Abstract
Surface Pre-reacted Glass-ionomer (S-PRG) filler is a bioactive filler produced by PRG technology, which has been applied to various dental materials. A S-PRG filler can release multiple ions from a glass-ionomer phase formed in the filler. In the present study, detailed inhibitory effects induced by S-PRG eluate (prepared with S-PRG filler) against Streptococcus mutans, a major pathogen of dental caries, were investigated. S-PRG eluate effectively inhibited S. mutans growth especially in the bacterium before the logarithmic growth phase. Microarray analysis was performed to identify changes in S. mutans gene expression in the presence of the S-PRG eluate. The S-PRG eluate prominently downregulated operons related to S. mutans sugar metabolism, such as the pdh operon encoding the pyruvate dehydrogenase complex and the glg operon encoding a putative glycogen synthase. The S-PRG eluate inhibited several in vitro properties of S. mutans relative to the development of dental caries especially prior to active growth. These results suggest that the S-PRG eluate may effectively inhibit the bacterial growth of S. mutans following downregulation of operons involved in sugar metabolism resulting in attenuation of the cariogenicity of S. mutans, especially before the active growth phase.
Introduction
Streptococcus mutans has been implicated as a primary causative agent of dental caries in humans1. Although the mechanisms of dental caries have been well investigated and the incidence of dental caries has been reduced in most developing countries, eradication of dental caries remains difficult2. Thus, novel dental products for prevention of dental caries are under development in various countries3–5.
Surface Pre-Reacted Glass-ionomer (S-PRG) fillers have been synthesized by PRG technology involving reaction between fluoroboroaluminosilicate glass and a polyacrylic acid solution6. The S-PRG filler is used in various dental materials including composite resins, bonding agents, cements, and resin sealants7,8. In addition, research focusing on the usefulness of S-PRG fillers in oral hygiene products such as mouthwashes to inhibit the bacteria or oral malodour has been reported9. S-PRG fillers can release six ions, fluoride (F−), sodium (Na+), borate (BO33−), aluminium (Al3+), silicate (SiO32−), and strontium (Sr2+), which have antimicrobial activity against various oral bacteria10. Although antimicrobial activity of S-PRG fillers against S. mutans has been reported11–13, the detailed inhibition mechanisms of S-PRG fillers remain unknown.
Dental caries development caused by S. mutans is induced by bacterial growth, survival and adhesion resulting in biofilm formation by microbial communities14. Sugar metabolism is an important factor for S. mutans growth and survival15,16, which is induced via the Embden-Meyerhof-Parnas pathway16. The sugar metabolic pathways including the Embden-Meyerhof-Parnas pathway are mainly observed in S. mutans during growth rather than in the stationary phase17.
In the present study, we investigated whether a S-PRG eluate prepared with a S-PRG filler can inhibit the bacterial growth of S. mutans. In addition, a molecular biological approach focusing on alterations in S. mutans gene expression in the presence of S-PRG eluate was performed using DNA microarray analysis. Furthermore, we analysed the inhibitory effects of the S-PRG eluate on several in vitro properties of S. mutans relavent to the development of dental caries.
Results
Inhibitory effects of S-PRG eluate on bacterial growth
S-PRG eluate was added at final concentrations of 0%, 6.3%, 12.5% and 25.0% in brain heart infusion (BHI) broth (Difco Laboratories, Detroit, MI, USA). Bacterial suspensions were adjusted in the BHI broth with or without S-PRG eluate at final concentrations ranging from 1.0 × 103 to 1.0 × 108 CFU/ml. After 18-h incubation at 37 °C, bacterial growth was measured at OD550 and bacterial suspensions were then streaked onto Mitis Salivarius agar plates (Difco Laboratories) containing bacitracin (0.2 U/ml; Sigma-Aldrich, St. Louis, MO, USA) and 15% (w/v) sucrose (MSB agar), which were anaerobically cultured at 37 °C for 48 h. S-PRG eluate added to bacterial suspensions (1.0 × 103 to 1.0 × 105 CFU/ml in BHI broth) prominently inhibited bacterial growth, even after incubation at 37 °C for 18 h. This inhibition was S-PRG-concentration-dependent for both OD550 densities and bacterial numbers (Fig. 1A,B). Although slightly lower OD550 values were observed in bacterial suspensions with concentrations ranging from 1.0 × 106 to 1.0 × 108 CFU/ml, the test strains at concentrations >1.0 × 106 CFU/ml did not show extensive reduction in cell numbers even when the S-PRG eluate was added at high concentrations. Thus, test strains adjusted to a final density of 1.0 × 107 CFU/ml were not growth inhibited even in the presence of 25% of S-PRG eluate after 18-h incubation at 37 °C and were mainly used in subsequent studies. Next, we monitored the kinetics of growth inhibition of 1.0 × 107 CFU/ml S. mutans in the presence of each concentration of S-PRG eluate before reaching the stationary phase. Bacterial growth of S. mutans without S-PRG eluate reached a plateau 7-h after incubation, and the lag times were lengthened in a dose dependent manner with the S-PRG eluate (Fig. 1C). The lag time for S. mutans to reach the stationary phase in the presence of 25% S-PRG eluate was approximately twice that in the absence of the S-PRG eluate. Furthermore, survival of 1.0 × 107 CFU/ml S. mutans in the presence of each concentration of S-PRG eluate after the stationary phase was monitored (Fig. 1D). The recovered bacterial numbers were not different among S. mutans in the presence of each concentration of S-PRG eluate after two days of incubation. However, the numbers of recovered S. mutans were reduced in a dose dependent manner with the S-PRG eluate and no bacteria were recovered in the presence of 25% S-PRG eluate 10 days after incubation.
Figure 1.
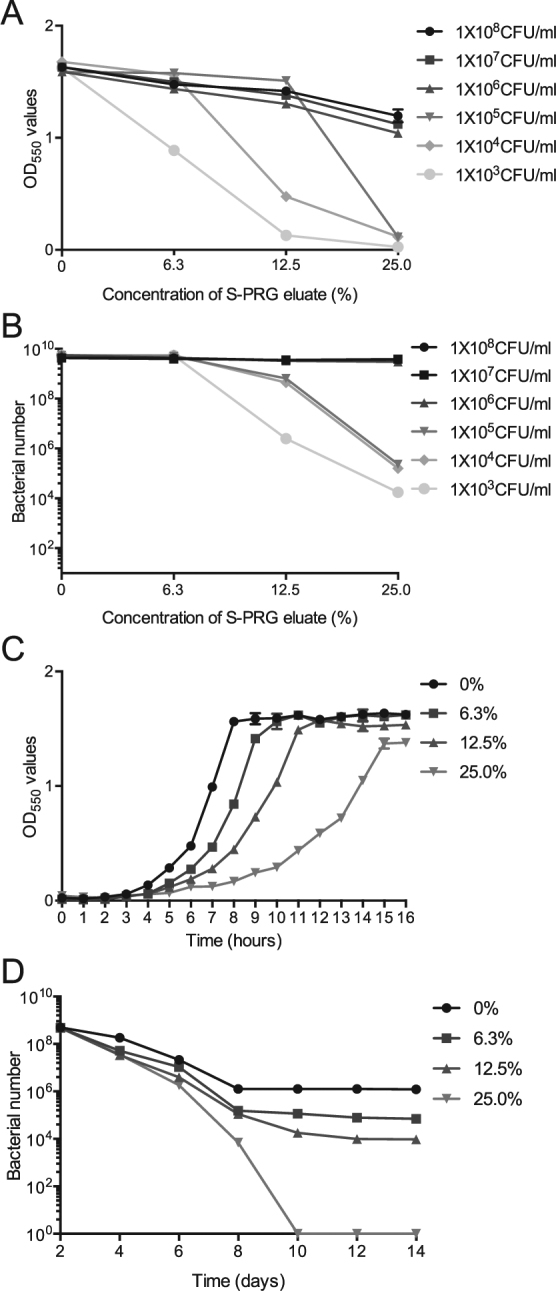
Inhibition on S. mutans MT8148 grown by the S-PRG eluate. (A,B) Bacterial growth by adding varying concentration of the eluate followed by 18 h incubation. Growth was determined by OD550 values in BHI broth (A) and recovered bacterial numbers on MSB plates (B). (C) Bacterial growth by adding 1 × 107 CFU/ml of S. mutans at multiple time points, which was determined by OD550 values in BHI broth. (D) Bacterial survival by adding 1 × 107 CFU/ml of S. mutans at multiple time points, which was determined by adding serial dilutions of the bacterial suspensions to MSB plates.
DNA microarray assays
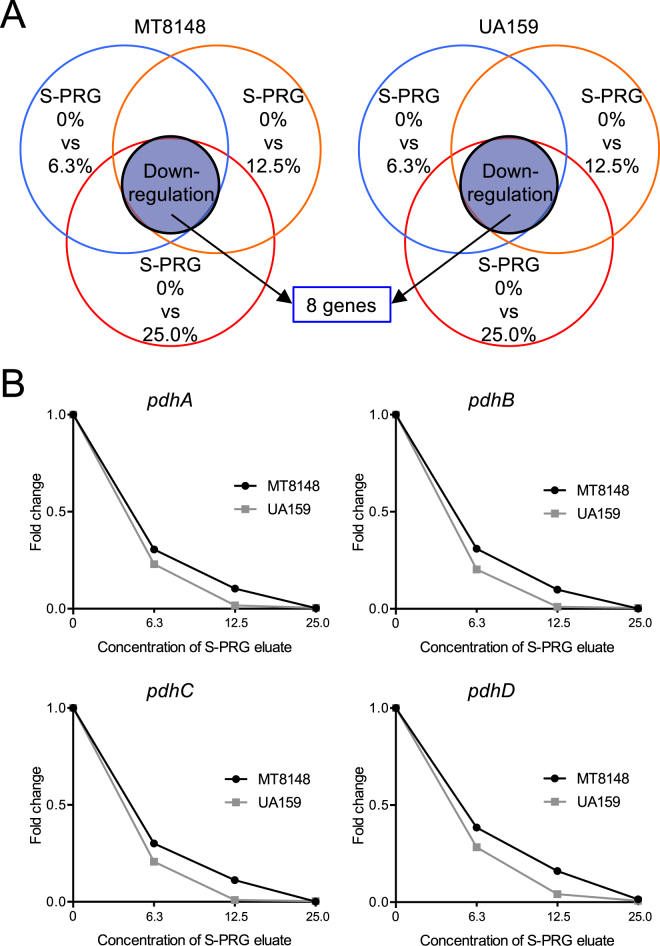
The S-PRG eluate could delay bacterial growth and inhibit bacterial survival of 1.0 × 107 CFU/ml S. mutans; thus, we decided to identify key genes of S. mutans affected by the S-PRG eluate. 1.0 × 107 CFU/ml of S. mutans MT8148 and UA159 in BHI broth were cultured with each specified concentration of S-PRG eluate at 37 °C for 18 h. Next, RNA samples were extracted from each sample for microarray analysis. In the microarray analysis, we evaluated three conditions at several concentrations of S-PRG eluate: 0% versus 6.3%, 0% versus 12.5%, and 0% versus 25.0%. From a list of genes, we selected those with increased or decreased changes of greater than 1.0 of Log2 ratio for comparison. First, we identified genes prominently regulated under all three conditions, which were identified in both S. mutans MT8148 and UA159, to reduce confounding effects of false signals (Fig. 2A). DNA microarray analysis revealed that eight genes were downregulated in all comparisons (Tables 1, 2). Among these genes, genes encoding the pyruvate dehydrogenase (PDH) complex, which plays an important role in S. mutans survival and is closely related to sugar metabolism15,18, were prominently downregulated. The PDH complex forms an operon containing four genes, pdhD, pdhA, pdhB, and pdhC15, all of which were downregulated by S-PRG eluate in a concentration-dependent manner in both MT8148 and UA159 (Fig. 2B).
Figure 2.
Altered expression of key genes in both S. mutans MT8148 and UA159 under three different conditions using DNA microarray analysis. (A) Detection of altered genes in the presence of S-PRG eluate. (B) Changes in the expression of genes in the pdh operon of MT8148 and UA159 in the presence of S-PRG eluate.
Table 1.
List of downregulated genes of MT8148 in the presence of S-PRG eluate, which were observed in both MT8148 and UA159 under three different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3%% | S-PRG12.5 | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| clp | 721354.1 | 10905.7 | 3934.7 | −1.47 | 0.361 | 2604.3 | −2.07 | 0.239 | 779.1 | −3.81 | 0.071 |
| gldA | 720934.1 | 79738.3 | 15155.0 | −2.40 | 0.190 | 4829.4 | −4.04 | 0.061 | 2716.1 | −4.88 | 0.034 |
| pdhA | 721781.1 | 157937.7 | 49355.6 | −1.68 | 0.312 | 17366.4 | −3.19 | 0.110 | 451.1 | −8.45 | 0.003 |
| pdhB | 721780.1 | 118457.0 | 38021.4 | −1.64 | 0.321 | 12313.0 | −3.26 | 0.104 | 232.9 | −9.05 | 0.002 |
| pdhC | 721779.1 | 147220.0 | 47212.9 | −1.64 | 0.321 | 17231.7 | −3.09 | 0.117 | 205.2 | −9.49 | 0.001 |
| pdhD | 721782.1 | 20637.6 | 8184.3 | −1.33 | 0.397 | 3493.2 | −2.56 | 0.169 | 332.1 | −5.98 | 0.016 |
| pfl2 | 720932.1 | 16001.1 | 5582.2 | −1.52 | 0.349 | 3688.0 | −2.12 | 0.231 | 798.1 | −4.33 | 0.050 |
| phsG | 721879.1 | 36063.9 | 13506.8 | −1.42 | 0.375 | 6031.0 | −2.58 | 0.167 | 1521.6 | −4.57 | 0.042 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal in each concentration of S-PRG eluate/signal without S-PRG eluate.
Table 2.
List of downregulated genes of UA159 in the presence of S-PRG eluate, which were observed in both MT8148 and UA159 under three different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3% | S-PRG12.5% | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| clp | 721354.1 | 18865.6 | 6619.2 | −1.51 | 0.351 | 2873.2 | −2.71 | 0.152 | 3817.9 | −2.30 | 0.202 |
| gldA | 720934.1 | 55312.5 | 21392.5 | −1.37 | 0.387 | 9103.4 | −2.60 | 0.165 | 2217.9 | −4.64 | 0.040 |
| pdhA | 721781.1 | 30983.4 | 7458.1 | −2.06 | 0.241 | 547.5 | −5.82 | 0.018 | 151.5 | −7.68 | 0.005 |
| pdhB | 721780.1 | 26341.2 | 5687.7 | −2.21 | 0.216 | 283.9 | −6.53 | 0.011 | 131.4 | −7.69 | 0.005 |
| pdhC | 721779.1 | 32996.4 | 7188.0 | −2.20 | 0.218 | 320.5 | −6.69 | 0.010 | 128.0 | −8.01 | 0.004 |
| pdhD | 721782.1 | 5817.6 | 1717.4 | −1.76 | 0.295 | 250.6 | −4.54 | 0.043 | 47.4 | −6.95 | 0.008 |
| pfl2 | 720932.1 | 11402.1 | 3593.0 | −1.67 | 0.315 | 2135.1 | −2.42 | 0.187 | 1288.9 | −3.14 | 0.113 |
| phsG | 721879.1 | 41481.6 | 17147.4 | −1.27 | 0.413 | 8112.9 | −2.35 | 0.196 | 1380.1 | −4.91 | 0.033 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal with each concentration of S-PRG eluate/signal without S-PRG eluate.
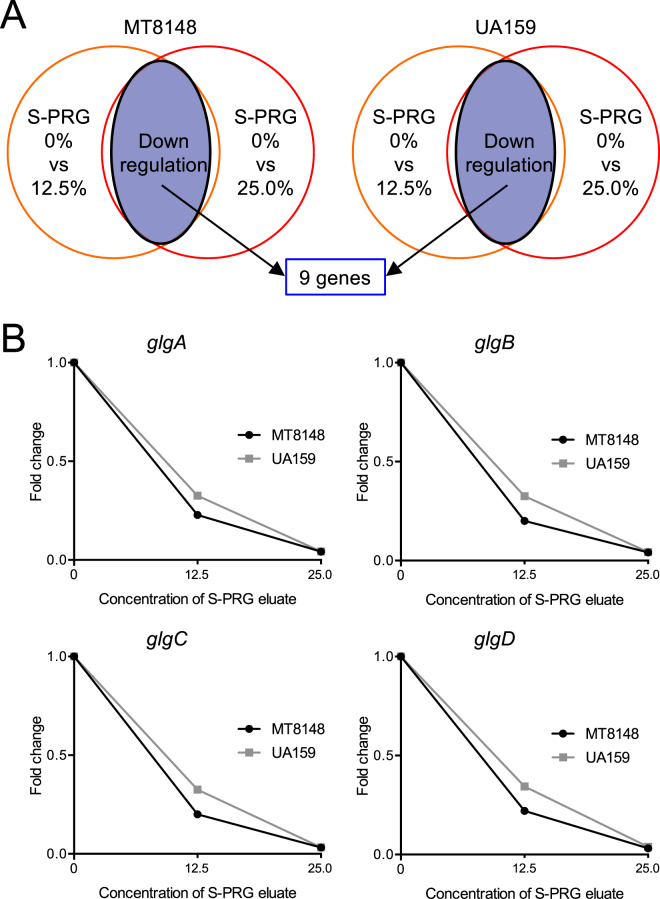
We also performed DNA microarray analyses under two different conditions focusing on S-PRG eluate concentrations of 0% versus 12.5% and 0% versus 25.0% (Fig. 3A), which revealed that nine genes were downregulated in both MT8148 and UA159 (Tables 3, 4). Among the downregulated genes, four genes encoding the putative glycogen synthase, named glgA, glgB, glgC, and glgD19, were downregulated by S-PRG eluate in a concentration-dependent manner (Fig. 3B). These genes are involved in glycogen synthesis and the glycogen produced is used for S. mutans survival under sugar-starved conditions19,20. We further analysed genes prominently regulated under the three conditions at several concentrations of S-PRG eluate: 6.3%, 12.5%, and 25.0%, which were identified in either MT8148 or UA159 (Fig. 4A). Among the 40 genes identified (8 and 32 genes identified in MT8148 and UA159, respectively) (Tables 5, 6), the lac operon (lacA, lacB, lacC, lacD, lacE, lacF, lacG), which is involved in galactose and lactose metabolism in S. mutans21, was prominently downregulated in UA159 in a concentration-dependent manner (Fig. 4B). Although the comY operon (comYA, comYB, comYC, comYD), which is associated with quorum sensing and biofilm formation22,23, was downregulated in UA159, the signals and inhibition of the comY operon were less prominent when compared to other operons.
Figure 3.
Altered expression of key genes in both S. mutans MT8148 and UA159 under two different conditions detected by DNA microarray analysis. (A) Detection of altered genes in the presence of S-PRG eluate. (B) Changes in the expression of genes in the glg operon of MT8148 and UA159 in the presence of S-PRG eluate.
Table 3.
List of downregulated genes of MT8148 in the presence of S-PRG eluate, which were observed in both MT8148 and UA159 under two different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3% | S-PRG12.5% | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| celR | 721935.1 | 34392.8 | 18718.7 | −0.88 | 0.545 | 7975.4 | −2.11 | 0.232 | 11089.6 | −1.63 | 0.323 |
| glgA | 721880.1 | 44037.6 | 25098.8 | −0.81 | 0.570 | 10246.4 | −2.10 | 0.233 | 1907.0 | −4.53 | 0.043 |
| glgB | 721883.1 | 29635.1 | 20348.1 | −0.54 | 0.687 | 6239.4 | −2.25 | 0.211 | 1451.3 | −4.35 | 0.049 |
| glgC | 721882.1 | 45034.7 | 29677.1 | −0.60 | 0.659 | 9715.7 | −2.21 | 0.215 | 1516.4 | −4.89 | 0.034 |
| glgD | 721881.1 | 210544.6 | 147984.5 | −0.51 | 0.703 | 48733.9 | −2.11 | 0.231 | 6864.6 | −4.94 | 0.033 |
| pfl | 720850.1 | 200380.8 | 202526.0 | 0.02 | 1.011 | 36209.8 | −2.47 | 0.181 | 24825.3 | −3.01 | 0.124 |
| ptcA | 721934.1 | 21838.2 | 12188.5 | −0.84 | 0.558 | 4817.3 | −2.18 | 0.221 | 6571.4 | −1.73 | 0.301 |
| ptcC | 721932.1 | 8470.6 | 4527.1 | −0.90 | 0.534 | 2615.5 | −1.70 | 0.309 | 2737.8 | −1.63 | 0.323 |
| wapA | 721382.1 | 3431.3 | 1740.2 | −0.98 | 0.507 | 1588.4 | −1.11 | 0.463 | 1652.3 | −1.05 | 0.482 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal with each concentration of S-PRG eluate/signal without S-PRG eluate.
Table 4.
List of downregulated genes of UA159 in the presence of S-PRG eluate, which were observed in both MT8148 and UA159 under two different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3% | S-PRG12.5% | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| celR | 721935.1 | 11936.0 | 1137.1 | −3.39 | 0.095 | 366.9 | −5.02 | 0.031 | 38.4 | −8.30 | 0.003 |
| glgA | 721880.1 | 30585.2 | 17989.6 | −0.77 | 0.588 | 10161.1 | −1.59 | 0.332 | 1366.7 | −4.48 | 0.045 |
| glgB | 721883.1 | 18468.0 | 16127.7 | −0.20 | 0.873 | 8708.1 | −1.08 | 0.471 | 932.6 | −4.31 | 0.050 |
| glgC | 721882.1 | 30765.2 | 23441.6 | −0.39 | 0.762 | 10508.8 | −1.55 | 0.342 | 1055.7 | −4.87 | 0.034 |
| glgD | 721881.1 | 134094.9 | 101989.3 | −0.39 | 0.761 | 48072.9 | −1.48 | 0.359 | 5395.6 | −4.64 | 0.040 |
| pfl | 720850.1 | 210979.6 | 166145.4 | −0.35 | 0.787 | 69347.0 | −1.61 | 0.329 | 9805.4 | −4.43 | 0.046 |
| ptcA | 721934.1 | 7508.6 | 633.3 | −3.57 | 0.084 | 207.5 | −5.19 | 0.027 | 17.7 | −8.74 | 0.002 |
| ptcC | 721932.1 | 2921.0 | 366.2 | −3.00 | 0.125 | 124.6 | −4.56 | 0.043 | 38.5 | −6.25 | 0.013 |
| wapA | 721382.1 | 3891.4 | 2323.7 | −0.76 | 0.590 | 1931.4 | −1.01 | 0.496 | 568.7 | −2.77 | 0.146 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal with each concentration of S-PRG eluate/signal without S-PRG eluate.
Figure 4.
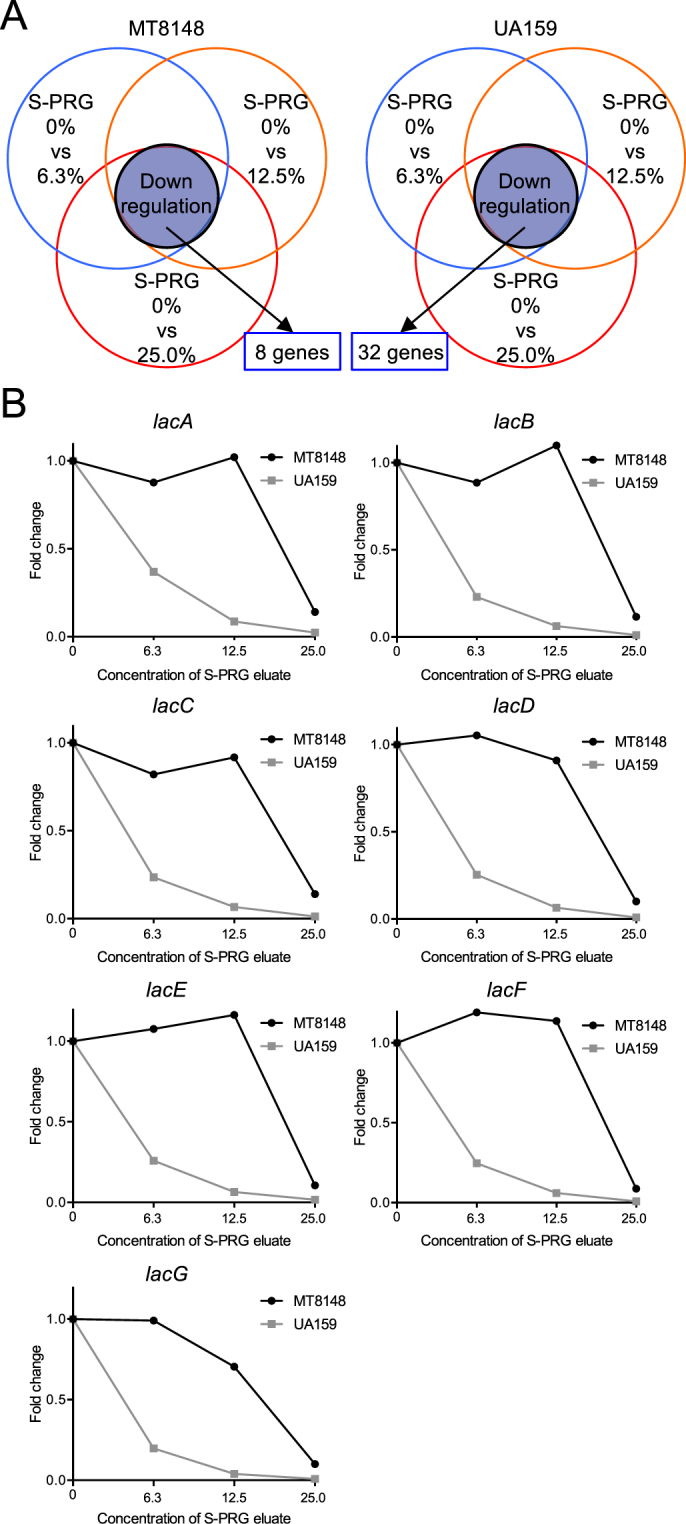
Altered expression of key genes in either S. mutans MT8148 or UA159 under three different conditions using DNA microarray analysis. (A) Detection of altered genes in the presence of S-PRG eluate. (B) Changes in the expression of genes in the lac operon of MT8148 and UA159 in the presence of S-PRG eluate.
Table 5.
List of downregulated genes of MT8148 in the presence of S-PRG eluate, which were observed in either MT8148 or UA159 under three different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3% | S-PRG12.5% | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| bcc | 721410.1 | 3656.2 | 2409.2 | −0.6 | 0.659 | 2386.3 | −0.61 | 0.653 | 1414.7 | −3.81 | 0.071 |
| celR | 721935.1 | 34392.8 | 18718.7 | −0.88 | 0.545 | 7975.4 | −2.11 | 0.232 | 11089.6 | −1.63 | 0.323 |
| cilA | 721415.1 | 2407.2 | 1780.9 | −0.43 | 0.741 | 2047.7 | −0.23 | 0.851 | 1084.1 | −1.15 | 0.451 |
| cilB | 721414.1 | 2040.5 | 1377.5 | −0.57 | 0.675 | 1407.6 | −0.53 | 0.69 | 719.7 | −1.5 | 0.353 |
| cilG | 721413.1 | 1982.2 | 1594.2 | −0.31 | 0.804 | 1700.8 | −0.22 | 0.858 | 719.2 | −1.46 | 0.363 |
| citC | 721405.1 | 350.4 | 534.4 | 0.61 | 1.524 | 757.7 | 1.11 | 2.161 | 165.1 | −1.09 | 0.47 |
| citG | 721406.1 | 471.1 | 806.7 | 0.78 | 1.713 | 1094.5 | 1.22 | 2.324 | 143.2 | −1.72 | 0.303 |
| citG2 | 721416.1 | 3123.7 | 2413.7 | −0.37 | 0.772 | 2832.3 | −0.14 | 0.907 | 1131.6 | −1.47 | 0.362 |
| coaA | 721513.1 | 186.1 | 250 | 0.43 | 1.344 | 329.3 | 0.82 | 1.767 | 79.9 | −1.22 | 0.429 |
| comEA | 721053.1 | 264.2 | 352.6 | 0.42 | 1.335 | 521.8 | 0.98 | 1.975 | 32.2 | −3.04 | 0.122 |
| comYA | 722285.1 | 151.4 | 221.1 | 0.55 | 1.463 | 343.8 | 1.19 | 2.276 | 22.1 | −2.78 | 0.145 |
| comYB | 722284.1 | 253.4 | 374 | 0.56 | 1.479 | 515.5 | 1.03 | 2.042 | 46.4 | −2.44 | 0.184 |
| comYC | 722283.1 | 276.9 | 452.1 | 0.71 | 1.635 | 684 | 1.31 | 2.473 | 69 | −2 | 0.249 |
| comYD | 722282.1 | 147.2 | 263.5 | 0.85 | 1.801 | 402.7 | 1.45 | 2.724 | 43.7 | −1.76 | 0.295 |
| galR | 721293.1 | 1196.1 | 1062.5 | −0.17 | 0.888 | 1628.9 | 0.44 | 1.36 | 347.9 | −1.78 | 0.291 |
| grpE | 720560.1 | 24097.8 | 10856.3 | −1.15 | 0.451 | 10952.4 | −1.14 | 0.454 | 3039.2 | −2.99 | 0.126 |
| hsdM | 721299.1 | 103.1 | 17.6 | −2.55 | 0.171 | 14.2 | −2.86 | 0.138 | 40.4 | −1.35 | 0.392 |
| lacA | 721844.1 | 503.8 | 491.8 | −0.04 | 0.975 | 691.5 | 0.46 | 1.371 | 77.4 | −2.71 | 0.153 |
| lacB | 721843.1 | 1420.2 | 1345.4 | −0.08 | 0.947 | 1602.7 | 0.17 | 1.129 | 174.2 | −3.03 | 0.123 |
| lacC | 721842.1 | 693.3 | 681.8 | −0.02 | 0.984 | 733.5 | 0.08 | 1.06 | 104.1 | −2.73 | 0.15 |
| lacD | 721841.1 | 791.7 | 947.8 | 0.24 | 1.184 | 786.9 | −0.01 | 0.994 | 88.4 | −3.16 | 0.112 |
| lacD2 | 720591.1 | 3352.2 | 1741.7 | −0.94 | 0.52 | 1563.4 | −1.1 | 0.466 | 103.9 | −5.01 | 0.031 |
| lacE | 721839.1 | 1851.5 | 2078.7 | 0.17 | 1.124 | 2250.3 | 0.28 | 1.216 | 210.6 | −3.14 | 0.114 |
| lacF | 721840.1 | 1250.6 | 1567.3 | 0.33 | 1.253 | 1506.8 | 0.27 | 1.205 | 127.4 | −3.3 | 0.102 |
| lacG | 721838.1 | 7506.5 | 7801.4 | 0.06 | 1.039 | 5603.4 | −0.42 | 0.747 | 783.6 | −3.26 | 0.104 |
| lacX | 721837.1 | 2089.4 | 2392.3 | 0.19 | 1.143 | 3630.3 | 0.8 | 1.737 | 211.3 | −3.31 | 0.101 |
| lytR | 721011.1 | 2102.5 | 1188.7 | −0.82 | 0.565 | 1104.9 | −0.93 | 0.526 | 507.7 | −2.05 | 0.241 |
| oadB | 721411.1 | 4735.5 | 3705.5 | −0.36 | 0.782 | 3979.2 | −0.25 | 0.84 | 2741.2 | −0.79 | 0.578 |
| pflC | 720930.1 | 968.8 | 781.5 | −0.31 | 0.806 | 904.2 | −0.1 | 0.933 | 645 | −0.59 | 0.664 |
| ptcA | 721934.1 | 21838.2 | 12188.5 | −0.84 | 0.558 | 4817.3 | −2.18 | 0.221 | 6571.4 | −1.73 | 0.301 |
| ptcB | 721936.1 | 1549.5 | 1220.1 | −0.35 | 0.786 | 1351.8 | −0.2 | 0.872 | 1410.3 | −0.14 | 0.91 |
| ptcC | 721932.1 | 8470.6 | 4527.1 | −0.9 | 0.534 | 2615.5 | −1.7 | 0.309 | 2737.8 | −1.63 | 0.323 |
| pycB | 721417.1 | 3315.9 | 2227.8 | −0.57 | 0.672 | 2493.8 | −0.41 | 0.752 | 2372.2 | −0.48 | 0.715 |
| rgpG | 720710.1 | 107407.9 | 3328.3 | −5.01 | 0.031 | 4296.3 | −4.64 | 0.04 | 4296.3 | −4.64 | 0.04 |
| scnE | 722129.1 | 47 | 23.3 | −1 | 0.5 | 14.1 | −1.74 | 0.3 | 4.2 | −3.47 | 0.09 |
| spaP | 721042.1 | 7581.6 | 3685 | −1.04 | 0.486 | 1858 | −2.03 | 0.245 | 1936 | −1.97 | 0.255 |
| ssb2 | 722266.1 | 673.6 | 980.5 | 0.54 | 1.459 | 1165.4 | 0.79 | 1.733 | 529 | −0.35 | 0.787 |
| trk | 721902.1 | 29059.7 | 13948.7 | −1.06 | 0.48 | 6713.2 | −2.11 | 0.231 | 5386.5 | −2.43 | 0.185 |
| trkB | 721901.1 | 21115.7 | 10376.4 | −1.03 | 0.491 | 4014.8 | −2.4 | 0.19 | 3970.1 | −2.41 | 0.188 |
| wapA | 721382.1 | 3431.3 | 1715.5 | −1 | 0.5 | 1588.4 | −1.11 | 0.463 | 1652.3 | −1.05 | 0.482 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal with each concentration of S-PRG eluate/signal without S-PRG eluate.
Table 6.
List of downregulated genes of UA159 in the presence of S-PRG eluate, which were observed in either MT8148 or UA159 under three different conditions using DNA microarray analysis.
| Gene | Accession number | S-PRG 0% | S-PRG 6.3% | S-PRG12.5% | S-PRG 25.0% | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Signal | Signal | Log2* | Change# | Signal | Log2* | Change# | Signal | Log2* | Change# | ||
| bcc | 721410.1 | 5980.6 | 1943.9 | −1.62 | 0.325 | 1876.8 | −1.67 | 0.314 | 1108.3 | −2.43 | 0.185 |
| celR | 721935.1 | 11936 | 1137.1 | −3.4 | 0.095 | 366.9 | −5.02 | 0.031 | 38.4 | −8.3 | 0.003 |
| cilA | 721415.1 | 5421.4 | 1804.4 | −1.59 | 0.332 | 1879 | −1.53 | 0.347 | 726.1 | −2.9 | 0.134 |
| cilB | 721414.1 | 3708.8 | 1459.6 | −1.34 | 0.394 | 1488.8 | −1.32 | 0.402 | 712.4 | −2.38 | 0.192 |
| cilG | 721413.1 | 4245.9 | 1734 | −1.29 | 0.408 | 1999.6 | −1.09 | 0.471 | 1059.9 | −2 | 0.249 |
| citC | 721405.1 | 1927.9 | 218.7 | −3.15 | 0.113 | 121.6 | −3.99 | 0.063 | 164.4 | −3.55 | 0.085 |
| citG | 721406.1 | 1543.6 | 282.1 | −2.45 | 0.183 | 138.2 | −3.48 | 0.089 | 183.1 | −3.1 | 0.117 |
| citG2 | 721416.1 | 5644.4 | 1906.9 | −1.56 | 0.338 | 1774.5 | −1.67 | 0.314 | 694.5 | −3.02 | 0.123 |
| coaA | 721513.1 | 126.2 | 39.1 | −1.69 | 0.31 | 49.8 | −1.34 | 0.395 | 54.6 | −1.21 | 0.433 |
| comEA | 721053.1 | 135.8 | 67.9 | −1 | 0.5 | 29.2 | −2.22 | 0.215 | 29.5 | −2.21 | 0.217 |
| comYA | 722285.1 | 231.2 | 37.6 | −2.65 | 0.159 | 17.9 | −3.74 | 0.075 | 28.8 | −3.02 | 0.124 |
| comYB | 722284.1 | 606.2 | 90 | −2.76 | 0.148 | 42 | −3.87 | 0.069 | 69 | −3.14 | 0.113 |
| comYC | 722283.1 | 903.7 | 109.7 | −3.05 | 0.121 | 45.5 | −4.34 | 0.049 | 102.2 | −3.14 | 0.113 |
| comYD | 722282.1 | 750.9 | 89.6 | −3.13 | 0.114 | 38 | −4.34 | 0.05 | 70.7 | −3.43 | 0.093 |
| galR | 721293.1 | 998.6 | 467 | −1.1 | 0.468 | 222.2 | −2.17 | 0.222 | 396.7 | −1.33 | 0.397 |
| grpE | 720560.1 | 40392.6 | 24381 | −0.72 | 0.603 | 15498.9 | −1.38 | 0.384 | 23261.7 | −0.8 | 0.576 |
| hsdM | 721299.1 | 236.4 | 230.2 | −0.04 | 0.974 | 233.2 | −0.02 | 0.986 | 693.4 | 1.55 | 2.933 |
| lacA | 721844.1 | 1232.9 | 487.2 | −1.34 | 0.395 | 115.2 | −3.42 | 0.093 | 34 | −5.19 | 0.027 |
| lacB | 721843.1 | 3958.7 | 974.1 | −2.02 | 0.246 | 263.3 | −3.91 | 0.066 | 59.5 | −6.09 | 0.015 |
| lacC | 721842.1 | 2894.5 | 707.6 | −2.03 | 0.245 | 203.3 | −3.83 | 0.07 | 44.2 | −6.07 | 0.015 |
| lacD | 721841.1 | 2529.6 | 976.9 | −1.37 | 0.386 | 173.4 | −3.88 | 0.068 | 25.2 | −6.65 | 0.01 |
| lacD2 | 720591.1 | 1990.2 | 422.6 | −2.24 | 0.212 | 162.4 | −3.61 | 0.082 | 39.2 | −5.67 | 0.02 |
| lacE | 721839.1 | 4352.6 | 1160.3 | −1.91 | 0.267 | 303.3 | −3.85 | 0.07 | 71.2 | −5.93 | 0.016 |
| lacF | 721840.1 | 3438.8 | 936 | −1.88 | 0.272 | 227.8 | −3.92 | 0.066 | 38.3 | −6.54 | 0.011 |
| lacG | 721838.1 | 30256.7 | 6224.5 | −2.28 | 0.206 | 1223.1 | −4.63 | 0.04 | 270.5 | −6.81 | 0.009 |
| lacX | 721837.1 | 2139.1 | 804.4 | −1.41 | 0.376 | 251.6 | −3.09 | 0.118 | 84.2 | −4.67 | 0.039 |
| lytR | 721011.1 | 1717.4 | 772.8 | −1.15 | 0.45 | 841.5 | −1.03 | 0.49 | 419.1 | −2.04 | 0.244 |
| oadB | 721411.1 | 11315.4 | 4350.1 | −1.38 | 0.384 | 4811.2 | −1.23 | 0.425 | 2660.8 | −2.09 | 0.235 |
| pflC | 720930.1 | 1164.2 | 478.1 | −1.28 | 0.411 | 391.9 | −1.57 | 0.337 | 544.7 | −1.1 | 0.468 |
| ptcA | 721934.1 | 7508.6 | 633.3 | −3.57 | 0.084 | 207.5 | −5.19 | 0.027 | 17.7 | −8.74 | 0.002 |
| ptcB | 721936.1 | 615.6 | 239.6 | −1.37 | 0.388 | 103.5 | −2.58 | 0.167 | 21.7 | −4.84 | 0.035 |
| ptcC | 721932.1 | 2921 | 366.2 | −3 | 0.125 | 124.6 | −4.56 | 0.043 | 38.5 | −6.25 | 0.013 |
| pycB | 721417.1 | 22423.4 | 5598.4 | −2 | 0.25 | 4989.7 | −2.17 | 0.223 | 2210.2 | −3.34 | 0.099 |
| rgpG | 720710.1 | 1621.9 | 1575.5 | −0.04 | 0.971 | 1983.1 | 0.29 | 1.223 | 4685.6 | 1.53 | 2.889 |
| scnE | 722129.1 | 324.3 | 412.9 | 0.35 | 1.273 | 291.9 | −0.15 | 0.9 | 284 | −0.19 | 0.876 |
| spaP | 721042.1 | 3168.9 | 3842.9 | 0.28 | 1.212 | 2127.1 | −0.58 | 0.671 | 2296.4 | −0.47 | 0.724 |
| ssb2 | 722266.1 | 769.7 | 369.5 | −1.06 | 0.48 | 271.3 | −1.51 | 0.352 | 308.6 | −1.32 | 0.401 |
| trk | 721902.1 | 14957.3 | 11522.8 | −0.38 | 0.77 | 10018.8 | −0.58 | 0.67 | 3609 | −2.05 | 0.241 |
| trkB | 721901.1 | 10436.8 | 7856.4 | −0.41 | 0.753 | 7320 | −0.51 | 0.701 | 2494.6 | −2.06 | 0.239 |
| wapA | 721382.1 | 3891.4 | 2323.7 | −0.74 | 0.597 | 1931.4 | −1.01 | 0.496 | 568.7 | −2.78 | 0.146 |
*Log2 means Log2 ratio. #Change means Fold change. The Log2 ratio and Fold change were calculated by signal with each concentration of S-PRG eluate/signal without S-PRG eluate.
No genes were upregulated in both MT8148 and UA159 under all three eluate conditions tested. DNA microarray analysis employing two different eluate concentrations (Supplemental Fig. 1A) showed only two genes were upregulated with lower fold changes in both MT8148 and UA159 (Supplemental Tables 1 and 2). We further identified six genes in either MT8148 or UA159 under the three different conditions tested (Supplemental Fig. 1B). However, these changes in expressions for all of the genes were relatively small (Supplemental Tables 3 and 4).
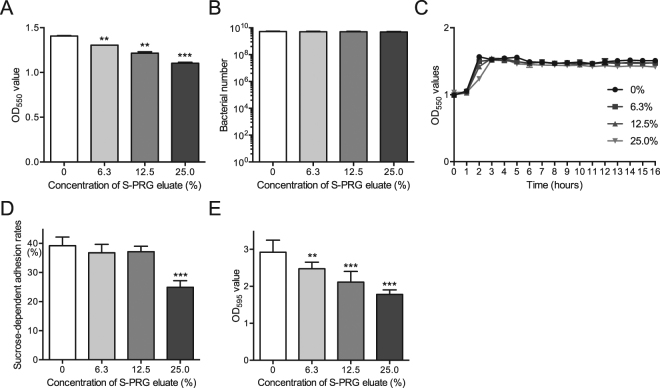
Inhibitory effects of S-PRG eluate in in vitro sucrose-dependent adhesion
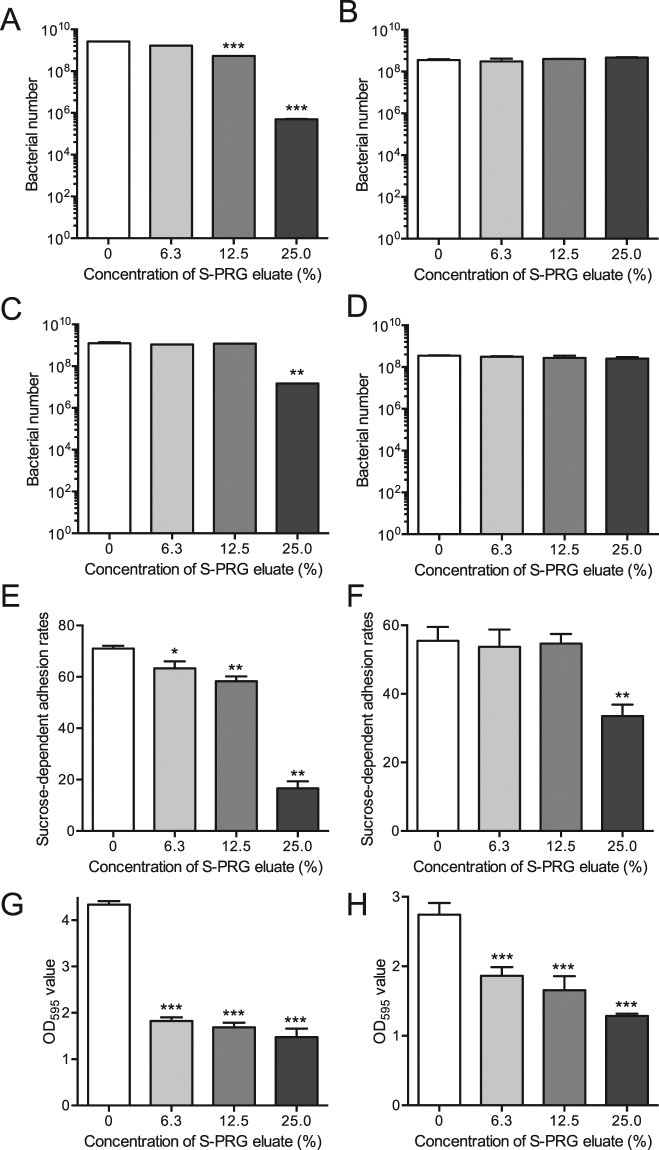
Bacterial suspensions were adjusted in BHI broth containing 1% sucrose to a final concentration of 1.0 × 107 CFU/ml S. mutans with or without S-PRG eluate. The bacterial suspensions were then cultured at 37 °C for 18 h at a 30° angle and sucrose-dependent adhesion analysis was performed as previously described24. Before the analysis, we confirmed that there were no differences in the total bacterial numbers tested (adhesive cells and non-adhesive cells) in the cultured bacteria among various concentration of S-PRG eluate (Fig. 5A,B). S-PRG eluate significantly inhibited sucrose-dependent adhesion of S. mutans in a concentration-dependent manner (P < 0.05) (Fig. 5C).
Figure 5.
Sucrose-dependent adhesion of S. mutans MT8148 in the presence of various concentrations of S-PRG eluate. Bacterial growth for cells used in the sucrose-dependent adhesion assay was determined by OD550 values (A) and recovered bacterial numbers on MSB plates (B). (C) Sucrose-dependent adhesion rates. Significant differences were determined using ANOVA with Bonferroni correction. *P < 0.05, **P < 0.01, and ***P < 0.001 versus no S-PRG eluate.
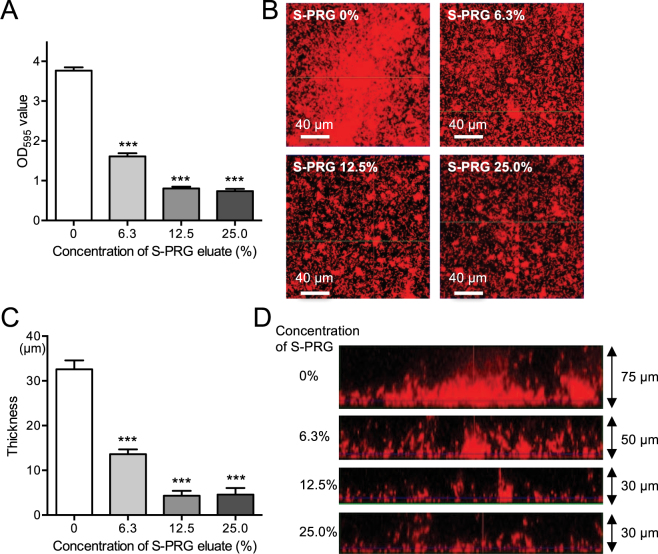
Inhibitory effects of S-PRG eluate on biofilm formation
Bacterial suspensions were adjusted in BHI broth containing 0%, 0.25% and 1% sucrose to a final concentration of 1.0 × 107 CFU/ml S. mutans in the presence or absence of S-PRG eluate. The bacterial suspensions were then added to saliva coated 96-well polystyrene microtiter plates. After incubation at 37 °C for 24 h, biofilms were quantified following staining with crystal violet and the structures of the biofilms were observed by confocal laser scanning microscopy. The quantity of formed biofilms was similar in BHI broth containing 0.25% and 1% sucrose, which were drastically reduced even at a low concentration of S-PRG eluate (Fig. 6A,B, Supplemental Fig. 2A). Both biofilm density and thickness were significantly reduced in the presence of S-PRG eluate, with significant differences (P < 0.001) (Fig. 6C,D). On the other hand, only weak biofilms were observed in the absence of sucrose regardless of the presence or absence of S-PRG eluate (Supplemental Fig. 2B).
Figure 6.
Biofilm formation by S. mutans MT8148 grown in BHI with 0.25% sucrose in the presence of various concentrations of S-PRG eluate. (A) Quantity of biofilm formation. (B) Representative images of formed biofilms using confocal scanning laser microscopy. (C) Biofilm thickness. (D) Representative images of biofilm thickness using confocal scanning laser microscopy. Significant differences were determined using ANOVA with Bonferroni correction. ***P < 0.001 versus no S-PRG eluate.
Inhibitory effects of S-PRG eluate on cellular hydrophobicity
Hydrophobic interactions involving S. mutans and tooth surfaces play a major role in the sucrose-independent initial adhesion of S. mutans to teeth25. We determined whether the S-PRG eluate inhibited the hydrophobic properties of S. mutans using an in vitro cellular hydrophobicity assay, which was assessed by the hydrophobic interaction of S. mutans and n-hexadecane26. A mixture of each concentration of S-PRG eluate and bacterial cultures adjusted to OD550 of 0.6 was vigorously vortexed for 1 min in the presence of n-hexadecane to induce hydrophobic interaction between n-hexadecane and the test strain. Cellular hydrophobicity rates were assessed via n-hexadecane partitioning by S. mutans in the presence of various concentrations of S-PRG eluate and showed no significant differences (Fig. 7A). Next, to examine the effects of incubation time on such interactions, we further analysed cellular hydrophobicity using S. mutans cells pretreated with S-PRG eluate for 18 h. The S-PRG eluate did not affect bacterial numbers (Fig. 7B), and significantly inhibited cellular hydrophobicity (P < 0.05, P < 0.01) (Fig. 7C).
Figure 7.
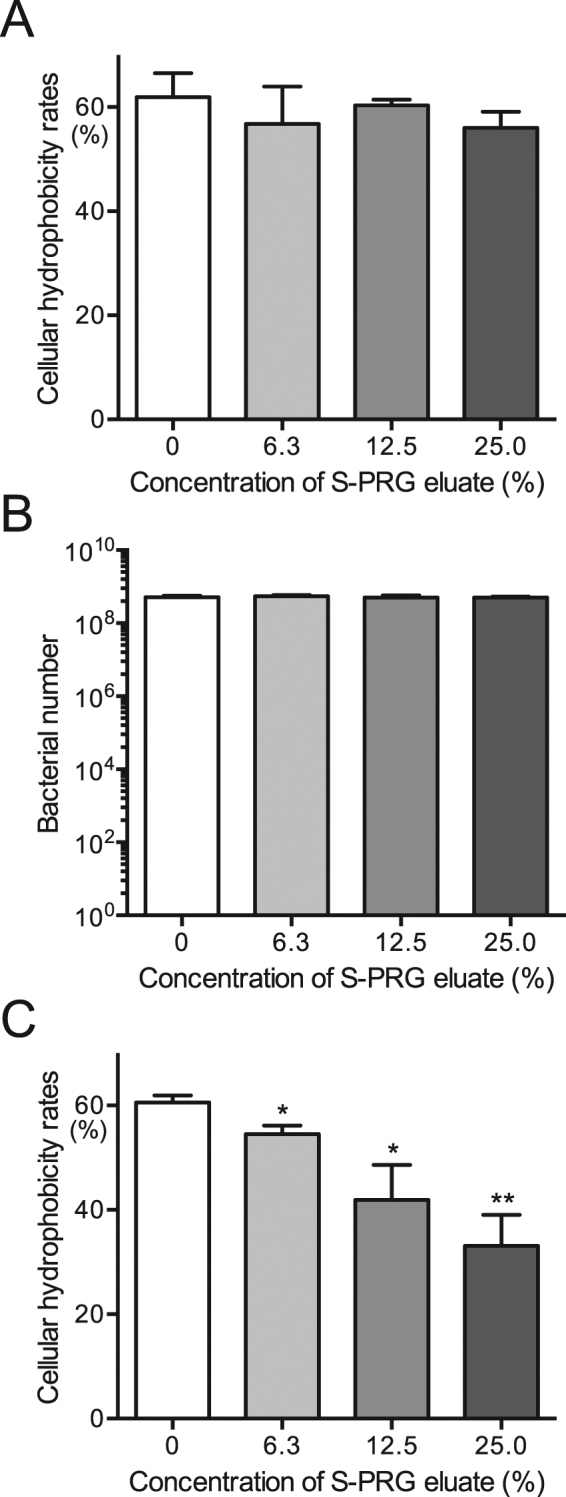
Cellular hydrophobicity of S. mutans MT8148 in the presence of various concentrations of S-PRG eluate. (A) Cellular hydrophobicity in the presence of S-PRG eluate. (B) Bacterial numbers used in the cellular hydrophobicity assay (C) Cellular hydrophobicity following the addition of S-PRG eluate 18 h before the assay. Significant differences were determined using ANOVA with Bonferroni correction. *P < 0.05 and **P < 0.01 versus no S-PRG eluate.
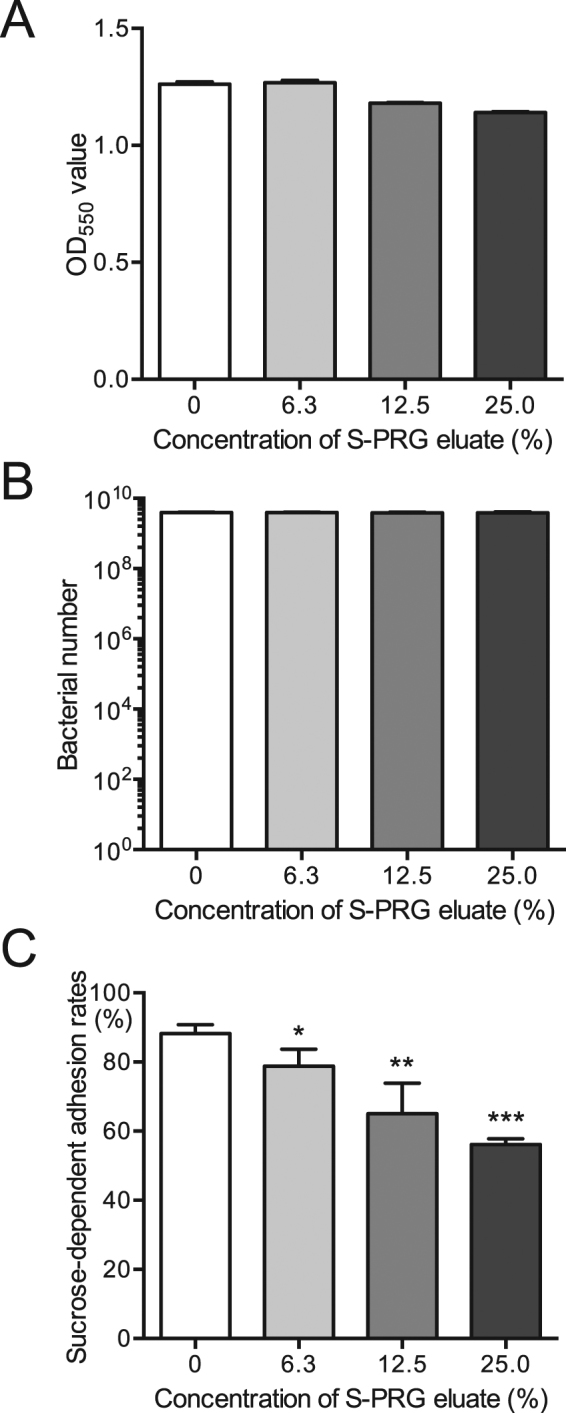
Inhibitory effects of S-PRG eluate against S. mutans in post-logarithmic phase
Bacteria in the stationary phase exhibit low sugar metabolic activity17. Thus, we investigated the effects of S-PRG eluate on bacterial growth, sucrose-dependent adhesion and biofilm formation using post-logarithmic phase 1.0 × 109 CFU/ml of S. mutans. Bacterial numbers were not affected by S-PRG eluate after 18-h incubation although the OD550 densities were lower at the higher concentrations of S-PRG eluate in a dose dependent manner (Fig. 8A,B). In addition, bacterial growth in the presence of each S-PRG eluate was monitored and reached the stationary phase within 3-h after incubation (Fig. 8C). Although sucrose-dependent adhesion and biofilm formation by S. mutans in the post-logarithmic phase were inhibited in the presence of S-PRG eluate, the inhibitory effects were not as prominent as those using 1.0 × 107 CFU/ml of S. mutans prior to the logarithmic growth phase (Fig. 8D,E).
Figure 8.
In vitro properties of S. mutans MT8148 in the late logarithmic phase in the presence of various concentrations of S-PRG eluate. (A, B) Bacterial growth by adding 1 × 109 CFU/ml of S. mutans with 18-h incubation, which was determined by OD550 values in BHI broth (A) and recovered bacterial numbers on MSB plates (B). (C) Bacterial growth by adding 1 × 109 CFU/ml of S. mutans at multiple time points, which was determined by OD550 values in BHI broth. (D) Sucrose-dependent adhesion rates. (E) Quantitation of biofilm formation. Significant differences were determined using ANOVA with Bonferroni correction. *P < 0.05, **P < 0.01, and ***P < 0.001 versus no S-PRG eluate.
Inhibitory effects of S-PRG eluate against other streptococci
S-PRG eluate prominently repressed the expression of genes encoding the pyruvate dehydrogenase (PDH) complex, and the complex is known to be important for bacterial growth and survival27. Thus, we analyzed the inhibitory effect of S-PRG eluate against other oral streptococci using two different types of oral streptococcal species; Streptococcus sobrinus B13 which is a S. mutans-related species with sucrose-dependent cariogenic properties28, and Streptococcus gordonii ATCC10558 lacking sucrose-dependent cariogenic properties29. S. sobrinus before reaching the logarithmic phase (adjusted to 1 × 105 CFU/ml) was inhibited by S-PRG eluate in a concentration-dependent manner (Fig. 9A), whereas inhibition was not observed using the bacteria in the post-logarithmic phase (adjusted to 1 × 109 CFU/ml) (Fig. 9B), similar to what was observed with S. mutans. The inhibitory effect on S. gordonii in the presence of S-PRG eluate was observed only with the bacterium before reaching the logarithmic growth phase (adjusted to 1 × 105 CFU/ml), though the inhibitory effect was lower than that observed in S. mutans and S. sobrinus (Fig. 9C,D). In addition, the S-PRG eluate significantly inhibited sucrose-dependent adhesion and biofilm formation by S. sobrinus before reaching the logarithmic growth phase (adjusted to 1 × 107 CFU/ml) (P < 0.05) (Fig. 9E–H).
Figure 9.
The in vitro properties of Streptococcus sobrinus B13 and Streptococcus gordonii ATCC10558 in the presence of various concentrations of S-PRG eluate. Bacterial growth using S. sobrinus before reaching the logarithmic phase (adjusted to 1 × 105 CFU/ml) (A) and in the post-logarithmic phase (adjusted to 1 × 109 CFU/ml) (B) following 18 h incubation, which was determined by recovered bacterial numbers on MSB plates. Bacterial growth using S. gordonii reaching the logarithmic phase (adjusted to 1 × 105 CFU/ml) (C) and in the post-logarithmic phase (adjusted to 1 × 109 CFU/ml) (D) with 18 h incubations, which was determined by recovered bacterial numbers on MSB plates. Sucrose-dependent adhesion rates using bacterial suspensions of S. sobrinus B13 adjusted before reaching the logarithmic phase (adjusted to 1 × 107 CFU/ml) (E) and in the post-logarithmic phase (adjusted to 1 × 109 CFU/ml) (F), respectively. Quantitation of biofilm formation using bacterial suspensions of S. sobrinus B13 adjusted to 1 × 107 CFU/ml (G) and 1 × 109 CFU/ml (H), respectively. Significant differences were determined using ANOVA with Bonferroni correction. *P < 0.05, **P < 0.01, and ***P < 0.001 versus no S-PRG eluate.
Discussion
Recent studies showed that a S-PRG filler could inhibit the growth and adherence of S. mutans12,13. However, these studies were performed for the purpose of developing dental products containing S-PRG filler, and not necessarily for antimicrobial activity. In the present study, we analysed the inhibitory effects of S-PRG eluate against S. mutans, focusing on the cariogenic properties and virulence genes of the bacterium, which are closely related to dental caries development.
S-PRG filler has been widely used in dental products, mainly contained in sealant and composite resins7,8. Although the effects of the presence of each ion component may be different from that in the presence of the other divalent and trivalent ions, releases of all ions from the S-PRG filler in composite resin could be confirmed even after overnight incubation13. In addition, the released ions were also identified in whole saliva12. Thus, inhibitory effects similar to those of S-PRG eluate observed in the present study may be expected for dental materials containing S-PRG filler, although the exact amounts of the released ions may be different for each clinical situation.
Multiple ions may affect the biological activity for many bacteria30. On the other hand, some of the bacteria such as Psuedomonas aeruginosa have developed a variety of mechanisms to escape the inhibitory effects of the ions. For example, P. aeruginosa can immobilize metal ions outside of the cell, reducing the inhibitory effect of the ions31. Conversely, the existence of multiple metal ions sometimes contributes to supporting biofilm formation by bacteria30,32. In the present study, a S- PRG eluate could effectively inhibit the growth and in vitro cariogenicity of S. mutans. In addition, a previous study showed that all of six ions released from a S- PRG filler could inhibit bacterial growth13. However, it was reported that some of the proteins of S. mutans encodes metal ion transport systems, which are considered as virulence factors for dental caries and infective endocarditis33,34. Therefore, further studies should be performed focusing on how the inhibitory effects against S. mutans induced by these ions alter bacterial virulence.
One of the most important factors for growth and survival of S. mutans is sugar metabolism. The sugar metabolism induced by S. mutans occurs through the Embden-Meyerhof-Parnas pathway, followed by acid production from pyruvate16. Inhibition of S. mutans growth in the presence of S-PRG eluate may result from interference with the Embden-Meyerhof-Parnas pathway or subsequent acid production pathways. To identify the key S. mutans genes closely related to the inhibition of growth and survival of the bacterium in the presence of S-PRG eluate, we performed DNA microarray analysis.
Based upon the results of the microarray analysis, S-PRG eluate appeared to strongly downregulate several S. mutans genes and the downregulation of some genes in the presence of S-PRG eluate was over 100-fold greater than in the absence of the eluate. Conversely, no gene was upregulated by more than 10-fold in the presence of S-PRG eluate compared with that in the absence of S-PRG eluate. These results indicated that the inhibitory effects on bacterial growth and survival of S. mutans in the presence of S-PRG eluate were primarily induced by downregulation of several S. mutans genes. Interestingly, three operons (pdh, glg, and lac), all of which play important roles in the survival and sugar metabolism of S. mutans, were downregulated in the presence of the S-PRG eluate in a concentration-dependent manner. Thus, these operons may be responsible for the inhibition of bacterial growth and survival observed in the presence of the S-PRG eluate.
Among the genes downregulated by the S-PRG eluate in the DNA microarray analysis, the PDH complex was prominently negatively regulated. The PDH complex converts pyruvate produced following sugar metabolism by S. mutans via the Embden-Meyerhof-Parnas pathway to acetyl-CoA and CO235. The PDH complex is encoded by an operon consisting of pdhD, pdhA, pdhB, and pdhC genes, and a pdhD defective mutant strain showed extremely reduced survival as compared with the parent strain15. Thus, S-PRG eluate likely affects the pdhD gene, resulting in reduced bacterial survival. In addition, the glg operon (glgB, glgC, glgD, glgA, glgP), which is involved in glycogen synthesis from glucose-1-phosphate via ADP-glucose21,35, was downregulated in the presence of high concentrations of S-PRG eluate. Although strong inhibition was only observed in UA159, the lac operon (lacA, lacB, lacC, lacD, lacF, lacE, lacG), which is involved in galactose and lactose metabolic pathways21, was also downregulated.
The S-PRG eluate could inhibit several in vitro properties related to the development of dental caries in S. mutans before the active growth phase. When we further analysed the inhibitory effects of S-PRG eluate against S. mutans in the post-logarithmic phase, the effects of the S-PRG eluate were less pronounced. These results indicated that a major role for the S-PRG eluate is the inhibition of S. mutans virulence and growth, which occurs before the active growth phase. Therefore, the inhibition of several in vitro properties of S. mutans before the growth phase in the presence of S-PRG eluate may be result from downregulation of genes such as those in the pdh operon and glg operons.
Another possible explanation for the inhibitory effects of S-PRG eluate on the in vitro cariogenic properties of S. mutans is that the S-PRG eluate may affect other S. mutans virulence genes since DNA microarray assays were performed under fixed incubation conditions. However, we did test various S-PRG eluate concentrations as well as different S. mutans strains. Therefore, further molecular biological analyses should be performed focusing on the inhibitory mechanisms of S-PRG eluate against other relevant in vitro cariogenic properties of S. mutans.
In the microarray analysis, S-PRG eluate most prominently repressed the expression of genes encoding the pyruvate dehydrogenase (PDH) complex, and the complex is known to be important for the bacterial growth and survival27. The PDH complex is also present in various bacterial species such as Mycobacterium tuberculosis and Escherichia coli36,37. Therefore, we thought that other oral streptococcal species may be growth inhibited by the S-PRG eluate prior to their active growth phase rather than following logarithmic growth phase. Based on this hypothesis, we have added the results of bacterial growth with or without S-PRG eluate using Streptococcus sobrinus (similar cariogenic bacteria to S. mutans) and Streptococcus gordonii (non-cariogenic bacteria), both of which were growth inhibited in the presence of S-PRG eluate, especially before the growth phase. On the other hand, some of the cariogenic properties such as sucrose-dependent colonization are specific for S. mutans and S. sobrinus. Thus, we analyzed the inhibitory effects on several in-vitro cariogenic properties of S. sobrinus in the presence of S-PRG eluate, which showed that the S-PRG eluate inhibited the cariogenic properties of S. sobrinus most prominently prior to active growth phase rather after logarithmic growth similar to that observed with S. mutans. These results may indicate that S-PRG eluate can inhibit S. mutans as well as various oral streptococci in a similar manner.
In summary, S-PRG eluates inhibited the bacterial growth of S. mutans and downregulated genes involved in sugar metabolism. In addition, the S-PRG eluate clearly inhibited several in vitro cariogenic properties of S. mutans. Taken together, our results suggest that the S-PRG eluate may be a useful tool for reducing dental caries due to its inhibitory effects on the bacterial growth and the sugar-dependent cariogenic properties of S. mutans.
Methods
S-PRG eluate preparations
S-PRG eluate was prepared as described previously38, and was provided by Shofu Inc. (Kyoto, Japan). Briefly, S-PRG filler was mixed with an equal amount of distilled water and mixed gently at room temperature for 24 h, followed by centrifugation at 3,000 × g, 23 °C for 6 h to separate the filler and the liquid. The supernatant was then filtered (pore size: 0.45μm) to remove any residual insoluble material and the resulting filtrate was used as the S-PRG eluate. The concentrations of ions released from S-PRG filler except for F− (i.e., Al3+, BO33−, Na+, SiO32−, and Sr2+) were measured by using an emission spectrophotometer (ICPS-8000, Shimadzu Co., Kyoto, Japan). In addition, the concentration of F− was confirmed with a F− electrode (Model 9609BNWP, Orion Research Inc., Beverly, MA, USA) using an ion selective electrode meter (Model 720 A, Orion Research Inc.). The ion concentrations of the S-PRG eluate were as follows: Al3+ = 19.6 ppm, BO33− = 1,656.5 ppm, Na+ = 618.5 ppm, SiO32− = 13.9 ppm, Sr2+ = 126.8 ppm, and F− = 141.0 ppm. The S-PRG eluate was diluted with brain heart infusion (BHI) broth (Difco Laboratories) at concentration of 25%, 12.5%, 6.3%, and 0%, respectively, and was used in subsequent studies.
Bacterial strains and growth condition
S. mutans strains MT8148 and UA159 were selected from the stock culture collection in our laboratory39–41. In addition, S. sobrinus B13 and S. gordonii ATCC10558 were also used28,29. Strains were confirmed to be S. mutans, S. sobrinus and S. gordonii based on their biochemical properties and observation of colony morphologies on MS with and without bacitracin agar plates. These strains were cultured in BHI broth at 37 °C for 18 h and used in subsequent studies.
Growth inhibition assay
The growth inhibition assays were performed according to methods described previously with some modification42. Cells from overnight cultures of S. mutans MT8148 grown in BHI broth were collected by centrifugation at 3,000 × g for 10 min. We confirmed that bacterial suspensions adjusted in BHI broth to OD550 = 1.0 were equal to 1 × 109 CFU/ml by adding serial dilutions of the bacterial suspensions onto MSB agar plates. Using the suspensions, bacterial dilutions with final concentrations ranging from 1.0 × 103 to 1.0 × 108 CFU/ml with or without S-PRG eluate were prepared. After 18-h incubation at 37 °C, bacterial growth was measured at OD550 and bacterial suspensions were then streaked onto MSB plates supplemented with 15% (w/v) sucrose and 0.2 U/ml of bacitracin and incubated anaerobically at 37 °C for 48 h. The numbers of colonies were counted after identifying the characteristic colony morphology of mutans streptococci. As for S. sobrinus B13 and S. gordonii ATCC10558, the inhibitory effects of S-PRG eluate on bacterial suspensions with concentrations of 1.0 × 105 CFU/ml were also analysed, since S. mutans was significantly inhibited by S-PRG eluate at this density. In addition, to monitor the kinetics of bacterial growth for MT8148, OD550 values of the bacterial suspensions were adjusted 1.0 × 107 CFU/ml with or without S-PRG eluate and monitored every 1 hour until the bacterial growth reached the stationary phase. In addition, survival of 1.0 × 107 CFU/ml of the bacteria added to BHI in the presence of each concentration of S-PRG eluate was monitored for 2 weeks by adding aliquots of the bacterial suspensions to MSB plates every 48 hours.
The growth of S. mutans MT8148 in late logarithmic phase was determined by a previously described method43. S. mutans MT8148, which was grown for 24 h to reach stationary phase, was adjusted to a final concentration of 1.0 × 109 CFU/ml in BHI broth with or without S-PRG eluate. After a further 18-h incubation at 37 °C, bacterial growth was measured at OD550 and bacterial suspensions were streaked onto MSB plates, followed by colony counting as described above. In addition, to monitor the kinetics of bacterial growth, the bacterial suspensions adjusted to a final concentration of 1.0 × 109 CFU/ml with or without S-PRG eluate were cultured at 37 °C and OD550 values of the bacterial suspensions were measured every hour until the bacteria entered the stationary phase. As for S. sobrinus B13 and S. gordonii ATCC10558, 1.0 × 109 CFU/ml of the bacteria were incubated with S-PRG eluate at 37 °C for 18 h and the bacterial suspensions were then streaked onto MSB plates, followed by colony counting as described above.
DNA microarray assays
A systematic analysis of gene alterations in S. mutans was performed using DNA microarrays to identify S. mutans gene expression affected by the S-PRG eluate, as described previously44. Briefly, 1.0 × 107 CFU/ml of S. mutans MT8148 and UA159 in BHI broth were cultured with each specified concentration of S-PRG eluate at 37 °C for 18 h. Amino-allyl amplified RNA was then obtained from total RNA using the Amino-allyl MessageAmp aRNA kit (Ambion, Inc., Austin, TX, USA). The purity, concentration, and quality of the RNA samples were confirmed with a NanoDrop One (Thermo Fisher Scientific) and Agilent 2100 bioanalyser (Agilent Technologies, Inc., Santa Clara, CA, USA). The purity and quality of RNA were assessed by nucleic acid absorbance at A260/A230 and A260/A280 of each sample and were more than 2.0, respectively. The concentrations of RNA of these samples ranged from 50 to 440 ng/μl. All samples were used for microarray assays after adjustment to a minimum concentration (50 ng/μl). The microarray assays were carried out by Takara Bio. Inc. (Otsu, Japan) using products for microarray analysis manufactured by Agilent Technologies, according to the manufacturer’s protocols. Briefly, Cy3 complimentary RNA was labelled with a Low Input Quick Amp Labeling Kit, One-Color (Agilent Technologies), and the Cy3-Labeled complimentary RNA was hybridized with the complete genome of S. mutans UA159 assembled with the Agilent Expression Array kit. After washing with Gene Expression Wash Buffers Pack (Agilent Technologies), the hybridization images were analysed using an Agilent Microarray Scanner (G2565CA) (Agilent Technologies). Quantitative data were obtained using Agilent Feature Extraction software (Agilent Technologies) and corrections for background signal intensity for the data were performed using a method, previously described45. Altered genes were identified using three different comparisons for each S. mutans strain and focused on the following S-PRG eluate concentrations: 0% versus 6.3%, 0% versus 12.5%, and 0% versus 25.0%. In addition, altered genes using two different comparisons for each S. mutans strain were also determined using S-PRG eluate concentrations of 0% versus 12.5% and 0% versus 25.0%. We selected genes with changes of increase or decrease greater than 1.0 of Log2 ratio in presence or absence of the indicated concentrations of eluate.
Sucrose-dependent adhesion
Sucrose-dependent adhesion to a glass surface was analysed as previously described with some modification24. Cells from overnight cultures of S. mutans MT8148 and S. sobrinus B13 were collected by centrifugation at 3,000 × g for 10 min, respectively. Cultures were adjusted in BHI broth containing 1% sucrose to a final concentration of 1.0 × 107 CFU/ml with or without S-PRG eluate. Bacterial suspensions were then cultured at 37 °C for 18 h at a 30° angle. After incubation, both adhesive and non-adhesive cells were measured by OD550 values and were streaked onto MSB plates to confirm the bacterial number in the culture tubes. In addition, culture tubes were also prepared other than those for the confirmation of the bacterial numbers were used for the sucrose-dependent adhesion assay. The culture tubes were vigorously vibrated with a vortex mixer for 3 s and non-adhesive cells were transferred to fresh tubes. Cells remaining on the glass surface (adhesive cells) were removed using a rubber scraper and suspended in 3 ml of water. Both adhesive and non-adhesive cells were dispersed by ultrasonication, and the cell masses were determined by densitometry at OD550. Total cells were calculated as OD550 (adhesive cells + non-adhesive cells), and the percent adherence was calculated as 100 × OD550 (adhesive cells)/OD550 (total cells).
Sucrose-dependent adhesion of S. mutans MT8148 and S. sobrinus B13 in the late logarithmic phase was determined according to a method described previously43. These bacteria were grown for 24 h to reach the stationary phase and were adjusted to a final concentration of 1.0 × 109 CFU/ml in BHI broth containing 1% sucrose with or without S-PRG eluate. Aliquots were cultured at 37 °C for 18 h at a 30° angle. Adherence rates were calculated as described above.
Biofilm assay
The quantity of formed biofilms was assessed as previously described with some modification46,47. Human saliva collected from a healthy volunteer was centrifuged at 12,000 × g for 10 min and the supernatant was filtered (pore size: 0.45 μm). The supernatant was diluted 1:3 with Milli-Q water to produce 25% saliva and coated 96-well polystyrene microtiter plates for 2 h. Cells from overnight cultures of S. mutans MT8148 or S. sobrinus B13 were collected by centrifugation at 3,000 × g for 10 min. Cultures were adjusted to 1.0 × 107 CFU/ml in BHI broth containing 0%, 0.25% and 1% sucrose with or without S-PRG eluate. Next, 200 µl of the bacterial suspensions were added to 96-well polystyrene microtiter plates coated with human saliva. After incubation at 37 °C for 24 h, the plates were washed three times with phosphate-buffered saline (PBS) to remove loosely bound bacteria. Biofilms were fixed with 25% formaldehyde for 10 min and stained with 1% crystal violet in water (Sigma-Aldrich) for 15 min at room temperature. Next, the plates were washed three times and dissolved in 95% ethanol before quantification of the absorbance at 595 nm with an enzyme-linked immunosorbent assay microplate reader (Thermo Fisher Scientific, Waltham, MA, USA).
Biofilm formation by S. mutans MT8148 and S. sobrinus B13 in late logarithmic phase was determined as follows. The bacteria were grown for 24 h to reach the stationary phase and adjusted in BHI broth containing 0.25% sucrose to a final concentration of 1.0 × 109 CFU/ml with or without S-PRG eluate. Next, 200 µl of the bacterial suspensions were added to 96-well plates coated with 25% saliva followed by incubation at 37 °C for 24 h. Analysis of biofilm formation was then performed as described above.
Microscopic observation of in vitro biofilms
Quantitative and structural analysis of biofilms by confocal laser scanning microscopy was performed as described previously with some modifications48. Cells from overnight cultures of S. mutans MT8148 were collected by centrifugation at 3,000 × g for 10 min. Bacterial cells were next resuspended in 1 ml of Milli-Q water with 5 µl of 10 mM hexidium iodide (Invitrogen, Carlsbad, CA, USA) and incubated in the dark for 15 min at room temperature. The bacterial suspension was adjusted in BHI broth containing 0.25% sucrose to each specified cell concentration with or without S-PRG eluate. Next, 200 µl of the bacterial suspension were added to a chambered cover glass system (CultureWellTM, Grace Bio Labs, Bend, OR, USA) coated with filtered human saliva. The chamber was then incubated at 37 °C for 18 h in the dark. At the end of the experimental period, non-attached S. mutans cells were washed with PBS and biofilms were observed by confocal scanning laser microscopy using a TCS-SP5 microscope (Leica Microsystems GmbH, Wetzlar, Germany) with reflected laser light at 488 nm, as well as a DMI6000 B fluorescence microscope (Leica) and a 63× oil immersion objective. Biofilm thickness was measured as follows: an image taken in the z-axis was divided into 10 sections and the thickness of the central portion was measured. The mean value and standard deviation for the thickness were then calculated.
Hydrophobic interaction
Cellular hydrophobicity was determined using n-hexadecane (Wako) as previously described with some modifications26. Overnight cultures of S. mutans MT8148 were collected by centrifugation at 3,000 × g for 10 min. Cultures were adjusted to OD550 of 0.6 with or without S-PRG eluate Next, 0.2 ml of n-hexadecane was added to 3 ml of bacterial cells and then uniformly agitated with a vortex mixer for 1 min to induce hydrophobic interaction between the test strain and n-hexadecane. The mixture was left to stand for 10 min at room temperature and the optical density of the aqueous phase was determined at OD550. The incorporation rate was calculated as follows: [1 − OD550 (aqueous phase of the tube containing the cell suspensions with added n-hexadecane)/OD550 (aqueous phase of the tube containing only cell suspensions)] × 100 (%)]. The results are shown as the mean ± SD from four independent experiments.
Cellular hydrophobicity using pre-logarithmic S. mutans cells pretreated with S-PRG eluate was also evaluated as follows. S. mutans MT8148 was added at a final concentration of 1.0 × 107 CFU/ml in BHI broth with or without S-PRG eluate. After the bacterial suspensions were incubated at 37 °C for 18 h, cultures were adjusted to OD550 of 0.6 and the bacterial suspensions were streaked onto MSB plates to confirm the bacterial numbers. Additional bacterial cultures adjusted to OD550 of 0.6 with or without S-PRG eluate were prepared to use for the following the cellular hydrophobicity assays, as described above.
Statistical analysis
Statistical analyses were conducted using GraphPad Prism 6 (GraphPad Software Inc., La Jolla, CA, USA). Intergroup differences were analysed using an analysis of variance (ANOVA) with Bonferroni correction. Results were considered to be significantly different at P < 0.05.
Electronic supplementary material
Acknowledgements
The authors thank Prof. Howard K. Kuramitsu, State University of New York at Buffalo for editing the manuscript. This work was supported by the Fund for Scientific Promotion of SHOFU Inc., Kyoto, Japan. All authors read and approved the final manuscript.
Author Contributions
R.N. designed the entire study under the supervision of K.N. R.N., Y.M. and S.M. performed the experiments, and data interpretation was conducted by R.N. Y.M., K.N., R.N. and K.N. wrote the manuscript, which all authors read and approved.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23354-x.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Hamada S, Slade HD. Biology, immunology, and cariogenicity of Streptococcus mutans. Microbiol Rev. 1980;44:331–384. doi: 10.1128/mr.44.2.331-384.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Touger-Decker R, van Loveren C. Sugars and dental caries. Am J Clin Nutr. 2003;78:881–892. doi: 10.1093/ajcn/78.4.881S. [DOI] [PubMed] [Google Scholar]
- 3.Randall RC, Wilson NH. Glass-ionomer restoratives: a systematic review of a secondary caries treatment effect. J Dent Res. 1999;78:628–637. doi: 10.1177/00220345990780020101. [DOI] [PubMed] [Google Scholar]
- 4.Imazato S. Bio-active restorative materials with antibacterial effects: new dimension of innovation in restorative dentistry. Dent Mater J. 2009;28:11–19. doi: 10.4012/dmj.28.11. [DOI] [PubMed] [Google Scholar]
- 5.Sicca C, Bobbio E, Quartuccio N, Nicolò G, Cistaro A. Prevention of dental caries: A review of effective treatments. J Clin Exp Dent. 2016;8:604–610. doi: 10.4317/jced.52890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ikemura K, et al. Optimizing filler content in an adhesive system containing pre-reacted glass-ionomer fillers. Dent Mater. 2003;19:137–146. doi: 10.1016/S0109-5641(02)00022-2. [DOI] [PubMed] [Google Scholar]
- 7.Shimazu K, Ogata K, Karibe H. Caries-preventive effect of fissure sealant containing surface reaction-type pre-reacted glass ionomer filler and bonded by self-etching primer. J Clin Pediatr Dent. 2012;36:343–347. doi: 10.17796/jcpd.36.4.n444r730r773un53. [DOI] [PubMed] [Google Scholar]
- 8.Ma S, et al. Effects of a coating resin containing S-PRG filler to prevent demineralization of root surfaces. Dent Mater J. 2012;31:909–915. doi: 10.4012/dmj.2012-061. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki N, et al. Effects of S-PRG eluate on oral biofilm and oral malodor. Arch Oral Biol. 2014;59:407–413. doi: 10.1016/j.archoralbio.2014.01.009. [DOI] [PubMed] [Google Scholar]
- 10.Ito S, et al. Effects of surface pre-reacted glass-ionomer fillers on mineral induction by phosphoprotein. J Dent. 2011;39:72–79. doi: 10.1016/j.jdent.2010.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Yoneda, M., et al Effect of S-PRG Eluate on Biofilm Formation and Enzyme Activity of Oral Bacteria. Int J Dent. 2012, ID 814913, 6 pages (2012). [DOI] [PMC free article] [PubMed]
- 12.Hotta M, Morikawa T, Tamura D, Kusakabe S. Adherence of Streptococcus sanguinis and Streptococcus mutans to saliva-coated S-PRG resin blocks. Dent Mater J. 2014;33:261–267. doi: 10.4012/dmj.2013-242. [DOI] [PubMed] [Google Scholar]
- 13.Miki S, et al. Antibacterial activity of resin composites containing surface pre-reacted glass-ionomer (S-PRG) filler. Dent Mater. 2016;32:1095–1102. doi: 10.1016/j.dental.2016.06.018. [DOI] [PubMed] [Google Scholar]
- 14.Klein, M. I., Hwang, G., Santos, P. H., Campanella, O. H. & Koo, H. Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Front Cell Infect Microbiol 5–10 (2015). [DOI] [PMC free article] [PubMed]
- 15.Busuioc M, Buttaro BA, Piggot PJ. The pdh operon is expressed in a subpopulation of stationary-phase bacteria and is important for survival of sugar-starved Streptococcus mutans. J Bacteriol. 2010;192:4395–4402. doi: 10.1128/JB.00574-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato Y, Okamoto-Shibayama K, Azuma T. Glucose-PTS Involvement in Maltose Metabolism by Streptococcus mutans. Bull Tokyo Dent Coll. 2015;56:93–103. doi: 10.2209/tdcpublication.56.93. [DOI] [PubMed] [Google Scholar]
- 17.Renye JA, Jr., Piggot PJ, Daneo-Moore L, Buttaro BA. Persistence of Streptococcus mutans in stationary-phase batch cultures and biofilms. Appl Environ Microbiol. 2004;70:6181–6187. doi: 10.1128/AEM.70.10.6181-6187.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korithoski B, Lévesque CM, Cvitkovitch DG. The involvement of the pyruvate dehydrogenase E1alpha subunit, in Streptococcus mutans acid tolerance. FEMS Microbiol Lett. 2008;289:13–19. doi: 10.1111/j.1574-6968.2008.01351.x. [DOI] [PubMed] [Google Scholar]
- 19.Busuioc M, Mackiewicz K, Buttaro BA, Piggot PJ. Role of intracellular polysaccharide in persistence of Streptococcus mutans. J Bacteriol. 2009;191:7315–7322. doi: 10.1128/JB.00425-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Asención Diez MD, Demonte AM, Guerrero S. A, Ballicora, M. A., Iglesias, A. A. The ADP-glucose pyrophosphorylase from Streptococcus mutans provides evidence for the regulation of polysaccharide biosynthesis in Firmicutes. Mol Microbiol. 2013;90:1011–1027. doi: 10.1111/mmi.12413. [DOI] [PubMed] [Google Scholar]
- 21.Zeng L, Das S, Burne RA. Utilization of lactose and galactose by Streptococcus mutans: transport, toxicity, and carbon catabolite repression. J Bacteriol. 2010;192:2434–2444. doi: 10.1128/JB.01624-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lunsford RD, Roble AG. comYA, a gene similar to comGA of Bacillus subtilis, is essential for competence-factor-dependent DNA transformation in Streptococcus gordonii. J Bacteriol. 1997;179:3122–3126. doi: 10.1128/jb.179.10.3122-3126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Merritt J, Qi F, Shi WA. A unique nine-gene comY operon in Streptococcus mutans. Microbiology. 2005;151:157–166. doi: 10.1099/mic.0.27554-0. [DOI] [PubMed] [Google Scholar]
- 24.Kawabata S, Hamada S. Studying biofilm formation of mutans streptococci. Methods Enzymol. 1999;310:513–523. doi: 10.1016/S0076-6879(99)10039-9. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg M, Judes H, Weiss E. Cell surface hydrophobicity of dental plaque microorganisms in situ. Infect Immun. 1983;42:831–834. doi: 10.1128/iai.42.2.831-834.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg M, Gutnick D, Rosenberg E. Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol Lett. 1980;9:29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x. [DOI] [Google Scholar]
- 27.Krzyściak W, et al. The usefulness of biotyping in the determination of selected pathogenicity determinants in Streptococcus mutans. BMC Microbiol. 2014;14:194. doi: 10.1186/1471-2180-14-194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi S, Koga K, Hayashida O, Nakano Y, Hasegawa Y. Glucan-binding domain of a glucosyltransferase from Streptococcus sobrinus: isolation of a 55-kilodalton peptide from a trypsin digest of glucosyltransferase prebound to insoluble glucan. Infect Immun. 1989;57:2210–2213. doi: 10.1128/iai.57.7.2210-2213.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nagata E, de Toledo A, Oho T. Invasion of human aortic endothelial cells by oral viridans group streptococci and induction of inflammatory cytokine production. Mol Oral Microbiol. 2011;26:78–88. doi: 10.1111/j.2041-1014.2010.00597.x. [DOI] [PubMed] [Google Scholar]
- 30.Abinaya S,P, Gautam P. Studies on the biofilm produced by Pseudomonas aeruginosa grown in different metal fatty acid salt media and its application in biodegradation of fatty acids and bioremediation of heavy metal ions. Can J Microbiol. 2017;63:61–73. doi: 10.1139/cjm-2015-0384. [DOI] [PubMed] [Google Scholar]
- 31.McLean RJ, Fortin D, Brown DA. Microbial metal-binding mechanisms and their relation to nuclear waste disposal. Can J Microbiol. 1996;42:392–400. doi: 10.1139/m96-055. [DOI] [Google Scholar]
- 32.Wloka M, Rehage H, Flemming H-C, Wingender J. Structure and rheological behaviour of the extracellular polymeric substance network of mucoid Pseudomonas aeruginosa biofilms. Biofilms. 2005;2:275–283. doi: 10.1017/S1479050506002031. [DOI] [Google Scholar]
- 33.Paik S, Brown A, Munro CL, Cornelissen CN, Kitten T. The sloABCR operon of Streptococcus mutans encodes an Mn and Fe transport systemrequired for endocarditis virulence and its Mn-dependent repressor. J Bacteriol. 2003;185:5967–5975. doi: 10.1128/JB.185.20.5967-5975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Binepal G, et al. Trk2 Potassium Transport System in Streptococcus mutans and Its Role in Potassium Homeostasis, Biofilm Formation, and Stress Tolerance. J Bacteriol. 2016;198:1087–1100. doi: 10.1128/JB.00813-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlsson J, Kujala U, Edlund MB. Pyruvate dehydrogenase activity in Streptococcus mutans. Infect Immun. 1985;49:674–678. doi: 10.1128/iai.49.3.674-678.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu S, Yang Y, Jin R, Zhang M, Wang H. Purification and characterization of a functionally active Mycobacterium tuberculosis prephenatedehydrogenase. Protein Expr Purif. 2006;49:151–158. doi: 10.1016/j.pep.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 37.Chen S, Vincent S, Wilson DB, Ganem B. Mapping of chorismate mutase and prephenate dehydrogenase domains in the Escherichia coli T-protein. Eur J Biochem. 2003;270:757–763. doi: 10.1046/j.1432-1033.2003.03438.x. [DOI] [PubMed] [Google Scholar]
- 38.Fujimoto Y, et al. Detection of ions released from S-PRG fillers and their modulation effect. Dent Mater J. 2010;29:392–397. doi: 10.4012/dmj.2010-015. [DOI] [PubMed] [Google Scholar]
- 39.Ooshima T, Izumitani A, Sobue S, Hamada S. Cariostatic effect of palatinose on experimental dental caries in rats. Jpn. J Med Sci Biol. 1983;36:219–223. doi: 10.7883/yoken1952.36.219. [DOI] [PubMed] [Google Scholar]
- 40.Miyatani F, et al. Relationship between Cnm-positive Streptococcus mutans and cerebral microbleeds in humans. Oral Dis. 2015;21:886–893. doi: 10.1111/odi.12360. [DOI] [PubMed] [Google Scholar]
- 41.Ajdić D, et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc Natl Acad Sci USA. 2002;99:14434–14439. doi: 10.1073/pnas.172501299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sasaki H, et al. Antibacterial activity of polyphenol components in oolong tea extract against Streptococcus mutans. Caries Res. 2004;38:2–8. doi: 10.1159/000073913. [DOI] [PubMed] [Google Scholar]
- 43.Ooshima T, et al. Contributions of three glycosyltransferases to sucrose-dependent adherence of Streptococcus mutans. J Dent Res. 2001;80:1672–1677. doi: 10.1177/00220345010800071401. [DOI] [PubMed] [Google Scholar]
- 44.Park, S. N. et al Antimicrobial mechanism of oleanolic and ursolic acids on Streptococcus mutans UA159. Curr Microbiol (in press). [DOI] [PubMed]
- 45.Zahurak M, et al. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattos-Graner RO, et al. Cloning of the Streptococcus mutans gene encoding glucan binding protein B and analysis of genetic diversity and protein production in clinical isolates. Infect. Immun. 2001;69:6931–6941. doi: 10.1128/IAI.69.11.6931-6941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ardin AC, et al. Identification and functional analysis of an ammonium transporter in Streptococcus mutans. PLoS One. 2014;9:e107569. doi: 10.1371/journal.pone.0107569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuboniwa M, et al. Streptococcus gordonii utilizes several distinct gene functions to recruit Porphyromonas gingivalis into a mixed community. Mol Microbiol. 2006;60:121–139. doi: 10.1111/j.1365-2958.2006.05099.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.