ABSTRACT
The yeast Candida parapsilosis is an increasingly common cause of systemic fungal infections among immunocompromised individuals, including premature infants. Adhesion to host surfaces is an important step in pathogenesis, but this process has not been extensively studied in this organism. A microfluidics assay was developed to test the ability of C. parapsilosis to adhere to immobilized host extracellular matrix proteins under physiological fluid shear conditions. Growth in mammalian tissue culture medium at 37°C for 3 to 6 h led to the induction of an adhesive phenotype at shear forces of 1 to 5 dynes/cm2 in some isolates of C. parapsilosis. Glutamic acid, proline, and calcium appeared to be the minimally necessary requirements for increased adhesion in these assays. To determine whether genes homologous to the ALS gene family of C. albicans were important for the adhesive phenotype, the expression levels of 5 homologous C. parapsilosis genes were quantified by using quantitative PCR (qPCR) under conditions leading to increased adhesion. CPAR2_404800 (CpALS7) and CPAR2_404780 showed increased expression levels compared to those in control yeast. The extent of adhesion was variable among different isolates, and linear regression identified the expression of CpALS7 but not CPAR2_404780 as having a strong positive correlation with adhesion. A homozygous CpALS7 deletion strain was deficient in adhesion, whereas the expression of CpALS7 in Saccharomyces cerevisiae resulted in increased adhesion. Together, these data provide strong evidence that CpAls7 aids in the adherence of C. parapsilosis to the extracellular matrix under shear forces and support its previously reported role in virulence.
KEYWORDS: adhesion, Candida parapsilosis, shear force, extracellular matrix
INTRODUCTION
Candida species are the leading cause of systemic, invasive fungal infections worldwide (1). Occurring primarily among immunocompromised individuals, these infections are initiated by a compromise of mucosal or skin barriers, access to the bloodstream, and subsequent escape from the blood to invade deeper tissues (1, 2). Given this sequence, mechanisms by which Candida can interact with and then breach blood vessel walls are likely important components of pathogenesis.
Candida albicans causes the majority of invasive infections (3) and is widely considered to be the most virulent species. The shift in morphology from its budding yeast form to a filamentous, hyphal form is well recognized as a key virulence determinant (4). Adhesion of C. albicans to host surfaces has been extensively studied. Although initial contact with epithelial and endothelial cells is likely made by the yeast form, initiation of hypha formation follows rapidly, and novel adhesins that have key roles in further adhesion are expressed on the hyphal surface (5).
The best-studied family of adhesins in C. albicans is the ALS (agglutinin-like sequence) gene family (6). This family is comprised of at least eight genes that encode large glycosylphosphatidylinositol (GPI)-linked glycoproteins on the cell surface (7). Several ALS genes have been shown to confer adhesion to mammalian host surfaces (8). Of note, ALS3 has been shown to mediate the binding of C. albicans to a variety of biotic matrices (6). It has also been implicated in binding to N-cadherin to initiate endocytosis by endothelial cells (9) and may also have a role in the penetration of the central nervous system (10). The induction of ALS3 occurs with hyphal growth (11), and its expression has been detected in yeast isolated from a murine model of disseminated candidiasis and from patients with oropharyngeal candidiasis (12).
Although the majority of cases of nosocomial candidiasis is attributed to C. albicans, the emergence of non-albicans Candida species has been a growing cause for concern (13, 14). In particular, Candida parapsilosis has been documented as the non-albicans Candida species most commonly isolated from blood cultures in multiple studies; in some populations, it has surpassed C. albicans as the most frequently isolated Candida species (15). Despite the increasing recognition of C. parapsilosis as a clinically significant opportunistic pathogen, it remains poorly characterized relative to C. albicans (16). Thus, there is a need to further investigate the pathobiology of C. parapsilosis, with an eye toward investigating virulence mechanisms, identifying potential drug targets, and developing novel therapeutics to better combat candidiasis caused by non-albicans Candida species.
Recent efforts have shed light on putative adhesins in the C. parapsilosis genome. Proteomic analysis of a cell wall extract from C. parapsilosis identified multiple proteins that bound to fibronectin, vitronectin, and/or laminin (17). Among these proteins were those encoded by CPAR2_404800 and CPAR2_404780, which bear significant identity to the ALS gene family (17). These same 2 gene products were purified from extracts of C. parapsilosis surface proteins based on their binding affinity for human plasminogen and high-molecular-mass kininogen (18). Furthermore, the site-specific deletion of CPAR2_404800 reduced the adhesion of C. parapsilosis to buccal epithelial cells and significantly attenuated virulence in a murine model of urinary tract infection (19). This gene was selected for targeting in this study because its product bears the highest sequence identity to C. albicans Als3. The authors of that study named this gene CpALS7 based on synteny with C. albicans ALS7 according to the Candida Gene Order database (http://cgob3.ucd.ie/) (20, 21). For consistency, this gene is referred to as CpALS7 throughout this study. These recent findings suggest that ALS homologues in C. parapsilosis carry an adhesive function and may be important players in the pathogenesis of invasive infection.
The objective of this study was to identify conditions that induce adhesion in C. parapsilosis and examine differential CpALS gene expression under adhesive and nonadhesive conditions, with the goal of further characterizing adhesins in C. parapsilosis. In the bloodstream, Candida spp. encounter fluid shear forces. In order to establish deep-seated infections, Candida in the bloodstream must adhere to vessel walls while resisting hemodynamic shear forces. To better model this milieu in vitro, we measured the adhesion of C. parapsilosis to immobilized substrates under physiological fluid shear using a microfluidics apparatus.
RESULTS
Effect of growth conditions on adhesion of C. parapsilosis to host proteins under shear.
The induction of hyphal growth in C. albicans leads to the expression of novel proteins that have a role in adhesion to host structures. To test whether similar growth conditions would increase the adhesion of C. parapsilosis, strain JMB81 was grown in mammalian tissue culture medium 199 (M199) for 3 h at 37°C. Of note, in this and all subsequent experiments, the morphology of all of the strains studied under any growth condition was uniformly the yeast form. No elongated yeast cells or pseudohyphae were observed under any conditions. The capacity of C. parapsilosis yeast cells “induced” under these conditions to adhere to immobilized bovine serum albumin (BSA) was tested in a microfluidics adhesion assay under various shear forces. C. parapsilosis cells induced in M199 showed greatly increased adhesion under fluid shear at 5 dynes/cm2 compared to control yeast cells (Fig. 1B and 2A). Under each shear force tested, the adhesion of induced yeast cells was significantly higher than that of control yeast cells, with peak adherence at 5 dynes/cm2 (Fig. 2A). Furthermore, increases in shear from 1 to 2 and from 2 to 5 dynes/cm2 resulted in significant increases in the adhesion of induced yeast cells (P < 0.001 in each case). In contrast, control yeast cells showed a significant decrease in adhesion with each increase in shear force above 1 dyne/cm2, as expected (P < 0.001). Similar results were obtained with four distinct clinical isolates of C. parapsilosis (22, 23), where induction resulted in greater adhesion at 5 dynes/cm2 than that at 1 dyne/cm2 (not shown). Adhesion to different substrates was compared between control and induced C. parapsilosis cells. Induction increased adhesion significantly toward BSA, fibronectin, collagen, and gelatin (Fig. 2B). In contrast, channels left uncoated with any substrate (Hanks' balanced salt solution [HBSS]) showed very low adhesion for both control and induced yeast cells.
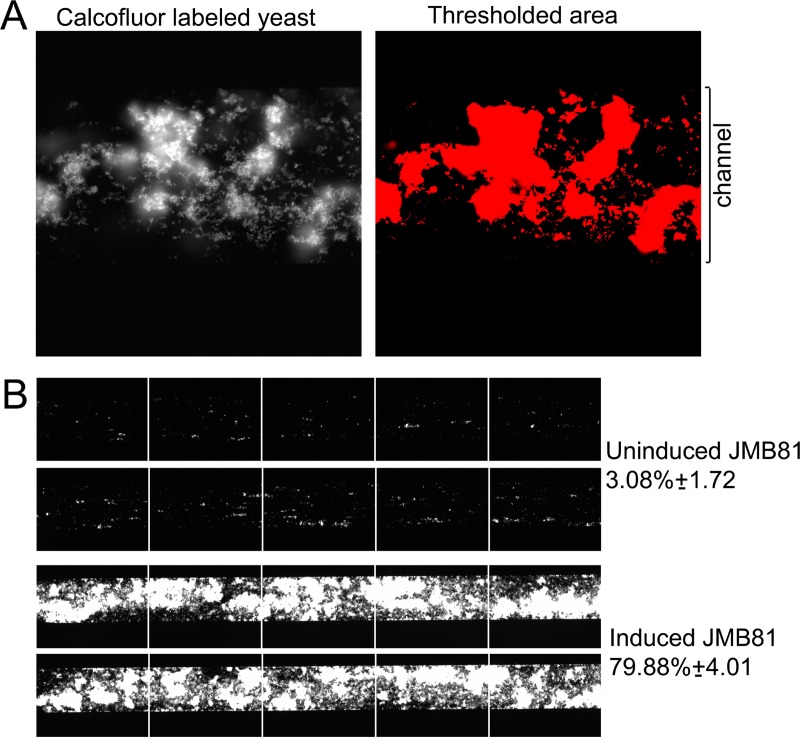
FIG 1.
C. parapsilosis adhesion under fluid shear. (A, left) Calcofluor-labeled fluorescent yeast cells were observed to adhere to the microchannel under specific conditions. (Right) Fluorescence-thresholded area. (B) Microchannels under each condition were run in duplicate and are represented as a series of adjacent images encompassing approximately 80% of the channel area. The top pair shows control C. parapsilosis cells adhering to immobilized BSA under fluid shear at 5 dynes/cm2. The bottom pair shows induced yeast cells showing a consistently increased adhesion index under these conditions. The fluid flow direction is from right to left. The adhesion index represents the area of the channel covered by yeast, expressed as a percentage ± the standard deviation, as indicated.
FIG 2.
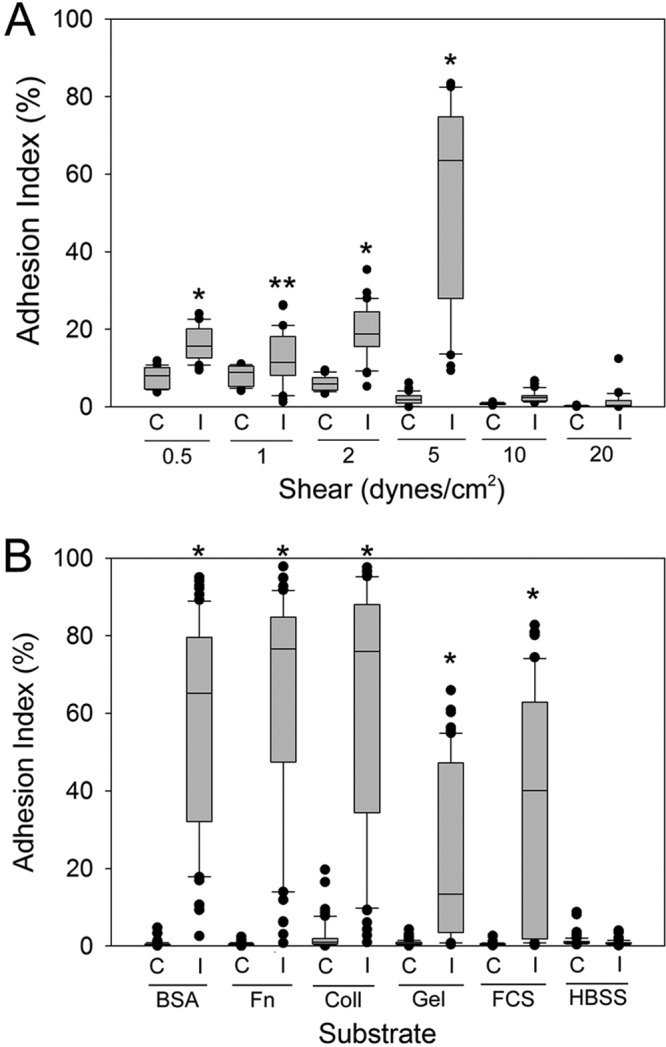
Effect of shear force and the substrate on C. parapsilosis adhesion. C. parapsilosis was induced by growth in M199 for 3 h at 37°C. (A) Adhesion assays were conducted in a microfluidics environment under various shear forces. The adhesion index was calculated from duplicate channels with a minimum of 3 biological replicates. Induced yeast cells (I) were compared to control yeast cells (C) at each shear force via a Mann-Whitney rank sum test. *, P < 0.001; **, P = 0.02 (for induced versus control yeast cells at each shear force). (B) Adhesion assays were conducted as described above, at 5 dynes/cm2, over microfluidics channels coated with the indicated substrates (BSA, bovine serum albumin; Fn, fibronectin; Coll, collagen; Gel, gelatin; FCS, fetal calf serum; HBSS, Hanks' balanced salt solution plus Ca2+/Mg+). *, P < 0.001.
To further define the components of M199 that may have led to increased adhesion, assays were conducted following induction in a variety of media. Parallel incubation of C. parapsilosis in yeast extract-peptone-dextrose (YPD) medium, water, or DPBS+ (Dulbecco's phosphate-buffered saline containing Ca2+ and Mg2+) did not result in increased adhesion; similarly, control yeast cells (grown in YPD medium) resuspended in M199 immediately prior to loading into the assay mixture did not show increased adhesion (not shown). Incubation in HBSS+ (HBSS containing calcium, magnesium, and 10 mM HEPES) also did not promote adhesion, so various components present in M199 but absent in HBSS+ were added individually or in combinations to HBSS+ during induction (Fig. 3). Small but significant increases in adhesion were observed when aspartic acid (D), glutamic acid (E), and proline (P) were added in combination to HBSS+. Assessment of each amino acid individually suggested that either glutamic acid or proline, but not aspartic acid, was sufficient to induce adhesion. Higher concentrations of these amino acids were tested and failed to further augment adhesion (not shown). Furthermore, the addition of EDTA to M199 after induction substantially reduced adhesion compared to that with M199 alone, suggesting that divalent cations are also required for yeast adhesion under flow.
FIG 3.

Components of M199 that contribute to adhesion. C. parapsilosis strain JMB81 was induced by growth in the indicated media for 3 h at 37°C and tested in adhesion assays. +/+, HBSS+; D, aspartic acid; E, glutamic acid; P, proline. The adhesion index was calculated from duplicate channels with a minimum of 3 biological replicates. Statistical analysis was performed by using Kruskal-Wallis analysis of variance on ranks, and between-group comparisons were made by using Dunn's method.
Induction in M199 medium resulted in increased adhesion to immobilized BSA, even when 10% fetal bovine serum was added during adhesion under shear (Fig. 4). Induction of yeast cells in 100% serum followed by shear flow in 100% serum also resulted in significantly increased adhesion compared to that of control yeast cells.
FIG 4.
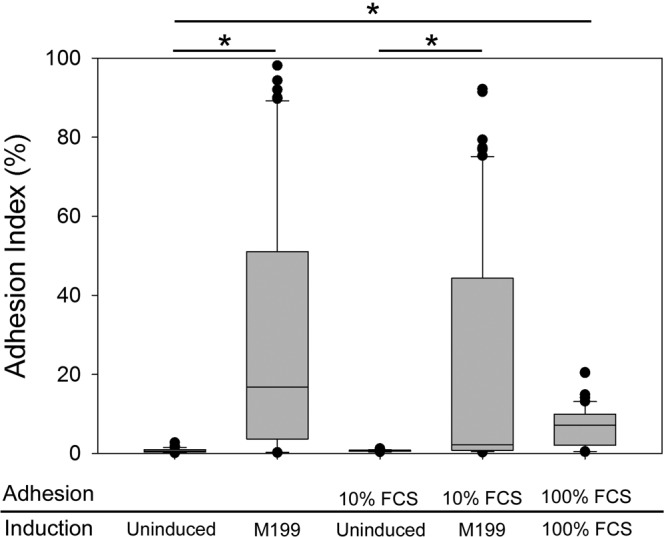
Effect of serum on adhesion. C. parapsilosis strain JMB81 was induced by growth for 3 h at 37°C in either M199 or 100% fetal calf serum (FCS) and tested in adhesion assays in the presence or absence of fetal calf serum, as indicated. The adhesion index was calculated from duplicate channels with a minimum of 3 biological replicates. Statistical analysis was performed by using Kruskal-Wallis analysis of variance on ranks, and between-group comparisons were made by using Dunn's method. *, P < 0.001.
Positive correlation of CpALS7 expression with adhesion.
To identify genes that may have a role in the increased adhesion of C. parapsilosis grown in M199, homologues of the ALS gene family of C. albicans (8, 24) were identified by using the Candida Genome Database (16). Five candidate genes were identified (see Table 2), and primers appropriate for quantitative PCR (qPCR) analysis of each gene transcript were designed and validated. Only 2 genes, CpALS7 and CPAR2_404780, demonstrated a significant increase in expression in M199 relative to that in YPD medium (Fig. 5). Among the ALS gene family homologues tested, the expression of CpALS7 showed by far the largest increase in expression when grown in M199. When additional C. parapsilosis isolates (Table 1) were tested under the same growth conditions, considerable variability in CpALS7 mRNA expression levels was observed. Because genomic heterogeneity among C. parapsilosis isolates has been noted (25), standard PCR was used with genomic DNA as the template to test for the presence of genomic sequences for each gene in each isolate. All of the wild-type isolates listed in Table 1 generated a PCR product for each gene, with the exception of strains JMB77 and Ro75. These two strains had a PCR product for all genes except CPAR2_404790. To test the hypothesis that the level of expression of CpALS7 is directly associated with adhesion, the C. parapsilosis isolates that showed variability in CpALS7 expression levels were tested in adhesion assays under shear at 5 dynes/cm2 (Fig. 6). Higher expression levels of CpALS7 showed a strong positive correlation with adhesion (R2 = 0.71; P < 0.001), suggesting that CpALS7 may have a direct role in the adhesion phenotype. None of the other ALS genes surveyed showed a positive or negative correlation with the adhesion index (see Fig. S1 in the supplemental material).
TABLE 2.
C. parapsilosis ALS and housekeeping gene homologues and primers
| Gene | Abbreviation or alternate gene name | qPCR primer | Primer sequence |
|---|---|---|---|
| CPAR2_500660 | 500660 | 500660_F | ATGGCTTGGATCAACCACTT |
| 500660_R | CAGAACTTACGGCAGGGGTA | ||
| CPAR2_404770 | 404770 | 404770_F | AACTGGCCATGAAAACAAGG |
| 404770_R | CCATAGCAGGTTTGCACAGA | ||
| CPAR2_404790 | 404790 | 404790_F | GGCGTCAAACACAGAATCAA |
| 404790_R | GCTGCTGTTGCTGTGACATT | ||
| CPAR2_404800 | CpALS7 | 404800_F | ACCACCACCGAGGTTACAAA |
| 404800_R | TGGATCTCCGGTTTTGAGTC | ||
| CPAR2_404780a | 404780 | 404780_F | CGCAAGTACTCCTTCCAACC |
| 404780_R | CCATTACCTTCGTTTCCATTG | ||
| CPAR2_201570 | CpACT1 | CpACT1_F | CGAACGTGGTTACGGTTTCT |
| CpACT1_R | TGACCATCTGGCAATTCGTA | ||
| CPAR2_502950 | CpPMA1 | CpPMA1_F | CGCCAACGAAGTTGTTCCTG |
| CpPMA1_R | AGATTCACCAGTGATGGCGG | ||
| CPAR2_806050 | CpRIP1 | CpRIP1_F | CCCAATTGGTGAAGCAGGTG |
| CpRIP1_R | CCAAGTTCAAAGGGGCTGGA | ||
| CPAR2_807570 | CpRPP2B | CpRPP2B_F | CTCCCCATCTGCTTCAGACA |
| CpRPP2B_R | TGAGGCCAATTTGGTGTTACCT | ||
| CPAR2_204740 | CpLSC2 | CpLSC2_F | TTGGACTGTGGTGGTACTGC |
| CpLSC2_R | TTGCGGCAATCAAACCCTTG | ||
| CPAR2_104580 | CpIMH3 | CpIMH3_F | TCACTGTTGGGGACGTCAAG |
| CpIMH3_R | GGATCGTCGTTGTTTTCGTGA | ||
| CPAR2_401060 | CpCPA1 | CpCPA1_F | CCCGAGAGCATGACTGATCC |
| CpCPA1_R | AGCAACATCACCAACGACGA | ||
| CPAR2_808670 | CpTDH3 | CpTDH3_F | ACCACCAACTGTTTGGCTCC |
| CpTDH3_R | ACCACCTCTCCAGTCCTTGT |
The primer set for CPAR2_404780 was noted to also recognize CPAR2_500660 during the review process. Analysis of qPCR data from samples containing these primer sets indicated that the expression level of CPAR2_500660 was extremely low and contributed ∼1% to the total mRNA levels at most. This observation is very consistent with previously reported data (19). Since the contribution of CPAR2_500660 to the calculations of CPAR2_404780 levels was negligible, the cross-hybridization of this primer set does not affect the data presented in a meaningful way.
FIG 5.
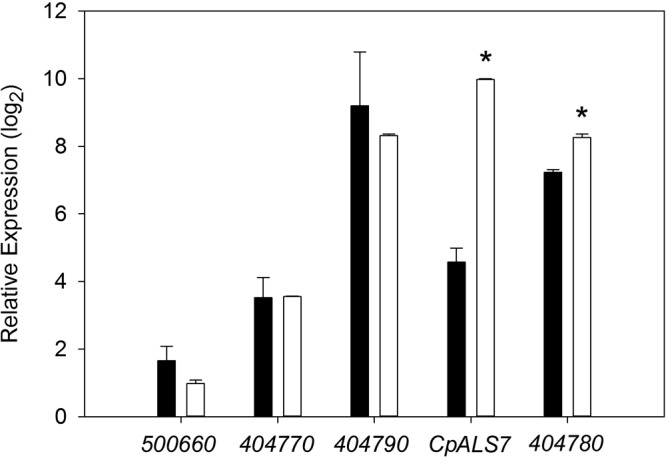
Expression of CpALS genes during growth in M199. C. parapsilosis strain JMB81 was induced by growth in M199 for 3 h at 37°C, and qPCR was performed to evaluate the transcription of ALS gene homologues. Gene names are abbreviated as listed in Table 2. The relative expression level is shown for each gene from control cells (black bars) and induced cells (white bars). Error bars represent standard deviations from duplicate samples with 3 biological replicates. Comparisons of expression levels for each gene under control and induced conditions were made by a 2-tailed t test. *, P < 0.001 for induced versus control cells.
TABLE 1.
Strains and plasmids used in this study
FIG 6.
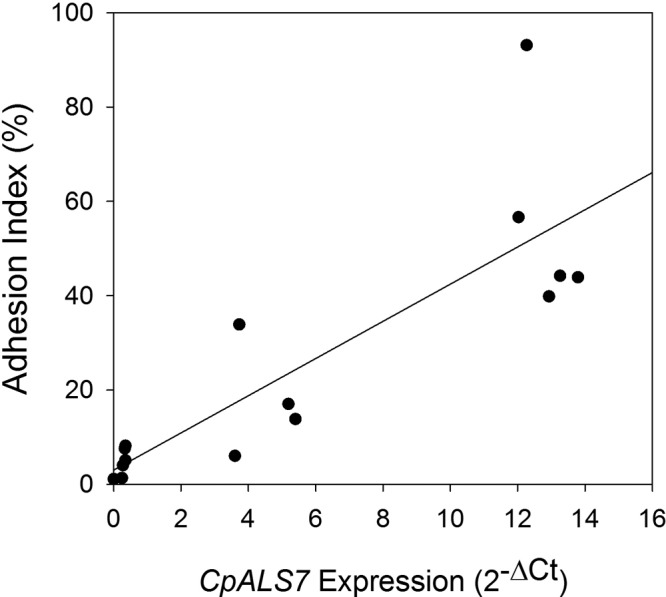
Association of CpALS7 expression with adhesion. C. parapsilosis isolates that showed various expression levels of CpALS7 when induced by growth in M199 for 3 h at 37°C were tested in adhesion assays as described in the text. The adhesion index was plotted against the expression level of CpALS7 measured by qPCR as described in the text. Linear regression analysis showed a strong positive correlation between CpALS7 expression and adhesion (R2 = 0.71; P < 0.001).
To test the relationship between CpALS7 expression and adhesion in a more direct manner, the gene was cloned and expressed in Saccharomyces cerevisiae under the control of a galactose-inducible promoter (Fig. 7A). The induction of CpALS7 expression in S. cerevisiae yeast cells led to significantly increased adhesion at 1 and 5 dynes/cm2 relative to that of yeast cells that were grown with glucose as a carbon source (P < 0.001). Yeast cells that contained the vector without CpALS7 showed minimal adhesion regardless of the carbon source. To further investigate the effect of CpALS7 on adhesion, a CpALS7 deletion strain (KO) (19) and its wild-type parent (ATCC 22019) were tested in the adhesion assay (Fig. 7B). Although the parent strain had low baseline levels of adhesion, induction in M199 medium resulted in a small increase. The induction of this strain also demonstrated a modest increase in the CpALS7 RNA expression level relative to those of the other strains tested (not shown). Nevertheless, the CpALS7 deletion strain had significantly reduced adherence in these assays. Taken together, these data suggest that CpALS7 expression is an important component of the adhesion of C. parapsilosis to immobilized albumin under shear.
FIG 7.
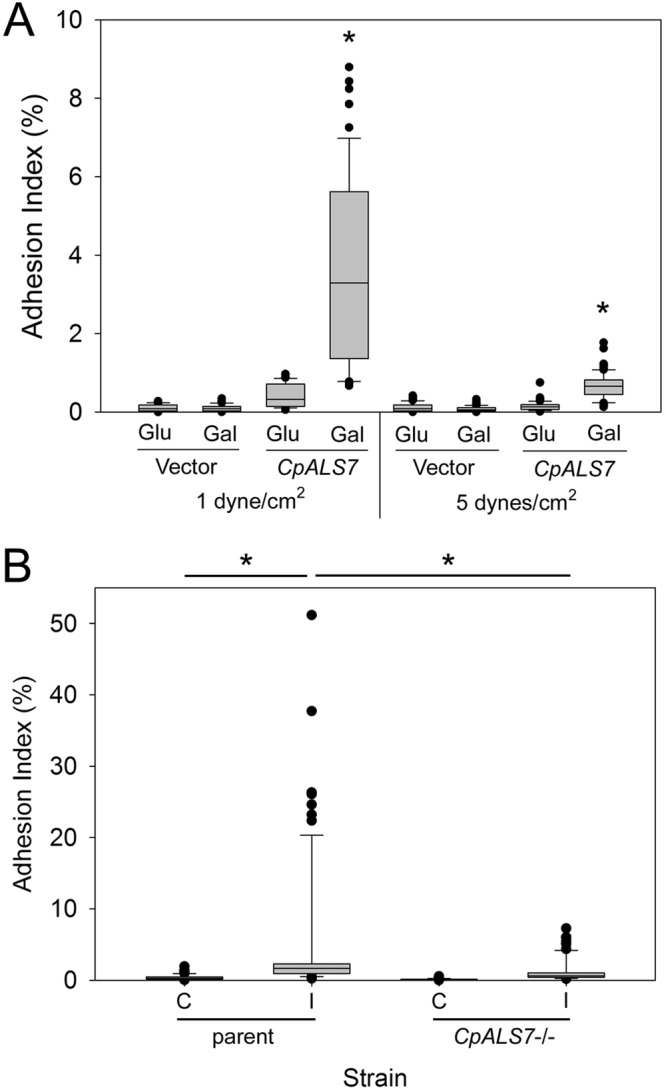
Expression of CpALS7 leads to increased adhesion. (A) CpALS7 was cloned into the S. cerevisiae expression vector pYES2 under the control of the galactose-inducible promoter. Yeast cells containing this plasmid or the vector only were grown with either glucose (Glu) (repressive) or galactose (Gal) (inductive) as the sole carbon source, and adhesion was measured in the microfluidics device with a shear force of 1 or 5 dynes/cm2. Data are from duplicate samples at each shear force with 5 biological replicates. Comparisons between cells grown in Glu and those grown in Gal were made by using a Mann-Whitney rank sum test. *, P < 0.001 for Gal versus Glu at each shear force. (B) A C. parapsilosis strain harboring a homozygous deletion of CpALS7 (CpALS7−/−) and the wild-type parent strain from which it was derived (19) were induced by growth in M199 for 3 h at 37°C and tested in adhesion assays as described above. The adhesion indices of induced (I) and control (C) yeast cells were calculated from duplicate channels with a minimum of 3 biological replicates. Statistical analysis was performed by using Kruskal-Wallis analysis of variance on ranks, and between-group comparisons were made by using Dunn's method. *, P < 0.001.
DISCUSSION
Adhesion of C. parapsilosis to host surfaces remains poorly characterized relative to that of C. albicans. Investigation into the mechanisms by which C. parapsilosis adheres to the extracellular matrix (ECM) offers insight into the organism's underlying pathobiology and may provide a route to disrupt adhesion and prevent subsequent invasive infection. The objective of this study was to identify adhesins in C. parapsilosis that mediate binding to the extracellular matrix under physiologically relevant shear flow.
Because members of the ALS gene family in C. albicans have a role in adhesion, we screened for differential gene expression of ALS homologues under adhesive and nonadhesive conditions to identify putative adhesins in C. parapsilosis. The expression of CpALS7 was markedly upregulated relative to other ALS gene homologues in strains that demonstrated inducible adhesion. A role for CpAls7 in adhesion was first suggested by a study showing that the protein was isolated from a fibronectin affinity column (17). Another study showed that a strain deficient in CpALS7 had reduced adhesion to buccal epithelial cells in vitro and a reduction in the fungal burden in a mouse model of urinary tract infection (19). Our study provides additional evidence to support a role for CpAls7 in adhesion. First, we utilized an inducible expression system in S. cerevisiae, a well-established strategy to identify putative adhesins in Candida spp. (26). The induction of CpALS7 expression in S. cerevisiae yielded a significant increase in adhesion in our assay system. However, the level of adhesion of the S. cerevisiae transformant remained relatively low compared to those of adhesive C. parapsilosis strains. It is possible that differences in codon usage contributed to the decreased functionality, as C. parapsilosis belongs to the CTG clade (where CTG is translated as serine rather than leucine as it is in S. cerevisiae). However, such discrepancies in adhesion are also likely to reflect inherent differences in the overall surface structures of C. parapsilosis and the generally nonadherent yeast S. cerevisiae.
Individual C. parapsilosis isolates displayed wide variability in the expression of CpALS7 mRNA under inducing conditions. The variability in CpALS7 expression and associated adhesion may reflect underlying genomic heterogeneity among strains of C. parapsilosis. Available genomic data collected from C. parapsilosis isolates have uncovered marked heterogeneity in general and variability in the presence and copy number of ALS genes in particular, reinforcing the understanding that C. parapsilosis is not clonal in its global distribution (25). Although we detected a PCR product for each ALS gene tested in each strain, except for CPAR2_404790 in 2 isolates, PCR analysis provides only a rough assessment of the genomic heterogeneity that could affect gene expression. The possible underlying genomic variability among the isolates in our study is the subject of ongoing investigation. We used this variability to probe whether variation in CpALS7 expression among different isolates could explain differences in the efficiency of adhesion to immobilized BSA in our flow adhesion assays. The strong positive correlation between the CpALS7 mRNA level and the adhesion index provides further evidence of its role in adhesion.
The ability of induced yeast cells to adhere to fibronectin, collagen, and BSA under shear suggests that CpAls7 binds a wide spectrum of substrates. Binding to various substrates has also been reported for several ALS genes in C. albicans (6, 12), and recent efforts have provided insight into the molecular mechanisms involved (27, 28). Als proteins in C. albicans consist of four domains: an N-terminal (NT) domain that is the most conserved among Als proteins, a threonine-rich (T) region, a central region with a variable number of 36-amino-acid repeats, and a C-terminal domain that varies in size and contains a GPI anchor attaching the protein to the cell wall (6). The NT domain confers the adhesive function and contains a peptide-binding cavity (PBC) that has been resolved by X-ray crystallography (27). The PBC contains an invariant lysine residue that is required to interact with the C-terminal carboxyl group of a peptide ligand, and the structure and molecular interactions within the PBC account for the broad substrate specificity of these proteins. Examination of the deduced primary sequence of CpAls7 confirms that this protein has the same domain structure as that of the C. albicans Als family and bears the conserved lysine residue as well as the 8 cysteine residues that are responsible for the conserved disulfide bonds. As such, the mechanism of ligand binding is likely similar to that of C. albicans.
The preferential binding of C. parapsilosis to the ECM under fluid shear is reminiscent of microbial “catch bonding,” a phenomenon in which tensile force applied to an environment increases the strength of bonds formed between microbes and/or between microbes and surfaces (29, 30). Recent work has established the involvement of C. albicans Als5 in catch bonding (31). Both wild-type C. albicans and S. cerevisiae expressing Als5 showed increased attachment as the shear force increased from 0.2 to 1.6 dynes/cm2. Higher shear force also led to more robust biofilm growth. The importance of the amyloid-forming region (AFR) of Als5 was demonstrated in this study. Additional work with Als3 has provided evidence that allows the separation of adhesion functions of Als3 (to protein ligands) from its role in the aggregation of fungal cells to each other (28). These data suggest that the AFR is more relevant for the latter. In our system, both adhesion to the ECM substrate and the aggregation of yeast cells to each other likely have a role in increasing the “adhesion index.” Structural similarities predicted by the similar domain structure of CpAls7 suggest that similar mechanisms are likely to be at play, but confirmation will require experimental validation. Regardless, the preferential binding of C. parapsilosis to the ECM at 5 dynes/cm2 may demonstrate the significance of this particular shear stress during adhesion to the ECM in the bloodstream or urinary tract. Given that physiological shear stress levels in capillaries and postcapillary venules are generally ≤5 dynes/cm2, these results suggest that initial adhesion is more likely to take place at these locales, as opposed to arterial fluid shear, which typically ranges from 10 to 30 dynes/cm2 (32, 33).
We hypothesize that M199, as a mammalian cell culture medium, may partially mimic the host environment and induce adaptive changes in C. parapsilosis, analogous to the yeast-to-hypha switch of C. albicans. It should be noted, however, that the expression levels of ALS gene products in C. albicans can vary considerably under in vitro and in vivo conditions, which may limit the applicability of these findings (34). Adhesion also required the presence of Ca2+ ions. Ca2+ is present in plasma and tissue extracellular fluid, and the mammalian integrin, cadherin, and selectin families of adhesion molecules are dependent on the presence of millimolar levels of divalent cations. The requirement for Ca2+ further suggests that CpAls7 may have evolved to provide adhesive functions in blood and tissues; however, it is not clear at present if divalent cations are required by the adhesin, the substrate, or both.
This study provides new and compelling evidence that CpAls7 aids in the adherence of C. parapsilosis to the ECM. CpAls7 may therefore represent an important inducible virulence factor for C. parapsilosis to adhere to blood vessel walls while resisting hemodynamic fluid shear and may play a role in the early stages of biofilm formation. Lessons learned not only are likely to be applicable to this specific host-pathogen interaction but also will better inform the pathogenic lifestyles of organisms that require a bloodstream phase.
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Table 1. C. parapsilosis strains were maintained in frozen stocks in 25% glycerol and recovered on YPD agar (1% yeast extract, 2% peptone, 2% dextrose). Parent strain ATCC 22019 and the homozygous als7Δ/als7Δ mutant (CpALS7 KO) were graciously provided by Arianna Tavanti (19). Cultures for adhesion assays and qPCR were grown overnight with vigorous agitation in YPD broth at 37°C. S. cerevisiae strain INVSc1 was maintained as described above, with incubations at 30°C.
The S. cerevisiae transformants JMB278 and JMB279 (see below) were grown overnight in SC-URA (synthetic complete yeast agar without uracil) at 30°C with vigorous agitation. Following culture overnight, cells were washed with sterile water and adjusted to an optical density (A600) of 0.4 in fresh SC-URA supplemented with either 2% galactose or 2% glucose (to induce or repress expression under the control of the GAL1 promoter, respectively). The cultures were incubated at 30°C for 24 h with vigorous agitation. Cultures were washed with HBSS+ and adjusted to a concentration of 3 × 106 yeast cells ml−1 before introduction into the microfluidics adhesion assay mixture.
C. parapsilosis strains were grown overnight in YPD broth at 37°C with vigorous agitation. To achieve the adhesive phenotype, cultures grown overnight were washed with sterile water, resuspended in medium 199 (catalogue number BW12-117Q; BioWhittaker/Lonza) at a concentration of 3 × 106 yeast cells ml−1, and incubated at 37°C for 3 to 6 h. This process is referred to as “induction.” Medium 199 was selected for induction because it efficiently induces hyphal growth and the expression of ALS genes important for adhesion in C. albicans. Medium 199 did not induce pseudohyphal growth of any C. parapsilosis strain studied under the conditions described here. Experiments involving amino acid supplements in HBSS+ were conducted with the following concentrations: 225 μM l-aspartic acid, 455 μM l-glutamic acid, and 347 μM l-proline.
Microfluidic adhesion assay.
Shear flow adhesion assays were conducted by using the Bioflux 200 system (Fluxion Biosciences) and Bioflux microchannel glass coverslip bottom plates at 37°C. This approach permitted running 24 samples in parallel under defined fluid shear and provided a medium-throughput assay for the simultaneous analysis of multiple conditions. Bioflux microchannels were coated with 2% BSA, 100 μg/ml fibronectin (R&D Systems), 800 μg/ml collagen type I (Sigma), or 1 mg/ml gelatin (Sigma) in HBSS+ for 48 h. Microchannels were then washed with HBSS+ at 2 dynes/cm2 for 1 to 2 h to remove the unbound substrate. Yeast suspensions were introduced into each microchannel, and fluid flow was initiated for the entire plate. After 30 min of adhesion under fluid flow, yeast cells were aspirated from inlet and outlet wells without disturbing the channel area, DPBS+ containing 5 μM calcofluor white (catalogue number F3543; Sigma) was added to microchannels, and fluid flow was resumed for another 10 min to wash away unbound yeast. Calcofluor is a fluorescent dye that binds to and stains yeast cell walls (Fig. 1A). The addition of calcofluor during washing (rather than during the adhesion step) reduced potential effects that the fluorescent dye binding may have on yeast adhesion. After washing, channels were imaged on a Nikon TE2000 inverted microscope using a Roper CoolSNAP HQ camera and Metavue 6.42r6 software (Molecular Devices). For some experiments, a Nikon Ti-E microscope with an Andor Zyla sCMOS camera and NIS-Elements software (Nikon) were used. On each microscope, a motorized stage was used to expedite the imaging of adjacent fields of view, permitting the programmed sequential capture of >80% of the channel length (Fig. 1B). Yeast adhesion was then calculated by using quantitative image analysis. Images were thresholded to differentiate brightly fluorescent yeast cells from dim background. A single threshold value was used per experiment so that all microchannels were analyzed with the same cutoff value. The thresholded area was measured by software and divided by the total imaged channel area to determine the percentage of the channel surface area covered by adherent yeast cells, defined as the adhesion index. This approach was developed because calcofluor fluorescence is dependent on pH and yeast strains. Measurement of the thresholded area minimized minor variations of the fluorescence intensity upon the calculation of adhesion efficiency. Microchannels were run in duplicate. The use of automated image capture and quantitative analysis reduced variability and contributed to the reproducible measurement of yeast adhesion. Data from a minimum of 3 independent experiments were pooled and were not normally distributed. Medians and percentile ranges were calculated, and comparisons between groups were made by a Mann-Whitney rank sum test or Kruskal-Wallis analysis of variance (ANOVA) on ranks, as indicated, with P values of <0.05 being considered significant. Statistical analysis was performed by using SigmaPlot version 13.0.
RNA extraction, reverse transcription, and quantitative PCR.
RNA was isolated from cell pellets grown under the specified conditions, flash frozen in liquid nitrogen, and promptly stored at −80°C. Total RNA was extracted and purified by using the RiboPure Yeast extraction kit (Ambion/Life Technologies) according to the manufacturer's instructions. Prior to reverse transcription (RT), RNA samples were treated with DNase (Ambion/Life Technologies) to digest genomic DNA. RNA integrity was routinely monitored by agarose gel electrophoresis. cDNA was synthesized from 1 μg total RNA with a blend of oligo(dT) and random hexamer primers using iScript reverse transcriptase (Bio-Rad). To verify the absence of genomic DNA, no-reverse-transcriptase controls were run in parallel with each sample.
Quantitative PCR was performed by using iTaq Universal SYBR green (Bio-Rad) and the ABI 7500 Fast system (Applied Biosystems). Primer sequences are listed in Table 2. All primers were used at a concentration of 500 nM. Primer specificity was determined via melting curve analysis and agarose gel electrophoresis. Controls lacking a template were routinely run to check for contamination or primer-dimer formation. Amplification efficiencies were assessed by using relative standard curves constructed from a cDNA dilution series. All efficiencies were estimated to be 92 to 100%. Given the sensitivity required for our experiments, we determined that the relative quantitation (2−ΔΔCT) method was suitable to calculate expression levels. The relative RNA expression level of each candidate gene was quantified by comparing cells grown in M199 (induced) to cells grown in YPD medium (control).
To identify a suitable reference gene for RT-qPCR analyses, a series of housekeeping gene homologues from C. albicans (ACT1, PMA1, RIP1, RPP2B, LSC2, IMH3, TDH3, and CPA1) was evaluated for expression stability (35). Relative transcript levels were examined under different growth conditions, as well as across different strains, by using RT-qPCR. According to the Normfinder algorithm (36), ACT1 displayed the most stable expression profile and was therefore selected to be the reference gene for our studies.
PCR amplification of CpALS genes.
To test for the presence of each ALS gene at the genomic level in the C. parapsilosis strains used in this study (Table 1), standard PCR was conducted by using genomic DNA from each strain as the template and the primers for each gene listed in Table 2. Reactions were carried out by using Illustra Ready-To-Go PCR beads (GE Healthcare) under conditions recommended by the manufacturer. PCR products were analyzed by agarose gel electrophoresis.
Cloning and expression of CpALS7 in S. cerevisiae.
A circular polymerase extension cloning (CPEC) strategy (37) was used to clone the C. parapsilosis CpALS7 gene under the control of the GAL1 promoter in the yeast expression vector pYES2 (Invitrogen). The gene was PCR amplified from genomic DNA of JMB81 by using Phusion high-fidelity polymerase (Thermo Scientific) according to the manufacturer's instructions, with the following CPEC forward and reverse primers, respectively: 5′-GGCCGCCAGTGTGCGCACGAATCCAATGGTCAAAC-3′ and 5′-GCCAGTGTGATGGATATCTGCAGTTAGAATAACAAGAAAAGACTCAAGAC-3′. The product was purified through a spin column, and 100 ng was added to a CPEC reaction mixture with 100 ng pYES2 plasmid DNA linearized with EcoRI, 400 μM deoxynucleoside triphosphates (dNTPs), and 1 U Phusion polymerase. The reaction mixture was heated in a thermocycler to 98°C for 3 min, followed by 10 cycles of 98°C for 30 s, 55°C for 30 s, and 72°C for 2.5 min, and the reaction was completed with a 10-min incubation at 72°C. The entire reaction mixture was transformed into competent Escherichia coli Top10 cells, and ampicillin-resistant transformants were screened with appropriate restriction digests. The expected sequence of the resulting plasmid (pBMJ44) was confirmed by DNA sequencing. pBMJ44 was transformed into S. cerevisiae INVSc1 by electroporation. Transformants containing an empty vector (JMB278) or CpALS7 (JMB279) were selected on SC-URA (Sigma) and subsequently cultured in SC-URA at 30°C.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Arianna Tavanti for providing the CpALS7 deletion strain.
Research reported in this publication was supported by an institutional development award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health (P30GM114750) and the Kilguss Research Core at Women & Infants Hospital of Rhode Island. It was also supported by the William and Mary Oh-William and Elsa Zopfi Professorship in Pediatrics for Perinatal Research (S.K.S. and J.M.B.).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00892-17.
REFERENCES
- 1.Kullberg BJ, Arendrup MC. 2015. Invasive candidiasis. N Engl J Med 373:1445–1456. doi: 10.1056/NEJMra1315399. [DOI] [PubMed] [Google Scholar]
- 2.Noyola DE, Fernandez M, Moylett EH, Baker CJ. 2001. Ophthalmologic, visceral, and cardiac involvement in neonates with candidemia. Clin Infect Dis 32:1018–1023. doi: 10.1086/319601. [DOI] [PubMed] [Google Scholar]
- 3.Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, Marr KA, Pfaller MA, Chang CH, Webster KM. 2009. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis 48:1695–1703. doi: 10.1086/599039. [DOI] [PubMed] [Google Scholar]
- 4.Thompson DS, Carlisle PL, Kadosh D. 2011. Coevolution of morphology and virulence in Candida species. Eukaryot Cell 10:1173–1182. doi: 10.1128/EC.05085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hofs S, Mogavero S, Hube B. 2016. Interaction of Candida albicans with host cells: virulence factors, host defense, escape strategies, and the microbiota. J Microbiol 54:149–169. doi: 10.1007/s12275-016-5514-0. [DOI] [PubMed] [Google Scholar]
- 6.Hoyer LL, Cota E. 2016. Candida albicans agglutinin-like sequence (Als) family vignettes: a review of Als protein structure and function. Front Microbiol 7:280. doi: 10.3389/fmicb.2016.00280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoyer LL. 2001. The ALS gene family of Candida albicans. Trends Microbiol 9:176–180. doi: 10.1016/S0966-842X(01)01984-9. [DOI] [PubMed] [Google Scholar]
- 8.Hoyer LL, Green CB, Oh SH, Zhao X. 2008. Discovering the secrets of the Candida albicans agglutinin-like sequence (ALS) gene family—a sticky pursuit. Med Mycol 46:1–15. doi: 10.1080/13693780701435317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE, Filler SG. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol 5:e64. doi: 10.1371/journal.pbio.0050064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y, Phan QT, Luo G, Solis NV, Liu Y, Cormack BP, Edwards JE Jr, Ibrahim AS, Filler SG. 2013. Investigation of the function of Candida albicans Als3 by heterologous expression in Candida glabrata. Infect Immun 81:2528–2535. doi: 10.1128/IAI.00013-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida RS, Brunke S, Albrecht A, Thewes S, Laue M, Edwards JE, Filler SG, Hube B. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog 4:e1000217. doi: 10.1371/journal.ppat.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu Y, Filler SG. 2011. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 10:168–173. doi: 10.1128/EC.00279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira GH, Muller PR, Szeszs MW, Levin AS, Melhem MS. 2010. Five-year evaluation of bloodstream yeast infections in a tertiary hospital: the predominance of non-C. albicans Candida species. Med Mycol 48:839–842. doi: 10.3109/13693780903580121. [DOI] [PubMed] [Google Scholar]
- 14.Pfaller MA, Diekema DJ. 2004. Rare and emerging opportunistic fungal pathogens: concern for resistance beyond Candida albicans and Aspergillus fumigatus. J Clin Microbiol 42:4419–4431. doi: 10.1128/JCM.42.10.4419-4431.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Trofa D, Gacser A, Nosanchuk JD. 2008. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev 21:606–625. doi: 10.1128/CMR.00013-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skrzypek MS, Binkley J, Binkley G, Miyasato SR, Simison M, Sherlock G. 2017. The Candida Genome Database (CGD): incorporation of assembly 22, systematic identifiers and visualization of high throughput sequencing data. Nucleic Acids Res 45:D592–D596. doi: 10.1093/nar/gkw924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozik A, Karkowska-Kuleta J, Zajac D, Bochenska O, Kedracka-Krok S, Jankowska U, Rapala-Kozik M. 2015. Fibronectin-, vitronectin- and laminin-binding proteins at the cell walls of Candida parapsilosis and Candida tropicalis pathogenic yeasts. BMC Microbiol 15:197. doi: 10.1186/s12866-015-0531-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karkowska-Kuleta J, Zajac D, Bras G, Bochenska O, Rapala-Kozik M, Kozik A. 2017. Binding of human plasminogen and high-molecular-mass kininogen by cell surface-exposed proteins of Candida parapsilosis. Acta Biochim Pol 64:391–400. doi: 10.18388/abp.2017_1609. [DOI] [PubMed] [Google Scholar]
- 19.Bertini A, Zoppo M, Lombardi L, Rizzato C, De Carolis E, Vella A, Torelli R, Sanguinetti M, Tavanti A. 2016. Targeted gene disruption in Candida parapsilosis demonstrates a role for CPAR2_404800 in adhesion to a biotic surface and in a murine model of ascending urinary tract infection. Virulence 7:85–97. doi: 10.1080/21505594.2015.1112491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguire SL, OhEigeartaigh SS, Byrne KP, Schroder MS, O'Gaora P, Wolfe KH, Butler G. 2013. Comparative genome analysis and gene finding in Candida species using CGOB. Mol Biol Evol 30:1281–1291. doi: 10.1093/molbev/mst042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzpatrick DA, O'Gaora P, Byrne KP, Butler G. 2010. Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11:290. doi: 10.1186/1471-2164-11-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bliss JM, Wong AY, Bhak G, Laforce-Nesbitt SS, Taylor S, Tan S, Stoll BJ, Higgins RD, Shankaran S, Benjamin DK Jr. 2012. Candida virulence properties and adverse clinical outcomes in neonatal candidiasis. J Pediatr 161:441.e2–447.e2. doi: 10.1016/j.jpeds.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bliss JM, Basavegowda KP, Watson WJ, Sheikh AU, Ryan RM. 2008. Vertical and horizontal transmission of Candida albicans in very low birth weight infants using DNA fingerprinting techniques. Pediatr Infect Dis J 27:231–235. doi: 10.1097/INF.0b013e31815bb69d. [DOI] [PubMed] [Google Scholar]
- 24.Butler G, Rasmussen MD, Lin MF, Santos MA, Sakthikumar S, Munro CA, Rheinbay E, Grabherr M, Forche A, Reedy JL, Agrafioti I, Arnaud MB, Bates S, Brown AJ, Brunke S, Costanzo MC, Fitzpatrick DA, de Groot PW, Harris D, Hoyer LL, Hube B, Klis FM, Kodira C, Lennard N, Logue ME, Martin R, Neiman AM, Nikolaou E, Quail MA, Quinn J, Santos MC, Schmitzberger FF, Sherlock G, Shah P, Silverstein KA, Skrzypek MS, Soll D, Staggs R, Stansfield I, Stumpf MP, Sudbery PE, Srikantha T, Zeng Q, Berman J, Berriman M, Heitman J, Gow NA, Lorenz MC, Birren BW, Kellis M, et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662. doi: 10.1038/nature08064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pryszcz LP, Nemeth T, Gacser A, Gabaldon T. 2013. Unexpected genomic variability in clinical and environmental strains of the pathogenic yeast Candida parapsilosis. Genome Biol Evol 5:2382–2392. doi: 10.1093/gbe/evt185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JE Jr. 2004. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem 279:30480–30489. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- 27.Salgado PS, Yan R, Taylor JD, Burchell L, Jones R, Hoyer LL, Matthews SJ, Simpson PJ, Cota E. 2011. Structural basis for the broad specificity to host-cell ligands by the pathogenic fungus Candida albicans. Proc Natl Acad Sci U S A 108:15775–15779. doi: 10.1073/pnas.1103496108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin J, Oh SH, Jones R, Garnett JA, Salgado PS, Rusnakova S, Matthews SJ, Hoyer LL, Cota E. 2014. The peptide-binding cavity is essential for Als3-mediated adhesion of Candida albicans to human cells. J Biol Chem 289:18401–18412. doi: 10.1074/jbc.M114.547877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Isberg RR, Barnes P. 2002. Dancing with the host; flow-dependent bacterial adhesion. Cell 110:1–4. doi: 10.1016/S0092-8674(02)00821-8. [DOI] [PubMed] [Google Scholar]
- 30.Thomas W. 2008. Catch bonds in adhesion. Annu Rev Biomed Eng 10:39–57. doi: 10.1146/annurev.bioeng.10.061807.160427. [DOI] [PubMed] [Google Scholar]
- 31.Chan CX, Lipke PN. 2014. Role of force-sensitive amyloid-like interactions in fungal catch bonding and biofilms. Eukaryot Cell 13:1136–1142. doi: 10.1128/EC.00068-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.dela Paz NG, D'Amore PA. 2009. Arterial versus venous endothelial cells. Cell Tissue Res 335:5–16. doi: 10.1007/s00441-008-0706-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shaik SS, Soltau TD, Chaturvedi G, Totapally B, Hagood JS, Andrews WW, Athar M, Voitenok NN, Killingsworth CR, Patel RP, Fallon MB, Maheshwari A. 2009. Low intensity shear stress increases endothelial ELR+ CXC chemokine production via a focal adhesion kinase-p38β MAPK-NF-κB pathway. J Biol Chem 284:5945–5955. doi: 10.1074/jbc.M807205200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Coleman DA, Oh SH, Manfra-Maretta SL, Hoyer LL. 2012. A monoclonal antibody specific for Candida albicans Als4 demonstrates overlapping localization of Als family proteins on the fungal cell surface and highlights differences between Als localization in vitro and in vivo. FEMS Immunol Med Microbiol 64:321–333. doi: 10.1111/j.1574-695X.2011.00914.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nailis H, Coenye T, Van Nieuwerburgh F, Deforce D, Nelis HJ. 2006. Development and evaluation of different normalization strategies for gene expression studies in Candida albicans biofilms by real-time PCR. BMC Mol Biol 7:25. doi: 10.1186/1471-2199-7-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen CL, Jensen JL, Orntoft TF. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 37.Quan J, Tian J. 2014. Circular polymerase extension cloning. Methods Mol Biol 1116:103–117. doi: 10.1007/978-1-62703-764-8_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.