Abstract
Copper is essential for eukaryotic life, and animals must acquire this nutrient through the diet and distribute it to cells and organelles for proper function of biological targets. Indeed, mutations in the central copper exporter ATP7A contribute to a spectrum of diseases, including Menkes disease, with symptoms ranging from neurodegeneration to lax connective tissue. As such, a better understanding of the fundamental impacts of ATP7A mutations on in vivo copper distrbutions is of relevance to those affected by these diseases. Here we combine metal imaging and optical imaging techniques at a variety of spatial resolutions to identify tissues and structures with altered copper levels in the Calamitygw71 zebrafish model of Menkes disease. Rapid profiling of tissue slices with LA-ICP-MS identified reduced copper levels in the brain, neuroretina, and liver of Menkes fish compared to control specimens. High resolution nanoSIMS imaging of the neuroretina, combined with electron and confocal microscopies, identified the megamitochondria of photoreceptors as loci of copper accumulation in wildtype fish, with lower levels of megamitochondrial copper observed in Calamitygw71 zebrafish. Interestingly, this localized copper decrease does not result in impaired photoreceptor development or altered megamitochondrial morphology, suggesting the prioritization of copper at sufficient levels for maintaining essential mitochondrial functions. Together, these data establish the Calamitygw71 zebrafish as an optically transparent in vivo model for the study of neural copper misregulation, illuminate a role for the ATP7A copper exporter in trafficking copper to the neuroretina, and highlight the utility of combining multiple imaging techniques for studying metals in whole organism settings with spatial resolution.
TOC image
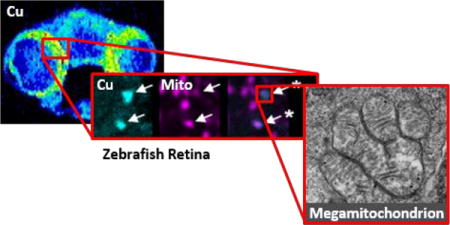
Coregistering quantitative high-resolution metal imaging with fluorescence imaging identifies copper accumulation in megamitochondria of photoreceptors within intact neuroretinal slices.
Introduction
Copper acquisition and trafficking are fundamental functions of eukaryotic organisms,1,2 with one major function for copper being an essential cofactor for cytochrome c oxidase, the terminal oxidase enzyme in the mitochondrial electron transport chain.3 Like other metal nutrients, copper must be obtained from the environment and trafficked to the correct tissues, cells, organelles, and proteins within the body. Indeed, dysregulation of copper trafficking can lead to mislocalisation of copper within an organism and contribute to disease.4–6 As such, identifying molecular sources and targets associated with copper dysregulation may reveal potential new targets for therapeutic benefit.7–10
In this context, ATP7A is the major copper export protein ubiquitously expressed across mammalian tissues.11 On a cellular level, ATP7A is responsible for exporting copper from the cell as well as loading copper into copper-dependent enzymes during Golgi processing.12 On a tissue level, ATP7A is required to mobilize copper from intestinal cells into the blood stream.13 Indeed, loss of ATP7A has been linked to accumulation of copper in the kidneys and intestines and severe copper deficiency in the other tissues of the body.14
In humans, mutations in the ATP7A protein are associated with a spectrum of diseases that span a wide range of severities.15 Distal motor neuropathy (DMN) is the least severe, manifesting in young-adulthood with the death of motor neurons and progressive muscle weakness.16 Occipital horn syndrome (OHS) typically appears in teenagers, with patients showing mild cognitive defects, failure of the autonomic nervous system, poor formation of connective tissue leading to lax skin and joints, as well as characteristic horns on the occipital bone at the base of the skull.17 Menkes disease is the most severe disease associated with ATP7A mutations, with symptoms typically manifesting within months of birth.15 Patients often have brain atrophy and seizures, poor formation of connective tissue, and coarse, light-coloured hair; Menkes disease is typically lethal during childhood. Neurological symptoms are common to all of these diseases, regardless of severity, leading to the hypothesis that the central nervous system is particularly sensitive to defects in copper metabolism.18,19 However, studying the neurological effects of low copper in these diseases can be complicated because neurological effects may be masked by other pathologies such as poor tissue structure or motor function.
Against this backdrop, we turned our attention to zebrafish as an optically transparent animal model of copper dysregulation. Specifically, the Calamitygw71 (Calgw71) allele in zebrafish contains an I1061S point mutation in ATP7A that causes impaired copper export.20 Interestingly, Calgw71 embryos are morphologically indistinguishable from their wildtype siblings but are sensitive to copper deprivation. The application of low levels of the copper chelator neocuproine impairs pigmentation in Calgw71 embryos at concentrations that have no effect on their wildtype siblings,20 suggesting that Calgw71 fish contain lower levels of copper than their wildtype siblings. We reasoned that if Calgw71 fish had lower copper levels than their wildtype siblings, the Calgw71 fish could provide an attractive model system to study the effects of reduced copper levels on neural function without the complications of gross morphological or functional defects typically associated with Menkes models.21 As copper concentrations in Calgw71 embryos across different tissues had not been measured directly, we were specifically interested in identifying whether neural tissue in particular within these fish was indeed copper deficient. Because zebrafish embryos are prohibitively small to subject to traditional dissection and bulk metal analysis methods such as atomic absorption (AA) or inductively coupled plasma mass spectrometry (ICP-MS), we turned to metal imaging methods to quantify the amount and location of copper within Calgw71 embryos.22 In this report, we employ a combination of laser ablation inductively coupled plasma mass spectrometry (LA-ICP-MS) to investigate the distribution and concentration of copper in Calgw71 embryos relative to wildtype along with nano-secondary ion mass spectrometry (nanoSIMS) to image the neuroretina with higher spatial resolution, revealing distinct copper puncta in this region. Electron microscopy provides evidence that the observed copper puncta represent the contents of the megamitochondria of photoreceptors. Additionally, generation of transgenic zebrafish with a fluorescent mitochondrial marker enables us to localize megamitochondria by confocal microscopy and subsequently quantify their Cu concentration by nanoSIMS.
Experimental
Materials
All chemicals were purchased from Sigma Aldrich, unless otherwise noted.
Zebrafish Husbandry
All zebrafish housing, care, and experiments were approved by the UC Berkeley Animal Care and Use Committee. Zebrafish were housed in the UC Berkeley Zebrafish Facility and kept at 28.5 °C on a 14-hr light/10-hr dark cycle. Embryos were produced by natural crosses and staged by hours post fertilization (hpf) or days post fertilization (dpf). To tightly control metal content in growing embryos, embryos were raised in E3 medium made from doubly-distilled (18 MΩ) water. Wildtype zebrafish were from the AB strain. Transgenic Calamitygw71 (Calgw71) fish were a gift from Prof. Jonathan Gitlin; genotyping of the Calgw71 line was conducted as described previously.20
Sample Preparation for LA-ICP-MS, nanoSIMS and Immunofluorescence
Zebrafish embryos were raised in E3 medium to 6 dpf. Zebrafish used for immunofluorescence were treated with 0.003% 1-phenyl-2-thiourea (PTU) beginning at 24 hours post fertilization (hpf) to limit pigmentation that might obscure fluorescent secondary antibodies. Embryos were euthanized in ice water and immediately embedded in Optimal Cutting Temperature (OCT) mounting media (Tissue Tek) in cryomolds (Tissue Tek). The embedded embryos were immediately frozen in a dry ice/isopentane bath and stored at −80 °C until sectioning. For 4–18 hours before sectioning, the embedded embryos were equilibrated to −20 °C. The embryos were sectioned into 20 μm slices using a Cryostat (Leica CM1950) and placed directly onto Superfrost PLUS slides (Thermo Fisher). For nanoSIMS analysis, before mounting slices on the slides, the slides were trimmed to 1.5 inches long using a diamond knife so that the slides could fit inside the nanoSIMS sample carrier. To avoid touching the surface of the trimmed slides during sample mounting, the slides were taped at the edges to standard-length slides using removable Scotch tape. The slices were air-dried and stored at room temperature until analysis.
To test the effect of fixation on metal imaging, some samples were fixed before mounting. In this case, 6 dpf embryos were euthanized in ice water and immediately submerged in 4% paraformaldehyde (PFA, Thermo Fisher) in phosphate buffered saline (PBS) overnight at 4 °C. The next day, embryos were washed three times in PBS, transferred to a solution of 30% sucrose in PBS for cryoprotection, and allowed to equilibrate until the embryos had sunk to the bottom of the tube (typically overnight). The fixed, cryoprotected embryos were mounted in OCT and processed as outlined above. Fixation perturbed metal localization and was not used in subsequent sample preparation (Figure S1). This fixation method was also used for samples prepared for immunofluorescence.
Laser Ablation Inductively Coupled Plasma Mass Spectrometry
Laser ablation was performed using an NWR213 laser with a TV2 sample chamber (ESI, Bozeman, MT) using the following parameters: Spot size: 6 μm; Fluence: 2.3 J cm−2; Stage speed: 15 μm s−1; Firing rate: 20 Hz; He flow: 800 mL min−1; Pattern spacing: 6 μm. Using these parameters, the tissue was fully ablated but the glass slide remained undamaged. The ablated material was introduced by helium gas flow into an iCAP-Qc ICP-MS (Thermo Fisher) and analyzed for 63Cu or 66Zn content using a 0.4 sec dwell time in standard acquisition mode. The resulting mass spectrometry traces and laser log files were processed in Igor Pro using the Iolite application. The Trace Elements data reduction scheme was used in Semi-quantitative mode using 63Cu or 66Zn as the reference trace and a custom matrix-matched standard to convert mass spectrometer counts to metal concentration. Quantitative metal maps were exported in csv format and imported into ImageJ for quantification of regions of interest.
Nano Secondary Ion Mass Spectrometry
Samples were prepared as outlined above and were dried for at least 24 hours before use in order to prevent sample distortion in the nanoSIMS vacuum chamber. To create a conductive surface, the samples were coated with 15–20 nm of gold using a Hummer sputter coater (Technics). During the initial optimization of sample preparation methods, carbon coating (12–15 nm, EMS 150T ES by EMS Quorum) was also tested. Both carbon and gold coating yielded excellent SIMS images (Figure S1). Due to the ease and consistency of gold coating in our facility, gold coating was used for all subsequent sample preparation.
The Cameca NanoSIMS 50 (Gennevilliers, France) at Lawrence Livermore National Lab was used to image the intracellular distribution of P, Ca, Cu, and Fe. All samples were first imaged by reflected light using an SMZ800 stereo microscope (Nikon) for slide mapping and a compound microscope mounted on an encoded X–Y stage (Leitz) for high magnification images. Fluorescence confocal microscopy was performed on dried tissue sections as described below (see Confocal Microscopy). Within the NanoSIMS 50, a reflected light camera was used to locate the correct sample on each slide, and total secondary ion imaging was used to locate specific target areas within each sample, based on morphological features of the tissue that had been previously identified using reflected light microscopy outside the NanoSIMS 50. Target areas were sputtered to a depth of ~60 nm23 before analysis with a focused 100 to 120 pA negative oxygen ion primary beam, which was scanned over 2500 μm2 rasters with 512 × 512 pixels to generate secondary ions. The secondary ion mass spectrometer was tuned for ~3000 and 12C+, 31P+, 40Ca+, 56Fe+, and 63Cu+ were detected simultaneously by electron multipliers in pulse counting mode. The correct metal ion peaks were identified using NBS610 glass (National Institute of Standards and Technology, USA), except for 12C+, which was located on the samples. Each analysis area was scanned 30 to 50 times with 500 ns pixel−1 dwell times to collect serial secondary ion images for quantification. For depth profiling, raster areas of 1250 μm2 were scanned up to 180 times using the conditions above. Each scan of the primary ion beam causes the ejection of secondary ions, removing a very thin layer of material from the sample surface. Sequential scanning thereby profiles ion concentrations in the z dimension through the sample.
The NanoSIMS ion image data were processed using custom software (LIMAGE, L.R. Nittler, Carnegie Institute of Washington, USA). The ion images were corrected for detector dead time and image shifts between scans, and then used to produce ion ratio images. For defined regions of interest, ion ratios were calculated for 31P+/12C+, 40Ca+/12C+, 56Fe+/12C+ and 63Cu+/12C+ by averaging the replicate scans. These data were quantified using matrix-matched standards (see below). In cases where fluorescence imaging had been performed before analysis, the samples were removed from the NanoSIMS and imaged again by reflected light microscopy on a compound microscope (Leitz); ion sputtering leaves a visible scar on the tissue that reveals the location of the analysis, allowing the metal maps and fluorescence images to be aligned (see Confocal Microscopy and Image Alignment).
Preparation of Matrix-Matched Standards for Quantitative Metal Maps
This protocol was adapted from published methods.24,25 Salmon muscle (30 mL of packed tissue) was digested by adding 10 mL of protease solution (0.25% trypsin, 10 mM EDTA, 0.1X PBS) and 48 μL of Collagenase P (100 mg mL−1 in HBSS) in a plastic tube. The solution was mixed with a plastic spatula (to minimize metal contamination) and incubated at 28° C for 4 hours with periodic mixing. The tissue was stored at 4°C overnight. The next day, the tissue was warmed to 28 °C for an additional 6 hours of digestion and homogenized in a Dounce homogenizer using 10 passes until the tissue was gooey and smooth. The tissue was separated into 500 μL aliquots in 1.5 mL tubes (Sarstedt) and frozen at −20 °C until metal addition. A solution of CuCl2, ZnCl2, Fe(citrate), CaCl2, MgCl2, and KCl (10,000 ppm each) was prepared in water. Dilutions of 5000, 1000, 500, 100, 50, and 10 ppm were made in water. Each dilution was mixed 1:10 with an aliquot of tissue (50 μL metal mixture per 500 μL tissue) and mixed with a hand-held mechanical homogenizer. To remove bubbles, the standards were centrifuged at 16,000 × g at room temperature for 2 hours. Any resulting supernatant was removed, and the vials were frozen in a dry ice/isopentane bath and stored at −80 °C until sectioning. Before sectioning, the standards were cut in half vertically. One half was sectioned into 20 μm slices using a Cryostat (Leica CM1950) and placed directly onto Superfrost PLUS slides (Thermo Fisher), air-dried, and stored at room temperature until analysis. The other half was divided into three parts for triplicate analysis by liquid ICP-MS. The samples for liquid analysis were weighed in 1.5 mL tubes (Sarstedt) and combined 1:1 (w/v) with concentrated nitric acid (BDH Aristar Ultra). After overnight incubation at room temperature, samples were diluted into 2% HNO3 (prepared from concentrated acid in milliQ water) and doped with a gallium internal standard (Inorganic Ventures, 20 ppb final concentration). The metal content was determined by measuring 63Cu and 66Zn using a Thermo Fisher iCAP-Qc ICP-MS in Kinetic Energy Discrimination (KED) mode with the He flow set to 4.426 mL min−1. Measurements were normalized to a standard curve of known metal concentrations doped with 20 ppb Ga. The standard curve was diluted from CMS-5 standard (Al, Cs, Co, Fe, Mg, Ni, Rb, Na, Zn, Ca, Cr, Cu, Li, Mn, K, Ag, and Sr in 2% nitric acid) with molybdenum, phosphorous and sulfur added (Inorganic Ventures). The calibration curves for LA-ICP-MS and nanoSIMS are shown in Figure S2.
Electron Microscopy
For ultrastructural analysis of the retina tissue, 6 dpf zebrafish embryos were euthanized in ice water and submerged in 0.1 M phosphate buffer containing 2% glutaraldehyde (Electron Microscopy Sciences). A fresh razor blade was used to remove the tail of each embryo in order to improve permeabilization with the fixative. Zebrafish were fixed for 1 hour at room temperature on a rotator and then left in fixative at 4 °C up to two weeks until embedding. To each tube containing zebrafish embryos in 1 mL of 2% glutaraldehyde in 0.1 M phosphate buffer was added 4 drops (~50 μL) of 4% OsO4 (Electron Microscopy Sciences) in water. The tubes were placed on a rocker for 15 minutes at room temperature. Osmium-containing fixative was removed, and the samples were washed three times with 0.1 M sodium cacodylate buffer (pH 7.4) for 5 minutes per wash. Sodium cacodylate buffer was removed, and embryos were dehydrated through a series of acetone:water washes (30%, 50%, 70%, 90%, 95% and 3× 100% acetone), 10 minutes per wash, at room temperature with rotation. Fresh resin was prepared: for 50 mL of resin, 23.5 g Eponate 12TM Resin (Ted Pella), 12.5 g dodecenylsuccinic anhydride (Ted Pella), and 14 g methyl-5-norbornene-2,3-dicarboxylic anhydride (Ted Pella) were combined and stirred thoroughly. Dehydrated embryos were infused with resin through a series of acetone:resin washes (3:1 acetone:resin, 1:1 acetone:resin, 1:3 acetone:resin), 20 minutes per wash at room temperature with rotation. The embryos were then treated with resin 2× 30 minutes and left in a fresh resin wash overnight at room temperature with rotation. The next day, benzyldimethylamine (BDMA, Ted Pella) accelerant was added to extra pre-mixed resin solution (0.75 mL BDMA per 50 mL resin) and stirred thoroughly for at least ten minutes. Bubbles were removed under vacuum for 15 minutes. Embryos were removed from tubes containing resin and placed in fresh 1.5 mL tubes. Resin containing accelerant was added to each tube containing samples, and the samples were careful stirred into suspension using a wooden toothpick. Samples were infused with accelerant by three washes with resin containing accelerant, at least 2 hours per wash, up to 12 hours total. Samples were mounted in resin containing accelerant in flat rubber molds (Pelco) and hardened in a 60 °C oven (Fisher Isotemp) for at least 48 hours.
Blocks containing samples were trimmed manually using a fresh razor blade rinsed with 95% ethanol. Trimmed blocks were faced using a glass knife and sectioned using a 3.0 ultra diamond knife (Diatome) on an UltraCut microtome (Reichert-Jung) to 70–100 nm and mounted on copper grids coated with formvar (Ted Pella). Grids were stained in a Pelco Grid Staining System with 2% aqueous uranyl acetate (Ted Pella) for 7 minutes, followed by six washes with distilled water. Grids were immediately stained for 5 minutes with Reynolds lead citrate and subsequently washed six times with distilled water. Grids were dried and stored at room temperature in the dark until imaging. Electron microscopy was performed on a Tecnai 120 KV (FEI by Thermo Scientific) in the UC Berkeley Electron Microscopy Facility.
Cloning
To locate the mitochondria in retinal cryosections, we expressed the mitochondrial marker TOM20 fused to the fluorescent protein mCherry under the actin promoter. The DNA construct for these experiments, pSCAC-69-pActin:TOM20-mCherry, was assembled by inserting the actin promoter and a gBlock coding for TOM20-iRFP into the pSCAC-69 backbone and subsequently replacing iRFP with mCherry due to a mismatch in laser lines on the stereoscope used for sorting zebrafish. The pSCAC-69 plasmid contains Tol2 transposable elements for insertion of DNA cargo into the genomic DNA. pSCAC-69 was a gift from Seok-Yong Choi (Addgene plasmid # 31241)26 and was amplified using the following primers: (FWD: cgttgagtataggtaatgactaggccGCCCCTTATTTGTGCTTGAT, REV: taggcctatttaggtgacactatagCCATGTCTGGACTTCTGAGG; Tm: 59 °C). The actin promoter was amplified from pMTB2-NLS-BirA-2A-mCherry_Ras, a gift from Tatjana Sauka-Spengler (Addgene plasmid # 80067)27 using the following primers: (FWD: ATAAGGGGCggcctagtcattacctatactcaacg, REV: CATGGctatagtgtcacctaaataggccta; Tm: 56 °C). The resulting vector and promoter PCR products were assembled by Gibson Assembly (NEB) to create the pSCAC-69-pActin plasmid. TOM20-iRFP was inserted into pSCAC-69-pActin by amplification of the plasmid backbone using the following primers (FWD: AGCTCGAATTAATTCATCGA, REV: GGCTATAGTGTCACCTAAATAGG; Tm: 57 °C) and Gibson Assembly of the backbone and a gBlock coding for TOM20-iRFP (Integrated DNA Technologies, see Supplemental Methods for gBlock sequence). To exchange iRFP for mCherry, the pSCAC-69-pActin:TOM20 vector was amplified using the following primers (FWD: AGAACCGCTGTTAGGATCT, REV: TAAAGCTCGAATTAATTCATCGA, Tm: 58 °C) and mCherry was amplified from an in-house vector (pcDNA3.1(+)-ePDZb-mCherry) using the following primers (FWD: AAACGACGGTCAGATCCTAACAGCGGTTCTATGGTGAGCAAGGGC, REV: TGGATCATCATCGATGAATTAATTCGAGCTTTACTTGTACAGCTCGTCCAT, Tm: 58 °C). The resulting vector and insert PCR products were assembled by Gibson Assembly (NEB) to create the final pSCAC-69-pActin:TOM20-mCherry construct. DNA used for injections was purified using the Zyppy Miniprep Kit (Zymo) to remove endotoxin.
Generation of Mosaic Tg(Actin:TOM20-mCherry) Zebrafish Embryos
Zebrafish embryos expressing the mitochondrial marker TOM20-mCherry were generated using Tol2 transposase activity, which inserts the DNA coding for the construct into genomic DNA. Zebrafish embryos at the 1- or 2-cell stage were injected with 1 nL of injection solution containing 25 ng μL−1 TolII RNA and 25 ng μL−1 pSCAC-69-pActin:TOM20-mCherry (see Cloning section, above) in water containing phenol red. Embryos were raised in E3 media and monitored daily for toxicity, but no toxicity was observed. At 6 dpf, embryos were prepared for nanoSIMS.
Immunofluorescence
To assess the abundance of photoreceptors in the retina of wiltype and Calgw71 embryos, immunofluorescence was performed as follows. Samples were prepared by PFA fixation, embedding, and cryosectioning, as outlined above. Slides were stored at −20 °C until use. Slides were warmed to room temperature and hydrated in dH2O for 3 minutes. Samples were equilibrated in 3× 5-minute washes with 0.5 M TBST (0.5 M NaCl, 0.05M Tris, 0.1% Tween-20, pH 7.4) and blocked for 1 hour at room temperature with gentle rocking in blocking solution (0.5 M TBST, 5% donkey serum (Jackson Labs), 2% non-fat dried milk). Primary antibody was applied in fresh blocking solution overnight at 4 °C without agitation. Primary antibodies were used at the following concentrations: Zpr-1 (ZIRC, 1:500), Zpr-2 (ZIRC, 1:500), Zpr-3 (ZIRC, 1:500), α-calretinin (EMD Millipore, 1:2000). The following day, samples were washed 3× 5 minutes in 0.5 M TBST, and secondary antibody was applied in blocking buffer for 2–4 hours at room temperature without agitation. Secondary antibodies (α-Mouse-AlexaFluor 647 and α-Rabbit-AlexaFluor 488) were purchased from Life Tech and used at 1:200. Samples were washed 3× 5 minutes in 0.5 M TBST and once for 3 minutes in tap water. Hoechst (10 mg mL−1, Life Tech) was diluted 1:2000 into PBS and applied for 3 minutes. Samples were washed 3× 5 minutes in PBS, dried briefly, and covered with coverslips using Fluoromount G. After 10 minutes, slides were sealed with clear nail polish (L. A. Colors). Slides were stored flat at 4 °C until imaging.
Confocal Microscopy and Image Alignment
All fluorescence imaging (of dried tissue slices to be analysed by NanoSIMS, as well as of fixed immunofluorescence samples) was performed on a Zeiss LSM 710 confocal microscope with a 20× air objective. Transmitted light was collected to visualize slice morphology. ZEN 2010 software was used to calculate the optimal scan rate and pixel dimensions for each region.
To align fluorescence images and metal maps, the morphological features of the tissue were used as fiducials. Fluorescence and transmitted light images taken on the confocal microscope were already aligned to each other, as they are taken at the same time. Metal maps were aligned to the scar left on the tissue by ion sputtering, which was visualized by refected light microscopy after NanoSIMS analysis. Then, the two image sets (fluorescence + transmitted light, and metal + reflected light) were aligned to each other using the morphological features of the tissue visible in both pre- and post-analysis transmitted and reflected light images. Following alignment, transmitted and reflected light images were removed, revealing aligned fluorescence images and metal maps.
Results
Laser ablation ICP-MS analysis of copper distributions in wildtype and Calgw71 zebrafish embryos
To assess whether Calgw71 embryos have altered copper levels relative to their wildtype (WT) siblings, we analysed tissue slices of Calgw71 and WT embryos by LA-ICP-MS. In this technique, each tissue slice is ablated by a laser beam rastering across its surface, and the metal concentration of the ablated material at each location is measured by ICP-MS, rendering a 2-dimensional metal map of the tissue.22 Flash frozen tissue was prepared in order to avoid the redistribution of metals that can occur during tissue fixation (Figure S1 and Experimental Methods).24,28 LA-ICP-MS was performed using a 6 μm diameter spot, which was sufficient to resolve distinct tissues within the zebrafish embryo and achieved, to our knowledge, the highest resolution images of endogenous metals acquired by laser ablation techniques. LA-ICP-MS revealed tissue-specific enrichment of copper in the neural tissue, liver, and heart of 6 days post-fertilization (dpf) wildtype embryos relative to the rest of the body (Figure 1, g-l). However, in Calgw71 embryos, copper accumulated in the kidneys and was largely excluded from the neural tissue (Figure 1, m-r). The difference in metal distribution between Calgw71 and WT embryos was specific to copper, as zinc distributions were similar between the two genotypes (Figure S3). Matrix-matched standards24 allowed quantification of these images, revealing significant differences in copper levels in the kidney, liver, and neural tissue of the two genotypes, but no differences in zinc levels (Figure 2, Figure S4).
Figure 1.
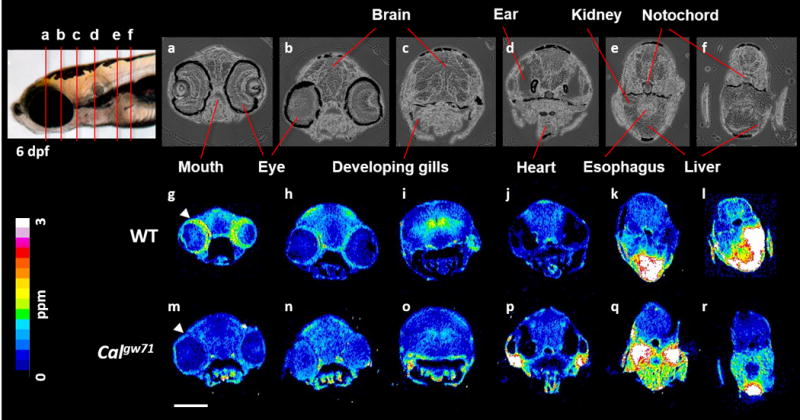
la-icp-ms images of wildtype (g-l) and calamitygw71 (m-r) embryos (6 dpf). transmitted light images (a-f) provide anatomical orientation. arrowheads (g, m) indicate the ring of copper observed around the outer edge of the retina of wildtype fish which is absent in calgw71 embryos. scale bar: 200 μm.
Figure 2.
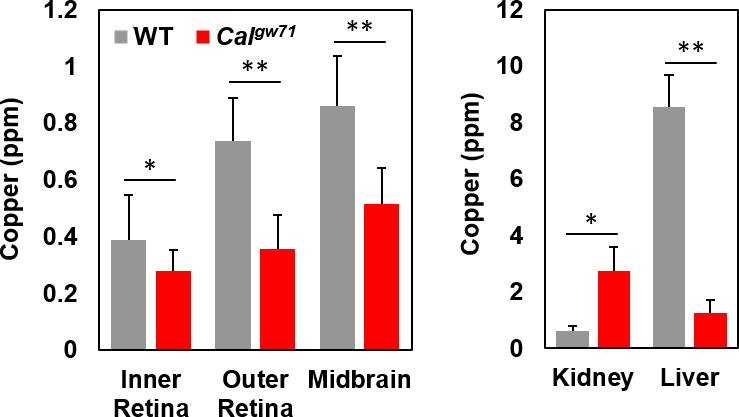
quantification of la-icp-ms images of wildtype and calgw71 embryos; n > 3 measurements per point. error bars represent standard deviation. *, p<0.05. **, p<0.001.
Multimodal copper mapping in zebrafish using LA-ICP-MS and nanoSIMS
Interested in further probing the differences in neural tissue copper levels between the two genotypes, we focused on the substantial difference in copper content in the outer retina of Calgw71 vs wildtype fish. Indeed, in wildtype fish, a bright ring of copper was observed along the outer edge of the retina, and this ring was absent in Calgw71 retinas (Figure 1g and 1m, arrowheads). The retina is a highly structured tissue, consisting of multiple concentric cell layers that radiate from the lens29,30 with each cell layer having a distinct function. As such, we sought to identify the retinal cell layer that contained these copper rings as a starting point. However, retinal cell layers in zebrafish embryos are ~5–30 μm wide each, making them difficult to distinguish with a 6 μm diameter ablation spot. Thus, we turned to the CAMECA NanoSIMS, a high-spatial-resolution imaging mass spectrometer that can achieve 100–200 nm spatial resolution metal maps of biological tissue.31,32 Interestingly, with this enhanced spatial resolution, nanoSIMS images resolved distinct copper puncta rather than revealing continuous rings of copper around the retinal edge (Figure 3). Using the phosphorous signal to identify cellular nuclei which distinguish retinal cell layers, the copper puncta were assigned to the outer nuclear layer (ONL), the retinal layer containing the photoreceptors. Additionally, to the best of our knowledge, these data represent the first implementation of matrix-matched standards of metals in biological tissue for nanoSIMS in order to quantify the copper content of these puncta. Copper puncta in Calgw71 embryos contained 60% of the copper content of the puncta in wildtype embryos (WT: 3.5 ± 0.7 ppm; Calgw71: 2.0 ± 0.3 ppm). Having mapped the copper rings observed by LA-ICP-MS to discrete puncta within the ONL of the retina, we next sought to identify the cellular structures containing this highly concentrated copper.
Figure 3.
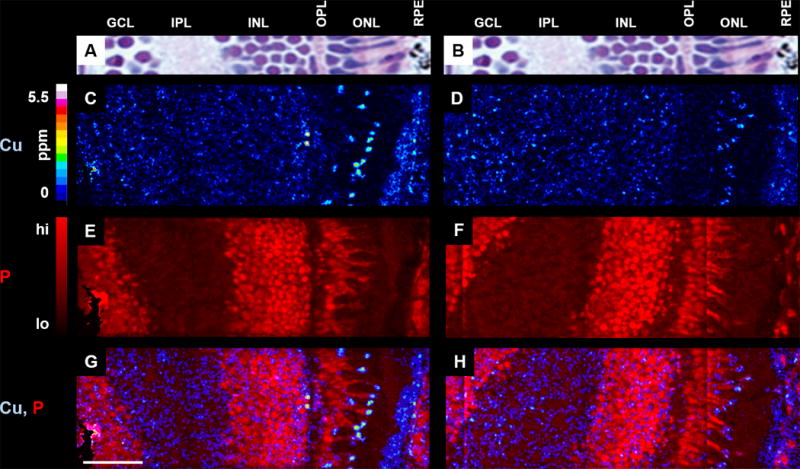
in the zebrafish embryo retina, copper is enriched in puncta in the outer nuclear layer (onl); calgw71 embryos (d, f, and h) contain less copper in this region than wt embryos (c, e, and g). representative nuclear staining provides anatomical orientation (a and b). nanosims images include copper (c and d), phosphorous (e and f) and an overlay of copper and phosphorous signals (g and h). gcl: ganglion cell layer; ipl: inner plexiform layer; inl: inner nuclear layer; opl: outer plexiform layer; onl: outer nuclear layer; rpe: retinal pigmented epithelium. scale bar 25 μm.
Photoreceptor megamitochondria are loci of retinal copper accumulation
Photoreceptor cells are highly complex neural cells responsible for sensing photons and generating the initial electrochemical signals that propagate through the neuroretina and are transmitted to the brain via the optic nerve.29,30 These cells are the most energy-demanding cells of the retina. In the dark, energy is devoted to maintaining ion gradients in the steady state; in the light, even more energy is required to mediate the turnover of each rhodopsin molecule that interacts with a photon.33–35 To provide the large quantity of ATP required for these processes, photoreceptors contain a large mass of mitochondria. In most mammals, many long, thin mitochondria are arranged in parallel within the inner segment of the photoreceptor cell.36,37 However, in other animals, including zebrafish38–40 and some tree shrews,41,42 large, bulky mitochondria with diameters exceeding 2 μm have been observed in the inner segments of photoreceptors. The unusual size of these mitochondria places them in a morphological class known as megamitochondria.43 Interestingly, megamitochondria are typically observed under conditions of cell stress.44–46 However, some megamitochondria have been observed in healthy cells,47,48 and the megamitochondria of photoreceptors appear to belong in this category. The role of megamitochondria in photoreceptors is unclear. Some groups have argued that megamitochondria provide the high ATP levels necessary for photoreceptor health.40 Others maintain that the main driver of megamitochondrial morphology is optical: the refractive index of megamitochondria is much higher than that of the rest of the retina, perhaps revealing a role for megamitochondria in collecting and focusing light.49,50 Nevertheless, we hypothesized that the observed copper puncta in photoreceptor cells might be localized to megamitochondria. Indeed, electron micrographs revealed that megamitochondria are located between the nuclei and the photoreceptor stacks in the ONL, a pattern that is strikingly similar to the pattern of the copper and phosphorous signals we observed in nanoSIMS images (Figure 4).
Figure 4.
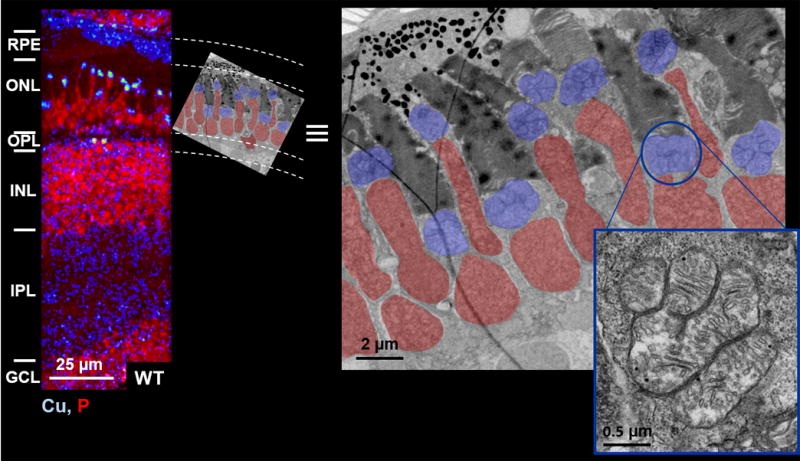
megamitochondria are located between nuclei and receptors in the outer nuclear layer, matching the pattern of copper puncta observed in the nanosims images. nanosims copper and phosphorous overlay (left) of a wildtype zebrafish retina and electron micrograph (right) of a similar region in another wildtype zebrafish retina. false color labels nuclei (red) and megamitochondria (blue). a copy of the electron micrograph is scaled and rotated to align with the nanosims image. inset: zoomed image of one megamitochondrion in gray scale. scale bar lengths are indicated. gcl: ganglion cell layer; ipl: inner plexiform layer; inl: inner nuclear layer; opl: outer plexiform layer; onl: outer nuclear layer; rpe: retinal pigmented epithelium.
In order to verify that copper puncta indeed correspond to megamitochondria, we tested the colocalization of a mitochondrial marker with the copper signal from a nanoSIMS image of the same tissue. To identify megamitochondria in these experiments, we avoided techniques that require fixation, such as electron microscopy and immunofluorescence, as we and others28 have shown that fixatives redistribute metals within tissue (Figure S1). Instead, we generated zebrafish embryos expressing Actin:TOM20-mCherry, an mCherry construct localized to the mitochondria and expressed under the actin promoter.51,52 Introduction of Actin:TOM20-mCherry DNA into the genomic DNA using Tol2 transposase yielded embryos expressing TOM20-mCherry in a mosaic pattern: some cells express the construct while others do not. In this case, a mosaic expression pattern worked to our advantage, allowing us to select retinal tissue where the labelled megamitochondria were spread apart and readily distinguished from one another. Using the fluorescence signal as a guide, we acquired nanoSIMS images of regions with distinct, labelled megamitochondria (Figure S5). Indeed, megamitochondria labelled with mCherry colocalized with copper puncta from nanoSIMS images of the same region (Figure 5). Interestingly, achieving 1:1 mapping of copper puncta to labelled megamitochondria required many scans of the nanoSIMS raster, as one optical slice by confocal microscopy covers a depth of 1–2 μm, while a single nanoSIMS scan analyses only a few nanometres of the tissue’s surface. With much higher resolution in the z dimension, nanoSIMS images provide multiple slices through megamitochondria and yield a profile of copper concentration through each megamitochondrion (Figure S6). Additionally, many copper puncta in the nanoSIMS images did not have corresponding labelled megamitochondria in the confocal images, as we intentionally selected images where the labelled megamitochondria were spread apart, with multiple unlabelled megamitochondria in the same frame.
Figure 5.
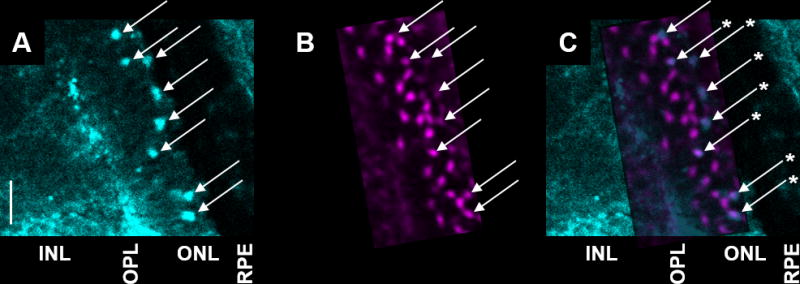
colocalization of megamitochondria with copper puncta. fluorescently labeled megamitochondria (a, cyan) overlap with copper puncta in nanosims images (b, magenta) of the same tissue. megamitochondria are indicated with arrows. asterisks denote colocalization of fluorescence and copper signals (c). inl: inner nuclear layer; opl: outer plexiform layer; onl: outer nuclear layer; rpe: retinal pigmented epithelium. scale bar: 5 μm.
Calgw71 zebrafish have sufficient copper to maintain megamitochondrial respiration to support photoreceptor growth
Having demonstrated that copper puncta observed in nanoSIMS images localize to megamitochondria, we sought to address whether there were any functional consequences of reduced copper in Calgw71 embryos relative to wildtype siblings. Photoreceptors with damaged or missing megamitochondria degrade due to the inability to produce sufficient ATP to support rhodopsin turnover.53 However, cone and rod photoreceptors were similarly abundant in Calgw71 and WT embryo retinas, as shown by immunofluorescence using Zpr-1 and Zpr-3, respectively (Figure 6). Additionally, electron micrographs revealed no gross morphological defects in photoreceptor or megamitochondrial architecture between wildtype and Calgw71 embryos (Figure S7). Together, these results suggest that the lower copper levels observed in megamitochondria of Calgw71 embryos relative to wildtype siblings do not disrupt megamitochondrial function to an extent that would cause photoreceptor degradation.
Figure 6.
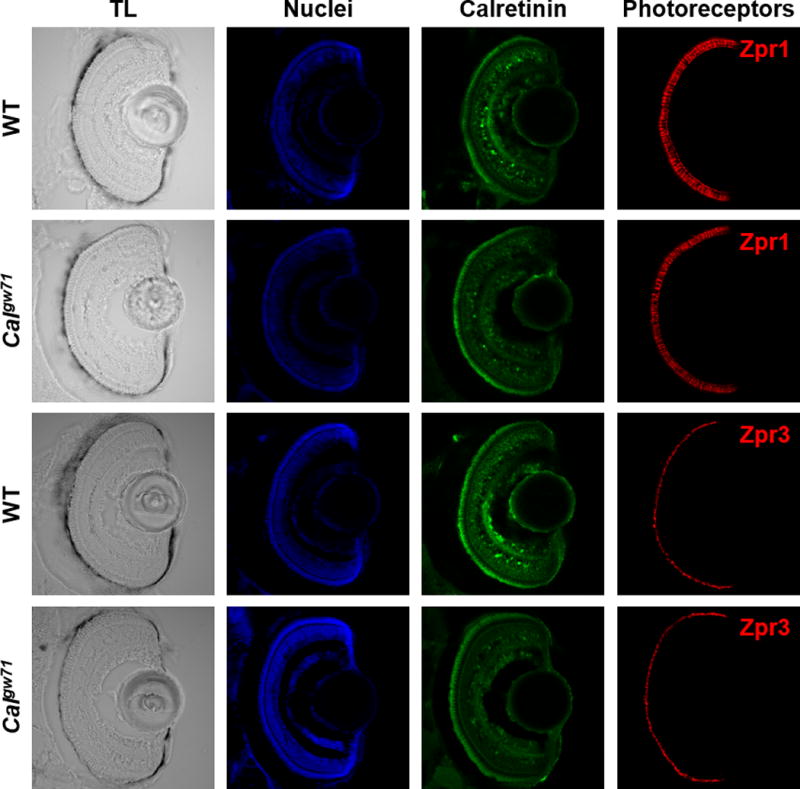
cone and rod photoreceptor abundance is similar in calgw71 and wt zebrafish. tl: transmitted light. nuclear staining (blue) and anti-calretinin (green) serve as controls for staining efficiency. the zpr1 antibody labels rods, while the zpr3 antibody labels cones (both red).
These results are in agreement with previous studies demonstrating the prioritization of copper for mitochondrial function.54 The majority of copper contained in mitochondria is not accounted for by enzyme cofactors55 and has been suggested to reside in the mitochondrial matrix as a CuL complex with unknown ligand L.56 Since the megamitochondria in Calgw71 embryos generate sufficient ATP to support photoreceptor health, we speculate that there is sufficient copper to support cytochrome c oxidase and that the observed decrease in copper may affect a different copper pool, such as the CuL reservoir. Indeed, cytochrome c oxidase activity in whole Calgw71 embryos or dissected Calgw71 embryo eyes was no different from that of WT embryos or embryo eyes (Figure S9). We do not currently have assays with appropriate sensitivity to measure cytochrome c oxidase activity or CuL levels of the megamitochondria in only the photoreceptor population, leaving the molecular origins of copper depletion in Calgw71 megamitochondria an open question for future investigation.
Discussion
The dynamic distribution of copper in an organism reflects the combined action of the copper import, export, and storage proteins and small molecules in each tissue. In this study, we find that mutation of the copper transporter ATP7A disrupts copper trafficking in zebrafish embryos. Copper is enriched in the kidneys while failing to reach normal levels in key tissues such as the liver and neural matter. This localization pattern suggests that copper is readily mobilized to the blood in Calgw71 embryos, precluding a role for ATP7A in mobilizing copper from the yolk; however, as copper is filtered from the blood by the kidneys, ATP7A fails to export copper into the urine, leading to copper accumulation in the kidney. This phenomenon has also been documented in the brindled mouse model of Menkes disease.14 In contrast to copper accumulation in the kidney, copper fails to be efficiently trafficked to the eyes, brain, and liver, suggesting that ATP7A may play an important role in moving copper out of the blood and into peripheral tissues.
Copper transport to neural tissue represents a special case of copper mobilization from blood, as the blood-brain barrier (BBB) is a highly regulated structure. In addition to the brain, the retina also lies behind the BBB, and molecular retinal composition reflects the ability of the BBB to transport nutrients into the retina. Our metal imaging data reveal accumulation of copper in the outer nuclear layer of wildtype neuroretina, specifically in the megamitochondria of photoreceptors, confirming that the BBB transports copper and that this copper is trafficked to specific organelles within cells. In contrast to wildtype embryos, Calgw71 embryos accumulate only about half as much copper in photoreceptor megamitochondria, suggesting that ATP7A plays an important role in mobilizing copper across the blood-brain barrier to these cells and organelles. This result supports the use of the Calgw71 zebrafish as a model of neural copper deprivation in fish that are otherwise morphologically healthy.
The role of copper in the retina and the impact of copper dyshomeostasis on retinal tissue is an area of active research. Studies of Menkes57,58 and Wilson diseases,59,60 copper metabolism disorders caused by mutations in the copper exporters ATP7A and ATP7B, respectively, have found significant optical pathologies including disrupted retinal signalling and loss of sight.61,62 Additionally, alterations in copper metabolism have been associated with age-related macular degeneration.63,64 More recently, the importance of peptidylglycine α-amidating monooxygenase (PAM), a copper-containing monoamine oxidase, in photoreceptor ciliogenesis has been identified,65,66 and copper has been found to be required for spontaneous firing in the retina.67 In this study, the roles of copper accumulation in the megamitochondria remain unclear. Despite significantly lower copper levels in Calgw71 megamitochondria relative to wildtype, these organelles continue to support the development of apparently healthy photoreceptors, suggesting compensation mechanisms that may prioritize metalation of essential mitochondrial components such as cytochrome c oxidase. Previous studies of copper shortages have described a hierarchy of metalation68 in which proteins with high copper affinity are metalated preferentially over proteins with lower-affinity sites.69 Whether these principles account for healthy photoreceptors in Calgw71 embryos is a subject for further study.
From a technical perspective, this study highlights the sequential use of multiple imaging techniques with increasing spatial resolution to identify tissues, cells, and subcellular organelles enriched in a metal of interest. Rapid profiling methods such as LA-ICP-MS are powerful because of their ability to scan many tissue slices over a short period of time, providing metal signatures for specific tissues and genotypes and identifying differences between them. After identifying regions of interest using LA-ICP-MS, techniques such as nanoSIMS may be used to obtain higher resolution images of metal distribution, identifying the specific cellular and subcellular context in which the metal is located. Coupling nanoSIMS with high resolution optical techniques such as confocal fluorescence microscopy, colocalization of optical markers with metal signals in the absence of tissue fixation may be readily achieved, allowing the unambiguous identification of subcellular compartments enriched in a metal.
As a final point, matrix-matched standards constitute a critical part of this study because they enable quantitative comparison of metal imaging data collected from multiple samples. These standards allow for direct correlation between LA-ICP-MS and nanoSIMS results, providing numerical readouts of metal content in tissues without dissection or digestion and facilitating the quantification of metal concentrations in structures that are too small to analyse by bulk techniques, such as a single megamitochondrion.
Conclusions
The combination of rapid metal profiling by LA-ICP-MS with high resolution metal imaging by nanoSIMS and electron and fluorescence confocal microscopies identified copper accumulation in megamitochondria of photoreceptors in wildtype zebrafish embryos. In contrast, megamitochondria of Calgw71 Menkes model zebrafish embryos contain ca. 60% of wildtype copper levels. Despite low megamitochondrial copper, cone and rod photoreceptors are equally numerous in Calgw71 embryos and wildtypes, and no differences in megamitochondrial morphology were observed. These results suggest that ATP7A is involved in the transport of copper across the blood-brain barrier. Additionally, as megamitochondria with low total copper support normal photoreceptor development, we suggest that copper sufficiency may be prioritized within mitochondria to ensure mitochondrial function. The retina is one of the most valuable sensory inputs in daily human life.29 Vision impairment due to eye disease affects over 30 million adults in the United States, with over 2 million cases due to macular degeneration,70 the degeneration of the central portion of the retina, which is currently considered to be incurable.71 The combined imaging results highlight the central importance of copper in this system and characterize a major cellular structure for copper accumulation.
As techniques for imaging metals in biological tissue continue to improve and the statistical methods applied to metal image analysis become more sophisticated,72,73 the power of metal imaging to identify biologically relevant differences between genotypes will become increasingly valuable. Indeed, the application of high-resolution metal imaging to highly structured tissues can potentially unravel unappreciated roles of metals in cellular subtypes within tissues that were previously invisible because bulk or low-resolution imaging techniques average across many cells within a tissue. Efforts along these lines are underway in our laboratory and others.
Supplementary Material
Significance to Metallomics.
Copper is an essential nutrient that must be acquired from the diet and properly distributed through the body. Genetic mutations that disrupt copper transport in humans contribute to many diseases, including Menkes disease. Here, we use zebrafish as an optically transparent model of Menkes disease to probe how copper distributions are altered in a whole animal setting. We combine multiple techniques for imaging metals in Menkes fish to identify reduced copper levels localized to retinal photoreceptor cells, illuminating a source of copper dysregulation in this disease.
Acknowledgments
Work at UC Berkeley, including LA-ICP-MS, confocal and electron microscopy, cloning, and zebrafish husbandry, was supported by the NIH (GM 79465 to C.J.C.). C.M.A. was supported by a Fannie and John Hertz Foundation Fellowship and an NIH Chemical Biology Interface Training Grant (T32 GM066698). C.J.C. is an Investigator with the Howard Hughes Medical Institute and a CIFAR Senior Fellow. NanoSIMS work at LLNL was performed under the auspices of the U.S. DOE under contract number DE-AC52-07NA27344 and was supported by the U.S. DOE Genomic Science Program under contract SCW1039.
We thank Christina Ramon for help with sample preparation for nanoSIMS analysis. We thank Prof. Jonathan Gitlin for sharing the Calgw71 zebrafish line. We thank Reena Zalpuri and the UC Berkeley Electron Microscope facility for instruction and assistance in preparing and imaging samples by electron microscopy. We thank Mel Boren, Kate Kliman and the staff at the UC Berkeley Zebrafish Facility for housing and feeding our zebrafish lines.
Footnotes
Electronic Supplementary Information (ESI) available online. See DOI: 10.1039/x0xx00000x
Conflicts of interest
There are no conflicts to declare.
References
- 1.Lutsenko S. Copper trafficking to the secretory pathway. Metallomics. 2016;8:840–852. doi: 10.1039/c6mt00176a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Öhrvik H, Thiele DJ. How copper traverses cellular membranes through the mammalian copper transporter 1, Ctr1. Annals of the New York Academy of Sciences. 2014;1314:32–41. doi: 10.1111/nyas.12371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hamza I, Gitlin JD. Copper chaperones for cytochrome c oxidase and human disease. Journal of Bioenergetics and Biomembranes. 2002;34:381–388. doi: 10.1023/a:1021254104012. [DOI] [PubMed] [Google Scholar]
- 4.Prasad AN, Ojha R. Menkes disease: what a multidisciplinary approach can do. Journal of Multidisciplinary Healthcare. 2016;9:371–385. doi: 10.2147/jmdh.s93454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lutsenko S. Modifying factors and phenotypic diversity in Wilson’s disease. Annals of the New York Academy of Sciences. 2014;1315:56–63. doi: 10.1111/nyas.12420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee J, Prohaska JR, Thiele DJ. Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proceedings of the National Academy of Sciences. 2001;98:6842–6847. doi: 10.1073/pnas.111058698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedera P. Update on the clinical management of Wilson's disease. The Application of Clinical Genetics. 2017;10:9–19. doi: 10.2147/tacg.s79121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies KM, Mercer JFB, Chen N, Double KL. Copper dyshomoeostasis in Parkinson’s disease: implications for pathogenesis and indications for novel therapeutics. Clinical Science. 2016;130:565–574. doi: 10.1042/cs20150153. [DOI] [PubMed] [Google Scholar]
- 9.Hordyjewska A, Popiołek Ł, Kocot J. The many “faces” of copper in medicine and treatment. BioMetals. 2014;27:611–621. doi: 10.1007/s10534-014-9736-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaler SG. Translational research investigations on ATP7A: an important human copper ATPase. Annals of the New York Academy of Sciences. 2014;1314:64–68. doi: 10.1111/nyas.12422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Linz R, Lutsenko S. Copper-transporting ATPases ATP7A and ATP7B: cousins, not twins. Journal of Bioenergetics and Biomembranes. 2007;39:403–407. doi: 10.1007/s10863-007-9101-2. [DOI] [PubMed] [Google Scholar]
- 12.Lutsenko S, LeShane ES, Shinde U. Biochemical basis of regulation of human copper-transporting ATPases. Archives of Biochemistry and Biophysics. 2007;463:134–148. doi: 10.1016/j.abb.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gupta A, Lutsenko S. Human copper transporters: mechanism, role in human diseases and therapeutic potential. Future Medicinal Chemistry. 2009;1:1125–1142. doi: 10.4155/fmc.09.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoshimura N, Kida K, Usutani S, Nishimura M. Histochemical localization of copper in various organs of brindled mice after copper therapy. Pathology International. 1995;45:10–18. doi: 10.1111/j.1440-1827.1995.tb03374.x. [DOI] [PubMed] [Google Scholar]
- 15.Kaler SG. ATP7A-related copper transport diseases— emerging concepts and future trends. Nature Reviews Neurology. 2011;7:15–29. doi: 10.1038/nrneurol.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaler SG. In: GeneReviews(R) Pagon RA, editor. 1993. [Google Scholar]
- 17.Tumer Z. An overview and update of ATP7A mutations leading to Menkes disease and occipital horn syndrome. Human Mutation. 2013;34:417–429. doi: 10.1002/humu.22266. [DOI] [PubMed] [Google Scholar]
- 18.Chang CJ. Bioinorganic life and neural activity: Toward a chemistry of consciousness? Accounts of Chemical Research. 2017;50:535–538. doi: 10.1021/acs.accounts.6b00531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ahuja A, Dev K, Tanwar RS, Selwal KK, Tyagi PK. Copper mediated neurological disorder: Visions into amyotrophic lateral sclerosis, Alzheimer and Menkes disease. Journal of Trace Elements in Medicine and Biology. 2015;29:11–23. doi: 10.1016/j.jtemb.2014.05.003. [DOI] [PubMed] [Google Scholar]
- 20.Madsen EC, Gitlin JD. Zebrafish mutants calamity and catastrophe define critical pathways of gene–nutrient interactions in developmental copper metabolism. PLoS Genetics. 2008;4:e1000261. doi: 10.1371/journal.pgen.1000261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lenartowicz M, et al. Mottled mice and non-mammalian models of menkes disease. Frontiers in Molecular Neuroscience. 2015;8 doi: 10.3389/fnmol.2015.00072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ackerman CM, Lee S, Chang CJ. Analytical methods for imaging metals in biology: From transition metal metabolism to transition metal signaling. Analytical Chemistry. 2017;89:22–41. doi: 10.1021/acs.analchem.6b04631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ghosal S, et al. Imaging and 3D Elemental characterization of intact bacterial spores by high-resolution secondary ion mass spectrometry. Analytical Chemistry. 2008;80:5986–5992. doi: 10.1021/ac8006279. [DOI] [PubMed] [Google Scholar]
- 24.Hare DJ, Lear J, Bishop D, Beavis A, Doble PA. Protocol for production of matrix-matched brain tissue standards for imaging by laser ablation-inductively coupled plasma-mass spectrometry. Analytical Methods. 2013;5:1915. doi: 10.1039/c3ay26248k. [DOI] [Google Scholar]
- 25.Becker JS, Zoriy MV, Pickhardt C, Palomero-Gallagher N, Zilles K. Imaging of copper, zinc, and other elements in thin section of human brain samples (hippocampus) by laser ablation inductively coupled plasma mass spectrometry. Analytical Chemistry. 2005;77:3208–3216. doi: 10.1021/ac040184q. [DOI] [PubMed] [Google Scholar]
- 26.Jung Kim M, Ho Kang K, Kim CH, Choi SY. Real-time imaging of mitochondria in transgenic zebrafish expressing mitochondrially targeted GFP. BioTechniques. 2008;45:331–334. doi: 10.2144/000112909. [DOI] [PubMed] [Google Scholar]
- 27.Trinh LA, et al. Biotagging of specific cell populations in zebrafish reveals gene regulatory logic encoded in the nuclear transcriptome. Cell Reports. 2017;19:425–440. doi: 10.1016/j.celrep.2017.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hackett MJ, et al. Chemical alterations to murine brain tissue induced by formalin fixation: implications for biospectroscopic imaging and mapping studies of disease pathogenesis. The Analyst. 2011;136:2941. doi: 10.1039/c0an00269k. [DOI] [PubMed] [Google Scholar]
- 29.Grossniklaus HE, Geisert EE, Nickerson JM. Introduction to the retina. 2015;134:383–396. doi: 10.1016/bs.pmbts.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 30.Stenkamp DL. Development of the vertebrate eye and retina. 2015;134:397–414. doi: 10.1016/bs.pmbts.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Herrmann AM, et al. Nano-scale secondary ion mass spectrometry — A new analytical tool in biogeochemistry and soil ecology: A review article. Soil Biology and Biochemistry. 2007;39:1835–1850. doi: 10.1016/j.soilbio.2007.03.011. [DOI] [Google Scholar]
- 32.Hong-Hermesdorf A, et al. Subcellular metal imaging identifies dynamic sites of Cu accumulation in Chlamydomonas. Nature Chemical Biology. 2014;10:1034–1042. doi: 10.1038/nchembio.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu DY, Cringle SJ. Oxygen distribution and consumption within the retina in vascularised and avascular retinas and in animal models of retinal disease. Progress in Retinal and Eye Research. 2001;20:175–208. doi: 10.1016/s1350-9462(00)00027-6. [DOI] [PubMed] [Google Scholar]
- 34.Linton JD, et al. Flow of energy in the outer retina in darkness and in light. Proceedings of the National Academy of Sciences. 2010;107:8599–8604. doi: 10.1073/pnas.1002471107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kooragayala K, et al. Quantification of oxygen consumption in retina ex vivo demonstrates limited reserve capacity of photoreceptor mitochondria. Investigative Opthalmology & Visual Science. 2015;56:8428. doi: 10.1167/iovs.15-17901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stone J, van Driel D, Valter K, Rees S, Provis J. The locations of mitochondria in mammalian photoreceptors: Relation to retinal vasculature. Brain Research. 2008;1189:58–69. doi: 10.1016/j.brainres.2007.10.083. [DOI] [PubMed] [Google Scholar]
- 37.Hoang QV, Linsenmeier RA, Chung CK, Curcio CA. Photoreceptor inner segments in monkey and human retina: mitochondrial density, optics, and regional variation. Vis Neurosci. 2002;19:395–407. doi: 10.1017/s0952523802194028. [DOI] [PubMed] [Google Scholar]
- 38.Tarboush R, Novales Flamarique I, Chapman GB, Connaughton VP. Variability in mitochondria of zebrafish photoreceptor ellipsoids. Visual Neuroscience. 2014;31:11–23. doi: 10.1017/s095252381300059x. [DOI] [PubMed] [Google Scholar]
- 39.Kim J, et al. The presence of megamitochondria in the ellipsoid of photoreceptor inner segment of the zebrafish retina. Anatomia, Histologia, Embryologia: Journal of Veterinary Medicine Series C. 2005;34:339–342. doi: 10.1111/j.1439-0264.2005.00612.x. [DOI] [PubMed] [Google Scholar]
- 40.Masuda T, Wada Y, Kawamura S. ES1 is a mitochondrial enlarging factor contributing to form mega-mitochondria in zebrafish cones. Scientific Reports. 2016;6 doi: 10.1038/srep22360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knabe W, Kuhn HJ. Morphogenesis of megamitochondria in the retinal cone inner segments of Tupaia belangeri (Scandentia) Cell and Tissue Research. 1996;285:1–9. doi: 10.1007/s004410050614. [DOI] [PubMed] [Google Scholar]
- 42.Lluch S, López-Fuster MJ, Ventura J. Cornea, retina, and lens morphology in five Soricidae species (Soricomorpha: Mammalia) Anatomical Science International. 2009;84:312–322. doi: 10.1007/s12565-009-0042-1. [DOI] [PubMed] [Google Scholar]
- 43.Wakabayashi T. Megamitochondria formation - physiology and pathology. Journal of Cellular and Molecular Medicine. 2002;6:497–538. doi: 10.1111/j.1582-4934.2002.tb00452.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoppel CL, Tandler B, Fujioka H, Riva A. Dynamic organization of mitochondria in human heart and in myocardial disease. The International Journal of Biochemistry & Cell Biology. 2009;41:1949–1956. doi: 10.1016/j.biocel.2009.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Caldwell SH, et al. NASH and cryptogenic cirrhosis: a histological analysis. Annals of Hepatology. 2009;8:346–352. [PMC free article] [PubMed] [Google Scholar]
- 46.Zsengellér ZK, et al. Methylmalonic acidemia: A megamitochondrial disorder affecting the kidney. Pediatric Nephrology. 2014;29:2139–2146. doi: 10.1007/s00467-014-2847-y. [DOI] [PubMed] [Google Scholar]
- 47.Tandler B, Nagato T, Phillips CJ. Megamitochondria in the serous acinar cells of the submandibular gland of the neotropical fruit bat, Artibeus obscurus. The Anatomical Record. 1997;248:13–17. doi: 10.1002/(SICI)1097-0185(199705)248:1<13::AID-AR2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 48.Spicer SS, Parmley RT, Boyd L, Schulte BA. Giant mitochondria distinct from enlarged mitochondria in secretory and ciliated cells of gerbil trachea and bronchioles. American Journal of Anatomy. 1990;188:269–281. doi: 10.1002/aja.1001880306. [DOI] [PubMed] [Google Scholar]
- 49.Knabe W, Skatchkov S, Kuhn HJ. “Lens mitochondria” in the retinal cones of the tree-shrew Tupaia belangeri. Vision Research. 1997;37:267–271. doi: 10.1016/s0042-6989(96)00199-x. [DOI] [PubMed] [Google Scholar]
- 50.Lluch S, López-Fuster MJ, Ventura J. Giant mitochondria in the retina cone inner segments of shrews of genusSorex(Insectivora, Soricidae) The Anatomical Record Part A: Discoveries in Molecular, Cellular, and Evolutionary Biology. 2003;272A:484–490. doi: 10.1002/ar.a.10066. [DOI] [PubMed] [Google Scholar]
- 51.Kanaji S, Iwahashi J, Kida Y, Sakaguchi M, Mihara K. Characterization of the signal that directs Tom20 to the mitochondrial outer membrane. The Journal of Cell Biology. 2000;151:277–288. doi: 10.1083/jcb.151.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noble S, Godoy R, Affaticati P, Ekker M. Transgenic zebrafish expressing mCherry in the mitochondria of dopaminergic neurons. Zebrafish. 2015;12:349–356. doi: 10.1089/zeb.2015.1085. [DOI] [PubMed] [Google Scholar]
- 53.Desplan C, et al. Impaired mitochondrial energy production causes light-induced photoreceptor degeneration independent of oxidative stress. PLOS Biology. 2015;13:e1002197. doi: 10.1371/journal.pbio.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dodani SC, Leary SC, Cobine PA, Winge DR, Chang CJ. A targetable fluorescent sensor reveals that copper-deficient SCO1 and SCO2 patient cells prioritize mitochondrial copper homeostasis. Journal of the American Chemical Society. 2011;133:8606–8616. doi: 10.1021/ja2004158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baker ZN, Cobine PA, Leary SC. The mitochondrion: a central architect of copper homeostasis. Metallomics. 2017;9:1501–1512. doi: 10.1039/c7mt00221a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cobine PA, Ojeda LD, Rigby KM, Winge DR. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. Journal of Biological Chemistry. 2004;279:14447–14455. doi: 10.1074/jbc.M312693200. [DOI] [PubMed] [Google Scholar]
- 57.Seelenfreund MH, Gartner S, Vinger PF. The ocular pathology of Menkes’ disease. (Kinky hair disease) Archives of Ophthalmology. 1968;80:718–720. doi: 10.1001/archopht.1968.00980050720007. [DOI] [PubMed] [Google Scholar]
- 58.Ferreira R. Menkes disease New ocular and electroretinographic findings. Ophthalmology. 1998;105:1076–1078. doi: 10.1016/s0161-6420(98)96010-9. [DOI] [PubMed] [Google Scholar]
- 59.Dingle J, Havener WH. Ophthalmoscopic changes in a patient with Wilson’s disease during long-term penicillamine therapy. Annals of Ophthalmology. 1978;10:1227–1230. [PubMed] [Google Scholar]
- 60.Satishchandra P, Ravishankar Naik K. Visual pathway abnormalities Wilson’s disease: an electrophysiological study using electroretinography and visual evoked potentials. Journal of the Neurological Sciences. 2000;176:13–20. doi: 10.1016/s0022-510x(00)00280-x. [DOI] [PubMed] [Google Scholar]
- 61.Waggoner DJ, Bartnikas TB, Gitlin JD. The role of copper in neurodegenerative disease. Neurobiology of Disease. 1999;6:221–230. doi: 10.1006/nbdi.1999.0250. [DOI] [PubMed] [Google Scholar]
- 62.Krajacic P, et al. Retinal localization and copper-dependent relocalization of the Wilson and Menkes disease proteins. Investigative Opthalmology & Visual Science. 2006;47:3129. doi: 10.1167/iovs.05-1601. [DOI] [PubMed] [Google Scholar]
- 63.Erie JC, Good JA, Butz JA, Pulido JS. Reduced zinc and copper in the retinal pigment epithelium and choroid in age-related macular degeneration. American Journal of Ophthalmology. 2009;147:276–282.e271. doi: 10.1016/j.ajo.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 64.Newsome DA, et al. Macular degeneration and elevated serum ceruloplasmin. Investigative Ophthalmology & Visual Science. 1986;27:1675–1680. [PubMed] [Google Scholar]
- 65.Kumar D, et al. Early eukaryotic origins for cilia-associated bioactive peptide-amidating activity. Journal of Cell Science. 2016;129:943–956. doi: 10.1242/jcs.177410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bachmann-Gagescu R, et al. The ciliopathy gene cc2d2a controls zebrafish photoreceptor outer segment development through a role in Rab8-dependent vesicle trafficking. Human Molecular Genetics. 2011;20:4041–4055. doi: 10.1093/hmg/ddr332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dodani SC, et al. Copper is an endogenous modulator of neural circuit spontaneous activity. Proceedings of the National Academy of Sciences. 2014;111:16280–16285. doi: 10.1073/pnas.1409796111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mendelsohn BA, et al. Atp7a determines a hierarchy of copper metabolism essential for notochord development. Cell Metabolism. 2006;4:155–162. doi: 10.1016/j.cmet.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 69.Banci L, et al. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 70.Prevalence of Adult Vision Impairment and Age-Related Eye Diseases in America. 2017 < https://nei.nih.gov/eyedata/adultvision_usa>.
- 71.Foundation, A. M. D. What is Macular Degeneration? 2017 < https://www.macular.org/what-macular-degeneration>.
- 72.James CJ, Hesse CW. Independent component analysis for biomedical signals. Physiological Measurement. 2005;26:R15–39. doi: 10.1088/0967-3334/26/1/r02. [DOI] [PubMed] [Google Scholar]
- 73.James SA, et al. φXANES: In vivo imaging of metal-protein coordination environments. Scientific Reports. 2016;6 doi: 10.1038/srep20350. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


