Abstract
Objective:
To investigate the association between CYP2C19 and ABCB1 polymorphisms and clopidogrel resistance (CR) in patients with cardiovascular disease in Beijing district.
Methods:
In total, 325 patients were enrolled in the study, including 101 experimental group patients and 224 control group patients. The experimental group was divided into CR group (n=30) and non-CR group (n=71) according to the adenosine diphosphate (ADP)-induced platelet inhibition rate in thromboelastography (TEG) (ADP-induced platelet inhibition rate of <30% was defined as CR and rate of 30%–100% was defined as non-CR). Genotypes, including CYP2C19*2, CYP2C19*3, CYP2C19*4, CYP2C19*5, CYP2C19*17, and ABCB1, were determined using time-of-flight mass spectrometry (Clin-TOF) and Sanger sequencing in all patients.
Results:
In the experimental group, carriers of CYP2C19 heterozygous (*1/*2, n=46; *1/*3, n=7), and mutation homozygous (*2/*2, n=7; *2/*3, n=3; *3/*3, n=0) genotypes showed significantly lower ADP-induced platelet inhibition rates than noncarriers (*1/*1, n=38; p=0.035 and 0.001, respectively); the carriage of mutant CYP2C19*2 or *3 allele was significantly associated with an increased risk of CR. In contrast, carriers of ABCB1 heterozygous (TC, n=50) showed significantly lower ADP-induced platelet inhibition rates than noncarriers (CC, n=39, p=0.097), and there was no significant correlation between ABCB1 genotypes and higher CR risk.
Conclusion:
The carriage of CYP2C19*2 or *3 mutant allele was significantly associated with attenuated platelet response to clopidogrel and increased CR risk. The carriage of ABCB1 mutant allele was not significantly associated with CR risk.
Keywords: CYP2C19, ABCB1 gene, polymorphism, clopidogrel resistance, adenosine diphosphate, induced platelet inhibition rate
Introduction
The use of clopidogrel in combination with aspirin is recommended for the prevention of ischemic events in cardiovascular patients with acute coronary syndrome (ACS) or in those undergoing percutaneous coronary intervention (PCI) according to current guidelines (1). Despite this standard treatment, there are still numerous adverse cardiovascular events, and attenuated responses to clopidogrel therapy and clopidogrel resistance (CR) are regarded to be the main reasons.
Clopidogrel is an inactive prodrug that is absorbed by regula-ting the P-glycoprotein encoded by ABCB1 gene in the intestine. This requires two bioconversion steps that are regulated by the hepatic cytochrome P450 (CYP) system to form active product, which targets and blocks adenosine diphosphate (ADP) P2Y12 receptor, and indirectly inhibits GPIIb/IIIa receptor-binding fib-rinogen, and then inhibits platelet aggregation (2).
Although the mechanism of CR has not been fully elucidated yet, a variety of factors, including external and internal factors, have been identified. External factors included patient compliance, drug dose, and drug interactions. Internal factors inclu-ded gene polymorphisms, activation of other platelet pathways, and increased release of ADP. Among these factors, hepatic cytochrome P4502C19 (CYP2C19) is one of the key enzymes of clopidogrel oxidative metabolism, which plays a role as the main metabolic enzyme of the CYP system. The main mutant alleles, CYP2C19*2 and CYP2C19*3, are the most common genotypes in Asian populations (frequency of 30%–50% and 5%–10%, respectively) (3). The presence of CYP2C19*2 or CYP2C19*3 mutant allele increases CR risk (4,5). The other two alleles, CYP2C19*4 and CYP2C19*5, are mainly distributed in Caucasian race and are associated with attenuated response to clopidogrel. Although platelet aggregation significantly decreases in patients with CYP2C19*17 mutant allele, the risk of ischemic events significantly increases (6). ABCB1 is a family of transmembrane transporters that primarily transport substrates to the plasma membrane u-sing the energy generated by ATP hydrolysis. It plays a leading role in the human multiple drug resistance gene family, the mutation influences the absorption of clopidogrel, but the correlation with CR has not reached a consensus.
Therefore, this study aimed to investigate the association between CYP2C19 and ABCB1 polymorphisms and CR risk in clopidogrel-treated Chinese patients to provide evidence for the early prediction of CR, to guide the rational use of drugs, and to reduce the incidence of adverse cardiovascular events.
Methods
Study populations
The patients were selected from May to October 2016. A total of 325 patients were enrolled in the study, including 101 experimental group patients and 224 control group patients. The exclusion criteria of the experimental group were as follows: (1) Patients with cardiovascular disease, including ACS and undergoing PCI; and (2) Consecutive clopidogrel on-treatment patients, including imported drugs, such as Plavix (a loading dose of 300 mg for first time, followed by 75 mg/day), and domestic drug, such as Taijia (50 mg/day), with platelet function testing. The exclusion criteria of the control group were as follows: The chest X-ray, physical examination, electrocardiogram, blood routine parameters, urine routine parameters, blood biochemical tests, medical history were no abnormalities, no chronic bleeding and bleeding tendency, coagulation index and platelet aggregation rate without exception. The study protocol complied with the Declaration of Helsinki and was approved by the Ethics Committee.
CYP2C19 genotyping by time-of-flight mass spectrometry (Clin-TOF) and Sanger sequencing
Genomic DNA was extracted from blood using the Blood genomic DNA Extraction Kit [DP348, Tiangen Biotech (Beijing) Co., Ltd.]. Primers were designed by Bioyong Technologies Inc., and synthesized by Generay Biotech (Shanghai) Co., Ltd. First, multiplex PCR reactions were performed with 5 µL of reaction mixture, comprising 1 µL genomic DNA, 2.78 µL PCR mixture I, 0.22 µL enzyme I, and 1 µL amplification primer. The PCR conditions were as follows: initial denaturation at 37°C for 10 min and 95°C for 10 min; 45 cycles of annealing at 95°C for 30 s, 56°C for 30 s, and 72°C for 60 s; and final extension at 72°C for 5 min. Then, SAP was used to eliminate uncombined dNTPs after amplification in order to make them unavailable for future reaction. Dephosphorylation conditions were 37°C for 40 min and 85°C for 5 min. Finally, single-base primer extension reactions were conducted using the following reaction mixture: 1.019 µL PCR mixture III, 0.041 µL enzyme III, and 0.940 µL amplification primer. The reactions were performed using a two-cycling loop program as follows: initial denaturation at 94°C for 30 s; 40 cycles of annealing at 94°C for 5 s, 56°C for 5 s, and 80°C for 5 s; and final extension at 72°C for 3 min. These annealing and extension steps were repeated five times.
Genotypes were determined using time-of-flight mass spectrometry (CLIN-TOF-II, Bioyong Technologies Inc.) and Sanger sequencing at Beijing Sunbiotech Co., Ltd.
The primer sequences used in DNA sequencing are as follows: CYP2C19*2-F: AACCAGAGCTTGGCATATTG; CYP2C19*2-R: TGCTTCTCAAGCATTACTCC; CYP2C19*3-F: TTCCAATCATTTAGCTTCACCCT; CYP2C19*3-R: GCATAAAATAAAGAACTTTGCCATC; CYP2C19*4-F: TGCATTGGAACCACTTGG; CYP2C19*4-R: TCCCTTACTGTTTACCCTCA; CYP2C19*5-F: CCTCCTATGATTCACCGAACAGT; CYP2C19*5-R: GGGTCAATCAGAGATTTCAGGTTA; CYP2C19*17-F: ACCAGGAGGTCAAGAAGC; CYP2C19*17-R: GAAGGCAGGAATTGTTAT; ABCB1-F: TCACAAGGAGGGTCAGG; and ABCB1-R: TTGGCAGTTTCAGTGTAAGA.
Platelet function testing
ADP-induced platelet inhibition rate was measured by TEG® 5000 Thrombelastograph® Hemostasis Analyzer system (Haemonetics Corporation). Volume of 2.7 mL venous blood anticoagulation with 3.2% sodium citrate and 4.0 mL venous blood anticoagulation with 14.7 U/mL lithium heparin was collected from patients received clopidogrel (75 mg, once daily) after 4 days, and completed test within 4 h. The analyzer has three channels; 20 µL 0.2 ml/L CaCl2 and 340 µL blood anticoagulation with citrate which mixed with Kaolin were added into 1-channel; 10 µL activator F and 360 µL blood anticoagulation with heparin were added into 2-channel; and 10 µL activator F, 10 µL ADP and 360 µL blood anticoagulation with heparin were added into 3-channel. Platelet inhibition rate was calculated by the instrument software, and the results were expressed as a percentage (%).
Clinical data collection
The demographic characteristics and baseline data, including age; gender; BMI; coronary risk factors (e.g., smoking, hypertension, and diabetes mellitus); and use of other medications [e.g., angiotensin-converting enzyme (ACE) inhibitor, angiotensin receptor blocker (ARB), and calcium-channel blocker], of 101 experimental group patients were collected and evaluated. These data were obtained through reviewing medical records of hospital.
Statistical methods
Data are expressed as number (percentage), mean±SD, or median with interquartile range (IQR). Continuous variables were analyzed using unpaired two-tailed t-test and ANOVA for >2 groups for Gaussian distribution and nonparametric tests for non-Gaussian distribution. Allele frequencies were analyzed using X2 test. The correlation between genotype and ADP-induced platelet inhibition rates was analyzed using Pearson correlation coefficient test. Other potential confounding factors between genotypes and the efficacy of clopidogrel treatment were excluded using multivariate logistic regression analysis. Statistical analyses were performed using the SPSS software, version 19.0 (SPSS, IL, USA). P<0.05 was considered statistically significant.
Results
Study population and CYP2C19*2 or *3 genotyping
A cohort of 101 experimental group patients was divided into three groups according to CYP2C19 genotype as follows: wild-type homozygotes (*1/*1, n=38), heterozygous (*1/*2 or *1/*3, n=53), and mutation homozygous (*2/*2, *2/*3 or *3/*3, n=10). Baseline characteristics of the study population according to their CYP2C19 genotype are summarized in Table 1.
Table 1.
Baseline characteristics of the experimental group patients according to the CYP2C19*2 and *3 genotype
| CYP2C19 | CYP2C19 | CYP2C19 | P (value) | |
|---|---|---|---|---|
| *1/*1 | *1/*2, *1/*3 | *2/*2, *2/*3, *3/*3 | ||
| (n=38) | (n=53) | (n=10) | ||
| Age, years | 69.71±13.44 | 69.62±11.20 | 64.30±15.38 | 0.440 |
| Male (%) | 8 (21.1%) | 17 (32.1%) | 3 (30.0%) | 0.512 |
| BMIa, Kg/m2 | 23.18±7.89 | 24.52±4.74 | 22.54±9.48 | 0.560 |
| Smoking status | 0.125 | |||
| Non-smoker | 30 (78.9%) | 48 (90.6%) | 10 (100%) | |
| Ex-smoker | 4 (10.5%) | 3 (5.7%) | 0 (0%) | |
| Habitual smoker | 4 (10.5%) | 2 (3.8%) | 0 (0%) | |
| Hypertension | 24 (63.2%) | 29 (54.7%) | 4 (40.0%) | 0.402 |
| Hyperlipidemia | 4 (10.5%) | 10 (18.9%) | 3 (30.0%) | 0.297 |
| Diabetes mellitus | 13 (34.2%) | 22 (41.5%) | 4 (40.0%) | 0.782 |
| ACEIb | 19 (50.0%) | 20 (37.7%) | 4 (40.0%) | 0.477 |
| ARBc | 28 (73.7%) | 39 (73.6%) | 7 (70.0%) | 0.935 |
| Calcium-channel blocker | 25 (65.8%) | 36 (67.9%) | 7 (70.0%) | 0.982 |
| β-receptor blocker | 30 (78.9%) | 46 (86.8%) | 8 (80.0%) | 0.579 |
| Statins | 35 (92.1%) | 51 (96.2%) | 10 (100.0%) | 0.546 |
| Proton-pump inhibitor | 22 (57.9%) | 30 (56.6%) | 4 (40.0%) | 0.538 |
| Aspirin | 10 (26.3%) | 13 (24.5%) | 3 (30.0%) | 0.941 |
- body mass index;
- angiotensin-converting enzyme inhibitor;
- angiotensin receptor blocker
Multivariate logistic regression analysis was performed to exclude other potential confounding factors between genotypes and the efficacy of clopidogrel treatment.
Demographic characteristics and baseline data between three genotypes of CYP2C19 groups were well balanced
Genotyping results
The genotyping results of the experimental group and the control group are shown in Table 2 and Table 3, respectively. There was no significant difference between the two groups (p=0.545).
Table 2.
Genotype frequency of the experimental group
| Percentage (%) Gene |
Wild-type homozygotes | Genotype Heterozygous |
Mutation homozygous |
|---|---|---|---|
| CYP2C19*2 | 45 (44.45) | 49 (48.51) | 7 (6.93) |
| CYP2C19*3 | 91 (90.10) | 10 (9.90) | 0 |
| CYP2C19*4 | 101 (100.00) | 0 | 0 |
| CYP2C19*5 | 101 (100.00) | 0 | 0 |
| CYP2C19*17 | 100 (99.01) | 1 (0.99) | 0 |
| ABCB1 | 39 (38.61) | 50 (49.50) | 12 (11.88) |
Table 3.
Genotype frequency of the control group
| Percentage (%) Gene |
Wild-type homozygotes | Genotype Heterozygous |
Mutation homozygous |
|---|---|---|---|
| CYP2C19*2 | 106 (47.32) | 101 (45.09) | 17 (7.59) |
| CYP2C19*3 | 208 (92.86) | 16 (7.14) | 0 |
| CYP2C19*4 | 223 (99.55) | 1 (0.45) | 0 |
| CYP2C19*5 | 224 (100.00) | 0 | 0 |
| CYP2C19*17 | 219 (97.77) | 5 (2.23) | 0 |
| ABCB1 | 81 (36.16) | 104 (46.43) | 39 (17.41) |
Allele frequencies of CYP2C19*2 and CYP2C19*3 (A) in the experimental group were 31.19% and 4.95%, respectively. Allele frequencies of CYP2C19*2 and CYP2C19*3 (A) in the control group were 30.4% and 4.7%, respectively.
The results of mass spectrometry of the genotype are shown in Figure 1.
Figure 1.
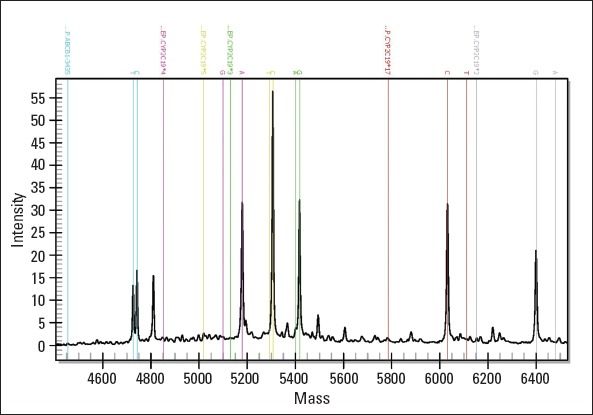
Mass spectrograms for genotypes CYP2C19*2, CYP2C19*3, CYP2C19*4, CYP2C19*5, CYP2C19*17, and ABCB1
Unextend primer (UEP) peak and two different alleles of one genotype are marked by dotted vertical lines in the same color
CYP2C19*2 or *3 genotypes and CR
As shown in Figure 2, the median value of ADP-induced platelet inhibition rate in the experimental group was 45.00% (IQR, 27.70%–66.75%). Median ADP-induced platelet inhibition rates among CYP2C19 genotypes were as follows: 57.60% (IQR, 34.78%–74.90%) for wild-type homozygous patients, 39.50% (IQR, 25.65%–62.20%) for heterozygous patients, and 23.45% (IQR, 8.45%–37.65%) for mutant homozygous patients. ADP-induced platelet inhibition rates were significantly different among groups (p=0.013, F=3.355), and the lowest rate was observed for mutant homozygous patients. Moreover, difference in ADP-induced platelet inhibition rates was examined between *1/*1, *1/*2, *1/*3, *2/*3, and *3/*3; difference between *1/*1 and *1/*2, *2/*2, *2/*3 was significant (p=0.30, 0.07, and 0.19, respectively; SD=5.58, 10.48, and 15.27, respectively), but that difference between *1/*1 and *1/*3 (p=0.563; SD=10.48) was not significant. Multivariate linear regression model revealed that CYP2C19*2 or *3 mutant allele was independently associated with ADP-induced platelet inhibition rate (p=0.001; F=11.285; R=0.320a).
Figure 2.
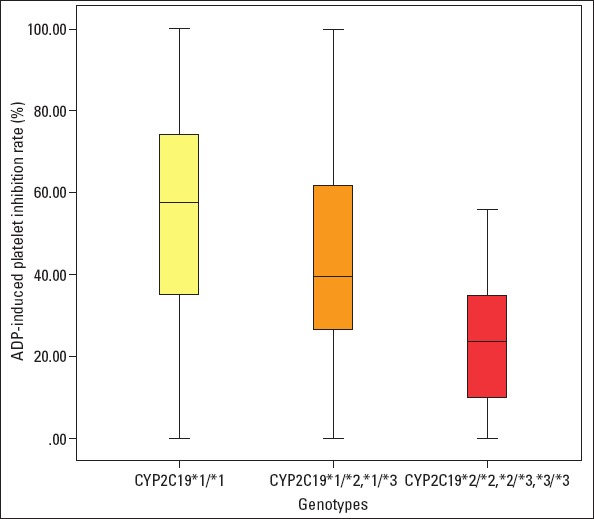
CYP2C19*2 or *3 genotypes and ADP- induced platelet inhibition rates
CR risk and presence of CYP2C19*2 or *3 genotypes in the experimental group are shown in Table 4; there was a statistically significant correlation between the genotypes and CR risk (p=0.028; Pearson Chi-square=7.123), and CR risk was the highest in mutant homozygous patients. Presence of CYP2C19*2 or *3 mutant alleles was identified to be an independent predictor of CR risk (p=0.026; t=2.270).
Table 4.
The relationship between CYP2C19*2 or *3 genotypes and CR
| Metabolizes | Number | Percentage of entire group | Number of CRa | Percentage of CR in entire group | |
|---|---|---|---|---|---|
| Wild-type homozygotes | *1/*1 | 38 | 37.62% | 7 | 18.42% |
| Heterozygous | *1/*2 | 46 | 45.54% | 15 | 32.61% |
| *1/*3 | 7 | 6.93% | 2 | 28.57% | |
| Total | 53 | 52.48% | 17 | 32.08% | |
| Mutation homozygous | *2/*2 | 7 | 6.93% | 4 | 57.14% |
| *2/*3 | 3 | 2.97% | 2 | 66.67% | |
| *3/*3 | 0 | 0.00% | 0 | 0.00% | |
| Total | 10 | 9.90% | 6 | 60.00% |
- clopidogrel resistance
CYP2C19*4, CYP2C19*5, CYP2C19*17 genotypes
Six patients showed CYP2C19*17 mutant allele; one patient showed CYP2C19*4 mutant allele; and no patient showed CYP2C19*5 mutant.
ABCB1 genotypes and CR
The frequency of ABCB1 mutant allele in the experimental group is shown in Table 2. ADP-induced platelet inhibition rates were not significantly different among the groups (p=0.176, F=1.771); this difference between the wild-type homozygous CC and heterozygous TC was significant (p=0.097; SD=5.65), but that between wild-type homozygous CC and mutant homozygous TT (p=0.854; SD=8.73) was not significant. Furthermore, there was no significant correlation between the ABCB1 genotypes and increased CR risk.
Time-of-flight mass spectrometry system technology (Clin-TOF) and Sanger sequencing technology
Among all 325 patients, 1 patient showed inconsistent sequencing results; the consistency rate between time-of-flight mass spectrometry system (Clin-TOF) and Sanger sequencing was 99.69%; thus, the inconsistency may be due to sample contamination during mass spectrometry.
Discussion
Clopidogrel is an oral prodrug that needs to be converted to an active metabolite to irreversibly bind to the P2Y12 receptor. Approximately 85% of clopidogrel is hydrolyzed to inactive metabolites by the contribution of CYP2C19, CYP3A4 or CYP3A5, CYP2C9, CYP1A2, and CYP2B6 isozymes, and the rest passes through two sequential steps that are dependent on cytochrome P450 (CYP) into active metabolites. CYP2C19 genotype affects both metabolic steps and is the most important determinant of the pharmacokinetic and pharmacodynamic responses to clopidogrel although it accounts for only approximately 12% of the reported variability (7-10). The presence of any dysfunctional CYP2C19 alleles (*2, *3, *4, *5) is associated with CR risk, ischemic events, and stent thrombosis (7, 8, 11, 12), whereas the presence of the CYP2C19 allele (*17) is associated with an increased risk of bleeding (13). Gene mutations that regulate clopidogrel absorption and excretion, such as gene encoding P-glycoprotein multidrug resistance spontaneous transporters, ABCB1 (14), may also affect the risk of CR and clinical events during treatment.
In the present study, we observed that the frequencies of CYP2C19*2 and CYP2C19*3 alleles (A) in the experimental group and in the control group were consistent with those reported by Zou et al. (30.14% and 3.57%) (2) and were higher than those reported in Swedish Caucasians (15) and Ethiopians (16). The allele frequencies of CYP2C19*4, CYP2C19*5, and CYP2C19*17 in our cohort were lower than those in other races (17-19), confirming that allelic variants exhibit ethnic and geographic diversity. This study aimed to investigate the association between CYP2C19 and ABCB1 polymorphisms and the antiplatelet effect of clopidogrel. We observed that the presence of CYP2C19*2 or *3 mutant allele has a significant effect on platelet function, resulting in significantly lower ADP-induced platelet inhibition rates than in wild-type homozygous. The carriers of CYP2C19*2 or *3 mutant allele showed attenuated response to clopidogrel therapy and increased risk of CR. These results are supported by the previous studies (4-6, 20). ABCB1 mutation resulted in a decreased ADP-induced platelet inhibition in mutant homozygous patients compared to wild-type patients, but there is no evidence regar-ding the association between the ABCB1 genotype and CR. This result was supported by that reported by Shuldiner et al. (21) in addition to a significant association between ABCB1 polymorphism and CR reported by other researchers (22,23).
Patients with clinical ACS, particularly those undergoing PCI treatment, are recommended to receive a dual antiplatelet stra-tegy with clopidogrel and aspirin, but the carriage of CYP2C19*2 or *3 mutant allele is associated with a higher risk of CR, leading to increased incidence of adverse cardiovascular events. The pharmacogenomic testing of CYP2C19 is recommended by the ACC/AHA guideline to better individualize drug dose for patients to get maximum benefits from an alternative strategy (24).
There are a variety of platelet functions testing methods, including platelet aggregation experiments, such as light transmittance aggregometry (LTA), TEG, vasodilator-stimulated phosphoprotein (VASP) phosphorylation method, and use of VerifyNow instrument. LTA has been used as the gold standard; however, this method has a large demand for platelet-rich plasma samples, which is time-consuming and has poor reproducibility. The TEG method is a dynamic depiction of blood coagulation by highly sensitive overhanging filaments, which can provide coagulation and fibrinolytic information as well as a comprehensive reflection of the coagulation cascade reaction and platelet interaction with good reproducibility; thus, we selected TEG to test platelet function in this study.
Currently, there are a variety of methods, including PCR-RFLP (polymerase chain reaction-restriction fragment length polymorphism) method, use of Taq-Man probes, ARMS (amplification refractory mutation system), genechip, Sanger sequencing, and mass spectrometry. In this study, CYP2C19 and ABCB1 gene polymorphisms were for the first time detected using time-of-flight mass spectrometry (Clin-TOF) in Beijing district; Mass spectrometry, which is a soft-ionization technology that allows the analysis of biomolecules, has been reported to be a useful technology for the detection of SNPs (25). Its advantages include the wide range of molecular weight that can be assuaged, the fast speed of scanning, the simple steps of operation, and particularly higher sequencing flux compared with that in Sanger sequencing. Mass spectrometry has a potential to become an applicable approach in clinical diagnosis and can be applied to genetic individualized therapy.
Study limitations
This study has some limitations that need to be discussed. First, we excluded the differences between groups and ruled out the interaction of concurrent medications and coronary risk factors, but we did not exclude the possible bias of genetic polymorphisms, such as CYP3A4, CYP3A5, or other genetic variation involved in the reaction of P2Y12 inhibitors, that may influence the pharmacokinetics, pharmacodynamics, and clinical efficacy of clopidogrel. Second, the number of clopidogrel-treated patients was small, and we did not collect the follow-up the data of major adverse cardiovascular events (MACE) in these patients, which highlights the need for further researches to include a larger cohort and conduct a more complete follow-up data of study patients.
Conclusion
In this study, we investigated the association between CYP2C19 and ABCB1 polymorphisms and CR in clopidogrel-treated Chinese patients. We observed that the carriage of CYP2C19*2 or *3 mutant allele was significantly associated with attenuated platelet response to clopidogrel and increased risk of CR, whereas the carriage of ABCB1 mutant allele was not significantly associated with the risk of CR. Our findings can provide an evidence for the early prediction of CR, guide the rational use of drugs, and reduce the incidence of adverse cardiovascular events in patients.
Footnotes
Conflict of interest: None declared.
Peer-review: Externally peer-reviewed.
Authorship contributions: Concept – Z.Z.; Design – Z.Z.; Supervision – X.Z.; Fundings – X.Z.; Materials – Z.Z., H.X., Y.M., H.G., J.Z.; Data collection &/or processing – Z.Z., H.X., Y.M., H.G., J.Z.; Analysis &/or interpretation – Z.Z., H.X., Y.M., H.G., J.Z.; Literature search – Z.Z., H.X., Y.L., C.L., Y.S.; Writing – Z.Z., H.X., Y.L., C.L., Y.S.; Critical review – X.Z.
References
- 1.Aradi D, Komócsi A, Vorobcsuk A, Serebruany VL. Impact of clopidogrel and potent P2Y 12 -inhibitors on mortality and stroke in patients with acute coronary syndrome or undergoing percutaneous coronary intervention:a systematic review and meta-analysis. Thromb Haemost. 2013;109:93–101. doi: 10.1160/TH12-06-0377. [DOI] [PubMed] [Google Scholar]
- 2.Zou JJ, Xie HG, Chen SL, Tan J, Lin L, Zhao YY, et al. Influence of CYP2C19 loss-of-function variants on the antiplatelet effects and cardiovascular events in clopidogrel-treated Chinese patients undergoing percutaneous coronary intervention. Eur J Clin Pharmacol. 2013;69:771–7. doi: 10.1007/s00228-012-1392-5. [DOI] [PubMed] [Google Scholar]
- 3.Xie HG, Zou JJ, Hu ZY, Zhang JJ, Ye F, Chen SL. Individual variability in the disposition of and response to clopidogrel:pharmacogeno-mics and beyond. Pharmacol Ther. 2011;129:267–89. doi: 10.1016/j.pharmthera.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 4.Hulot JS, Bura A, Villard E, Azizi M, Remones V, Goyenvalle C, et al. Cytochrome P450 2C19 loss-of-function polymorphism is a major determinant of clopidogrel responsiveness in healthy subjects. Blood. 2006;108:2244–7. doi: 10.1182/blood-2006-04-013052. [DOI] [PubMed] [Google Scholar]
- 5.Brandt JT, Close SL, Iturria SJ, Payne CD, Farid NA, Ernest CS, et al. Common polymorphisms of CYP2C19 and CYP2C9 affect the pharmacokinetic and pharmacodynamic response to clopidogrel but not prasugrel. J Thromb Haemost. 2007;5:2429–36. doi: 10.1111/j.1538-7836.2007.02775.x. [DOI] [PubMed] [Google Scholar]
- 6.Baldwin RM, Ohlsson S, Pedersen RS, Mwinyi J, Ingelman-Sundberg M, Eliasson E, et al. Increased omeprazole metabolism in carriers of the CYP2C19*17 allele;a pharmacokinetic study in healthy volunteers. Br J Clin Pharmacol. 2008;65:767–74. doi: 10.1111/j.1365-2125.2008.03104.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mega JL, Close SL, Wiviott SD, Shen L, Hockett RD, Brandt JT, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N Engl J Med. 2009;360:354–62. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 8.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varenhorst C, James S, Erlinge D, Brandt JT, Braun OO, Man M, et al. Genetic variation of CYP2C19 affects both pharmacokinetic and pharmacodynamic responses to clopidogrel but not prasugrel in aspirin-treated patients with coronary artery disease. Eur Heart J. 2009;30:1744–52. doi: 10.1093/eurheartj/ehp157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varenhorst C, James S, Erlinge D, Braun OO, Brandt JT, Winters KJ, et al. Assessment of P2Y12 inhibition with the point-of-care device VerifyNow P2Y12 in patients treated with prasugrel or clopidogrel coadministered with aspirin. Am Heart J. 2009;157:562.e1–9. doi: 10.1016/j.ahj.2008.11.021. [DOI] [PubMed] [Google Scholar]
- 11.Collet JP, Hulot JS, Pena A, Villard E, Esteve JB, Silvain J, et al. Cytochrome P450 2C19 polymorphism in young patients treated with clopidogrel after myocardial infarction:a cohort study. Lancet. 2009;373:309–17. doi: 10.1016/S0140-6736(08)61845-0. [DOI] [PubMed] [Google Scholar]
- 12.Hulot JS, Collet JP, Silvain J, Pena A, Bellemain-Appaix A, Barthélémy O, et al. Cardiovascular risk in clopidogrel-treated patients according to cytochrome P450 2C19*2 loss-of-function allele or proton pump inhibitor coadministration:a systematic meta-analysis. J Am Coll Cardiol. 2010;56:134–43. doi: 10.1016/j.jacc.2009.12.071. [DOI] [PubMed] [Google Scholar]
- 13.Sibbing D, Koch W, Gebhard D, Schuster T, Braun S, Stegherr J, et al. Cytochrome 2C19*17 allelic variant, platelet aggregation, blee-ding events, and stent thrombosis in clopidogrel-treated patients with coronary stent placement. Circulation. 2010;121:512–8. doi: 10.1161/CIRCULATIONAHA.109.885194. [DOI] [PubMed] [Google Scholar]
- 14.Taubert D, von Beckerath N, Grimberg G, Lazar A, Jung N, Goeser T, et al. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006;80:486–501. doi: 10.1016/j.clpt.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 15.Chang M, Dahl ML, Tybring G, Götharson E, Bertilsson L. Use of omeprazole as a probe drug for CYP2C19 phenotype in Swedish Caucasians:comparison with S-mephenytoin hydroxylation phenotype and CYP2C19 genotype. Pharmacogenetics. 1995;5:358–63. doi: 10.1097/00008571-199512000-00004. [DOI] [PubMed] [Google Scholar]
- 16.Persson I, Aklillu E, Rodrigues F, Bertilsson L, Ingelman-Sundberg M. S-mephenytoin hydroxylation phenotype and CYP2C19 genotype among Ethiopians. Pharmacogenetics. 1996;6:521–6. doi: 10.1097/00008571-199612000-00005. [DOI] [PubMed] [Google Scholar]
- 17.Ferguson RJ, De Morais SM, Benhamou S, Bouchardy C, Blaisdell J, Ibeanu G, et al. A new genetic defect in human CYP2C19:mutation of the initiation codon is responsible for poor metabolism of S-mephenytoin. J Pharmacol Exp Ther. 1998;284:356–61. [PubMed] [Google Scholar]
- 18.Ibeanu GC, Blaisdell J, Ghanayem BI, Beyeler C, Benhamou S, Bouchardy C, et al. An additional defective allele, CYP2C19*5, contributes to the S-mephenytoin poor metabolizer phenotype in Caucasians. Pharmacogenetics. 1998;8:129–35. doi: 10.1097/00008571-199804000-00006. [DOI] [PubMed] [Google Scholar]
- 19.Ghasemi Z, Hashemi M, Ejabati M, Ebrahimi SM, Kheiri Manjili H, Sharafi A, et al. Development of a High-Resolution Melting Analysis Method for CYP2C19*17 Genotyping in Healthy Volunteers. Avicenna J Med Biotechnol. 2016;8:193–9. [PMC free article] [PubMed] [Google Scholar]
- 20.Chen BL, Zhang W, Li Q, Li YL, He YJ, Fan L, et al. Inhibition of ADP-induced platelet aggregation by clopidogrel is related to CYP2C19 genetic polymorphisms. Clin Exp Pharmacol Physiol. 2008;35:904–8. doi: 10.1111/j.1440-1681.2008.04915.x. [DOI] [PubMed] [Google Scholar]
- 21.Shuldiner AR, O'Connell JR, Bliden KP, Gandhi A, Ryan K, Horenstein RB, et al. Association of cytochrome P450 2C19 genotype with the antiplatelet effect and clinical efficacy of clopidogrel therapy. JAMA. 2009;302:849–57. doi: 10.1001/jama.2009.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mega JL, Close SL, Wiviott SD, Shen L, Walker JR, Simon T, et al. Genetic variants in ABCB1 and CYP2C19 and cardiovascular outcomes after treatment with clopidogrel and prasugrel in the TRITON-TIMI 38 trial:a pharmacogenetic analysis. Lancet. 2010;376:1312–9. doi: 10.1016/S0140-6736(10)61273-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taubert D, von Beckerath N, Grimberg G, Lazar A, Jung N, Goeser T, et al. Impact of P-glycoprotein on clopidogrel absorption. Clin Pharmacol Ther. 2006;80:486–501. doi: 10.1016/j.clpt.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 24.Wright RS, Anderson JL, Adams CD, Bridges CR, Casey DE Jr, Ettinger SM, et al. 2011 ACCF/AHA focused update of the Guidelines for the Management of Patients with Unstable Angina/Non-ST-Elevation Myocardial Infarction (updating the 2007 guideline):a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines developed in collaboration with the American College of Emergency Physicians, Society for Cardiovascular Angiography and Interventions, and Society of Thoracic Surgeons. J Am Coll Cardiol. 2011;57:1920–59. doi: 10.1016/j.jacc.2011.02.009. [DOI] [PubMed] [Google Scholar]
- 25.Ragoussis J, Elvidge GP, Kaur K, Colella S. Matrix-assisted laser desorption/ionisation, time-of-flight mass spectrometry in genomics research. PLoS Genet. 2006;2:e100. doi: 10.1371/journal.pgen.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]


