ABSTRACT
The pneumococcal capsular serotype is an important determinant of complement resistance and invasive disease potential, but other virulence factors have also been found to contribute. Pneumococcal surface protein C (PspC), a highly variable virulence protein that binds complement factor H to evade C3 opsonization, is divided into two subgroups: choline-bound subgroup I and LPxTG-anchored subgroup II. The prevalence of different PspC subgroups in invasive pneumococcal disease (IPD) and functional differences in complement evasion are unknown. The prevalence of PspC subgroups in IPD isolates was determined in a collection of 349 sequenced strains of Streptococcus pneumoniae isolated from adult patients. pspC deletion mutants and isogenic pspC switch mutants were constructed to study differences in factor H binding and complement evasion in relation to capsule thickness. Subgroup I pspC was far more prevalent in IPD isolates than subgroup II pspC. The presence of capsule was associated with a greater ability of bound factor H to reduce complement opsonization. Pneumococcal subgroup I PspC bound significantly more factor H and showed more effective complement evasion than subgroup II PspC in isogenic encapsulated pneumococci. We conclude that variation in the PspC subgroups, independent of capsule serotypes, affects pneumococcal factor H binding and its ability to evade complement deposition.
KEYWORDS: complement, factor H, PspC, Streptococcus pneumoniae, immune evasion, invasive disease
INTRODUCTION
Streptococcus pneumoniae is an important human pathogen that colonizes the upper respiratory tract. The pathogen is also an important cause of invasive diseases, such as pneumonia, sepsis, and meningitis. The pneumococcal polysaccharide capsule affects complement resistance and protects against phagocytic killing (1, 2). Epidemiological studies found that particular capsular serotypes are dominant in invasive disease whereas others are associated with nasopharyngeal carriage (3, 4). Besides the important role of the pneumococcal capsule, the genotype also affects complement resistance. Within the same serotype, significant differences in complement C3 deposition between isolates have been observed, indicating that the genetic background of the strain also affects complement resistance (5). A recent study suggests that within the same serotype and clonal complex, genetic differences in virulence genes encoding pneumococcal surface proteins A and C (PspA and PspC) affect the invasive disease potential (6). It is therefore of interest to gain more insight into how genetic variation in these genes affects complement resistance and contributes to pneumococcal virulence.
The complement system is an essential component of the host defense against S. pneumoniae (7). Complement activation by one of three pathways—the classical, lectin, and alternative pathways—leads to opsonization of the bacterial surface with the C3 activation products C3b and iC3b. These opsonins mediate phagocytosis mainly through complement receptors CR1 and CR3. Importantly, the alternative pathway amplifies the initial complement activation (8). C3b deposited on the bacterial surface is formed into an alternative-pathway C3 convertase cleaving more C3, which enhances C3b opsonization (9). The importance of the alternative pathway in complement activation is emphasized by the fact that many pathogens possess mechanisms to inhibit alternative-pathway activation by binding of the host alternative-pathway inhibitor factor H (10–14).
S. pneumoniae binds human factor H by PspC, also referred to as CbpA, SpsA, PbcA, and Hic (15–19). Factor H binding by PspC is a mechanism to evade complement deposition. In addition, PspC acts as an adhesion molecule by interacting with the secretory component of human IgA and the epithelial polymeric immunoglobulin receptor (pIgR) and binding to the laminin receptor on vascular endothelial cells, which facilitates adhesion and invasion (16, 17, 20–25). In vitro studies using human serum have demonstrated that factor H binding by S. pneumoniae strains is dependent on the presence of PspC but that the level of binding is influenced by the capsular serotype (1, 26).
The pspC gene shows large allelic variation. Eleven different types of pspC have been identified based on clusters of sequence homology. PspC consists of a C-terminal repeat region, a proline-rich domain, and N-terminal α-helical domains, also called R1 and R2 (27, 28). A factor H binding region of 121 amino acids (residues 38 to 158), containing multiple epitopes for factor H binding, has been identified in the N-terminal region (21). In the C-terminal region, a major difference in anchor sequence has been identified, dividing pspC into two subgroups: allelic types with a choline binding domain (classical; subgroup I) or an LPxTG-anchoring domain (nonclassical; subgroup II) (27).
The prevalences and distributions of the different PspC subgroups and types in invasive disease or carriage isolates have not been characterized thoroughly, although Iannelli et al. demonstrated a predominance of subgroup I PspC (74%) in a collection of 43 strains containing randomly chosen clinical isolates, standard laboratory strains, and American Type Culture Collection strains (27). However, it is not known whether variation in PspC type, independent of capsule differences, affects pneumococcal factor H binding and its ability to evade complement deposition. Here, we describe a far greater prevalence of choline-bound subgroup I PspC types than of LPxTG-anchored subgroup II PspC types in invasive pneumococcal disease (IPD) isolates. In addition, using isogenic pspC switch mutants, we demonstrate that subgroup I PspC is more effective in complement evasion than subgroup II PspC. These findings indicate that PspC-specific differences contribute to intraserotype variation in complement resistance.
RESULTS
PspC subgroup I is most prevalent in invasive pneumococcal disease isolates.
Analysis of the 349 invasive disease strains demonstrated that PspC subgroup I was present in 298 isolates (85.4%) and in an additional 19 isolates (5.4%) that contained both subgroup I and subgroup II PspC. Only 22 of the isolates (6.3%) contained subgroup II, and 10 isolates (2.9%) had no pspC (Table 1). Strains containing two pspC genes were mainly of serotypes 2, 6A, 6B, 19A, and 19F. Strains containing subgroup II were mostly serotype 3 with sequence type (ST) 180 (18 out of 22). Within a serotype, we found large variations in PspC types, whereas some serotypes contained only a single PspC type. Most sequence types were associated with a specific PspC type, though variation in PspC types within sequence types was also observed (see Table S2 in the supplemental material). We observed no association between PspC subgroups and disease outcomes.
TABLE 1.
PspC subgroup I is most prevalent in invasive pneumococcal disease isolatesa
| Subgroup | No. of isolates (n = 349) | % of total |
|---|---|---|
| PspC subgroup I (choline binding domain) | 298 | 85.4 |
| PspC subgroup II (LPxTG binding anchor) | 22 | 6.3 |
| PspC subgroups I and II | 19 | 5.4 |
| No PspC | 10 | 2.9 |
Capsule expression and PspC-mediated factor H binding contribute to complement evasion.
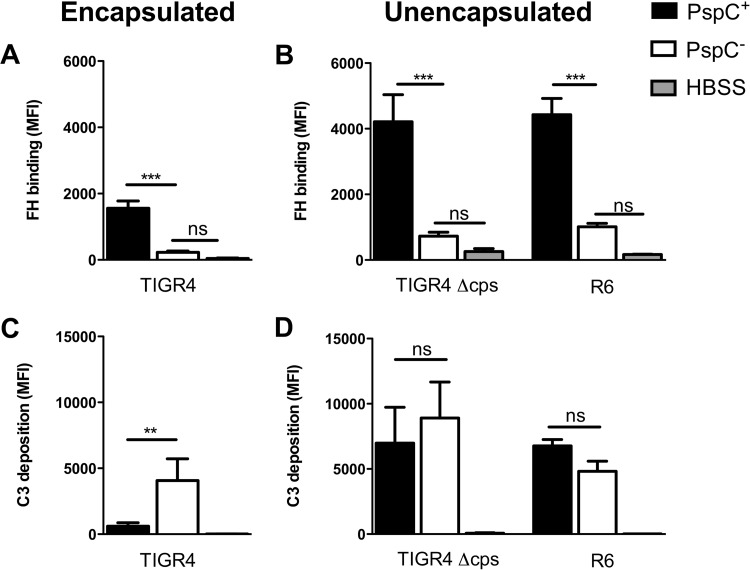
Capsular polysaccharide is known to inhibit complement opsonization (29). In addition, factor H binding by PspC is influenced by the capsular serotype (1, 2). We assessed the effect of the pneumococcal capsule on factor H binding to PspC. We compared factor H binding to the wild type and a pspC deletion (ΔpspC) mutant in encapsulated S. pneumoniae strain TIGR4, unencapsulated mutant TIGR4 (TIGR4 Δcps), and unencapsulated strain R6. In all backgrounds, deletion of pspC resulted in significantly reduced factor H binding as measured by flow cytometry following incubation in pooled human sera (Fig. 1A and B), demonstrating that PspC is the main factor H binding protein. Factor H binding to unencapsulated TIGR4 or R6 was higher than that to encapsulated TIGR4 (Fig. 1A and B).
FIG 1.
The effect of factor H (FH) binding on complement deposition is dependent on the presence of capsule. Factor H binding and C3 deposition on the surfaces of encapsulated S. pneumoniae strain TIGR4, unencapsulated TIGR4 (Δcps), and unencapsulated R6 and its pspC deletion mutants (PspC−) were measured by flow cytometry following incubation of the bacteria for 30 min at 37°C in 10% pooled human serum diluted in HBSS. (A and B) Factor H binding. (C and D) C3 deposition. MFI, mean fluorescence intensity. The bars represent means and standard deviations for results obtained from three or four separate experiments. Comparisons between strains were performed using ANOVA for repeated measurements with a Bonferroni correction for multiple comparisons. **, P < 0.01; ***, P < 0.001; ns, not significant.
We assessed the effects of pneumococcal capsule and pspC expression on complement resistance by comparing C3 opsonization of encapsulated TIGR4 and the pspC deletion mutant and of unencapsulated TIGR4 and R6. Encapsulated TIGR4 showed significantly reduced C3 opsonization compared to the ΔpspC mutant (Fig. 1C). To exclude the possibility that other factors besides binding of factor H affected bacterial C3 opsonization, we performed this experiment with mouse serum because mouse C3 is able to opsonize the bacterial surface (30) but mouse factor H is not able to bind PspC (31). With the use of mouse serum, no differences in C3 opsonization between encapsulated TIGR4 and the ΔpspC mutant were observed (see Fig. S1 in the supplemental material). This demonstrates that for encapsulated TIGR4, factor H binding contributes to complement evasion by reducing C3 opsonization. Loss of pspC had no effect on C3 opsonization for unencapsulated TIGR4 and the unencapsulated R6 strain (Fig. 1D). This indicated that factor H binding to PspC of unencapsulated TIGR4 and R6 did not contribute to complement resistance.
PspC subgroup-specific differences in factor H binding and complement resistance.
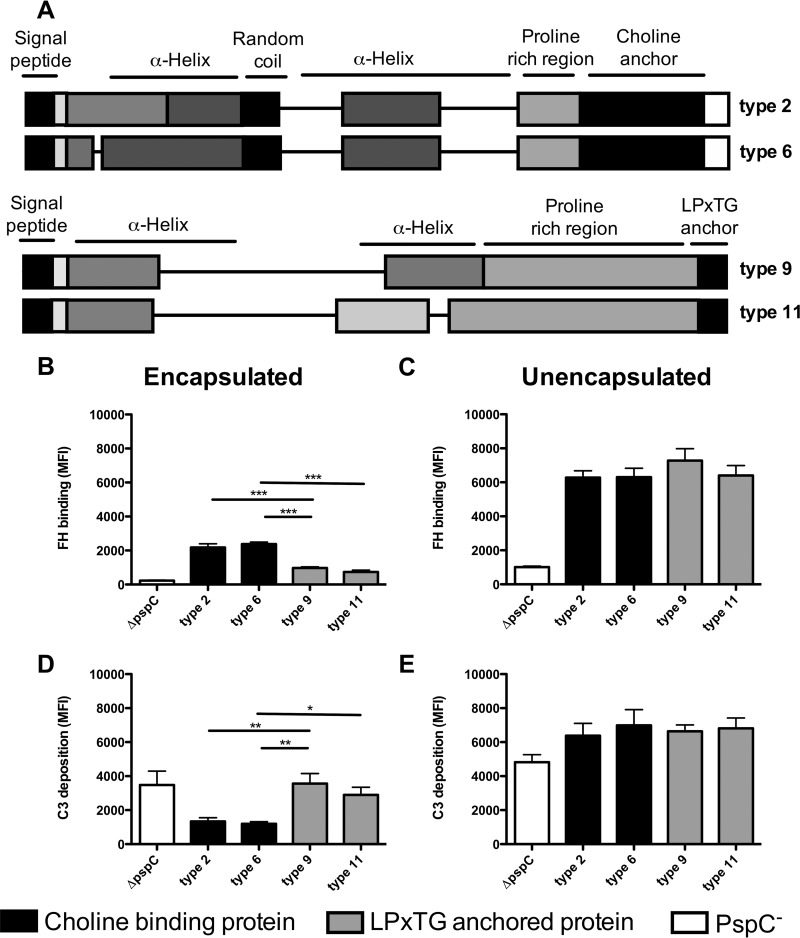
PspC types 2, 6, 9, and 11 were selected because PspC types 2 and 6 (subgroup I) have a choline binding domain and PspC types 9 and 11 (subgroup II) have an LPxTG binding anchor, and these types are heterogeneous in their factor H binding regions (Fig. 2A). The four pspC types were cloned into TIGR4, replacing the original pspC gene, in order to study PspC type-specific differences in complement evasion within the same genetic background. Quantitative PCR confirmed that the pspC gene expression levels of the 4 different PspC types within TIGR4 or R6 were equal (see Fig. S2 in the supplemental material). Due to the large amino acid diversity between the different PspC types, detection of protein expression by Western blot analysis using polyclonal mouse serum was not successful.
FIG 2.
PspC subgroup-specific differences affect factor H binding and complement resistance in encapsulated TIGR4, but not in unencapsulated R6. (A) Schematic representation of pspC type 2, 6, 9, and 11 domains based on the classification by Iannelli et al. (27). (B to E) Factor H binding and C3 deposition on the surface of S. pneumoniae TIGR4 ΔpspC (white bars) or TIGR4 containing pspC types 2 and 6 of subgroup I (black bars) or pspC types 9 and 11 of subgroup II (gray bars). Factor H (FH) binding and C3 deposition were measured by flow cytometry following incubation of the bacteria for 30 min at 37°C in 10% pooled human sera diluted in HBSS. (B and C) Factor H binding. (D and E) C3 deposition. The bars represent means and standard deviations for results obtained from three separate experiments. Comparisons between pspC types were performed using ANOVA for repeated measurements with a Bonferroni correction for multiple comparisons. *, P < 0.05; **, P < 0.01, ***, P < 0.001.
We found significantly enhanced factor H binding to pneumococci expressing subgroup I PspC types (2 and 6) compared to subgroup II PspC types (9 and 11) in the TIGR4 genetic background (Fig. 2B). Consistently, factor H binding was inversely correlated with C3 deposition on the bacterial surface, and thus, subgroup I PspC types 2 and 6 showed significantly reduced C3 deposition compared to subgroup II PspC types 9 and 11 in the encapsulated mutants (Fig. 2D). This demonstrated that PspC-specific differences affected factor H binding and complement resistance in encapsulated pneumococci. In an unencapsulated background, no difference in factor H binding between the various PspC types of subgroups I and II was found (Fig. 2C). In addition, no difference in C3 opsonization was found between the subgroup I and subgroup II PspC types in the unencapsulated R6 strains (Fig. 2E).
DISCUSSION
In this study, we found that subgroup I PspC types were more prevalent in IPD isolates than subgroup II PspC types. Expression of different PspC types in isogenic encapsulated pneumococci showed that subgroup I PspC types were more effective in complement evasion than subgroup II PspC types. In addition, we found that capsule affects PspC-mediated complement evasion. Previous studies examining the role of PspC in complement evasion have been performed in encapsulated strains (1, 26). Different serotypes showed large variation in the effect on complement deposition upon loss of PspC, even though all serotypes were shown to bind factor H via PspC (26). In line with this, we found that pneumococcal factor H binding is mainly PspC dependent, although some residual factor H binding was observed. This may be explained by factor H binding to Pht family proteins (32) or elongation factor Tu (Tuf), which was recently found to bind human factor H (33). In this study, we demonstrated that absence of a capsule within the same genetic background voids the ability of factor H binding to reduce complement deposition. These findings are in line with previous studies demonstrating that unencapsulated strains are more sensitive for complement deposition (2, 29). This might be explained by the fact that the absence of a capsule enhances antibody binding to subcapsular antigens and increases binding of complement mediators, such as C-reactive protein, both known to activate the complement classical pathway (2, 34). Enhanced classical-pathway activation may minimize the effect of alternative-pathway inhibition by factor H binding on overall complement deposition. This may explain why factor H binding had no effect on complement C3 deposition for the unencapsulated strains (2, 29).
The current study demonstrates that capsule affects factor H binding and its role in complement evasion. Even though encapsulated TIGR4 bound less factor H than unencapsulated TIGR4, the binding of factor H resulted in reduced complement deposition, suggesting that complement evasion by factor H binding is mainly important in the presence of capsule. This is further supported by the observation that thickly encapsulated, opaque phase variants are often found in invasive infections, in which pneumococcal complement evasion is vital for survival (29, 34–36).
Our findings indicate that factor H binding to unencapsulated pneumococci does not reduce C3 deposition on the bacterial surface. Unencapsulated pneumococci are rarely seen to cause disease (37) but are found in 4 to 19% of colonizing S. pneumoniae isolates (38, 39). During nasopharyngeal carriage, other functions of PspC have been indicated by previous reports showing its importance in adherence (16, 17, 20–23, 40). In addition, our observation that PspC binding of factor H by unencapsulated strains had no effect on complement resistance may help in understanding the loss of PspC in naturally occurring unencapsulated S. pneumoniae strains (41, 42). Other gene products in the naturally occurring unencapsulated strains, such as the PspC-like protein PspK, were shown to play a role in colonization and adherence but did not bind factor H (43–45).
We demonstrated that genetic differences in pspC affect complement resistance. Previous studies found differences in pneumococcal factor H binding between clinical isolates (1). However, this could not be attributed to differences in pspC alone, since other factors, such as the capsule type, also varied (1, 21, 26). In the current study, four PspC types selected from a cohort with IPD isolates were cloned into an isogenic background, isolating the effects of PspC type differences on factor H binding and complement resistance. Remarkably, in the absence of capsule, we found no differences in factor H binding between strains expressing the various PspC types, even though the sequences of the previously defined 121-amino-acid-long factor H binding region varied extensively (21). This indicates that all four PspC types have the same ability to bind factor H but that PspC in combination with the serotype 4 capsule resulted in differences in factor H binding. The lengths of the coding sequences vary between the PspC types, as pspC types 2 and 6 are 2,082 bp and 2,046 bp, respectively, whereas pspC groups 9 and 11 are 1,458 bp and 1,245 bp, respectively. These differences in length may explain the observed differences in factor H binding in combination with capsule. Some clinical isolates contain both PspC types 6 and 9. Our findings demonstrate that both are able to bind factor H for unencapsulated strains, but in the presence of capsule, PspC type 6 binds more factor H and decreases C3 deposition on the bacterial surface to a greater extent than PspC type 9.
Another important difference between the high and low factor H binding PspC types in our study is the PspC C-terminal domain, as PspC types 2 and 6 (subgroup I) have a choline binding domain and PspC types 9 and 11 (subgroup II) have an LPxTG binding anchor. Interestingly, the IPD isolate cohort described in this paper consists mainly of PspC types of subgroup I, whereas a much lower percentage of the PspC types belong to subgroup II. Analysis of strain collections including carriage and invasive isolates from the same region and period may help to further dissect the contribution of PspC subgroups and types to invasive disease potential. Additionally, more insight into epidemiological differences in PspC type prevalence and their contributions to invasive disease may have implications for vaccine design, because PspC is an important vaccine candidate (46–48). Our study demonstrates PspC subgroup and type distributions in invasive disease analogous to serotype distribution in invasive disease. No comparative data from a large set of clinical invasive disease isolates have been published to date, although Iannelli et al. demonstrated a predominance of subgroup I PspC (74%) in a collection of 43 strains containing randomly chosen clinical isolates, standard laboratory strains, and American Type Culture Collection strains (27). Thus, we cannot exclude the possibility that subgroup I PspC may also be more prevalent and may provide a selective advantage in mucosal infection and colonization. From our present study and the available data, we can conclude only that subgroup I PspC is more prevalent in invasive disease than subgroup II PspC, which correlates with increased complement resistance.
A strength of our study is that we studied various PspC types in an isogenic background. Others have demonstrated that the capsule type affects factor H binding to PspC and complement evasion in capsule switch mutants (1). Our findings are complementary to these data and demonstrate that genetic differences in pspC, using pspC switch mutants, affect complement resistance in the presence of polysaccharide capsule. This is in line with a previous pediatric study in which genetic variation in pspC was suggested to explain differences in invasiveness within the same serotype and clonal complex (6).
In conclusion, we found a higher prevalence of subgroup I PspC types than subgroup II PspC types in IPD isolates. Expression of different PspC types in isogenic encapsulated pneumococci showed that subgroup I PspC types are more effective in complement evasion than subgroup II PspC types. In addition, we showed that capsule thickness affects PspC-mediated complement evasion. These findings indicate that PspC type-specific differences contribute to intraserotype variation in complement resistance.
MATERIALS AND METHODS
Pneumococcal strain collection, sequence typing, and PspC typing.
The prevalences of various pspC types were studied in 349 sequenced S. pneumoniae isolates from IPD patients in Nijmegen, The Netherlands (2001 to 2011). All the bacteremia isolates were from patients with pneumonia (n = 312), meningitis (n = 30), endocarditis (n = 4), arthritis (n = 1), peritonitis (n = 1), and sinusitis (n = 1) (49, 50). The median age was 67 years (interquartile range, 53 to 78 years), and the male/female ratio was 0.86. This observational cohort study was approved by the local medical ethics committees of both participating hospitals. Genome sequences, serotypes, and sequence types were obtained from Cremers et al. (49). PspC-coding genes were identified from the genome sequences by aligning the 40-amino-acid conserved N terminus of PspC with the protein-coding sequences of all the genomes using BLASTP (51). PspC types were determined by aligning the PspC-coding genes with the known PspC gene sequences (27) using BLASTP, and the BLAST results were manually inspected.
Construction of pspC deletion mutants and isogenic pspC switch mutants.
Four different pspC types were selected from the bacteremia cohort and constructed into an isogenic background. The pspC types have been described by Iannelli et al. (27). Based on this description, we selected two PspC types with a choline binding anchor and two PspC types with an LPxTG binding anchor in the C-terminal domain with differences in the N-terminal factor H binding domain. To obtain these PspC types, we searched our strain collection and found isolates containing pspC genes belonging to the previously described PspC types. pspC type 2 was obtained from strain PBCN0094 (10050_2#53) and was 100% identical to the previously described pspC 2.2 (27), also found in TIGR4. pspC types 6 and 9 were both obtained from strain PBCN0133 (10050_2#83) and were cloned separately into an isogenic background. pspC type 9 showed 91% identity to the previously described pspC type 9.1, with all differences in the repeat region, and pspC type 6 showed 100% identity to the previously described pspC type 6.1. pspC type 11, also known as Hic, was found in strain PBCN0031 (10050_2#18) and showed 97% identity with the previously described pspC type 11.3. The pspC types found in these strains were cloned into an encapsulated TIGR4 strain and an unencapsulated R6 strain.
The various pspC types were cloned into the pspC deletion mutants to avoid recombination with the original pspC gene. Again, a megaprimer PCR product was constructed but contained, besides the pspC-flanking regions and a spectinomycin cassette, the selected pspC type originating from the above-mentioned clinical isolates. The spectinomycin cassette and the right flank were PCR amplified from TIGR4 ΔpspC containing the spectinomycin resistance cassette. The megaprimer PCR product was used to transform R6 ΔpspC (kanamycin resistant). Directed mutants were obtained by selective plating (using spectinomycin) and were checked for correct integration of the antibiotic resistance cassette and pspC-type gene by PCR using control primers located inside the genes. A PCR product from the left flank to the right flank containing the inserted pspC type and the spectinomycin cassette was used to transform TIGR4 ΔpspC (kanamycin resistant). Mutants were obtained by selective plating and were checked for correct integration of the antibiotic resistance cassette and pspC-type gene by PCR using control primers located inside the genes and sequencing. The primer sequences (obtained from Biolegio, Nijmegen, The Netherlands) are listed in Table S1 in the supplemental material. Correct integration of the pspC gene was confirmed by sequencing.
TIGR4 ΔpspC and R6 ΔpspC deletion mutants were constructed by allelic replacement of the target gene with an antibiotic resistance marker as described previously (52).
Bacterial strains and culture conditions.
Bacteria were grown on Columbia blood agar plates (Becton Dickinson) and in Todd-Hewitt broth supplemented with 5 g/liter yeast extract (THY) at 37°C and 5% CO2 to an optical density at 620 nm (OD620) of 0.2. The number of CFU per milliliter was determined by plating serial 10-fold dilutions on blood agar plates.
Factor H binding and C3 deposition assay.
Wild-type and mutant TIGR4 and R6 strains (1 × 107 CFU) were pelleted in a 96-well plate and resuspended in 10% (vol/vol) pooled normal human sera (GTI Diagnostic) or 20% (vol/vol) C57BL/6 mouse serum (Innovative Research) in Hanks balanced salt solution (HBSS) containing Ca2+ and Mg2+ to a total volume of 100 μl. The bacterial suspension was incubated for 30 min at 37°C in 5% CO2. After incubation, the bacteria were washed and labeled with polyclonal sheep anti-human factor H (Abcam), fluorescein isothiocyanate (FITC)-conjugated goat anti-human C3 (Cappel), or FITC-conjugated goat anti-mouse C3 (Cappel) diluted in phosphate-buffered saline (PBS) plus 2% bovine serum albumin (BSA). After 30 min of incubation, the bacteria were washed and the anti-factor H antibody was labeled with FITC-donkey anti-sheep IgG antibody (Jackson Immunoresearch). The bacteria were fixed with 2% paraformaldehyde. Factor H binding and C3 deposition were measured using an LSR II flow cytometer (BD Biosciences). The data were analyzed using FlowJo v10.1.
Quantitative-PCR analysis.
RNA from overnight-grown strains of TIGR4 and R6 expressing pspC type 2, 6, 9, or 11 was extracted using an RNeasy minikit (Qiagen), and DNA was removed with Turbo-DNase (Ambion). The RNA (1 μg) was reverse transcribed into cDNA using an iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed using SYBR green chemistry (Bio-Rad) and PspC-specific primers on a CFX-96 real-time PCR machine (Bio-Rad) (see Table S1 in the supplemental material). GyrA was used as a reference gene. Quantification cycle (Cq) values and relative expression values were calculated using CFX Manager software (Bio-Rad).
Statistics.
Differences between strains were analyzed using repeated-measures analysis of variance (ANOVA) with Bonferroni corrections for multiple comparisons. Differences were considered statistically significant when the P value was <0.05.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by the European Union's Seventh Framework Program (EC-GA no. 279185), the European Childhood Life-Threatening Infectious Diseases Study (EUCLIDS). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00010-18.
REFERENCES
- 1.Hyams C, Trzcinski K, Camberlein E, Weinberger DM, Chimalapati S, Noursadeghi M, Lipsitch M, Brown JS. 2013. Streptococcus pneumoniae capsular serotype invasiveness correlates with the degree of factor H binding and opsonization with C3b/iC3b. Infect Immun 81:354–363. doi: 10.1128/IAI.00862-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. 2010. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun 78:704–715. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brueggemann AB, Griffiths DT, Meats E, Peto T, Crook DW, Spratt BG. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J Infect Dis 187:1424–1432. doi: 10.1086/374624. [DOI] [PubMed] [Google Scholar]
- 4.Melin M, Trzcinski K, Antonio M, Meri S, Adegbola R, Kaijalainen T, Kayhty H, Vakevainen M. 2010. Serotype-related variation in susceptibility to complement deposition and opsonophagocytosis among clinical isolates of Streptococcus pneumoniae. Infect Immun 78:5252–5261. doi: 10.1128/IAI.00739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyams C, Opel S, Hanage W, Yuste J, Bax K, Henriques-Normark B, Spratt BG, Brown JS. 2011. Effects of Streptococcus pneumoniae strain background on complement resistance. PLoS One 6:e24581. doi: 10.1371/journal.pone.0024581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browall S, Norman M, Tangrot J, Galanis I, Sjostrom K, Dagerhamn J, Hellberg C, Pathak A, Spadafina T, Sandgren A, Battig P, Franzen O, Andersson B, Ortqvist A, Normark S, Henriques-Normark B. 2014. Intraclonal variations among Streptococcus pneumoniae isolates influence the likelihood of invasive disease in children. J Infect Dis 209:377–388. doi: 10.1093/infdis/jit481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ram S, Lewis LA, Rice PA. 2010. Infections of people with complement deficiencies and patients who have undergone splenectomy. Clin Microbiol Rev 23:740–780. doi: 10.1128/CMR.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harboe M, Mollnes TE. 2008. The alternative complement pathway revisited. J Cell Mol Med 12:1074–1084. doi: 10.1111/j.1582-4934.2008.00350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harboe M, Ulvund G, Vien L, Fung M, Mollnes TE. 2004. The quantitative role of alternative pathway amplification in classical pathway induced terminal complement activation. Clin Exp Immunol 138:439–446. doi: 10.1111/j.1365-2249.2004.02627.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beernink PT, Leipus A, Granoff DM. 2006. Rapid genetic grouping of factor H-binding protein (genome-derived neisserial antigen 1870), a promising group B meningococcal vaccine candidate. Clin Vaccine Immunol 13:758–763. doi: 10.1128/CVI.00097-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haapasalo K, Vuopio J, Syrjanen J, Suvilehto J, Massinen S, Karppelin M, Jarvela I, Meri S, Kere J, Jokiranta TS. 2012. Acquisition of complement factor H is important for pathogenesis of Streptococcus pyogenes infections: evidence from bacterial in vitro survival and human genetic association. J Immunol 188:426–435. doi: 10.4049/jimmunol.1102545. [DOI] [PubMed] [Google Scholar]
- 12.Sharp JA, Echague CG, Hair PS, Ward MD, Nyalwidhe JO, Geoghegan JA, Foster TJ, Cunnion KM. 2012. Staphylococcus aureus surface protein SdrE binds complement regulator factor H as an immune evasion tactic. PLoS One 7:e38407. doi: 10.1371/journal.pone.0038407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Langereis JD, de Jonge MI, Weiser JN. 2014. Binding of human factor H to outer membrane protein P5 of non-typeable Haemophilus influenzae contributes to complement resistance. Mol Microbiol 94:89–106. doi: 10.1111/mmi.12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rosadini CV, Ram S, Akerley BJ. 2014. Outer membrane protein P5 is required for resistance of nontypeable Haemophilus influenzae to both the classical and alternative complement pathways. Infect Immun 82:640–649. doi: 10.1128/IAI.01224-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dave S, Pangburn MK, Pruitt C, McDaniel LS. 2004. Interaction of human factor H with PspC of Streptococcus pneumoniae. Indian J Med Res 119(Suppl):66–73. [PubMed] [Google Scholar]
- 16.Hammerschmidt S, Talay SR, Brandtzaeg P, Chhatwal GS. 1997. SpsA, a novel pneumococcal surface protein with specific binding to secretory immunoglobulin A and secretory component. Mol Microbiol 25:1113–1124. doi: 10.1046/j.1365-2958.1997.5391899.x. [DOI] [PubMed] [Google Scholar]
- 17.Rosenow C, Ryan P, Weiser JN, Johnson S, Fontan P, Ortqvist A, Masure HR. 1997. Contribution of novel choline-binding proteins to adherence, colonization and immunogenicity of Streptococcus pneumoniae. Mol Microbiol 25:819–829. doi: 10.1111/j.1365-2958.1997.mmi494.x. [DOI] [PubMed] [Google Scholar]
- 18.Cheng Q, Finkel D, Hostetter MK. 2000. Novel purification scheme and functions for a C3-binding protein from Streptococcus pneumoniae. Biochemistry 39:5450–5457. doi: 10.1021/bi992157d. [DOI] [PubMed] [Google Scholar]
- 19.Janulczyk R, Iannelli F, Sjoholm AG, Pozzi G, Bjorck L. 2000. Hic, a novel surface protein of Streptococcus pneumoniae that interferes with complement function. J Biol Chem 275:37257–37263. doi: 10.1074/jbc.M004572200. [DOI] [PubMed] [Google Scholar]
- 20.Hava DL, Camilli A. 2002. Large-scale identification of serotype 4 Streptococcus pneumoniae virulence factors. Mol Microbiol 45:1389–1406. doi: 10.1046/j.1365-2958.2002.03106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hammerschmidt S, Agarwal V, Kunert A, Haelbich S, Skerka C, Zipfel PF. 2007. The host immune regulator factor H interacts via two contact sites with the PspC protein of Streptococcus pneumoniae and mediates adhesion to host epithelial cells. J Immunol 178:5848–5858. doi: 10.4049/jimmunol.178.9.5848. [DOI] [PubMed] [Google Scholar]
- 22.Elm C, Rohde M, Vaerman JP, Chhatwal GS, Hammerschmidt S. 2004. Characterization of the interaction of the pneumococcal surface protein SpsA with the human polymeric immunoglobulin receptor (hpIgR). Indian J Med Res 119(Suppl):61–65. [PubMed] [Google Scholar]
- 23.Zhang JR, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, Tuomanen E. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827–837. doi: 10.1016/S0092-8674(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 24.Iovino F, Molema G, Bijlsma JJ. 2014. Streptococcus pneumoniae interacts with pIgR expressed by the brain microvascular endothelium but does not co-localize with PAF receptor. PLoS One 9:e97914. doi: 10.1371/journal.pone.0097914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orihuela CJ, Mahdavi J, Thornton J, Mann B, Wooldridge KG, Abouseada N, Oldfield NJ, Self T, Ala'Aldeen DA, Tuomanen EI. 2009. Laminin receptor initiates bacterial contact with the blood brain barrier in experimental meningitis models. J Clin Invest 119:1638–1646. doi: 10.1172/JCI36759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuste J, Khandavilli S, Ansari N, Muttardi K, Ismail L, Hyams C, Weiser J, Mitchell T, Brown JS. 2010. The effects of PspC on complement-mediated immunity to Streptococcus pneumoniae vary with strain background and capsular serotype. Infect Immun 78:283–292. doi: 10.1128/IAI.00541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iannelli F, Oggioni MR, Pozzi G. 2002. Allelic variation in the highly polymorphic locus pspC of Streptococcus pneumoniae. Gene 284:63–71. doi: 10.1016/S0378-1119(01)00896-4. [DOI] [PubMed] [Google Scholar]
- 28.Luo R, Mann B, Lewis WS, Rowe A, Heath R, Stewart ML, Hamburger AE, Sivakolundu S, Lacy ER, Bjorkman PJ, Tuomanen E, Kriwacki RW. 2005. Solution structure of choline binding protein A, the major adhesin of Streptococcus pneumoniae. EMBO J 24:34–43. doi: 10.1038/sj.emboj.7600490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Q, Li YX, Douthitt K, Stahl GL, Thurman JM, Tong HH. 2012. Role of the alternative and classical complement activation pathway in complement mediated killing against Streptococcus pneumoniae colony opacity variants during acute pneumococcal otitis media in mice. Microbes Infect 14:1308–1318. doi: 10.1016/j.micinf.2012.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Maten E, Westra D, van Selm S, Langereis JD, Bootsma HJ, van Opzeeland FJ, de Groot R, Ruseva MM, Pickering MC, van den Heuvel LP, van de Kar NC, de Jonge MI, van der Flier M. 2016. Complement factor H serum levels determine resistance to pneumococcal invasive disease. J Infect Dis 213:1820–1827. doi: 10.1093/infdis/jiw029. [DOI] [PubMed] [Google Scholar]
- 31.Lu L, Ma Z, Jokiranta TS, Whitney AR, DeLeo FR, Zhang JR. 2008. Species-specific interaction of Streptococcus pneumoniae with human complement factor H. J Immunol 181:7138–7146. doi: 10.4049/jimmunol.181.10.7138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogunniyi AD, Grabowicz M, Mahdi LK, Cook J, Gordon DL, Sadlon TA, Paton JC. 2009. Pneumococcal histidine triad proteins are regulated by the Zn2+-dependent repressor AdcR and inhibit complement deposition through the recruitment of complement factor H. FASEB J 23:731–738. doi: 10.1096/fj.08-119537. [DOI] [PubMed] [Google Scholar]
- 33.Mohan S, Hertweck C, Dudda A, Hammerschmidt S, Skerka C, Hallstrom T, Zipfel PF. 2014. Tuf of Streptococcus pneumoniae is a surface displayed human complement regulator binding protein. Mol Immunol 62:249–264. doi: 10.1016/j.molimm.2014.06.029. [DOI] [PubMed] [Google Scholar]
- 34.Kim JO, Romero-Steiner S, Sorensen UB, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun 67:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Briles DE, Hollingshead SK, Paton JC, Ades EW, Novak L, van Ginkel FW, Benjamin WH Jr. 2003. Immunizations with pneumococcal surface protein A and pneumolysin are protective against pneumonia in a murine model of pulmonary infection with Streptococcus pneumoniae. J Infect Dis 188:339–348. doi: 10.1086/376571. [DOI] [PubMed] [Google Scholar]
- 36.Weiser JN. 1998. Phase variation in colony opacity by Streptococcus pneumoniae. Microb Drug Resist 4:129–135. doi: 10.1089/mdr.1998.4.129. [DOI] [PubMed] [Google Scholar]
- 37.Park IH, Geno KA, Sherwood LK, Nahm MH, Beall B. 2014. Population-based analysis of invasive nontypeable pneumococci reveals that most have defective capsule synthesis genes. PLoS One 9:e97825. doi: 10.1371/journal.pone.0097825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chewapreecha C, Harris SR, Croucher NJ, Turner C, Marttinen P, Cheng L, Pessia A, Aanensen DM, Mather AE, Page AJ, Salter SJ, Harris D, Nosten F, Goldblatt D, Corander J, Parkhill J, Turner P, Bentley SD. 2014. Dense genomic sampling identifies highways of pneumococcal recombination. Nat Genet 46:305–309. doi: 10.1038/ng.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sulikowska A, Grzesiowski P, Sadowy E, Fiett J, Hryniewicz W. 2004. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolated from the nasopharynges of asymptomatic children and molecular analysis of S. pneumoniae and H influenzae strain replacement in the nasopharynx. J Clin Microbiol 42:3942–3949. doi: 10.1128/JCM.42.9.3942-3949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quin LR, Onwubiko C, Moore QC, Mills MF, McDaniel LS, Carmicle S. 2007. Factor H binding to PspC of Streptococcus pneumoniae increases adherence to human cell lines in vitro and enhances invasion of mouse lungs in vivo. Infect Immun 75:4082–4087. doi: 10.1128/IAI.00474-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Keller LE, Thomas JC, Luo X, Nahm MH, McDaniel LS, Robinson DA. 2013. Draft genome sequences of five multilocus sequence types of nonencapsulated Streptococcus pneumoniae. Genome Announc 1:e00520-. doi: 10.1128/genomeA.00520-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavares DA, Simoes AS, Bootsma HJ, Hermans PW, de Lencastre H, Sa-Leao R. 2014. Non-typeable pneumococci circulating in Portugal are of cps type NCC2 and have genomic features typical of encapsulated isolates. BMC Genomics 15:863. doi: 10.1186/1471-2164-15-863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Valentino MD, McGuire AM, Rosch JW, Bispo PJ, Burnham C, Sanfilippo CM, Carter RA, Zegans ME, Beall B, Earl AM, Tuomanen EI, Morris TW, Haas W, Gilmore MS. 2014. Unencapsulated Streptococcus pneumoniae from conjunctivitis encode variant traits and belong to a distinct phylogenetic cluster. Nat Commun 5:5411. doi: 10.1038/ncomms6411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park IH, Kim KH, Andrade AL, Briles DE, McDaniel LS, Nahm MH. 2012. Nontypeable pneumococci can be divided into multiple cps types, including one type expressing the novel gene pspK. mBio 3:e00035-. doi: 10.1128/mBio.00035-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keller LE, Jones CV, Thornton JA, Sanders ME, Swiatlo E, Nahm MH, Park IH, McDaniel LS. 2013. PspK of Streptococcus pneumoniae increases adherence to epithelial cells and enhances nasopharyngeal colonization. Infect Immun 81:173–181. doi: 10.1128/IAI.00755-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brooks-Walter A, Briles DE, Hollingshead SK. 1999. The pspC gene of Streptococcus pneumoniae encodes a polymorphic protein, PspC, which elicits cross-reactive antibodies to PspA and provides immunity to pneumococcal bacteremia. Infect Immun 67:6533–6542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vadesilho CF, Ferreira DM, Gordon SB, Briles DE, Moreno AT, Oliveira ML, Ho PL, Miyaji EN. 2014. Mapping of epitopes recognized by antibodies induced by immunization of mice with PspA and PspC. Clin Vaccine Immunol 21:940–948. doi: 10.1128/CVI.00239-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moreno AT, Oliveira ML, Ho PL, Vadesilho CF, Palma GM, Ferreira JM Jr, Ferreira DM, Santos SR, Martinez MB, Miyaji EN. 2012. Cross-reactivity of antipneumococcal surface protein C (PspC) antibodies with different strains and evaluation of inhibition of human complement factor H and secretory IgA binding via PspC. Clin Vaccine Immunol 19:499–507. doi: 10.1128/CVI.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cremers AJ, Mobegi FM, de Jonge MI, van Hijum SA, Meis JF, Hermans PW, Ferwerda G, Bentley SD, Zomer AL. 2015. The post-vaccine microevolution of invasive Streptococcus pneumoniae. Sci Rep 5:14952. doi: 10.1038/srep14952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cremers AJ, Meis JF, Walraven G, Jongh CE, Ferwerda G, Hermans PW. 2014. Effects of 7-valent pneumococcal conjugate 1 vaccine on the severity of adult 2 bacteremic pneumococcal pneumonia. Vaccine 32:3989–3994. doi: 10.1016/j.vaccine.2014.04.089. [DOI] [PubMed] [Google Scholar]
- 51.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burghout P, Bootsma HJ, Kloosterman TG, Bijlsma JJ, de Jongh CE, Kuipers OP, Hermans PW. 2007. Search for genes essential for pneumococcal transformation: the RADA DNA repair protein plays a role in genomic recombination of donor DNA. J Bacteriol 189:6540–6550. doi: 10.1128/JB.00573-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.