ABSTRACT
Chlamydia pecorum is an important intracellular bacterium that causes a range of diseases in animals, including a native Australian marsupial, the koala. In humans and animals, a gamma interferon (IFN-γ)-mediated immune response is important for the control of intracellular bacteria. The present study tested the hypotheses that C. pecorum can escape IFN-γ-mediated depletion of host cell tryptophan pools. In doing so, we demonstrated that, unlike Chlamydia trachomatis, C. pecorum is completely resistant to IFN-γ in human epithelial cells. While the growth of C. pecorum was inhibited in tryptophan-deficient medium, it could be restored by the addition of kynurenine, anthranilic acid, and indole, metabolites that could be exploited by the gene products of the C. pecorum tryptophan biosynthesis operon. We also found that expression of trp genes was detectable only when C. pecorum was grown in tryptophan-free medium, with gene repression occurring in response to the addition of kynurenine, anthranilic acid, and indole. When grown in bovine kidney epithelial cells, bovine IFN-γ also failed to restrict the growth of C. pecorum, while C. trachomatis was inhibited, suggesting that C. pecorum could use the same mechanisms to evade the immune response in vivo in its natural host. Highlighting the different mechanisms triggered by IFN-γ, however, both species failed to grow in murine McCoy cells treated with murine IFN-γ. This work confirms previous hypotheses about the potential survival of C. pecorum after IFN-γ-mediated host cell tryptophan depletion and raises questions about the immune pathways used by the natural hosts of C. pecorum to control the widespread pathogen.
KEYWORDS: Chlamydia; IFN-γ; tryptophan synthase; indoleamine 2,3-dioxygenase (IDO1); in vitro IFN-γ
INTRODUCTION
Chlamydiae are significant obligate intracellular bacterial pathogens of humans and animals. An important characteristic of chlamydiae is their biphasic developmental cycle consisting of the extracellular infectious elementary body (EB) and the metabolically active reticulate body (1). At the beginning of infection, elementary bodies are taken up by the cell into a membranous vacuole. Within the vacuole, the elementary bodies transform into reticulate bodies, which then undergo replication. The reticulate bodies eventually transform back into infectious elementary bodies and are then released via host cell lysis to infect neighboring cells (2). Another well-characterized feature of the chlamydial cell cycle is the in vitro ability to convert to a viable but noninfectious aberrant form (3–5) as one phenotype of the chlamydial stress response (6). This stress response can be induced by different factors, including gamma interferon (IFN-γ), penicillin exposure, and chlamydia phage infection, as well as amino acid or iron deprivation (7–10). The first of these, IFN-γ, is an important cytokine secreted by T cells and is involved with the host defense against Chlamydia infection (11).
While IFN-γ induces many host cellular responses, including the activation of p47 GTPases in murine cell lines (12, 13), one of the most important responses in terms of antichlamydial activity in human epithelial cells is the IFN-γ-mediated induction of the indoleamine 2,3-dioxygenase (IDO1) enzyme, which catalyzes l-tryptophan to l-kynurenine, resulting in inhibition of chlamydial growth and replication (14). In turn, one of the primary ways that chlamydial species appear to be able to resist IFN-γ exposure is via the presence and activity of the tryptophan biosynthesis operon in their genomes (15, 16). As in other bacteria, some chlamydial species have a complete or incomplete tryptophan biosynthesis operon regulated by an aporepressor, TrpR (17). When tryptophan is available, it enables TrpR to bind its cognate operator and repress the operon, while under tryptophan-limiting conditions, TrpR is unable to form the complex with the operator, resulting in tryptophan biosynthesis gene expression.
Genetic variation in the presence and completeness of the operon between chlamydial species, and indeed between strains of the same species, appears to be closely linked to the ability to survive IFN-γ-mediated tryptophan depletion (15, 16). Chlamydia trachomatis genital strains carry an incomplete tryptophan synthase gene (trpBA), which enables them to synthesize tryptophan only from indole (18, 19). In contrast, Chlamydia pneumoniae is highly sensitive to IFN-γ, as it does not have any tryptophan-biosynthetic genes in its genome (20–22). Chlamydia caviae contains an almost complete tryptophan operon (trpRDFCAB), kynU, and prsA (23) and, in HeLa cells, is completely resistant to IFN-γ, suggesting the presence of functional tryptophan biosynthesis enzymes that are capable of utilizing a range of metabolites, such as kynurenine, anthranilate, and indole, to synthesize tryptophan and restore growth under tryptophan-limiting conditions (24). While this situation appears to be consistent across most of the chlamydial species, there are exceptions: for instance, Chlamydia muridarum appears to resist the antichlamydial effects of IFN-γ in mouse cells via a different mechanism. IFN-γ priming induces p47 GTPases, not IDO1, in mouse epithelial cells (12). A large cytotoxin produced by C. muridarum was originally proposed to circumvent and escape the bactericidal effects of p47 GTPases (13); however, this has been refuted more recently following mutational studies (25). The genetic basis of C. muridarum IFN-γ resistance in murine cells thus remains unknown.
Chlamydia pecorum is recognized as an important pathogen that has been associated with diseases in livestock (cattle, sheep, goats, and pigs) and wild animals, including the koala (26–28). Clinical manifestations in livestock include polyarthritis, keratoconjunctivitis, encephalomyelitis, and abortion (29, 30). Chlamydial polyarthritis can appear rapidly and typically affects growing lambs from 4 to 8 months of age (28). Affected animals may develop debilitating arthritis that interferes with grazing and leads to weight loss and growth retardation (31). Beyond these reports, perhaps the most common outcome of C. pecorum infection is the absence of disease signs, with growing evidence that these subclinical infections may nevertheless impact the growth of affected animals, particularly dairy cattle (32, 33). C. pecorum is also a major driver of declines in the numbers of a native Australian marsupial, the koala (27). While much is still to be learned about the biology of the pathogen, recent breakthroughs in chlamydial genomics have revealed that the genome of the pathogen is largely conserved across different species and that if biological differences exist, they are influenced by only a limited number of single nucleotide polymorphisms (34, 35).
Unlike most of the other chlamydial species except C. caviae, C. pecorum carries an almost complete tryptophan biosynthesis operon containing the trpRDFCAB genes (35, 36). Notably, unlike in the other chlamydial species, this operon is not found within the chlamydial plasticity zone, a genetically variable region of chlamydial genomes normally found to contain virulence factors. In their place in this region, C. pecorum instead encodes several predicted cytotoxins (35), similar to those previously thought to be involved in C. muridarum IFN-γ resistance (25). Based on the presence of these genetic loci, we hypothesized that C. pecorum is resistant to IFN-γ exposure in vitro. Building on recent research that has begun to characterize the stress response of this important veterinary pathogen (37, 38), in the current study, we investigated the impact of IFN-γ on growth inhibition of C. pecorum in a range of different cell lines to explore the different mechanisms that C. pecorum may potentially use in vivo to avoid this important antichlamydial activity of the host.
RESULTS
Impact of IFN-γ treatment on the growth of C. pecorum in HEp-2 cells.
Previous studies of other chlamydial species have investigated the chlamydial response to IFN-γ-mediated depletion of host cell tryptophan pools in various cell lines (7, 22, 39, 40). In order to understand how C. pecorum responds to this phenomenon, the growth characteristics of the C. pecorum strains (IPA and E58) were measured in human epithelial cells treated with IFN-γ. These characteristics were compared to those of C. trachomatis D grown under similar conditions. Previous studies also revealed that the most significant growth inhibition occurred when the host cells were pretreated with IFN-γ (13). Accordingly, HEp-2 cells were preexposed to various concentrations (0, 200, 500, and 1,000 U/ml) of IFN-γ for 24 h prior to infection. IFN-γ was replenished every 24 h until 36 h postinfection (hpi) to deplete intracellular tryptophan pools, as previously described (22).
Under these conditions, viability experiments to assess the production of infectious C. pecorum EBs revealed no significant effect on the growth of C. pecorum cultures treated with IFN-γ (Fig. 1), with minor reductions in detectable C. pecorum inclusion-forming units (IFU) observed at 1,000 U IFN-γ/ml. At similar and lower concentrations, IFN-γ treatment resulted in a significant decrease in the growth of C. trachomatis D in comparison to the control group supplied with normal Dulbecco's modified Eagle's medium (DMEM) (P < 0.001). In the presence of 500 U IFN-γ/ml, the number of C. trachomatis D IFU per milliliter was reduced from that in the control infection by approximately 50%, whereas treatment with 1,000 U of IFN-γ/ml resulted in no detectable remaining infectious C. trachomatis EBs (Fig. 1).
FIG 1.
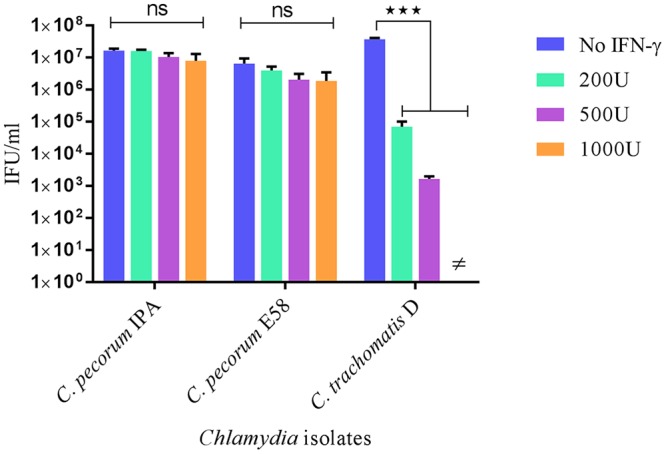
Viable infectious yields of C. pecorum IPA, C. pecorum E58, and C. trachomatis D strains after treatment with different concentrations of IFN-γ. Shown are the yields of C. pecorum IPA, C. pecorum E58, and C. trachomatis D generated after IFN-γ treatment of HEp-2 cell cultures. The HEp-2 cultures were pretreated with 0, 200, 500, and 1,000 U of IFN-γ/ml for 24 h and infected at an MOI of 0.5. The cultures were harvested at 36 hpi, and the numbers of infectious progeny were determined by measuring inclusion-forming units that formed infections in a new monolayer of host cells. ≠, no IFU were detected. The error bars indicate SD of the mean of three replicates (n = 9). Statistics were determined using unpaired two-tailed t tests. ***, P < 0.001 versus untreated controls; ns, not significant.
IDO1 expressed in IFN-γ-treated HEp2 cells infected by Chlamydia.
IFN-γ treatment of human cells induces IDO1, leading to catalysis of l-tryptophan and subsequent reduction of chlamydial growth (41). In order to confirm activation of IDO1, we infected HEp-2 cells preexposed to 1,000 U IFN-γ/ml with both C. pecorum and C. trachomatis D isolates and measured the IFN-γ-induced IDO1 activation by (i) expression of the IDO1 gene and (ii) Western blotting to monitor the presence of IDO1 in cultures. As shown in Fig. 2A, the level of IDO1 gene expression was significantly increased in C. pecorum IPA, C. pecorum E58, and C. trachomatis D cultures treated with IFN-γ compared to untreated cultures, with peak IDO1 mRNA gene expression detected in HEp-2 cells treated with 1,000 U/ml (P < 0.001). To confirm translation of the IDO1 protein, we also detected IDO1 expression by Western blotting using anti-IDO1 rabbit monoclonal antibody (Fig. 2B). Consistent with the IDO1 gene expression detected, IDO1 protein was expressed when Chlamydia-infected cultures were exposed to IFN-γ. Taken together, these observations confirmed that C. pecorum-infected HEp-2 cells expressed IDO1 following IFN-γ exposure and that IDO1 expression was not inhibited by C. pecorum IPA, C. pecorum E58, or C. trachomatis D infection.
FIG 2.
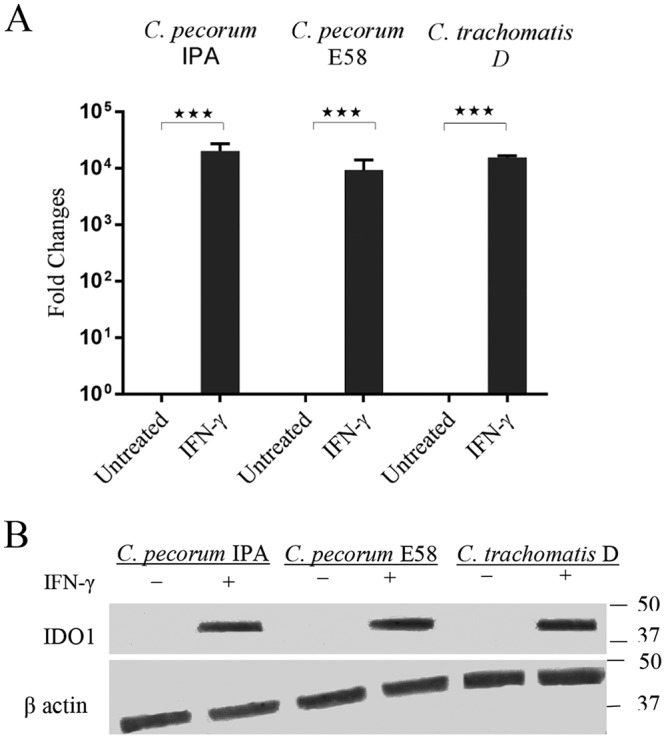
Presence of IDO1 protein in IFN-γ-treated Chlamydia cultures. (A) IDO1 gene expression in HEp-2 cells stimulated with IFN-γ and infected with C. pecorum IPA, C. pecorum E58, or C. trachomatis D. Total RNA was collected from untreated (DMEM) and treated (1,000 U IFN-γ/ml) groups at 20 hpi. The results for IDO1 were normalized against β-actin. The data are expressed as the fold increase relative to the untreated group. Statistics were determined using unpaired two-tailed t tests. ***, P < 0.001 versus untreated controls. (B) IDO1 protein expression of IFN-γ (1,000 U/ml)-treated HEp-2 cell lysate probed with anti-IDO1 monoclonal antibody. The sizes of relevant molecular mass markers in kDa are indicated on the right, and the Western blot band identities are indicated on the left.
C. pecorum can utilize the intermediate substrates of tryptophan metabolism.
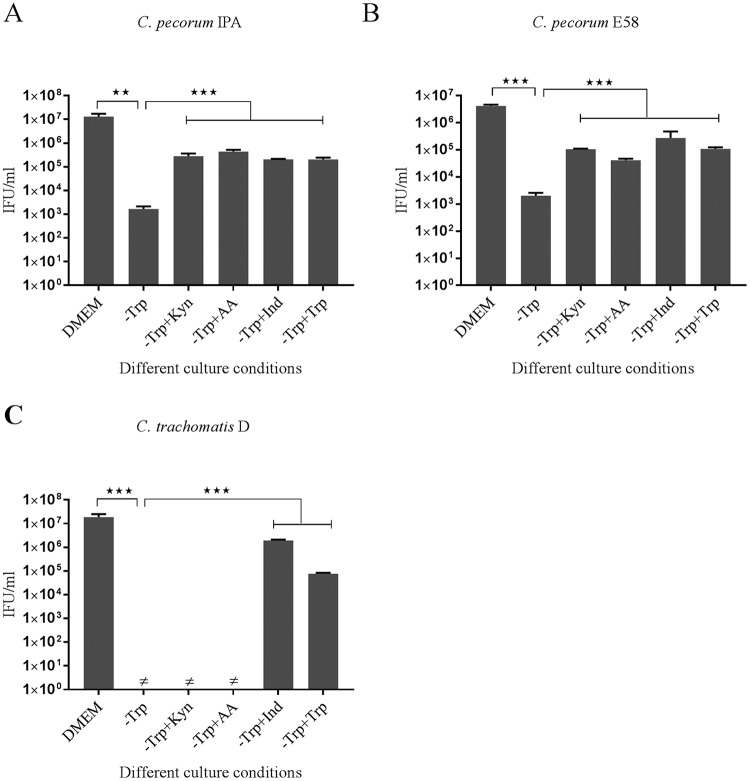
The success of C. pecorum strains in avoiding inhibition of growth in IFN-γ-treated cultures led us to develop an alternate hypothesis that, as revealed in the genome sequences of the pathogen, C. pecorum may respond to tryptophan depletion through the activity of the encoded products of the tryptophan synthase operon. In the next experiments, we tested a hypothesis that C. pecorum can utilize kynurenine, anthranilic acid (AA), and indole to synthesize l-tryptophan through the activity of the intact tryptophan synthase operon. To do so, we infected HEp-2 cells with C. pecorum IPA, C. pecorum E58, and C. trachomatis D strains under the following conditions: (i) in DMEM, (ii) in tryptophan-free DMEM, (iii) in tryptophan-free DMEM supplemented with l-kynurenine, (iv) in tryptophan-free DMEM supplemented with anthranilic acid, and (v) in tryptophan-free medium supplemented with indole. Cultures grown in tryptophan-free medium and rescued by the addition of l-tryptophan were also prepared as controls.
A significant decrease in viable infectious yields for C. pecorum IPA and C. pecorum E58 was observed when they were grown in tryptophan-free medium compared to the growth of the isolates in DMEM (P < 0.005) (Fig. 3A and B). No infectious progeny were observed for C. trachomatis D isolates grown in tryptophan-free medium (Fig. 3C). For the C. pecorum IPA isolate cultured in tryptophan-free medium, supplementation with alternative tryptophan substrates resulted in significantly increased yields of C. pecorum (P < 0.001). A similar trend was also observed for the C. pecorum E58 strains grown by the addition of kynurenine, anthranilic acid, and indole to the tryptophan-free medium (P < 0.001). In contrast, significant restoration of C. trachomatis D growth was achieved only when the isolates grown in tryptophan-free medium were supplemented with indole (P < 0.001). When the growth of all cultures in tryptophan-free medium was supplemented by exogenous tryptophan (fresh DMEM containing 16 mg/ml tryptophan according to the manufacturer's instructions), we were able to demonstrate rescue of all strains by observation of detectable IFU. For C. pecorum, this resulted in 103 to 105 IFU/ml recovered, while C. trachomatis experienced significant recovery of up to 104 IFU/ml (P < 0.001) (Fig. 3). Collectively, these results provide strong evidence that the capacity of C. pecorum to avoid the IFN-γ immune invasion in vitro is mainly due to the recycling abilities of different tryptophan precursors.
FIG 3.
Viable infectious yield of C. pecorum IPA, C. pecorum E58, and C. trachomatis D after supplementation with alternative substrates of tryptophan biosynthesis. HEp-2 cells infected with C. pecorum IPA, C. pecorum E58, or C. trachomatis D were grown at an MOI of 0.5 in DMEM; tryptophan-free medium (−Trp); or tryptophan-free culture rescued by l-kynurenine (−Trp+Kyn), anthranilic acid (−Trp+AA), indole (−Trp+Ind), or tryptophan (−Trp+Trp). The cultures were harvested at 36 hpi, and the infectious progeny were determined by measuring inclusion-forming units that formed infections in a new monolayer of host cells. The error bars represent SD of the means of three replicates (n = 9). Statistics were determined using unpaired two-tailed t tests. **, P < 0.005; ***, P < 0.001 versus untreated controls.
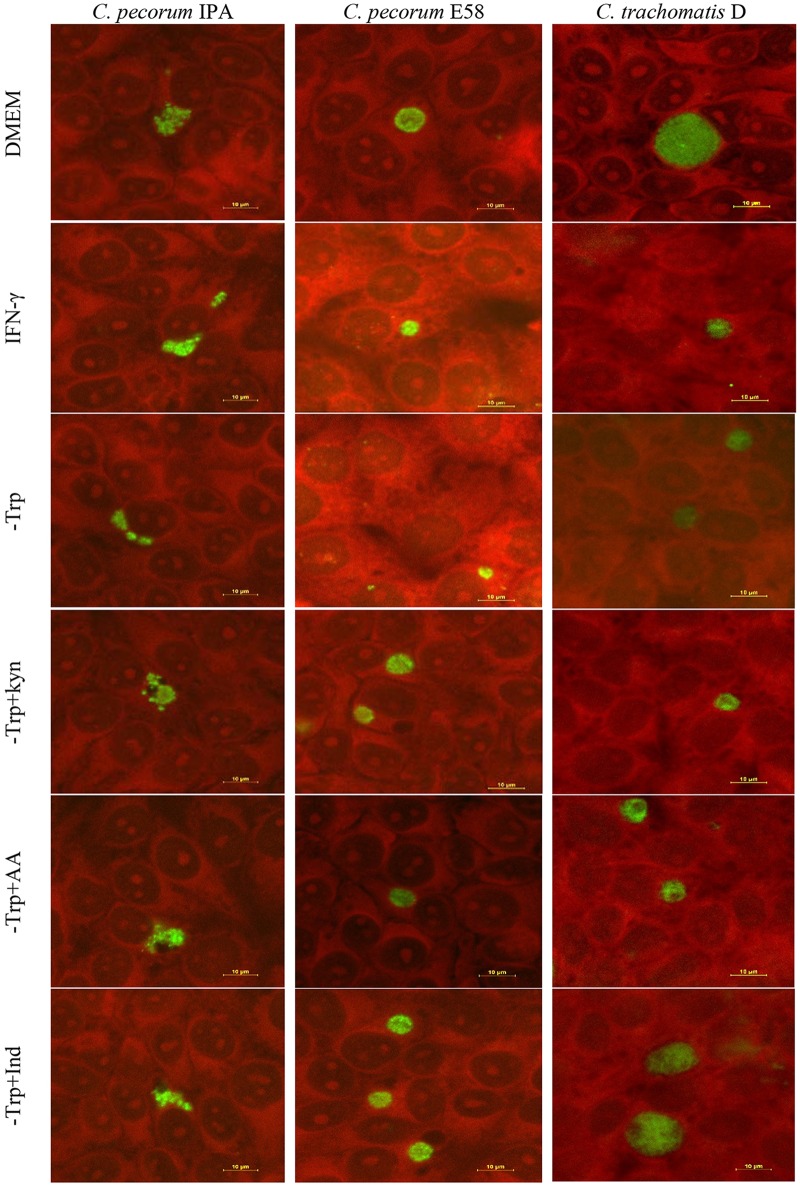
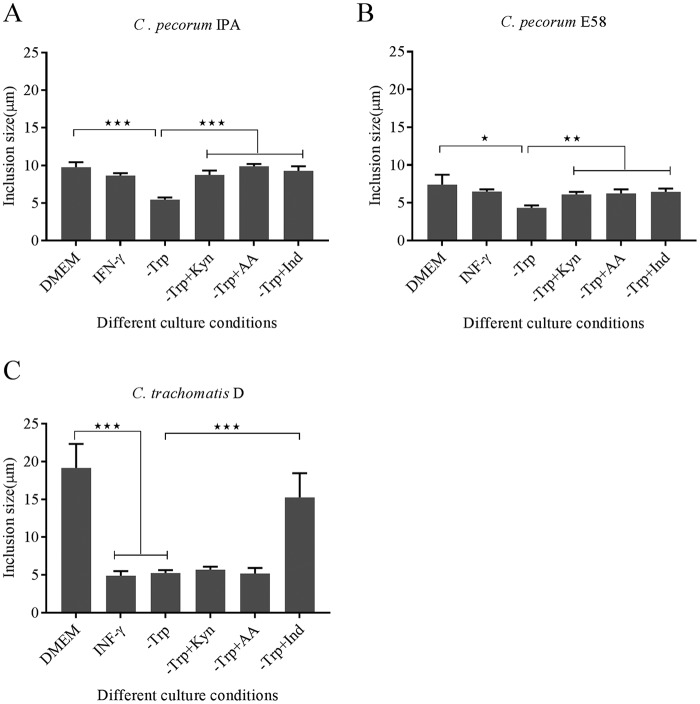
The chlamydial stress response induced by IFN-γ and other factors is typically characterized by the presence of aberrant reticulate bodies and a smaller number of inclusions with reduced size in vitro (21). Therefore, cultures under the above-described conditions were examined by chlamydial immunofluorescence and confocal microscopy to examine chlamydial inclusion morphology at 30 hpi. C. pecorum IPA and C. pecorum E58 inclusion morphology was varied in shape, with the former most often irregularly crescent shaped and the latter round (Fig. 4). In addition, C. trachomatis inclusions were generally round (Fig. 4). With the exception of the tryptophan-free-medium cultures (mean inclusion diameter, 5.45 ± 0.22 μm for IPA and 4.36 ± 0.24 μm for E58), C. pecorum inclusion sizes remained relatively the same under all other conditions tested (Fig. 5A and B). In contrast, for C. trachomatis D, significantly smaller inclusions were observable under IFN-γ exposure (4.9 ± 0.49 μm), in tryptophan-free medium (5.26 ± 0.26 μm), and in tryptophan-free medium supplemented with kynurenine (5.69 ± 0.32 μm) and anthranilic acid (5.2 ± 0.58 μm) culture conditions than in the control cultures (19.17 ± 2.59 μm) (P < 0.001) (Fig. 5C). The inclusion size was restored only when the C. trachomatis cultures were supplemented with indole (14.58 ± 2.62 μm) (Fig. 5C).
FIG 4.
Representative confocal microscopic images of C. pecorum IPA, C. pecorum E58, and C. trachomatis D isolates grown in HEp-2 cells. The culture conditions are indicated on the left. The cultures were grown at an MOI of 0.5 in DMEM, IFN-γ-treated (1,000 U/ml) medium; tryptophan-free medium (−Trp); or tryptophan-free culture rescued by l-kynurenine (−Trp+Kyn), anthranilic acid (−Trp+AA), or indole (−Trp+Ind) and stained with Chlamydia cell LPS at 30 hpi. Distinct inclusions (green) were detected under each condition within the host cell (red). Scale bars, 10 μm.
FIG 5.
Inclusion size measurement in untreated DMEM; IFN-γ-treated (1,000 U/ml) medium; tryptophan-free medium (−Trp); or tryptophan-free culture rescued by l-kynurenine (−Trp+Kyn), anthranilic acid (−Trp+AA), or indole (−Trp+Ind) at 30 hpi. The inclusion sizes were measured under each condition from three independent coverslips. The error bars represent SD of the means of three replicates (n = 20). Statistics were determined using unpaired two-tailed t tests. *, P < 0.05; **, P < 0.005; ***, P < 0.001 versus untreated controls.
Regulation of tryptophan gene expression in C. pecorum.
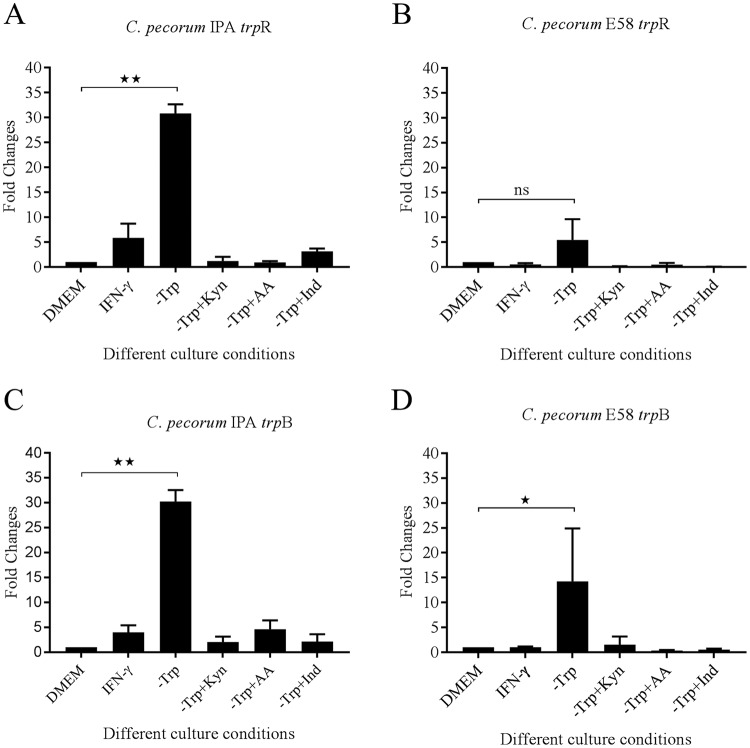
The trpR gene, considered the tryptophan-dependent regulator of tryptophan metabolism, regulates the expression of the chlamydial tryptophan biosynthesis genes (17). To understand trpR gene regulation in the C. pecorum response to tryptophan depletion, we investigated trpR gene expression in C. pecorum-infected HEp-2 cells upon exposure to IFN-γ or growth in tryptophan-free medium or following supplementation with tryptophan precursors by quantitative reverse transcription (qRT)-PCR. Compared to cultures grown in DMEM with tryptophan, we observed strong relative upregulation (>5-fold) of trpR gene expression for C. pecorum E58 cultured in tryptophan-free medium (Fig. 6B). This upregulation was even more significantly pronounced (>29-fold) for the C. pecorum IPA isolate grown in tryptophan-free medium (P < 0.005) (Fig. 6A). When we investigated trpR gene expression following addition of tryptophan precursors, such as l-kynurenine, anthranilic acid, and indole in the same cultures, trpR gene expression returned to baseline levels, similar to what was observed for the C. pecorum cultures grown in control DMEM. Similarly, when tryptophan was depleted via treatment with IFN-γ, the relative trpR transcript levels of C. pecorum IPA and E58 were similar to those of untreated DMEM cultures.
FIG 6.
trpR and trpB gene expression in HEp-2 cells. Hep-2 cells were infected with C. pecorum IPA and C. pecorum E58 strains in DMEM (control group); DMEM with IFN-γ; tryptophan-deficient medium (−Trp); or tryptophan-free medium supplemented with l-kynurenine (−Trp+Kyn), anthranilic acid (−Trp+AA), or indole (−Trp+Ind). Total RNA was isolated from infected cultures at 20 hpi. The trpR and trpB results were then normalized with 16S rRNA transcripts. The results are expressed as fold changes relative to the untreated DMEM group. (A) trpR gene expression of C. pecorum IPA in Hep-2 cells (n = 2; means and SD). (B) trpR gene expression of C. pecorum E58 in Hep-2 cells (n = 2; means and SD). (C) trpB gene expression of C. pecorum IPA in Hep-2 cells (n = 2; means and SD). (D) trpB gene expression of C. pecorum E58 in Hep-2 cells (n = 2; means and SD). Statistics were determined using unpaired two-tailed t tests. *, P < 0.05; **, P < 0.005; ns, not significant versus untreated controls.
We also tested trpB gene expression under all the conditions tested for C. pecorum trpR expression to evaluate cotranscription of the genes within the operon (Fig. 6C and D). In a pattern similar to that of trpR, and as expected, trpB gene expression was significantly upregulated in the C. pecorum cultures grown in tryptophan-free medium (P < 0.05). The relative expression of trpB was also observed to be similar to that of trpR for C. pecorum IPA and E58 when grown in tryptophan-free medium supplemented with tryptophan precursors.
C. pecorum strains grown in bovine epithelial cells are resistant to IFN-γ exposure.
As humans are not the natural host of C. pecorum, we were interested to examine the response of C. pecorum strains E58 (bovine origin) and IPA (ovine origin) to bovine IFN-γ. Previous reports suggested that bovine cells expressed tryptophan-degrading enzyme, IDO1, after stimulation with bovine IFN-γ in vitro (42, 43). Therefore, in order to assess whether the bovine cells treated with IFN-γ inhibit the growth of C. pecorum in vitro, we pretreated bovine kidney epithelial (BK) cells with 1,000 U/ml bovine IFN-γ 24 h prior to infection with either C. pecorum E58, C. pecorum IPA, or C. trachomatis D. IFN-γ-treated cultures of C. pecorum IPA yielded 1.9 × 107 IFU/ml, while cultures with DMEM yielded 4.2 × 107 IFU/ml (Fig. 7). A similar pattern was observed for the C. pecorum E58 strain, demonstrating only a minor detectable loss of IFU when grown in IFN-γ-exposed cultures. In contrast, IFN-γ-treated cultures significantly inhibited C. trachomatis D strain growth (no detectable viable yield) compared to DMEM cultures (P < 0.001) (Fig. 7). Based on these results, we concluded that all the strains in this experiment could grow in BK cells and that IFN-γ stimulation did not inhibit C. pecorum growth.
FIG 7.
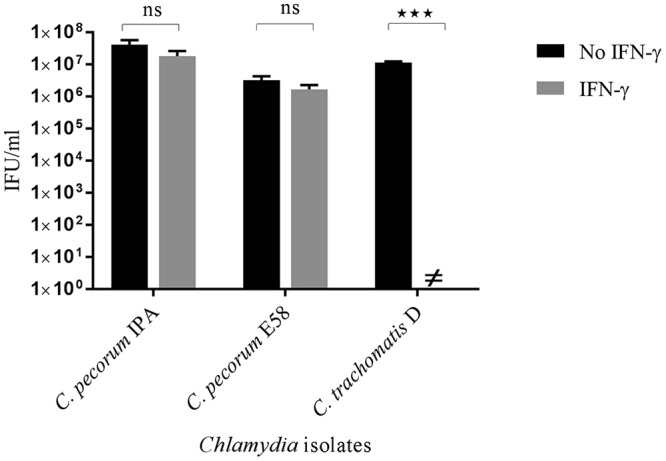
Effect of bovine IFN-γ on the growth of C. pecorum IPA, C. pecorum E58, and C. trachomatis D. BK cell monolayers pretreated with bovine IFN-γ (1,000 U/ml) were infected with the three strains at an MOI of 0.5. The cultures were harvested at 36 hpi, and the viable yield was determined by measuring inclusion-forming units that formed infections in a new monolayer of BK cells. ≠, no IFU were detected. The error bars represent SD of the means of three replicates (n = 9). Statistics were determined using unpaired two-tailed t tests. ***, P < 0.001; ns, not significant versus untreated controls.
C. pecorum strains are sensitive to IFN-γ in mouse cells.
Unlike IDO1 induction by IFN-γ in human and bovine cells, mouse epithelial cells induce p47 GTPases when stimulated by mouse IFN-γ (13, 44). Therefore, in order to determine whether mouse IFN-γ had an effect on the growth of C. pecorum strains grown in McCoy cells, C. pecorum and C. trachomatis isolates were used to infect McCoy cell monolayers exposed to IFN-γ. Interestingly, C. pecorum IPA infection in mouse cells treated with IFN-γ resulted in a significant, approximately 4 log10-fold reduction in infectious progeny at 36 hpi (P < 0.05) (Fig. 8). Similarly, the growth of C. pecorum E58 in IFN-γ-treated cultures was reduced significantly, from 3.0 × 107 IFU/ml in the DMEM control group to 3.8 × 103 IFU/ml. Similar results were also observed in C. trachomatis D-infected cultures following IFN-γ treatment, with significant reductions in detectable yields of EBs observed in comparison to their untreated counterparts (P < 0.001).
FIG 8.
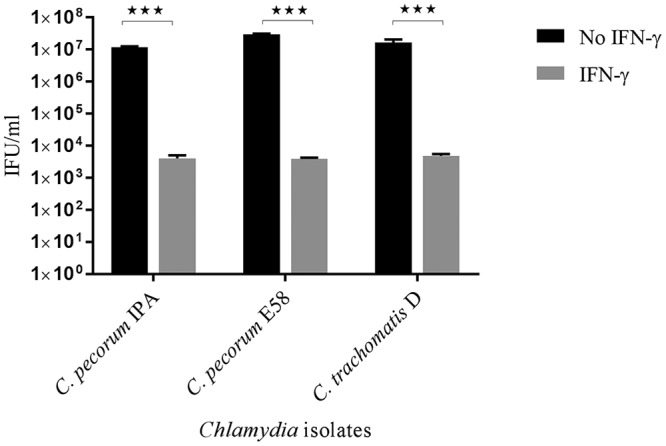
Effect of murine IFN-γ on the growth of C. pecorum IPA, C. pecorum E58, and C. trachomatis D in mouse cells. McCoy cell monolayers pretreated with murine IFN-γ (1,000 U/ml) were infected with the three strains at an MOI of 0.5. The cultures were harvested at 36 hpi, and the viable yields were determined by measuring inclusion-forming units that formed infections in a new monolayer of McCoy cells. The error bars represent SD of the means of three replicates (n = 9). Statistics were determined using unpaired two-tailed t tests. ***, P < 0.001 versus untreated controls.
DISCUSSION
Tryptophan is an important amino acid essential for the normal development of Chlamydia. Chlamydial tryptophan synthesis or lack thereof is linked to tissue tropism (15, 17), species specificity (22), virulence (14, 45), and the chlamydial stress response (6). IFN-γ-induced IDO1-depleted tryptophan catabolism is a well-described method for tryptophan limitation in host cells in vitro. However, the response to IFN-γ is not universal among Chlamydia species and may be associated with different virulence characteristics. It has been shown that the variability in IFN-γ sensitivity is mainly due to the presence of different suites of genes in the tryptophan operon (45). C. caviae has an almost complete operon including all the genes for tryptophan biosynthesis except trpE (23). In contrast, C. trachomatis has only trpB and trpA genes in the tryptophan operon (15) and C. pneumoniae has no genes (46). C. trachomatis and C. pneumoniae have been well documented to produce a chlamydial stress response to IFN-γ exposure (7, 47). Surprisingly, no relevant studies have been conducted on the C. pecorum–IFN-γ relationship, although C. pecorum comparative genomics revealed that C. pecorum possesses an almost complete tryptophan operon, trpRDCAB (35, 36).
In this study, we noted that livestock C. pecorum isolates can escape the IFN-γ effect via tryptophan depletion in human epithelial cells. Both C. pecorum isolates showed very little sensitivity to IFN-γ, whereas human C. trachomatis strains showed complete loss of infectious progeny following confirmation of IDO1 expression in the cell culture. In human cells, IFN-γ induces IDO1, which catalyzes the intracellular tryptophan pools to kynurenine (7, 48). In the present study, we observed that in the absence of tryptophan, C. pecorum growth could be restored in human epithelial cells by supplementation with kynurenine, anthranilic acid, and indole. This rescue of C. pecorum in tryptophan-deficient medium is congruent with the presence of an almost complete tryptophan operon including the trpRDFCAB, kynU, and prsA genes. This phenotype is also consistent with that of the related chlamydial species C. caviae, which was previously demonstrated to be completely resistant to IFN-γ treatment in HeLa cells due to its capacity to reuse kynurenine from the host cell as a substrate to synthesize tryptophan (24) thanks to trpRDFCAB, kynU, and prsA in its tryptophan operon. Our results also revealed that, as expected, the expression of this operon in tryptophan-depleted medium was dependent on the presence of tryptophan following the addition of kynurenine, anthranilic acid, and indole to the cultures. It was previously demonstrated that repression of trpR, the transcriptional regulator of the operon, is dependent on the tryptophan concentration and that repression does not commence until the ratio of tryptophan to TrpR approaches 100:1 (17). These results provide overwhelming evidence that adequate amounts of tryptophan can be synthesized by C. pecorum from these substrates and that this tryptophan can then bind to its cognate operator to repress transcription of the tryptophan operon.
While measuring the C. pecorum response to IFN-γ treatment in human cells provided an interesting comparison to C. trachomatis, the studies of C. pecorum resistance in other cell lines (i) highlighted differences in immune evasion strategies between C. pecorum and other veterinary chlamydiae (i.e., C. muridarum) and (ii) provided more information on the potential in vivo growth strategies utilized by the animal pathogen. Examination of whether C. pecorum can survive IFN-γ exposure in murine McCoy cells revealed that, like other chlamydial species except C. muridarum (12, 13, 22), C. pecorum growth was inhibited. We showed, however, that when grown in BK cells, C. pecorum isolates were resistant to bovine IFN-γ. In contrast, IFN-γ-treated BK cells inhibited C. trachomatis growth, possibly by an IDO1 depletion mechanism. Based on our previous data, we could then hypothesize that C. pecorum may avoid such an immune response due to the compensatory mechanisms available to the pathogen and courtesy of its nearly complete tryptophan biosynthesis operon. There is a reasonable amount of in vivo evidence to suggest that C. pecorum may indeed be able to do this in sheep and cattle. Subclinical C. pecorum gastrointestinal tract infections are common (28), with animals able to shed C. pecorum in fecal samples for extended periods (49). While evidence of aberrant chlamydial forms associated with in vivo infections are limited only to Chlamydia suis thus far (50), it is tempting to speculate that the IFN-γ resistance provided by the C. pecorum tryptophan biosynthesis operon may be an important virulence mechanism in the species to avoid mucosal responses in the gastrointestinal tracts of livestock hosts. In mouse models, experimental infections have revealed that downregulation of the gut immunity results in persistent subclinical infection with C. muridarum in the gastrointestinal tract with fecal shedding (51, 52). Studies of Mycobacterium avium subsp. paratuberculosis infection have also revealed that despite IDO1 gene and protein expression in infected gut tissues in a clinically infected sheep, the local adaptive immune response to the infection was inhibited, allowing the bacteria to survive and contributing to the chronicity of the infection (43). C. pecorum genetic experiments combined with in vivo infection studies will be required to verify this hypothesis. If true, however, it may have significance in influencing the future design of a C. pecorum vaccine, as vaccines for C. pecorum (53, 54) and other chlamydial species have, in part, focused on inducing pathogen-specific IFN-γ responses (55). The potential for IDO-independent effects of IFN-γ on chlamydial growth in bovine epithelial cells, as has been recently noted in both human (56) and murine (57) epithelial cells, as well as potential mechanisms that C. pecorum might use to evade them, requires additional investigation.
In summary, the results presented here demonstrate that C. pecorum produces complete resistance to IFN-γ-mediated tryptophan depletion due to its capacity to scavenge tryptophan from its metabolites from the host cell. Thus, the ability to synthesize tryptophan is likely to allow this intracellular pathogen to more readily adapt to the different environments within its natural host as a strategy to avoid host defenses. This study also provides further evidence that the relationship between IFN-γ and chlamydiae is host specific in nature. Further insights into the pathogenesis of C. pecorum will be needed for control strategies to be developed successfully for this enigmatic pathogen.
MATERIALS AND METHODS
Chlamydia isolates and culture conditions.
The Chlamydia isolates used in this study included an ovine polyarthritis isolate, C. pecorum IPA (VR-629; American Type Culture Collection [ATCC]); a calf sporadic bovine encephalomyelitis isolate, C. pecorum E58 (kindly provided by Bernhard Kaltenboeck, Auburn University); and C. trachomatis D (VR-885; ATCC). All the isolates were routinely cultured in the HEp-2 cell line (CCL-23; ATCC) at 37°C and 5% CO2 in DMEM growth medium (Gibco, Australia) supplemented with 5% heat-inactivated fetal calf serum (FCS) (Life Technologies, Australia), 120 μg streptomycin ml−1 (Sigma-Aldrich, Australia), and 50 μg gentamicin ml−1 (Gibco, Australia). Bovine kidney (BK) cells (CCL-22; ATCC) and McCoy mouse fibroblasts (CRL-1696; ATCC) were routinely grown in DMEM with 5% FCS, 50 μg gentamicin ml−1, and100 μg streptomycin ml−1. All experiments were conducted in 48-well plates (Sigma-Aldrich, Australia) at a multiplicity of infection (MOI) of 0.5 unless otherwise stated.
For IFN-γ experiments, 30,000 cells/well were seeded in a 48-well plate 24 h before infection with different concentrations of human IFN-γ (0, 200, 500, or 1,000 U/ml; Sigma-Aldrich, Australia), 1,000 U bovine IFN-γ/ml (Life Technologies, Australia), or 1,000 U murine IFN-γ/ml (Peprotech, USA) in HEp-2, BK, and McCoy cells, respectively. For the tryptophan-free experiment, 50,000 cells/well were seeded 24 h prior to infection in 48-well plates in 500 μl growth medium. Tryptophan precursors were added to the cultures at the following concentrations: 50 μM l-kynurenine (Sigma-Aldrich, Australia), 50 μM AA (Sigma-Aldrich, Australia), 50 μM indole (Sigma-Aldrich, Australia), and 16 μg/ml tryptophan (fresh DMEM [Gibco, Australia] according to the manufacturer's instructions). In all experiments, the indicated supplements were replenished at 24 hpi. Control cultures with no treatment were included in each experiment. The cultures were harvested at 36 hpi in sucrose-phosphate-glutamate (SPG) medium prior to storage at −80°C. The SPG medium consisted of 218 mM sucrose (Sigma-Aldrich, Australia), 3.76 mM KH2PO4 (Sigma-Aldrich, Australia), 7.1 mM K2HPO4 (Sigma-Aldrich, Australia), and 5 mM glutamic acid (Sigma-Aldrich, Australia), pH 7.4.
For Western blot analysis, 3 × 105 cells/well were seeded in a six-well plate (Sigma-Aldrich, Australia) in 3 ml growth medium 24 h prior to infection with IFN-γ. The cells were harvested at 36 hpi for subsequent use in Western blot analysis.
Determination of viable infectious yield.
The viable infectious yield of Chlamydia was evaluated by calculating the number of IFU per milliliter in HEp-2, BK, and McCoy cells. Confluent host cells were infected with harvested cell lysate in serial dilutions in 96-well cell culture plates (Sigma-Aldrich, Australia). At 30 hpi, the host cell monolayer was washed with phosphate-buffered saline (PBS) and fixed with 100% methanol for 10 min. The fixed cells were permeabilized with Triton X-100 for 15 min and then blocked with 1% bovine serum albumin (BSA) (Sigma-Aldrich, Australia) in PBS. Primary antibody (anti-Chlamydia HtrA) (10, 58) was incubated for 1 h at 1:500 dilution in 1% BSA, followed by three washes in 0.2% Tween 20 in PBS. Secondary antibody [goat anti-rabbit IgG(H+L)-Alexa Fluor 488; Invitrogen, Australia] was added at 1:600 dilution in 1% BSA for 1 h, followed by 4 washes in 0.2% Tween 20 in PBS. Triplicate samples at appropriate serial dilutions were counted in nine fields of view per dilution using fluorescence microscopy (Nikon Eclipse TiS fluorescence microscope). The viable infectious yield was determined as previously described (22). Graphical images were made and statistical analysis was conducted using GraphPad Prism (version 7).
Confocal microscopy.
Chlamydia-infected HEp-2 cell monolayers cultured on 8-mm coverslips in a 48-well plate were used for confocal laser scanning microscopy. At 30 hpi, the coverslip cultures were fixed with 100% methanol for 10 min and washed with PBS. The coverslips were stained with fluorescein isothiocyanate (FITC)-labeled Chlamydia cell lipopolysaccharide (LPS) (Cell Labs, Australia) and mounted on slides using Prolong Gold antifade (Life Technologies, Australia). Confocal images were obtained using a Nikon Eclipse TiS fluorescence microscope with a 60× oil objective. Images were processed for presentation using the NIS software suite. Sizes of inclusions at 30 hpi were measured to determine the effect of IFN-γ and the addition of kynurenine, anthranilic acid, and indole to tryptophan-deficient medium using ImageJ.
Western blotting.
Mock-infected cells and Chlamydia-infected cell cultures in 6-well plates were harvested at 36 hpi and washed with PBS. The cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (Sigma-Aldrich, Australia) and protease inhibitor cocktail (Sigma-Aldrich, Australia) with sonication. Twenty microliters of cell lysates was diluted in 2× Laemmli sample buffer (Bio-Rad, Australia) supplemented with 5% beta-mercaptoethanol, incubated at 95°C for 5 min, and centrifuged at 10,000 × g for 2 min. Samples were loaded into 10% mini-Protean TGX precast gels (Bio-Rad, Australia) at 150 V for 1 h. Precision Plus protein dual-color standards (10 to 250 kDa; Bio-Rad, Australia) and HeLa cell lysate plus IFN-γ (Santa Cruz Biotechnology, USA) were used as positive controls. Proteins were separated by SDS-PAGE following transfer onto a nitrocellulose membrane. The blots were blocked with 1× PBS plus 5% skim milk for 1 h at room temperature and probed with an anti-IDO1 rabbit primary monoclonal antibody (Abcam, Australia) at 1:1,000 dilution or anti β-actin rabbit polyclonal antibody at 1:2,000 dilution overnight at 4°C. The blots were washed three times in a rocker for 5 min each time with 0.2% Tween 20 and 1× PBS. The blots were then probed with horseradish peroxidase (HRP)-conjugated anti-rabbit secondary antibody (Abcam, Australia) at 1:5,000 dilution for 1 h at room temperature. The nitrocellulose membranes were washed four times in a rocker for 5 min each time with 0.2% Tween 20 and 1× PBS. The blot was analyzed with an Odyssey CLx imaging system.
Reverse transcriptase qRT-PCR for IDO1 and trpR genes.
C. pecorum-infected cell cultures were harvested in RNAlater (Sigma-Aldrich, Australia) at 20 hpi. Total RNA was extracted from the harvested cells using the RNeasy minikit (Qiagen, Australia) according to the manufacturer's instructions. The purity and concentration of the RNA were determined using a Nanodrop spectrophotometer. Following digestion of residual DNA by treatment with DNase, cDNA synthesis from RNA was performed using a Roche reverse transcription kit (Roche, Australia) according to the manufacturer's instructions. For host cell IDO1 gene expression, β-actin was used as a reference gene. The primers used for IDO1 (59) and β-actin (60) are listed in Table 1. The 16S rRNA gene was used as a reference gene for the chlamydial trpR and trpB gene expression studies. The primers used for trpR, trpB, and 16S rRNA are also listed in Table 1. All reactions were carried out on a Rotor-GeneQ (Qiagen, Australia) real-time PCR machine in a 20-μl final volume, using a Quantitect SYBR green PCR kit (Qiagen, Australia). The PCR conditions were 95°C for 5 min and 35 cycles of 25 s at 95°C, 30 s at 58°C, and 35 s at 72°C. The results are presented as normalized values of 2−ΔΔCT using the following equations (where CT is threshold cycle): ΔCT = (CT of the IDO1 or trpR or trpB gene − CT of the β-actin or 16S rRNA gene) and ΔΔCT = (ΔCT of the treated sample − ΔCT of the calibrator sample) (61). To test for DNA contamination, we used eight randomly selected DNase-treated RNA samples from the above-mentioned set as templates for conventional PCR targeting a 204-bp fragment of the chlamydial 16S rRNA gene (62) and a 250-bp fragment of the human β-actin gene. Amplification was not achieved in any of the samples, confirming the digestion of residual DNA in our RNA samples.
TABLE 1.
Primers used in this study
| Gene target | Primer orientation | Sequence | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| Human IDO1 | Forward | 5′-AGAAGTGGGCTTTGCTCTGC-3′ | 230 | 59 |
| Reverse | 5′-TGGCAAGACCTTACGGACATCTC-3′ | |||
| Human β-actin | Forward | 5′-CATGTACGTTGCTATCCAGGC-3′ | 250 | 60 |
| Reverse | 5′-CTCCTTAATGTCACGCACGAT-3′ | |||
| trpR | Forward | 5′-GATGGCTGGAATGCGTTTGT-3′ | 176 | This study |
| Reverse | 5′-CTCACACCATATGCCTCAG-3′ | |||
| trpB | Forward | 5′-AGACAGGAGCAGGACAAC-3′ | 198 | This study |
| Reverse | 5′-GGAGAGCCTCATTTACGG-3′ | |||
| 16S rRNA | Forward | 5′-AGTCGAACGGAATAATGGCT-3′ | 204 | 62 |
| Reverse | 5′-CCAACAAGCTGATATCCCAC-3′ |
Statistical analysis.
Statistical significance was analyzed using an unpaired two-tailed t test in GraphPad Prism (version 7). A P value of <0.05 was considered significant, and results are presented as means and standard deviations (SD).
ACKNOWLEDGMENTS
We thank Tomer Ventura, Sankhya Bommana, and Noa Ziklo for assistance with the Chlamydia cell culture and Western blot experiments.
This project was funded by an Australian Research Council Linkage Grant Project (LP140100315) awarded to A.P. and P.T.
We declare that we have no conflict of interest.
REFERENCES
- 1.Hackstadt T, Fischer ER, Scidmore MA, Rockey DD, Heinzen RA. 1997. Origins and functions of the chlamydial inclusion. Trends Microbiol 5:288–293. doi: 10.1016/S0966-842X(97)01061-5. [DOI] [PubMed] [Google Scholar]
- 2.Moulder JW. 1991. Interaction of chlamydiae and host cells in vitro. Microbiol Rev 55:143–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borel N, Pospischil A, Hudson AP, Rupp J, Schoborg RV. 2014. The role of viable but non-infectious developmental forms in chlamydial biology. Front Cell Infect Microbiol 4:97. doi: 10.3389/fcimb.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogan RJ, Mathews SA, Mukhopadhyay S, Summersgill JT, Timms P. 2004. Chlamydial persistence: beyond the biphasic paradigm. Infect Immun 72:1843–1855. doi: 10.1128/IAI.72.4.1843-1855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wyrick PB. 2010. Chlamydia trachomatis persistence in vitro: an overview. J Infect Dis 201(Suppl 2):S88–S95. doi: 10.1086/652394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bavoil PM. 2014. What's in a word: the use, misuse, and abuse of the word “persistence” in Chlamydia biology. Front Cell Infect Microbiol 4:27. doi: 10.3389/fcimb.2014.00027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beatty WL, Byrne GI, Morrison RP. 1993. Morphologic and antigenic characterization of interferon gamma-mediated persistent Chlamydia trachomatis infection in vitro. Proc Natl Acad Sci U S A 90:3998–4002. doi: 10.1073/pnas.90.9.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsia R, Ohayon H, Gounon P, Dautry-Varsat A, Bavoil PM. 2000. Phage infection of the obligate intracellular bacterium, Chlamydia psittaci strain guinea pig inclusion conjunctivitis. Microbes Infect 2:761–772. doi: 10.1016/S1286-4579(00)90356-3. [DOI] [PubMed] [Google Scholar]
- 9.Eickhoff M, Thalmann J, Hess S, Martin M, Laue T, Kruppa J, Brandes G, Klos A. 2007. Host cell responses to Chlamydia pneumoniae in gamma interferon-induced persistence overlap those of productive infection and are linked to genes involved in apoptosis, cell cycle, and metabolism. Infect Immun 75:2853–2863. doi: 10.1128/IAI.01045-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huston WM, Theodoropoulos C, Mathews SA, Timms P. 2008. Chlamydia trachomatis responds to heat shock, penicillin induced persistence, and IFN-gamma persistence by altering levels of the extracytoplasmic stress response protease HtrA. BMC Microbiol 8:190. doi: 10.1186/1471-2180-8-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morrison SG, Su H, Caldwell HD, Morrison RP. 2000. Immunity to murine Chlamydia trachomatis genital tract reinfection involves B cells and CD4(+) T cells but not CD8(+) T cells. Infect Immun 68:6979–6987. doi: 10.1128/IAI.68.12.6979-6987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roshick C, Wood H, Caldwell HD, McClarty G. 2006. Comparison of gamma interferon-mediated antichlamydial defense mechanisms in human and mouse cells. Infect Immun 74:225–238. doi: 10.1128/IAI.74.1.225-238.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson DE, Virok DP, Wood H, Roshick C, Johnson RM, Whitmire WM, Crane DD, Steele-Mortimer O, Kari L, McClarty G, Caldwell HD. 2005. Chlamydial IFN-gamma immune evasion is linked to host infection tropism. Proc Natl Acad Sci U S A 102:10658–10663. doi: 10.1073/pnas.0504198102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beatty WL, Byrne GI, Morrison RP. 1994. Repeated and persistent infection with Chlamydia and the development of chronic inflammation and disease. Trends Microbiol 2:94–98. doi: 10.1016/0966-842X(94)90542-8. [DOI] [PubMed] [Google Scholar]
- 15.Fehlner-Gardiner C, Roshick C, Carlson JH, Hughes S, Belland RJ, Caldwell HD, McClarty G. 2002. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem 277:26893–26903. [DOI] [PubMed] [Google Scholar]
- 16.Xie G, Bonner CA, Jensen RA. 2002. Dynamic diversity of the tryptophan pathway in chlamydiae: reductive evolution and a novel operon for tryptophan recapture. Genome Biol 3:research0051. doi: 10.1186/gb-2002-3-9-research0051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akers JC, Tan M. 2006. Molecular mechanism of tryptophan-dependent transcriptional regulation in Chlamydia trachomatis. J Bacteriol 188:4236–4243. doi: 10.1128/JB.01660-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morrison RP. 2000. Differential sensitivities of Chlamydia trachomatis strains to inhibitory effects of gamma interferon. Infect Immun 68:6038–6040. doi: 10.1128/IAI.68.10.6038-6040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Caldwell HD, Wood H, Crane D, Bailey R, Jones RB, Mabey D, Maclean I, Mohammed Z, Peeling R, Roshick C, Schachter J, Solomon AW, Stamm WE, Suchland RJ, Taylor L, West SK, Quinn TC, Belland RJ, McClarty G. 2003. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 111:1757–1769. doi: 10.1172/JCI17993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Byrne GI, Ouellette SP, Wang Z, Rao JP, Lu L, Beatty WL, Hudson AP. 2001. Chlamydia pneumoniae expresses genes required for DNA replication but not cytokinesis during persistent infection of HEp-2 cells. Infect Immun 69:5423–5429. doi: 10.1128/IAI.69.9.5423-5429.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantoja LG, Miller RD, Ramirez JA, Molestina RE, Summersgill JT. 2001. Characterization of Chlamydia pneumoniae persistence in HEp-2 cells treated with gamma interferon. Infect Immun 69:7927–7932. doi: 10.1128/IAI.69.12.7927-7932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chacko A, Barker CJ, Beagley KW, Hodson MP, Plan MR, Timms P, Huston WM. 2014. Increased sensitivity to tryptophan bioavailability is a positive adaptation by the human strains of Chlamydia pneumoniae. Mol Microbiol 93:797–813. doi: 10.1111/mmi.12701. [DOI] [PubMed] [Google Scholar]
- 23.Read TD, Myers GSA, Brunham RC, Nelson WC, Paulsen IT, Heidelberg J, Holtzapple E, Khouri H, Federova NB, Carty HA, Umayam LA, Haft DH, Peterson J, Beanan MJ, White O, Salzberg SL, Hsia R, McClarty G, Rank RG, Bavoil PM, Fraser CM. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res 31:2134–2147. doi: 10.1093/nar/gkg321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wood H, Roshick C, McClarty G. 2004. Tryptophan recycling is responsible for the interferon-gamma resistance of Chlamydia psittaci GPIC in indoleamine dioxygenase-expressing host cells. Mol Microbiol 52:903–916. doi: 10.1111/j.1365-2958.2004.04029.x. [DOI] [PubMed] [Google Scholar]
- 25.Rajaram K, Giebel AM, Toh E, Hu S, Newman JH, Morrison SG, Kari L, Morrison RP, Nelson DE. 2015. Mutational analysis of the Chlamydia muridarum plasticity zone. Infect Immun 83:2870–2881. doi: 10.1128/IAI.00106-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lenzko H, Moog U, Henning K, Lederbach R, Diller R, Menge C, Sachse K, Sprague L. 2011. High frequency of chlamydial co-infections in clinically healthy sheep flocks. BMC Vet Res 7:29. doi: 10.1186/1746-6148-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Polkinghorne A, Hanger J, Timms P. 2013. Recent advances in understanding the biology, epidemiology and control of chlamydial infections in koalas. Vet Microbiol 165:214–223. doi: 10.1016/j.vetmic.2013.02.026. [DOI] [PubMed] [Google Scholar]
- 28.Walker E, Lee EJ, Timms P, Polkinghorne A. 2015. Chlamydia pecorum infections in sheep and cattle: a common and under-recognised infectious disease with significant impact on animal health. Vet J 206:252–260. doi: 10.1016/j.tvjl.2015.09.022. [DOI] [PubMed] [Google Scholar]
- 29.DeGraves FJ, Gao D, Hehnen H-R, Schlapp T, Kaltenboeck B. 2003. Quantitative detection of Chlamydia psittaci and C. pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J Clin Microbiol 41:1726–1729. doi: 10.1128/JCM.41.4.1726-1729.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reinhold P, Sachse K, Kaltenboeck B. 2011. Chlamydiaceae in cattle: commensals, trigger organisms, or pathogens? Vet J 189:257–267. doi: 10.1016/j.tvjl.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Walker E, Moore C, Shearer P, Jelocnik M, Bommana S, Timms P, Polkinghorne A. 2016. Clinical, diagnostic and pathologic features of presumptive cases of Chlamydia pecorum-associated arthritis in Australian sheep flocks. BMC Vet Res 12:193. doi: 10.1186/s12917-016-0832-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Poudel A, Elsasser TH, Rahman KS, Chowdhury EU, Kaltenboeck B. 2012. Asymptomatic endemic Chlamydia pecorum infections reduce growth rates in calves by up to 48 percent. PLoS One 7:e44961. doi: 10.1371/journal.pone.0044961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reinhold P, Jaeger J, Liebler-Tenorio E, Berndt A, Bachmann R, Schubert E, Melzer F, Elschner M, Sachse K. 2008. Impact of latent infections with Chlamydophila species in young cattle. Vet J 175:202–211. doi: 10.1016/j.tvjl.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 34.Bachmann N, Fraser T, Bertelli C, Jelocnik M, Gillett A, Funnell O, Flanagan C, Myers GS, Timms P, Polkinghorne A. 2014. Comparative genomics of koala, cattle and sheep strains of Chlamydia pecorum. BMC Genomics 15:667. doi: 10.1186/1471-2164-15-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jelocnik M, Bachmann NL, Kaltenboeck B, Waugh C, Woolford L, Speight KN, Gillett A, Higgins DP, Flanagan C, Myers GS, Timms P, Polkinghorne A. 2015. Genetic diversity in the plasticity zone and the presence of the chlamydial plasmid differentiates Chlamydia pecorum strains from pigs, sheep, cattle, and koalas. BMC Genomics 16:893. doi: 10.1186/s12864-015-2053-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sait M, Livingstone M, Clark EM, Wheelhouse N, Spalding L, Markey B, Magnino S, Lainson FA, Myers GS, Longbottom D. 2014. Genome sequencing and comparative analysis of three Chlamydia pecorum strains associated with different pathogenic outcomes. BMC Genomics 15:23. doi: 10.1186/1471-2164-15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Leonard CA, Schoborg RV, Borel N. 2017. Productive and penicillin-stressed Chlamydia pecorum infection induces nuclear factor kappa B activation and interleukin-6 secretion in vitro. Front Cell Infect Microbiol 7:180. doi: 10.3389/fcimb.2017.00180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leonard CA, Dewez F, Borel N. 2016. Penicillin G-induced chlamydial stress response in a porcine strain of Chlamydia pecorum. Int J Microbiol 2016:3832917. doi: 10.1155/2016/3832917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlson JH, Hughes S, Hogan D, Cieplak G, Sturdevant DE, McClarty G, Caldwell HD, Belland RJ. 2004. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun 72:7063–7072. doi: 10.1128/IAI.72.12.7063-7072.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ziklo N, Huston WM, Taing K, Katouli M, Timms P. 2016. In vitro rescue of genital strains of Chlamydia trachomatis from interferon-gamma and tryptophan depletion with indole-positive, but not indole-negative Prevotella spp. BMC Microbiol 16:286. doi: 10.1186/s12866-016-0903-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taylor MW, Feng GS. 1991. Relationship between interferon-gamma, indoleamine 2,3-dioxygenase, and tryptophan catabolism. FASEB J 5:2516–2522. doi: 10.1096/fasebj.5.11.1907934. [DOI] [PubMed] [Google Scholar]
- 42.Spekker K, Czesla M, Ince V, Heseler K, Schmidt SK, Schares G, Daubener W. 2009. Indoleamine 2,3-dioxygenase is involved in defense against Neospora caninum in human and bovine cells. Infect Immun 77:4496–4501. doi: 10.1128/IAI.00310-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plain KM, de Silva K, Earl J, Begg DJ, Purdie AC, Whittington RJ. 2011. Indoleamine 2,3-dioxygenase, tryptophan catabolism, and Mycobacterium avium subsp. paratuberculosis: a model for chronic mycobacterial infections. Infect Immun 79:3821–3832. doi: 10.1128/IAI.05204-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson DE, Taylor LD, Shannon JG, Whitmire WM, Crane DD, McClarty G, Su H, Kari L, Caldwell HD. 2007. Phenotypic rescue of Chlamydia trachomatis growth in IFN-gamma treated mouse cells by irradiated Chlamydia muridarum. Cell Microbiol 9:2289–2298. doi: 10.1111/j.1462-5822.2007.00959.x. [DOI] [PubMed] [Google Scholar]
- 45.Shaw AC, Christiansen G, Roepstorff P, Birkelund S. 2000. Genetic differences in the Chlamydia trachomatis tryptophan synthase alpha-subunit can explain variations in serovar pathogenesis. Microbes Infect 2:581–592. doi: 10.1016/S1286-4579(00)00368-3. [DOI] [PubMed] [Google Scholar]
- 46.Kalman S, Mitchell W, Marathe R, Lammel C, Fan J, Hyman RW, Olinger L, Grimwood J, Davis RW, Stephens R. 1999. Comparative genomes of Chlamydia pneumoniae and C. trachomatis. Nat Genet 21:385–389. doi: 10.1038/7716. [DOI] [PubMed] [Google Scholar]
- 47.Borel N, Summersgill JT, Mukhopadhyay S, Miller RD, Ramirez JA, Pospischil A. 2008. Evidence for persistent Chlamydia pneumoniae infection of human coronary atheromas. Atherosclerosis 199:154–161. doi: 10.1016/j.atherosclerosis.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 48.Boehm U, Klamp T, Groot M, Howard JC. 1997. Cellular responses to interferon-gamma. Annu Rev Immunol 15:749–795. doi: 10.1146/annurev.immunol.15.1.749. [DOI] [PubMed] [Google Scholar]
- 49.Jelocnik M, Frentiu FD, Timms P, Polkinghorne A. 2013. Multi-locus sequence analysis provides insights into the molecular epidemiology of Chlamydia pecorum infections in Australian sheep, cattle and koalas. J Clin Microbiol 51:2625–2632. doi: 10.1128/JCM.00992-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pospischil A, Borel N, Chowdhury EH, Guscetti F. 2009. Aberrant chlamydial developmental forms in the gastrointestinal tract of pigs spontaneously and experimentally infected with Chlamydia suis. Vet Microbiol 135:147–156. doi: 10.1016/j.vetmic.2008.09.035. [DOI] [PubMed] [Google Scholar]
- 51.Yeruva L, Spencer N, Bowlin AK, Wang Y, Rank RG. 2013. Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 68:88–95. doi: 10.1111/2049-632X.12052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rank RG, Yeruva L. 2014. Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to “persistence” in human genital infections. Infect Immun 82:1362–1371. doi: 10.1128/IAI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Desclozeaux M, Jelocnik M, Whitting K, Saifzadeh S, Bommana S, Potter A, Gerdts V, Timms P, Polkinghorne A. 2017. Safety and immunogenicity of a prototype anti-Chlamydia pecorum recombinant protein vaccine in lambs and pregnant ewes. Vaccine 35:3461–3465. doi: 10.1016/j.vaccine.2017.03.091. [DOI] [PubMed] [Google Scholar]
- 54.Khan SA, Desclozeaux M, Waugh C, Hanger J, Loader J, Gerdts V, Potter A, Polkinghorne A, Beagley K, Timms P. 2016. Antibody and cytokine responses of koalas (Phascolarctos cinereus) vaccinated with recombinant chlamydial major outer membrane protein (MOMP) with two different adjuvants. PLoS One 11:e0156094. doi: 10.1371/journal.pone.0156094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cohen CR, Koochesfahani KM, Meier AS, Shen C, Karunakaran K, Ondondo B, Kinyari T, Mugo NR, Nguti R, Brunham RC. 2005. Immunoepidemiologic profile of Chlamydia trachomatis infection: importance of heat-shock protein 60 and interferon-gamma. J Infect Dis 192:591–599. doi: 10.1086/432070. [DOI] [PubMed] [Google Scholar]
- 56.Haldar AK, Piro AS, Finethy R, Espenschied ST, Brown HE, Giebel AM, Frickel EM, Nelson DE, Coers J. 2016. Chlamydia trachomatis is resistant to inclusion ubiquitination and associated host defense in gamma interferon-primed human epithelial cells. mBio 7:e01417–. doi: 10.1128/mBio.01417-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haldar AK, Foltz C, Finethy R, Piro AS, Feeley EM, Pilla-Moffett DM, Komatsu M, Frickel EM, Coers J. 2015. Ubiquitin systems mark pathogen-containing vacuoles as targets for host defense by guanylate binding proteins. Proc Natl Acad Sci U S A 112:E5628–E5637. doi: 10.1073/pnas.1515966112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huston WM, Swedberg JE, Harris JM, Walsh TP, Mathews SA, Timms P. 2007. The temperature activated HtrA protease from pathogen Chlamydia trachomatis acts as both a chaperone and protease at 37°C. FEBS Lett 581:3382–3386. doi: 10.1016/j.febslet.2007.06.039. [DOI] [PubMed] [Google Scholar]
- 59.Odemuyiwa SO, Ghahary A, Li Y, Puttagunta L, Lee JE, Musat-Marcu S, Ghahary A, Moqbel R. 2004. Cutting edge: human eosinophils regulate T cell subset selection through indoleamine 2,3-dioxygenase. J Immunol 173:5909–5913. doi: 10.4049/jimmunol.173.10.5909. [DOI] [PubMed] [Google Scholar]
- 60.Jiang F, Meng D, Weng M, Zhu W, Wu W, Kasper D, Walker WA. 2017. The symbiotic bacterial surface factor polysaccharide A on Bacteroides fragilis inhibits IL-1beta-induced inflammation in human fetal enterocytes via Toll receptors 2 and 4. PLoS One 12:e0172738. doi: 10.1371/journal.pone.0172738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 62.Marsh J, Kollipara A, Timms P, Polkinghorne A. 2011. Novel molecular markers of Chlamydia pecorum genetic diversity in the koala (Phascolarctos cinereus). BMC Microbiol 11:77. doi: 10.1186/1471-2180-11-77. [DOI] [PMC free article] [PubMed] [Google Scholar]