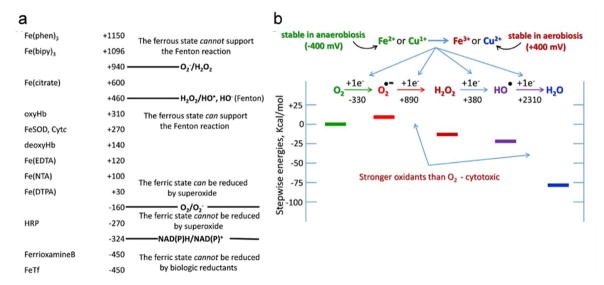
Fig. 1.
Iron and oxygen redox scales. (a) One-electron reduction potentials (in mV) of various iron complexes bracket the one-electron reduction potentials of oxygen and its 1e− reduction intermediates. The thermodynamically probable electron transfer events are indicated. Although not noted, typical copper complexes have potentials less than 100 mV and thus support these 1e− oxygen reduction reactions. (b) A thermodynamic scale of oxygen and its one-election reduction products. This diagram emphasizes the increasingly robust oxidation potential of oxygen’s 1e− reduced species.
The Reasoning
The potential intracellular cytotoxicity of O2 is reflected in the presence of a thiol-disulfide redox control system in all cells, anaerobes and aerobes alike; the thiolate anion is a kinetically and thermodynamically competent 1e− donor in dioxygen reduction to superoxide19, 24. In general, aerobes maintain this control via the mM GSH/GSSG redox buffer that, given the 100:1 ratio of these species in a resting cell, sets the intracellular reduction potential ≅-250 mV14, 25–29. A cell, whether free-living or organismal, deals also with extracellular redox stress which has the potential of damaging the cytoplasmic membrane and/or ecto-domains of proteins tethered to it. To the extent that the pro-oxidant reactivity of Fe2+ and Cu1+ contribute to this stress, limiting their concentration at the cell surface would be beneficial to an emerging aerobe. Multi-copper metallo-oxidases possess the enzymic activity to do just that.
Multi-copper oxidases (MCOs) contain, at least, 4 prosthetic copper atoms and thus in their fully reduced state provide the 4-electrons required to fully reduce dioxygen to 2H2O by-passing all of the intermediate 1e− reduced products collectively known as oxygen radicals1, 30, 31. All MCOs can be reduced by low-valent aromatic “hydroquinones” (phenols) and amines, e.g. para-phenylene diamine. These are “laccase” substrates, reflecting the role that some MCOs play in the formation and degradation of phenolic polymers found in spore coats and plant cell walls30, 32, 33. A relatively small cohort of MCOs express an additional activity towards Cu1+ (cuprous oxidases, e.g. bacterial CueO)34–37 or Fe2+ and Cu+1; although these latter MCOs long have been regarded as ferroxidases, a more descriptive common name is metallo-oxidase given their reactivity with both metal ions, a dual activity that has been appreciated only recently16, 30, 38, 39. The generic MCO reaction scheme and the role of the copper prosthetic groups in it is illustrated in Fig. 2.
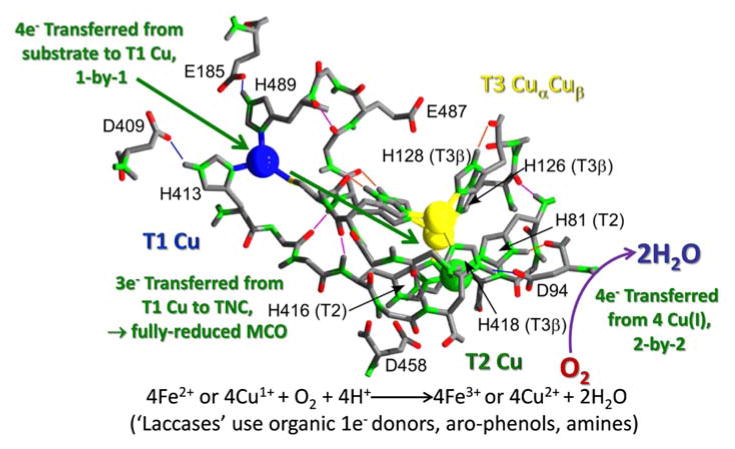
Fig. 2.
Electron transfer pathways in a multi-copper oxidase. Intermolecular, outersphere e− transfer from the reductant occurs at the T1 Cu into a lowest unoccupied molecular orbital that spans the protein’s four Cu atoms. Four consecutive 1e− substrate oxidations results in a fully reduced MCO. These 4e− are transferred, 2-by-2, to dioxygen bound at the trinuclear cluster (TNC) via a bound peroxy-intermediate1. The structure used here is Fet3 from S. cerevisiae (PDB 1ZPU)22.
There are two conceptual objections to the contention that metallo-oxidases represent a component of extracellular redox stress tool box; these objections can be phrased as questions: 1) are metallo-oxidases found at the extra-cytoplasmic surface of the cell membrane; and 2) is there a relatively high steady-state flux of Fe2+ and Cu1+ at this surface? A third and related objection is: 3) given the pro-oxidant behavior of these low valent metal ions, what would be the selective advantage of having such a flux at the cell’s surface in the first place?
These objections have been answered. First, all MCOs are either secreted, soluble proteins, or Type 1a membrane proteins with their catalytic sites found in the proteins’ carboxyl-terminal ectodomains40. Second, there is a flux of Fe2+ and Cu1+ at this surface irrespective of environmental aerobiosis due to the activity of cell surface metal reductases with activity towards both Fe3+ and Cu2+. The reducing equivalents for these reductases typically come from cytoplasmic reduced pyridine nucleotide co-factors41, 42 although some evidence has supported a direct role for dihydroascorbic acid as outlined below.
This raises the third objection: what was the selective advantage afforded by the extracellular production of a strong pro-oxidant, a potential cytotoxin? The answer? Having evolved in an anaerobic geochemical environment, the proto-aerobe had developed transport mechanisms for accumulation of these two essential trace elements in their low valent states, Fe2+ and Cu1+, i.e. ferrous and cuprous metal ion transporters. For a nascent aerobe to thrive in a geochemical milieu that now favored Fe3+ and Cu2+, a metal reductase was needed to supply the endogenous metal transporters with their ligands. The trade-off, however, was the potential oxidative stress posed by these low valent ions, a stress that could be managed by the coupled activity of a metallo-oxidase, an activity that like superoxide dismutases evolved and proliferated as aerobiosis spread from selective niches to the global environment.
The Evidence
Among eukaryotes, the universal copper uptake transporter conducts cuprous ion across the plasma membrane; in humans, this protein, hCTR1, is the product of the SLC31A1 locus43, 44. Similarly, the most ubiquitous iron uptake transporters expressed by eukaryotes are specific for ferrous iron. Again, in humans, there are two expressed by most if not all tissues, DMT1 and ZIP8 (and its orthologue, ZIP14) that in addition to Fe2+, transport other divalent, first-row transition metals2, 4, 45–48. Thus, for eukaryotes, their choice of ligand for iron and copper accumulation is one whose availability is diminishingly small in an environment in equilibrium with an atmosphere consisting of 21% O2 and at roughly neutral pH. A reasonable premise is that this choice is essentially vestigial, reflecting what was abundant when iron and copper acquisition pathways first evolved.
Eukaryotes encode and express a plethora of metalloreductases, e.g. fungi express the gene products of FRE loci49, 50 while humans express three genetically unrelated reductases, Dcytb (CYBRD1)51, 52, the Steap proteins (STEAP1-4)53–55, and SDR2 (FRRS1)56. All are Type III, multi-pass integral membrane proteins and most exhibit (some) residence in the plasma membrane. All except Steap1 and SDR2 have been shown to have turnover reductase activity towards both Fe3+ and Cu2+ 52, 55, 57. [Purified, reduced Steap1 reacts stoichiometrically with Fe3+ and Cu1+ but does not exhibit turnover kinetics with any oxidant58.] This metallo-reduction has been highlighted as “an essential biological process”59. In short, the low valent forms of these two metal ions are produced at the exocytoplasmic face of the eukaryotic plasma membrane. The iron and copper trafficking between reductase and permease in yeast and mammalian cells is illustrated in Fig. 3.
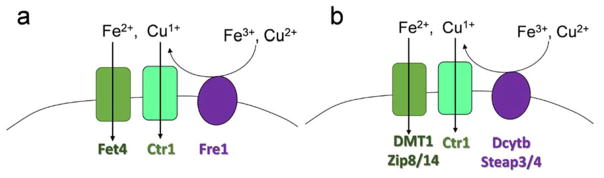
Fig. 3.
Eukaryotic metalloreductases and low valent copper and iron permeases. (a) The dominant plasma membrane metallo-reductase in fungi is a FRE protein, Fre1 in S. cerevisiae. Fungi express a canonical cuprous copper transporter, Ctr1, and a low-affinity ferrous iron uptake protein, Fet4. (b) Mammals express at least two distinct metalloreductases, Dcytb and a least two members of the Steap protein family, Steap3 and 4. The fungal and mammalian proteins share little homology but all contain inter-membrane porphyrins that function in shuttling electrons from cytosol to the exoplasm. For uptake, mammals express a canonical Ctr protein as well as two distinct families of ferrous iron transporters represented by DMT1 and Zip8/14.
This metallo-reduction itself is linked to cell redox status; this process drains reducing equivalents from the cytosol, in effect oxidizing it. There have been two suggestions as to the source of these electrons: reduced pyridine nucleotides and dihydroascorbic acid. Indeed, this cell surface reductase activity originally was referred to as an NAD(P)H oxidase one and was thought to play a role in cellular redox regulation and signaling41, 60, 61. On the other hand, some evidence has been interpreted to indicate ascorbate supplied the reducing equivalents 62–65. To be sure, sequence analysis did suggest that the fungal reductase, Fre1, was a heme protein homologous to the human NADPH oxidase66, 67, an inference confirmed by reverse genetic analysis67. These data suggested that the protein’s bis-heme prosthetic groups were an essential part of the intramolecular electron transfer pathway that shuttled reducing equivalents from the cytosol to the cell surface. These experiments did not examine the source of those equivalents, however. Thus, with the exception of Steap3 there are no structural nor structure-function data on which to base a specific reduction half-reaction mechanism. In the case of this member of the Steap family of reductase, N-terminal, cytosolic ‘reductase’ domain (PDB 2VQ3) contains a NADPH/flavin binding domain in which the binding of NADPH could be mapped 68.
Indeed, the most thoroughly examined metallo-reductase, oxidase pair is the Fre1, Fet3 one found in the plasma membrane of Saccharomyces cerevisiae and most other budding and filamentous fungi69–73. Log phase cultures of a common lab strain of S. cerevisiae exhibit a ferric reductase activity of 1.5 × 104 nmol/min/106 cells which is 103—104 fold greater than the rate of reduction of standard 1e− acceptors like MTT and TTC. The cupric reductase activity is somewhat less robust, but over 102 times greater than the reaction with any organic reductant73. These values, however, were quantified at super, saturating reductant concentrations so likely do not reflect typical physiologic velocities. Nonetheless, these measured metal ion substrate turnover efficiencies are ~102-fold greater than the respective metallo-oxidation by Fet3 of the reductase products Fe2+ and Cu1+. Consequently, even with the action of Fet3, there would be a steady-state flux of these low-valent pro-oxidants at the yeast plasma membrane albeit one that was tempered by metallo-oxidase action. This flux represents the ligand pool of low valent ionic species for transport into the cytoplasm by ferrous and cuprous ion transporters as illustrated in Fig. 3 above.
That this metallo-reductase, metallo-oxidase balancing act was homeostatic in nature is indicated by the following experiment in which the contribution Fet3 makes to yeast copper resistance was evaluated16. A parental strain of yeast is resistant to up to 5 mM cupric ion; in contrast, a fet3Δ strain exhibits growth arrest at less than 1 mM metal ion. Importantly, this sensitivity is not suppressed by concurrent deletion of the Ctr1 high-affinity Cu1+ transporter indicating that the cytotoxicity observed is a reflection of cell damage at the exo-cytoplasmic face of the plasma membrane and not due to intracellular insults. This origin model of copper toxicity and the role of a eukaryotic multicopper metallo-oxidase in it conforms to the likely mechanism by which the CueO MCO and its homologues contribute to copper resistance in bacteria35, 36. The function of the Fet3 ferroxidase in managing fungal copper resistance is illustrated in Fig. 4.
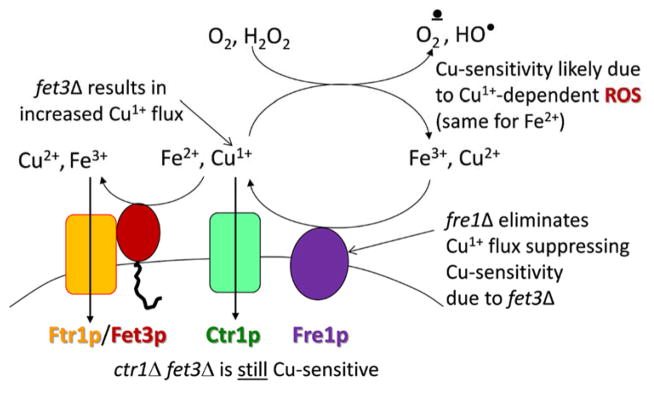
Fig. 4.
In yeast, deletion of FET3 leads to copper sensitivity. Common lab strains of S. cerevisiae exhibit normal growth up 10 mM copper. Growth of a fet3Δ strain is inhibited >0.5 mM copper, a sensitivity suppressed by deletion of FRE1. In contrast, the copper sensitivity of the fet3Δ strain is not suppressed by deletion of CTR1 indicating that the molecular trigger of the induced cytotoxicity is extracellular cuprous copper likely supporting redox cycling of dioxygen 1e− reduction products 16.
An obvious question arises, however, in regards to the presumed selective advantage metallo-reduction plays in support of low valent iron and copper uptake. For example, in yeast, with a substantial reoxidation of Fe2+ 39, what fraction of the reductase-generated ferrous iron substrate is available for uptake by the ferrous iron transporter, Fet474, 75? The adaptation to this apparent dilemma made by fungi, among a few other free-living eukaryotes, was to pair the ferroxidase with a ferric iron permease (an Ftr protein, for ferric iron transporter)76–78. Fet3 and Ftr1 form a FRET-detectable complex in the yeast plasma membrane in which the ferroxidase-generated Fe3+ is metabolically channeled to Ftr1 for permeation; that is, Fet3-generated Fe3+ is trafficked to Ftr1 by an associative, not dissociative mechanism12, 79. The FRET ‘image’ of this Fe-trafficking complex collected by confocal laser fluorescence microscopy is shown in Fig. 4a12. The tight, structural and mechanistic coupling of these proteins is indicated by the fact that exogenous, aqueous ferric ion is not substrate for Ftr1 iron permeation 79. Thus, the Fet/Ftr pattern reflects an iron uptake adaptation to the inherent cytotoxicity of Fe2+ under air. It is instructive to place the presumed vestigial ferrous iron permease, Fet4 and the aerobiosis-adapted Fet, Ftr pathway in their geochemical niches. Fet4, arising in a milieu awash with aqueous ionic ferrous iron, is a low-affinity transporter with KM ~40 μM 74; in this context, Fet4 can be compared functionally to the Fet, Ftr high-affinity system and its KM ~0.2 μM 79, appropriately adapted to iron metabolism in a ‘rusty’ environment.
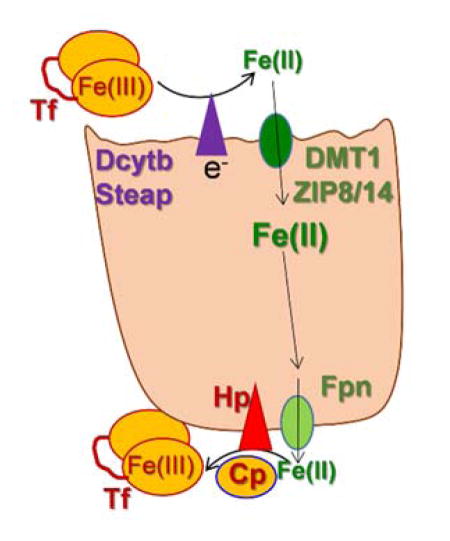
The tight metabolic coupling between ferroxidation and iron permeation in fungi is recapitulated in mammals by the ferrous iron exporter, ferroportin (Fpn) and the multi-copper ferroxidase, hephaestin (Hp) (and/or the orthologue, ceruloplasmin, Cp)2, 4, 5, 80–82. Like the fungal Fet, Ftr pair, the Hp, Fpn one also forms a FRET-detectable complex in the plasma membrane (Fig. 5b). However, rather than a ferroxidase to ferric ion permease iron trafficking pathway as in the Fet3, Ftr1 case, the Fpn-Hp scheme is a ferrous ion permeation to ferroxidation one. Again, however, the ferroxidation is the regulator of transmembrane iron trafficking: without the ‘terminal’ ferroxidation of the Fpn-transported Fe2+, the iron gets ‘stuck’ in Fpn, triggering permease protein retrieval from the plasma membrane82. The ferroxidase-dependent efflux via Fpn is illustrated in Fig. 6.
Fig. 5.
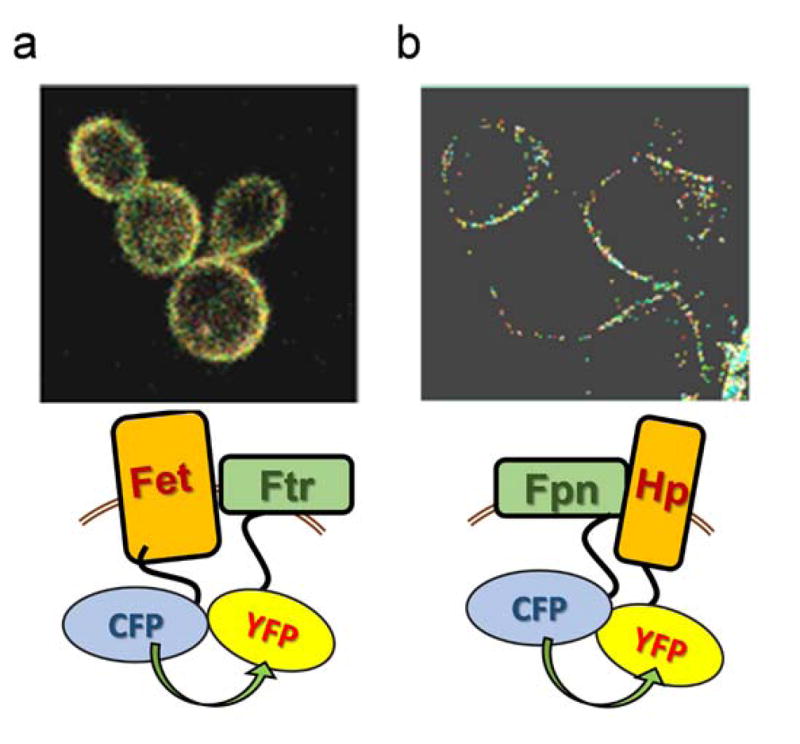
FRET images of ferroxidase, iron permease pairs. (a) Fluorescently-tagged Fet3 and Ftr1 were heterologously expressed in fet3Δftr1Δ S. cerevisiae. The FRET image shown reflects the increase in donor fluorescence upon photobleaching the acceptor. The FRET efficiency was 13.5% 12. (b) Fluorescently-tagged Fpn and Hp were heterologously expressed in HEK293T cells. The FRET image was generated as before. The efficiency was 9.8% which likely under-reports the biophysical energy transfer because it is uncorrected for donor fluor (Fpn-CFP) not in complex with Hp-YFP (Dlouhy and Kosman, unpublished).
Fig. 6.
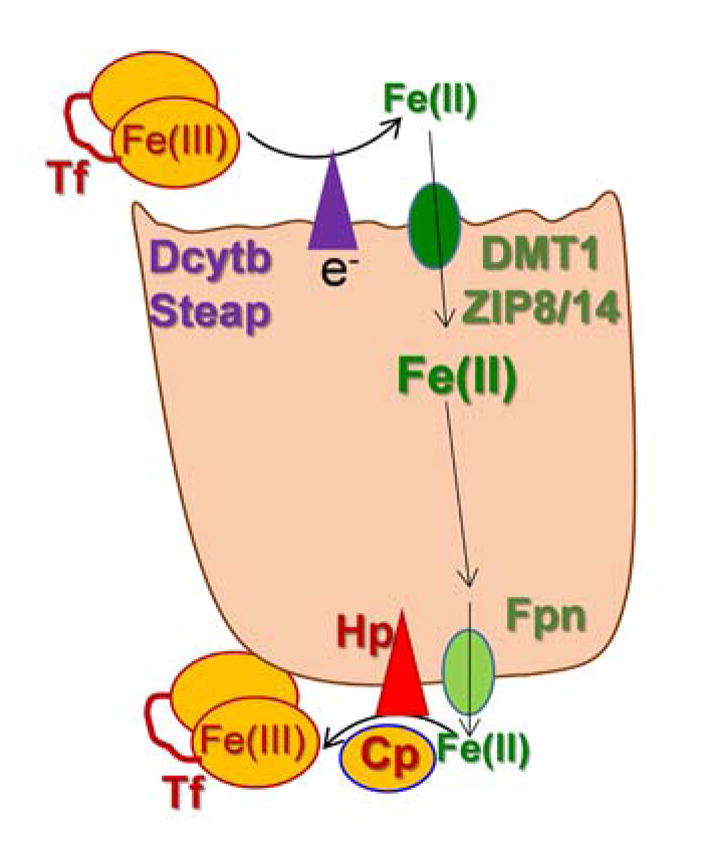
The ferroxidase-dependent step in mammalian iron trafficking: managing iron on the way out. Presented within the context of either an epithelial or endothelial cell (e.g. enterocyte or brain microcapillary), managing the efflux step relies on hephaestin (Hp) or ceruloplasmin (Cp) ferroxidation of the Fpn-trafficked Fe2+. In one model of this step, the Fe3+ product is channeled to a recipient ferric iron chelator, e.g. apo-transferrin (Tf) in an associative mechanism comparable to the fungal Fet, Ftr uptake pathway.
Summarizing the argument
One can appreciate the two ferroxidase-dependent iron trafficking mechanisms expressed by Eukarya in the context of the premise that the metallo-oxidase, at the least, tempers the flux of low valent metal ion pro-oxidants. In the case of fungal Fet3-Ftr1, by coupling ferroxidation (which manages the pro-oxidant) to permeation (which provides the essential trace element) both physiologic ends are achieved. In the Fpn-Hp system, Fpn transports Fe2+ out of the reducing environment of the cytosol but only when the ferrous iron pro-oxidant potential is ‘quenched’ by ferroxidation prior to or coincident with its release into the higher potential extra-cellular milieu. Indeed, this view provides a teleological context for the fact of the ferric iron specificity of transferrin. Indeed, one might also consider the possibility that like the ‘hand-off’ of Fe3+ from Fet3 to Ftr in fungal iron uptake, a similar transfer of Fe3+ occurs between Fpn/Hp and apo-Tf as the terminal step in mammalian cell iron efflux. In short, these two iron trafficking systems share the same intrinsic teleos, to get iron where it needs to go without causing any mischief.
Rebuttal: What’s missing from this argument?
There is little doubt about the progressive increase in the oceans’ level of dissolved oxygen starting ~ 1Gy BCE; the increase at that time-point is attributed, at least in part, to the disappearance of the ferrous iron that had buffered the O2 produced by photosynthetic bacteria starting ~2.5 Gy BCE83, 84. What more recently has been demonstrated is that concurrent with this oxygenation there was a sharp increase in dissolved trace elements, including copper85, 86. This is the geochemical context for the argument presented here.
Given this corresponding one-electron oxidation of the environmental Fe2+ and Cu1+, the selective advantage of a metallo-reductase to maintain the status quo, while arguable, is certainly is open to argument. While such an activity does maintain a source of low valent species for cell uptake, in the presence of dissolved O2 this same metal ion pool is a ready source of electrons for ROS generation. This is the very line of thinking provided as rationale for the metallo-oxidase side of this story. Indeed, given that the geologic oxidation event, itself, would have produced a steady flux of ROS from the oxidation of low valent metal ions, enzyme activities consistent with the suppression of the pathophysiologic consequences of that redox cycling would have been under strong selection87.
So, what is missing here is an explicit examination of the correspondence between the appearance, diversification and transfer of the genetic information encoding activities with selective advantage in this evolving geochemical milieu. To date, cladistics of this sort have focused on a specific component of the cellular response to this oxidation event. Thus, the bacterial copper homeostasis (and silver resistance) ‘island’ has been examined in detail (CHASRI), work that highlights the copper resistance advantage provided by the pco gene cluster including pcoA encoding the bacterial periplasmic cuprous oxidase88. On the other hand, a similarly thorough examination of the dispersal of the family of reductases that include metallo-reductases, the FRD superfamily, has shown that those members containing a ferric reductase domain (Pfam: PF01794) are found not only in eukaryotes, as is emphasized here, but also in bacteria, albeit at a relatively low probability (~12%)89. Note that this protein family is distinct from the one involved in dissimilatory metal reduction. The only ‘overlapping’ information provided by these two studies is the fact that the CHASRI that has been shared ubiquitously among bacteria does not encode a metallo-reductase88. On the other hand, the argument here posits that metallo-reductase activity has been selected for in support of metal accumulation, so the absence of a gene encoding such activity on a genetically mobile Cu-resistance element is not remarkable.
In this regard, except for studies on bacterial methanobactin90 and yersinibactin91 relatively little emphasis has been placed on how bacteria access nutrient copper; the emphasis is on how prokaryotes limit it. A similar disparity is found in reviewing the literature on how bacteria recover the iron bound to the siderophores they secrete to scavenge ferric iron from their environment. While there are numerous reports of a bacterial enzyme activity recovered from the extracellular milieu to the cytoplasm and everywhere in between that supports an NAD(P)H-dependent ferric reductase activity92, none of this activity has been linked to a gene or growth phenotype. While a cuprous oxidase activity has been described in the eukaryotic ferroxidases, yeast Fet3, human ceruloplasmin and algal Fox1, no ferroxidase activity has been described for any of the bacterial cuprous oxidases. This raises a question about the specific cellular and/or lifestyle elements that have selected for the iron and copper trafficking pathways outlined in this Perspective. One obvious conclusion is that the connections posited in this discussion between reductase and oxidase activities relate solely to eukaryotes and, possibly, to cell systems that typically or solely proliferate in a communal setting. This lifestyle is common to fungi and alga and, of course, to all multicellular organisms. On the other hand, bacteria, too, thrive in both planktonic (dispersed) and communal (bio-film) settings; might there be a difference in how this metal redox story plays out in these disparate growth habits? At least as far as anti-oxidant defense and antibiotic resistance is concerned, there are differences; bio-films are more resistant to both stressors93, 94. Certainly food for thought, and for further exploration.
Acknowledgments
The work reflected in the content of this Perspective has been supported in the author’s laboratory by the National Institutes of Health via grant awards RO1 DK053820, RO3 NS095063 and RO1 NS102337.
Abbreviations
- Tf
Transferrin
- mV
Millivolt
- GSH/GSSG
Reduced, oxidized glutathione
- ROS
Reactive oxygen species
- MCO
Multi-copper oxidase
- TNC
Tri-nuclear cluster
- ROS
Reactive oxygen species
- FRET
Fluorescence resonance energy transfer
- C(Y)FP
Cyan(Yellow) fluorescent protein
- Hp
Hephaestin
- Cp
Ceruloplasmin
- Fpn
Ferroportin
Footnotes
Conflicts of interest
The author has no conflict of interest.
References
- 1.Solomon EI. Dioxygen Binding, Activation, and Reduction to H2O by Cu Enzymes. Inorg Chem. 2016;55:6364–6375. doi: 10.1021/acs.inorgchem.6b01034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Knutson MD. Iron transport proteins: Gateways of cellular and systemic iron homeostasis. J Biol Chem. 2017;292:12735–12743. doi: 10.1074/jbc.R117.786632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lutsenko S. Copper trafficking to the secretory pathway. Metallomics. 2016;8:840–852. doi: 10.1039/c6mt00176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duck KA, Connor JR. Iron uptake and transport across physiological barriers. Biometals. 2016;29:573–591. doi: 10.1007/s10534-016-9952-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Potdar AA, Sarkar J, Das NK, Ghosh P, Gratzl M, Fox PL, Saidel GM. Computational modeling and analysis of iron release from macrophages. PLoS Comput Biol. 2014;10:e1003701. doi: 10.1371/journal.pcbi.1003701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maryon EB, Molloy SA, Zimnicka AM, Kaplan JH. Copper entry into human cells: progress and unanswered questions. Biometals. 2007;20:355–364. doi: 10.1007/s10534-006-9066-3. [DOI] [PubMed] [Google Scholar]
- 7.Bertini I, Cavallaro G, McGreevy KS. Cellular copper management-a draft user’s guide. Coordin Chem Rev. 2010;254:506–524. [Google Scholar]
- 8.Gawryluk RMR, Kamikawa R, Stairs CW, Silberman JD, Brown MW, Roger AJ. The Earliest Stages of Mitochondrial Adaptation to Low Oxygen Revealed in a Novel Rhizarian. Curr Biol. 2016;26:2729–2738. doi: 10.1016/j.cub.2016.08.025. [DOI] [PubMed] [Google Scholar]
- 9.Stairs CW, Leger MM, Roger AJ. Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos Trans R Soc Lond B Biol Sci. 2015;370:20140326. doi: 10.1098/rstb.2014.0326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hampl V, Stairs CW, Roger AJ. The tangled past of eukaryotic enzymes involved in anaerobic metabolism. Mob Genet Elements. 2011;1:71–74. doi: 10.4161/mge.1.1.15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Staerck C, Gastebois A, Vandeputte P, Calenda A, Larcher G, Gillmann L, Papon N, Bouchara JP, Fleury MJJ. Microbial antioxidant defense enzymes. Microb Pathog. 2017;110:56–65. doi: 10.1016/j.micpath.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 12.Singh A, Severance S, Kaur N, Wiltsie W, Kosman DJ. Assembly, activation, and trafficking of the Fet3p.Ftr1p high affinity iron permease complex in Saccharomyces cerevisiae. J Biol Chem. 2006;281:13355–13364. doi: 10.1074/jbc.M512042200. [DOI] [PubMed] [Google Scholar]
- 13.Sies H. Oxidative stress: a concept in redox biology and medicine. Redox Biol. 2015;4:180–183. doi: 10.1016/j.redox.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 15.Betteridge DJ. What is oxidative stress? Metabolism. 2000;49:3–8. doi: 10.1016/s0026-0495(00)80077-3. [DOI] [PubMed] [Google Scholar]
- 16.Shi X, Stoj C, Romeo A, Kosman DJ, Zhu Z. Fre1p Cu2+ reduction and Fet3p Cu1+ oxidation modulate copper toxicity in Saccharomyces cerevisiae. J Biol Chem. 2003;278:50309–50315. doi: 10.1074/jbc.M307019200. [DOI] [PubMed] [Google Scholar]
- 17.Adam FI, Bounds PL, Kissner R, Koppenol WH. Redox properties and activity of iron-citrate complexes: evidence for redox cycling. Chem Res Toxicol. 2015;28:604–614. doi: 10.1021/tx500377b. [DOI] [PubMed] [Google Scholar]
- 18.Qian SY, Buettner GR. Iron and dioxygen chemistry is an important route to initiation of biological free radical oxidations: an electron paramagnetic resonance spin trapping study. Free Radic Biol Med. 1999;26:1447–1456. doi: 10.1016/s0891-5849(99)00002-7. [DOI] [PubMed] [Google Scholar]
- 19.Koppenol WH, Butler j. Energetics of interconversion reactions of oxyradicals. Adv Free Rad Biol Med. 1985;1:91–131. [Google Scholar]
- 20.Aust SD, Morehouse LA, Thomas CE. Role of metals in oxygen radical reactions. J Free Radic Biol Med. 1985;1:3–25. doi: 10.1016/0748-5514(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 21.Halliwell B, Gutteridge JM. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem J. 1984;219:1–14. doi: 10.1042/bj2190001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor AB, Stoj CS, Ziegler L, Kosman DJ, Hart PJ. The copper-iron connection in biology: structure of the metallo-oxidase Fet3p. Proc Natl Acad Sci U S A. 2005;102:15459–15464. doi: 10.1073/pnas.0506227102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Halliwell B. Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 2006;141:312–322. doi: 10.1104/pp.106.077073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nauser T, Koppenol WH, Schoneich C. Protein thiyl radical reactions and product formation: a kinetic simulation. Free Radic Biol Med. 2015;80:158–163. doi: 10.1016/j.freeradbiomed.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McBean GJ. Cysteine, glutathione, and thiol redox balance in astrocytes. Antioxidants (Basel) 2017;6:62. doi: 10.3390/antiox6030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morgan B. Reassessing cellular glutathione homoeostasis: novel insights revealed by genetically encoded redox probes. Biochem Soc Trans. 2014;42:979–984. doi: 10.1042/BST20140101. [DOI] [PubMed] [Google Scholar]
- 27.Kirlin WG, Cai J, Thompson SA, Diaz D, Kavanagh TJ, Jones DP. Glutathione redox potential in response to differentiation and enzyme inducers. Free Radic Biol Med. 1999;27:1208–1218. doi: 10.1016/s0891-5849(99)00145-8. [DOI] [PubMed] [Google Scholar]
- 28.Lopez-Mirabal HR, Winther JR. Redox characteristics of the eukaryotic cytosol. Biochim Biophys Acta. 2008;1783:629–640. doi: 10.1016/j.bbamcr.2007.10.013. [DOI] [PubMed] [Google Scholar]
- 29.Go YM, Jones DP. Redox compartmentalization in eukaryotic cells. Biochim Biophys Acta. 2008;1780:1273–1290. doi: 10.1016/j.bbagen.2008.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kosman DJ. Multicopper oxidases: a workshop on copper coordination chemistry, electron transfer, and metallophysiology. J Biol Inorg Chem. 2010;15:15–28. doi: 10.1007/s00775-009-0590-9. [DOI] [PubMed] [Google Scholar]
- 31.Solomon EI, Sundaram UM, Machonkin TE. Multicopper oxidases and oxygenases. Chem Rev. 1996;96:2563–2605. doi: 10.1021/cr950046o. [DOI] [PubMed] [Google Scholar]
- 32.Reiss R, Ihssen J, Richter M, Eichhorn E, Schilling B, Thony-Meyer L. Laccase versus laccase-like multi-copper oxidase: a comparative study of similar enzymes with diverse substrate spectra. PLoS One. 2013;8:e65633. doi: 10.1371/journal.pone.0065633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enguita FJ, Marcal D, Martins LO, Grenha R, Henriques AO, Lindley PF, Carrondo MA. Substrate and dioxygen binding to the endospore coat laccase from Bacillus subtilis. J Biol Chem. 2004;279:23472–23476. doi: 10.1074/jbc.M314000200. [DOI] [PubMed] [Google Scholar]
- 34.Djoko KY, Chong LX, Wedd AG, Xiao Z. Reaction mechanisms of the multicopper oxidase CueO from Escherichia coli support its functional role as a cuprous oxidase. J Am Chem Soc. 2010;132:2005–2015. doi: 10.1021/ja9091903. [DOI] [PubMed] [Google Scholar]
- 35.Tree JJ, Kidd SP, Jennings MP, McEwan AG. Copper sensitivity of cueO mutants of Escherichia coli K-12 and the biochemical suppression of this phenotype. Biochem Biophys Res Commun. 2005;328:1205–1210. doi: 10.1016/j.bbrc.2005.01.084. [DOI] [PubMed] [Google Scholar]
- 36.Singh SK, Grass G, Rensing C, Montfort WR. Cuprous oxidase activity of CueO from Escherichia coli. J Bacteriol. 2004;186:7815–7817. doi: 10.1128/JB.186.22.7815-7817.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grass G, Rensing C. CueO is a multi-copper oxidase that confers copper tolerance in Escherichia coli. Biochem Biophys Res Commun. 2001;286:902–908. doi: 10.1006/bbrc.2001.5474. [DOI] [PubMed] [Google Scholar]
- 38.Stoj CS, Augustine AJ, Solomon EI, Kosman DJ. Structure-function analysis of the cuprous oxidase activity in Fet3p from Saccharomyces cerevisiae. J Biol Chem. 2007;282:7862–7868. doi: 10.1074/jbc.M609766200. [DOI] [PubMed] [Google Scholar]
- 39.Stoj C, Kosman DJ. Cuprous oxidase activity of yeast Fet3p and human ceruloplasmin: implication for function. FEBS Lett. 2003;554:422–426. doi: 10.1016/s0014-5793(03)01218-3. [DOI] [PubMed] [Google Scholar]
- 40.Kosman DJ. Encylopedia of Inorganic and Bioinorganic Chemistry. John Wiley and Sons; New York: 2017. [DOI] [Google Scholar]
- 41.Low H, Crane FL, Morre DJ. Putting together a plasma membrane NADH oxidase: a tale of three laboratories. Int J Biochem Cell Biol. 2012;44:1834–1838. doi: 10.1016/j.biocel.2012.06.032. [DOI] [PubMed] [Google Scholar]
- 42.Ly JD, Lawen A. Transplasma membrane electron transport: enzymes involved and biological function. Redox Rep. 2003;8:3–21. doi: 10.1179/135100003125001198. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan JH, Maryon EB. How mammalian cells acquire copper: An essential but potentially toxic metal. Biophys J. 2016;110:7–13. doi: 10.1016/j.bpj.2015.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuo YM, Zhou B, Cosco D, Gitschier J. The copper transporter CTR1 provides an essential function in mammalian embryonic development. Proc Natl Acad Sci U S A. 2001;98:6836–6841. doi: 10.1073/pnas.111057298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coffey R, Knutson MD. The plasma membrane metal-ion transporter ZIP14 contributes to nontransferrin-bound iron uptake by human beta-cells. Am J Physiol Cell Physiol. 2017;312:C169–C175. doi: 10.1152/ajpcell.00116.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nam H, Wang CY, Zhang L, Zhang W, Hojyo S, Fukada T, Knutson MD. ZIP14 and DMT1 in the liver, pancreas, and heart are differentially regulated by iron deficiency and overload: implications for tissue iron uptake in iron-related disorders. Haematologica. 2013;98:1049–1057. doi: 10.3324/haematol.2012.072314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang CY, Jenkitkasemwong S, Duarte S, Sparkman BK, Shawki A, Mackenzie B, Knutson MD. ZIP8 is an iron and zinc transporter whose cell-surface expression is up-regulated by cellular iron loading. J Biol Chem. 2012;287:34032–34043. doi: 10.1074/jbc.M112.367284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pinilla-Tenas JJ, Sparkman BK, Shawki A, Illing AC, Mitchell CJ, Zhao N, Liuzzi JP, Cousins RJ, Knutson MD, Mackenzie B. Zip14 is a complex broad-scope metal-ion transporter whose functional properties support roles in the cellular uptake of zinc and nontransferrin-bound iron. Am J Physiol Cell Physiol. 2011;301:C862–871. doi: 10.1152/ajpcell.00479.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saikia S, Oliveira D, Hu G, Kronstad J. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect Immun. 2014;82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Philpott CC. Iron uptake in fungi: a system for every source. Biochim Biophys Acta. 2006;1763:636–645. doi: 10.1016/j.bbamcr.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 51.Lane DJ, Bae DH, Merlot AM, Sahni S, Richardson DR. Duodenal cytochrome b (DCYTB) in iron metabolism: an update on function and regulation. Nutrients. 2015;7:2274–2296. doi: 10.3390/nu7042274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wyman S, Simpson RJ, McKie AT, Sharp PA. Dcytb (Cybrd1) functions as both a ferric and a cupric reductase in vitro. FEBS Lett. 2008;582:1901–1906. doi: 10.1016/j.febslet.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 53.Kleven MD, Dlakic M, Lawrence CM. Characterization of a single b-type heme, FAD, and metal binding sites in the transmembrane domain of six-transmembrane epithelial antigen of the prostate (STEAP) family proteins. J Biol Chem. 2015;290:22558–22569. doi: 10.1074/jbc.M115.664565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Knutson MD. Steap proteins: implications for iron and copper metabolism. Nutr Rev. 2007;65:335–340. doi: 10.1111/j.1753-4887.2007.tb00311.x. [DOI] [PubMed] [Google Scholar]
- 55.Ohgami RS, Campagna DR, McDonald A, Fleming MD. The Steap proteins are metalloreductases. Blood. 2006;108:1388–1394. doi: 10.1182/blood-2006-02-003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sparla F, Preger V, Pupillo P, Trost P. Characterization of a novel NADH-specific, FAD-containing, soluble reductase with ferric citrate reductase activity from maize seedlings. Arch Biochem Biophys. 1999;363:301–308. doi: 10.1006/abbi.1998.1085. [DOI] [PubMed] [Google Scholar]
- 57.Gauss GH, Kleven MD, Sendamarai AK, Fleming MD, Lawrence CM. The crystal structure of six-transmembrane epithelial antigen of the prostate 4 (Steap4), a ferri/cuprireductase, suggests a novel interdomain flavin-binding site. J Biol Chem. 2013;288:20668–20682. doi: 10.1074/jbc.M113.479154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim K, Mitra S, Wu G, Berka V, Song J, Yu Y, Poget S, Wang DN, Tsai AL, Zhou M. Six-transmembrane epithelial antigen of prostate 1 (STEAP1) has a single b heme and is capable of reducing Metal ion complexes and oxygen. Biochemistry. 2016;55:6673–6684. doi: 10.1021/acs.biochem.6b00610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pierre JL, Fontecave M, Crichton RR. Chemistry for an essential biological process: the reduction of ferric iron. Biometals. 2002;15:341–346. doi: 10.1023/a:1020259021641. [DOI] [PubMed] [Google Scholar]
- 60.Morre DJ, Davidson M, Geilen C, Lawrence J, Flesher G, Crowe R, Crane FL. NADH oxidase activity of rat liver plasma membrane activated by guanine nucleotides. Biochem J. 1993;292(Pt 3):647–653. doi: 10.1042/bj2920647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morre DJ, Brightman AO. NADH oxidase of plasma membranes. J Bioenerg Biomembr. 1991;23:469–489. doi: 10.1007/BF00771015. [DOI] [PubMed] [Google Scholar]
- 62.Asard H, Barbaro R, Trost P, Berczi A. Cytochromes b561: ascorbate-mediated trans-membrane electron transport. Antioxid Redox Signal. 2013;19:1026–1035. doi: 10.1089/ars.2012.5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su D, Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273:3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 64.Griesen D, Su D, Berczi A, Asard H. Localization of an ascorbate-reducible cytochrome b561 in the plant tonoplast. Plant Physiol. 2004;134:726–734. doi: 10.1104/pp.103.032359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Asard H, Horemans N, Caubergs RJ. Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett. 1992;306:143–146. doi: 10.1016/0014-5793(92)80986-q. [DOI] [PubMed] [Google Scholar]
- 66.Shatwell KP, Dancis A, Cross AR, Klausner RD, Segal AW. The FRE1 ferric reductase of Saccharomyces cerevisiae is a cytochrome b similar to that of NADPH oxidase. J Biol Chem. 1996;271:14240–14244. doi: 10.1074/jbc.271.24.14240. [DOI] [PubMed] [Google Scholar]
- 67.Finegold AA, Shatwell KP, Segal AW, Klausner RD, Dancis A. Intramembrane bis-heme motif for transmembrane electron transport conserved in a yeast iron reductase and the human NADPH oxidase. J Biol Chem. 1996;271:31021–31024. doi: 10.1074/jbc.271.49.31021. [DOI] [PubMed] [Google Scholar]
- 68.Sendamarai AK, Ohgami RS, Fleming MD, Lawrence CM. Structure of the membrane proximal oxidoreductase domain of human Steap3, the dominant ferrireductase of the erythroid transferrin cycle. Proc Natl Acad Sci U S A. 2008;105:7410–7415. doi: 10.1073/pnas.0801318105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kosman DJ. Redox cycling in iron uptake, efflux, and trafficking. J Biol Chem. 2010;285:26729–26735. doi: 10.1074/jbc.R110.113217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kosman DJ. Molecular mechanisms of iron uptake in fungi. Mol Microbiol. 2003;47:1185–1197. doi: 10.1046/j.1365-2958.2003.03368.x. [DOI] [PubMed] [Google Scholar]
- 71.Hassett RF, Romeo AM, Kosman DJ. Regulation of high affinity iron uptake in the yeast Saccharomyces cerevisiae. Role of dioxygen and Fe. J Biol Chem. 1998;273:7628–7636. doi: 10.1074/jbc.273.13.7628. [DOI] [PubMed] [Google Scholar]
- 72.de Silva DM, Askwith CC, Kaplan J. Molecular mechanisms of iron uptake in eukaryotes. Physiol Rev. 1996;76:31–47. doi: 10.1152/physrev.1996.76.1.31. [DOI] [PubMed] [Google Scholar]
- 73.Hassett R, Kosman DJ. Evidence for Cu(II) reduction as a component of copper uptake by Saccharomyces cerevisiae. J Biol Chem. 1995;270:128–134. doi: 10.1074/jbc.270.1.128. [DOI] [PubMed] [Google Scholar]
- 74.Dix D, Bridgham J, Broderius M, Eide D. Characterization of the FET4 protein of yeast. Evidence for a direct role in the transport of iron. J Biol Chem. 1997;272:11770–11777. doi: 10.1074/jbc.272.18.11770. [DOI] [PubMed] [Google Scholar]
- 75.Dix DR, Bridgham JT, Broderius MA, Byersdorfer CA, Eide DJ. The FET4 gene encodes the low affinity Fe(II) transport protein of Saccharomyces cerevisiae. J Biol Chem. 1994;269:26092–26099. [PubMed] [Google Scholar]
- 76.Ramanan N, Wang Y. A high-affinity iron permease essential for Candida albicans virulence. Science. 2000;288:1062–1064. doi: 10.1126/science.288.5468.1062. [DOI] [PubMed] [Google Scholar]
- 77.Askwith C, Kaplan J. An oxidase-permease-based iron transport system in Schizosaccharomyces pombe and its expression in Saccharomyces cerevisiae. J Biol Chem. 1997;272:401–405. doi: 10.1074/jbc.272.1.401. [DOI] [PubMed] [Google Scholar]
- 78.Stearman R, Yuan DS, Yamaguchi-Iwai Y, Klausner RD, Dancis A. A permease-oxidase complex involved in high-affinity iron uptake in yeast. Science. 1996;271:1552–1557. doi: 10.1126/science.271.5255.1552. [DOI] [PubMed] [Google Scholar]
- 79.Kwok EY, Severance S, Kosman DJ. Evidence for iron channeling in the Fet3p-Ftr1p high-affinity iron uptake complex in the yeast plasma membrane. Biochemistry. 2006;45:6317–6327. doi: 10.1021/bi052173c. [DOI] [PubMed] [Google Scholar]
- 80.McCarthy RC, Kosman DJ. Ferroportin and exocytoplasmic ferroxidase activity are required for brain microvascular endothelial cell iron efflux. J Biol Chem. 2013;288:17932–17940. doi: 10.1074/jbc.M113.455428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yeh KY, Yeh M, Glass J. Interactions between ferroportin and hephaestin in rat enterocytes are reduced after iron ingestion. Gastroenterology. 2011;141:292–299. 299e291. doi: 10.1053/j.gastro.2011.03.059. [DOI] [PubMed] [Google Scholar]
- 82.De Domenico I, Ward DM, di Patti MC, Jeong SY, David S, Musci G, Kaplan J. Ferroxidase activity is required for the stability of cell surface ferroportin in cells expressing GPI-ceruloplasmin. EMBO J. 2007;26:2823–2831. doi: 10.1038/sj.emboj.7601735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gacesa R, Dunlap WC, Barlow DJ, Laskowski RA, Long PF. Rising levels of atmospheric oxygen and evolution of Nrf2. Sci Rep. 2016;6:27740. doi: 10.1038/srep27740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lyons TW, Reinhard CT, Planavsky NJ. The rise of oxygen in Earth’s early ocean and atmosphere. Nature. 2014;506:307–315. doi: 10.1038/nature13068. [DOI] [PubMed] [Google Scholar]
- 85.Large RR, Halpin JA, Danyushevsky LV, Maslennikov VV, Bull SW, Long JA, Gregory DD, Lounejeva E, Lyons TW, Sack PJ, McGoldrick PJ, Calver CR. Trace element content of sedimentary pyrite as a new proxy for deep-time ocean-atmosphere evolution. Earth Planet Sc Lett. 2014;389:209–220. [Google Scholar]
- 86.Tribovillard N, Algeo TJ, Lyons T, Riboulleau A. Trace metals as paleoredox and paleoproductivity proxies: An update. Chem Geol. 2006;232:12–32. [Google Scholar]
- 87.Halliwell B. Antioxidant defence mechanisms: from the beginning to the end (of the beginning) Free Radic Res. 1999;31:261–272. doi: 10.1080/10715769900300841. [DOI] [PubMed] [Google Scholar]
- 88.Staehlin BM, Gibbons JG, Rokas A, O’Halloran TV, Slot JC. Evolution of a heavy metal homeostasis/resistance island reflects increasing copper stress in Enterobacteria. Genome Biol Evol. 2016;8:811–826. doi: 10.1093/gbe/evw031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhang X, Krause KH, Xenarios I, Soldati T, Boeckmann B. Evolution of the ferric reductase domain (FRD) superfamily: modularity, functional diversification, and signature motifs. PLoS One. 2013;8:e58126. doi: 10.1371/journal.pone.0058126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dassama LM, Kenney GE, Ro SY, Zielazinski EL, Rosenzweig AC. Methanobactin transport machinery. Proc Natl Acad Sci U S A. 2016;113:13027–13032. doi: 10.1073/pnas.1603578113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Koh EI, Robinson AE, Bandara N, Rogers BE, Henderson JP. Copper import in Escherichia coli by the yersiniabactin metallophore system. Nat Chem Biol. 2017;13:1016–1021. doi: 10.1038/nchembio.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Schroder I, Johnson E, de Vries S. Microbial ferric iron reductases. FEMS Microbiol Rev. 2003;27:427–447. doi: 10.1016/S0168-6445(03)00043-3. [DOI] [PubMed] [Google Scholar]
- 93.de la Fuente-Nunez C, Reffuveille F, Fernandez L, Hancock RE. Bacterial biofilm development as a multicellular adaptation: antibiotic resistance and new therapeutic strategies. Curr Opin Microbiol. 2013;16:580–589. doi: 10.1016/j.mib.2013.06.013. [DOI] [PubMed] [Google Scholar]
- 94.Aiassa V, Barnes AI, Albesa I. Resistance to ciprofloxacin by enhancement of antioxidant defenses in biofilm and planktonic Proteus mirabilis. Biochem Biophys Res Commun. 2010;393:84–88. doi: 10.1016/j.bbrc.2010.01.083. [DOI] [PubMed] [Google Scholar]