ABSTRACT
Ehrlichia chaffeensis has a group of well-characterized type I secreted tandem repeat protein (TRP) effectors that have moonlighting capabilities. TRPs modulate various cellular processes, reprogram host gene transcription as nucleomodulins, function as ubiquitin ligases, and directly activate conserved host cell signaling pathways to promote E. chaffeensis infection. One TRP-interacting host target is polycomb group ring finger protein 5 (PCGF5), a member of the polycomb group (PcG) protein family and a component of the polycomb repressive complex 1 (PRC1). The current study demonstrates that during early infection, PCGF5 strongly colocalizes with TRP120 in the nucleus and later dramatically redistributes to the ehrlichial vacuole along with other PCGF isoforms. Ectopic expression and immunoprecipitation of TRP120 confirmed the interaction of TRP120 with multiple different PCGF isoforms. At 48 h postinfection, a dramatic redistribution of PCGF isoforms from the nucleus to the ehrlichial vacuole was observed, which also temporally coincided with proteasomal degradation of PCGF isoforms and TRP120 expression on the vacuole. A decrease in PRC1-mediated repressive chromatin mark and an altered transcriptional activity in PRC1-associated Hox genes primarily from HOXB and HOXC clusters were observed along with the degradation of PCGF isoforms, suggesting disruption of the PRC1 in E. chaffeensis-infected cells. Notably, small interfering RNA (siRNA)-mediated knockdown of PCGF isoforms resulted in significantly increased E. chaffeensis infection. This study demonstrates a novel strategy in which E. chaffeensis manipulates PRC complexes through interactions between TRP120 and PCGF isoforms to promote infection.
KEYWORDS: Ehrlichia chaffeensis, PCGF, polycomb repressive complex, tandem repeat protein, Hox gene
INTRODUCTION
Ehrlichia chaffeensis is a Gram-negative, obligately intracellular bacterium that exhibits tropism for mononuclear phagocytes and causes the emerging tick-borne disease, human monocytotropic ehrlichiosis (HME) (1). E. chaffeensis has evolved strategies to evade innate host defenses of the mononuclear phagocyte, where it replicates in membrane-bound cytoplasmic vacuoles and avoids destruction (2, 3). During infection, E. chaffeensis significantly alters the transcriptional activity of genes encoding host cell proteins involved in various processes such as apoptosis, cellular differentiation, signal transduction, cytokine production, and membrane trafficking (4–7). The underlying molecular mechanisms responsible for these changes in gene expression during ehrlichial infection are not fully understood but are mediated in part by pathogen effector-directed host transcriptional modulation involving direct and epigenetic mechanisms.
Eukaryotic gene transcription is regulated by many different mechanisms and often involves single or multiple chemical modifications on a specific stretch of DNA and/or histones (8). Histone posttranslational modifications (HPTMs), like acetylation, phosphorylation, methylation, ubiquitination, and sumoylation, play a major role in regulating chromatin conformation and dictate the accessibility of DNA to its transcriptional machinery. Thus, HPTMs catalyzed by different chromatin-modifying enzymes like histone acetyltransferase, histone deacetylase, histone methyltransferase, and ubiquitin ligases are essential regulators of eukaryotic gene expression (9, 10).
Other intracellular bacteria, such as Anaplasma phagocytophilum and Legionella pneumophila, have been shown to manipulate host gene expression through effector-mediated host chromatin modifications (11, 12). In addition, E. chaffeensis tandem repeat protein (TRP) effectors interact with different chromatin-modifying proteins, like histone methylases and demethylases, protein components of the SWI/SNF chromatin remodeling complex, and polycomb group (PcG) proteins (e.g., polycomb group ring finger protein 5 [PCGF5]) (13). The E. chaffeensis effector, TRP120, strongly interacts with the RING domain of PCGF5 (14), a component of the polycomb repressive complex 1 (PRC1), which is a repressive regulator of various eukaryotic genes, with Hox genes being the most studied targets (15). Moreover, we have recently demonstrated that TRP120 has HECT E3 ubiquitin ligase activity resulting in ubiquitination and a subsequent decrease of PCGF5 in infected cells (16).
Polycomb repressive complexes (PRCs) are multisubunit protein complexes and are broadly divided into two groups (PRC1 and PRC2) (15, 17). PRC1 is responsible for monoubiquitination of histone 2A (H2A) at lysine 119 (H2AK119Ub), and PRC2 is involved in trimethylation of histone 3 (H3) at lysine 27 (H3K27Me3). Both PRC1- and PRC2-mediated posttranslational histone modifications result in changes in chromatin conformation and transcriptional inactivation of eukaryotic genes; thus, these HPTMs are considered to be repressive marks (18, 19).
PRC complexes are well-characterized Hox gene regulators that function by the addition of repressive chromatin marks (20). The Hox genes encode homeobox-containing transcription factors involved in cellular differentiation and proliferation of various cell types, including cells of hematopoietic lineage (21–23). In mammals, 39 Hox genes are usually found in four Hox gene clusters (A to D) which are located on four different chromosomes, at 7p15, 17p21, 12q13, and 2q31, respectively. Based on sequence similarity and position within the cluster, mammalian Hox genes have been assigned to 13 paralogous groups, and each cluster has 9 to 11 members (24).
E. chaffeensis TRP120 interacts with the PCGF component of PRC1, and a previous study demonstrated that knockdown of PCGF5 enhances ehrlichial infection (25). Thus, we investigated the functional relevance of this interaction to better understand the role of PcGs and PRC-associated functions during E. chaffeensis infection. We determined that E. chaffeensis TRP120 promotes intracellular infection by exploiting PcG proteins, resulting in altered PRC1-mediated repressive histone marks and Hox gene expression.
RESULTS
TRP120 interacts with PCGF5 in the host cell nucleus during early stages of infection.
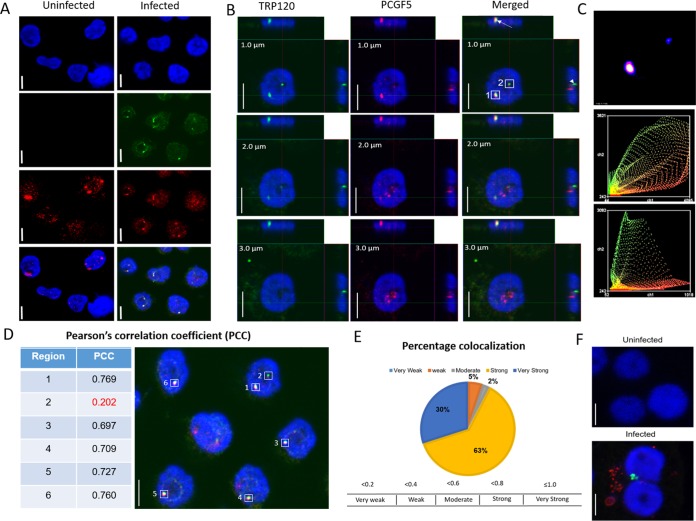
We have previously demonstrated that E. chaffeensis TRP120 interacts with PCGF5. Moreover, TRP120 is a nucleomodulin that translocates to the nucleus and binds to host DNA (26). Thus, we investigated the possibility of nuclear interaction of TRP120 with PCGF5 during E. chaffeensis infection. We dual-stained E. chaffeensis-infected THP-1 cells with PCGF5 and TRP120 antibodies and performed confocal laser microscopy. The time points were selected based on a previous Western blot analysis of the nuclear fraction of E. chaffeensis-infected THP-1 cells showing a significant amount of TRP120 present in host cell nuclei (26). TRP120 was detected as green fluorescence in the nucleus at 24 h postinfection (hpi) (Fig. 1A). At 24 hpi, we did not observe any remarkable differences in nuclear localization of PCGF5 in infected cells in comparison to uninfected controls as determined by corrected total cell florescence (CTCF) (see Fig. S5 in the supplemental material); however, the merged confocal laser micrographs of infected THP-1 cells showed colocalization of TRP120 with PCGF5 in the nucleus of infected THP-1 cells (Fig. 1B, arrow). Based on Pearson's correlation coefficient, the intensity of green fluorescing TRP120 exhibited a strong positive correlation with red fluorescing PCGF5 in more than 90% of the colocalizations examined, suggesting an interaction between these two proteins in the nucleus (Fig. 1D and E). TRP120 was not detected in the nucleus of uninfected THP-1 cells (Fig. 1A). Anti-Dsb (an ehrlichial periplasmic protein) antibody was used to confirm the infection 24 h postexposure to cell-free E. chaffeensis, as TRP120 is was not detectable on ehrlichiae at this time point (Fig. 1F).
FIG 1.
E. chaffeensis TRP120 interacts with PCGF5 in the nucleus during early (24 h) infection. E. chaffensis-infected or uninfected THP-1 cells were immunostained with TRP120 and PCGF5 antibodies (24 hpi) and examined using confocal laser microscopy. (A) Colocalization of punctate TRP120 (green) with PCGF5 (red) in the host cell nucleus (blue). Uninfected cells were treated similarly (left panels). (B) Z-stack images (section depth at the top left on each image) of a single nucleus showing strong (box 1) and weak (box 2) colocalization of TRP120 with PCGF5. Z-projections in the X-to-Z direction are presented at the top and in the Y-to-Z direction are presented at the right side of each image. (C) Intensity correlation analysis of a 1-μm section of the E. chaffeensis-infected cell in panel B demonstrating a positive PDM [product of the differences from the mean = (red intensity − mean red intensity) × (green intensity − mean green intensity)] value (top), which correlated to a strong and a weak colocalization of PCGF5 and TRP120 in region 1 and region 2, respectively. Intensity scatter of region 1 (middle) and region 2 (bottom) demonstrating strong fluorescence intensity for both the red and the green channel. (D) Table showing Pearson's correlation coefficient (PCC) of the highlighted regions shown in E. chaffeensis-infected cell (right panel). (E) Pie chart based on PCCs of multiple regions showing percentage colocalization of TRP120 with PCGF5 in the nucleus. N (number of images analyzed) = 6; n (total number of regions analyzed) = 38. (F) Composite images of confocal laser micrographs demonstrating the presence of E. chaffeensis Dsb (red) and TRP120 (green) in infected cells, confirming infection and nuclear transport of TRP120, at 24 hpi; scale bar, 10 μm.
PCGF isoforms redistribute to the E. chaffeensis vacuole during late stages of infection.
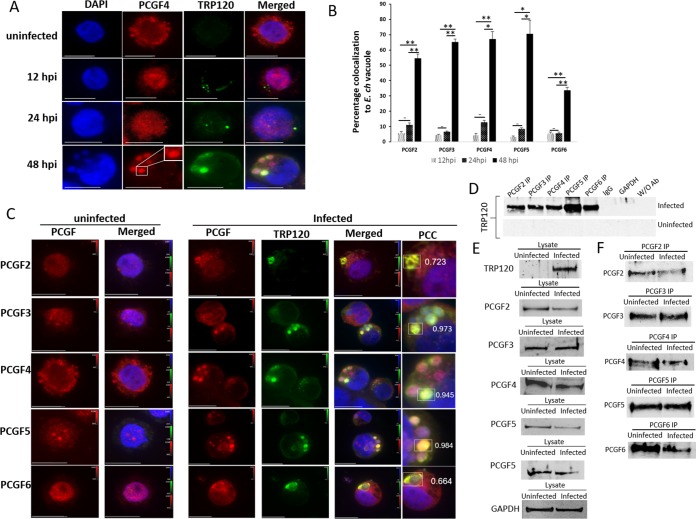
In a previous study, we demonstrated that the ectopically expressed TR region of TRP120 interacts with the RING domain of PCGF5 (13). The paralogous members of the PCGF family share a high similarity in their RING domain (>80% similarity), suggesting that isoforms in addition to PCGF5 may also interact with TRP120. To confirm that E. chaffeensis TRP120 explicitly colocalizes with different PCGFs, we ectopically expressed green fluorescent protein (GFP)-tagged wild-type (WT) TRP120 in HeLa cells, immunostained with PCGF isoform-specific antibody, and examined cells by immunofluorescence microscopy. Strong colocalization of native PCGF isoforms with ectopically expressed GFP-tagged TRP120 suggests an interaction between TRP120 and PCGFs in transfected HeLa cells (see Fig. S1 in the supplemental material). During infection, to observe colocalization of TRP120 and PCGF isoforms, we examined cells immunostained with TRP120 and PCGF isoform-specific antibodies at 12, 24, and 48 hpi using immunofluorescence microscopy (Fig. 2A). A significant redistribution of PCGF isoforms from the nucleus to the cytoplasmic compartment of infected cells was observed at 48 hpi (Fig. 2B). Based on Pearson's correlation coefficient, strong colocalization of TRP120 with PCGF isoforms was observed at the ehrlichial vacuole (Fig. 2C). To confirm the redistribution of these PCGF paralogs to the ehrlichial vacuole, confocal laser microscopy was performed on samples and images were captured in 0.5-μm increments (Fig. S2). The immunofluorescence and the confocal micrographs confirmed the recruitment of PCGF paralogs to the ehrlichial vacuole at 48 hpi (Fig. 2A and C; Fig. S2). There was no significant redistribution of PCGF isoforms to the ehrlichial vacuole during early stages (<24 h) of infection (Fig. S3). Interactions between TRP120 and PCGF isoforms were further reconfirmed using coimmunoprecipitation (co-IP) (Fig. 2D).
FIG 2.
PCGF isoforms colocalize with cytoplasmic E. chaffeensis vacuoles during the late stage (48 h) of infection. (A) Immunofluorescence analysis of E. chaffeensis-infected THP-1 cells. Immunofluorescence micrographs demonstrate distribution of PCGF4 isoforms in uninfected and E. chaffeensis-infected cells at 12, 24, and 48 hpi. PCGF4 was mainly localized in the nucleus of an uninfected cell or during early infection (12 and 24 hpi) but was distributed to the ehrlichial vacuole (rectangle) at a late stage (48 hpi). Scale bar, 10 μm. (B) Bar graph depicting percentages of PCGF-TRP colocalized cells for individual PCGF isoforms (n = 3; Student's two-tailed t test; *, P ≤ 0.05; **, P ≤ 0.005). (C) Immunofluorescence micrographs demonstrating colocalization (infected, rectangles) of E. chaffeensis expressing TRP120 (green) with PCGF2, -3, -4, -5, and -6 isoforms (red) at the ehrlichial vacuole (arrow) at 48 hpi compared to uninfected cells. The signal intensities of red, green, and blue fluorescence channels are shown on each image using a table (a.u., arbitrary units) to cross-reference index numbers to output values. The color map is used to look up the actual colors corresponding to each index number. The Pearson's correlation coefficients shown next to the regions of interest (rectangles) indicate a strong/very strong colocalization for PCGF2, -3, -4, -5, and -6 isoforms with TRP120 in the ehrlichial vacuole. Scale bar, 10 μm. (D) Coimmunoprecipitation using PCGF2, -3, -4, -5, and -6 antibodies and chemiluminescence detection of TRP120 using TRP120-specific antibody from E. chaffeensis-infected and uninfected THP-1 cell lysate. (E) Western blot analysis of E. chaffeensis-infected and uninfected input lysates using TRP120, PCGF2, PCGF3, PCGF4, PCGF5, and PCGF6 antibodies; GAPDH was used as a loading control. (F) Western blot analysis of immunoprecipitation eluates using PCGF2, -3, -4, -5, and -6 isoform-specific antibodies.
Redistribution of PCGFs to the ehrlichial vacuole results in decreased PCGF isoforms in E. chaffeensis-infected cell.
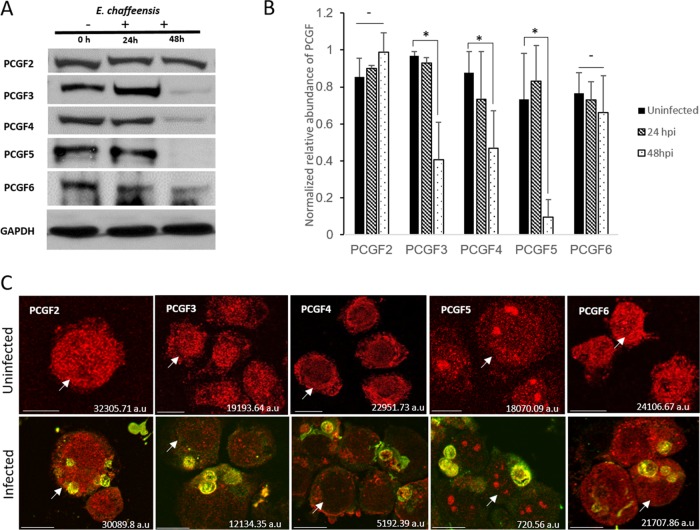
Since we observed a strong correlation between the localization of E. chaffeensis TRP120 and host PCGFs early in the nucleus and later in the cytosolic compartment, we investigated the fate of these host epigenetic regulators in uninfected and E. chaffeensis-infected THP-1 cells. Western blot analysis of the whole-cell lysate using PCGF antibodies demonstrated a significant decrease in the total levels of PCGF3, PCGF4, and PCGF5 in E. chaffeensis-infected cells at 48 hpi (Fig. 3A and B). There were no significant changes in PCGF2 or PCGF6 levels. PCGF1 was not detected in uninfected or infected cells using commercially available antibodies. The Western blot results were confirmed by immunofluorescence microscopy using CTCF to quantify fluorescence intensity (Fig. 3C).
FIG 3.
PCGFs redistribute to the ehrlichial vacuole during infection and result in a decreased total cellular level of PCGF isoforms. (A) Western blot analysis of THP-1 (E. chaffeensis-infected and uninfected) whole-cell lysates (0, 24, and 48 hpi) using isoform-specific anti-PCGF antibodies. Whole-cell lysates were subjected to SDS-PAGE, and the amount of each PCGF isoform was detected using chemiluminescence; GAPDH was used as a loading control. (B) GAPDH-normalized relative abundance of PCGF isoforms in uninfected and E. chaffeensis-infected THP-1 cells at 24 and 48 hpi. Error bars indicate standard deviations between experiments (n = 3; Student's two-tailed t test; *, P ≤ 0.05). (C) Confocal laser micrographs showing decrease in PCGF isoforms (red fluorescence) in E. chaffeensis-infected cells at 48 hpi (bottom panel) compared to those in uninfected cells (top panel); scale bar, 10 μm. Arrows indicate specific cells for which the total cell fluorescence was calculated and which is shown on the bottom right corner of the image.
The decrease in PCGF isoforms is due to degradation of these proteins in E. chaffeensis-infected cells.
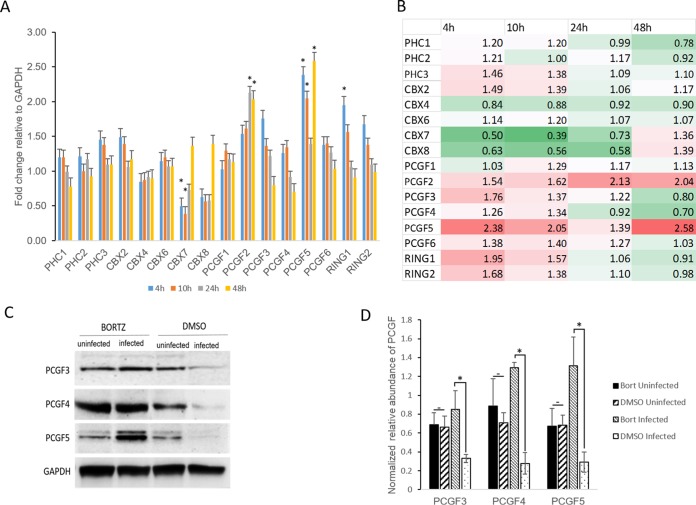
To confirm that the reduction in PCGF isoforms was not caused by a decrease in the transcriptional activity of these genes, the transcript levels of known PRC1 core components, including PCGF isoforms, were quantified using reverse transcription-quantitative PCR (RT-qPCR) in E. chaffeensis-infected cells at 4, 10, 24, and 48 hpi. Biologically significant changes (≥2-fold increase or ≤0.5 decrease) in transcriptional activity were not observed for most of these genes except CBX7, PCGF2, and PCGF5. The gene CBX7 showed a significant decrease in transcriptional activity at 4 and 10 hpi, PCGF2 showed an increase in transcriptional activity at 24 and 48 hpi, and PCGF5 exhibited increased gene transcription at 4, 10, and 48 hpi compared to uninfected controls (Fig. 4A and B). None of the PCGF genes showed a biologically significant decrease in transcriptional activity at 48 hpi. These data suggest that the reduction in PCGF isoforms in infected THP-1 cells at 48 hpi was due to the degradation of PCGF proteins and was not caused by a decrease in PCGF transcriptional activity during E. chaffeensis infection.
FIG 4.
Decrease in PCGF isoforms occurs via proteosomal degradation in E. chaffeensis-infected cells. (A) Graph representing fold changes in PRC1 core component gene expression in E. chaffeensis-infected cells at 4, 10, 24, and 48 hpi; n = 3. Biologically significant (approximately ≤2-fold increase or decrease) and statistically significant (Student's two-tailed t test; *, P ≤ 0.05) changes in gene transcription were observed for CBX7, PCGF2, PCGF5, and RING1 genes. (B) Heat map showing relative expression levels of PRC1 core component genes at 4, 10, 24, and 48 hpi. Each position in the heat map represents an individual gene (listed next to the heat map). The colors indicate differential expression (red indicates induction, green indicates repression, and white represents no significant change) from the average gene expression level in uninfected cells. The intensity of the color represents the amplitude of induction/repression. (C) Western blot analysis of bortezomib (BORTZ)- or DMSO-treated THP-1 (E. chaffeensis-infected and uninfected) whole-cell lysates using isoform-specific anti-PCGF antibodies. E. chaffeensis-infected or uninfected THP-1 cells were treated with 100 nM bortezomib 38 h postinfection for 10 h. The whole-cell lysates were then subjected to SDS-PAGE separation, and the relative abundance of PCGF isoforms was determined using chemiluminescence. (D) Graph representing GAPDH-normalized relative abundance of PCGF isoforms in bortezomib- or DMSO-treated E. chaffeensis-infected or uninfected THP-1 cell lysates. Error bars indicate standard deviations between experiments (n = 3; Student's two-tailed t test; *, P ≤ 0.05).
To further confirm that the decrease in PCGF isoforms was due to the degradation of PCGF proteins, the E. chaffeensis-infected cells were treated with bortezomib, a 26S proteasome inhibitor, and the whole-cell lysates were harvested for Western blot analysis with PCGF isoform-specific antibodies (Fig. 4C). The band intensity of individual PCGF isoforms was measured using densitometry, normalized with GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and represented as the normalized relative abundance of PCGF (Fig. 4D). E. chaffeensis-infected bortezomib-treated cells had a higher relative abundance of PCGF protein than infected dimethyl sulfoxide (DMSO)-treated cells, suggesting that PCGF degradation occurs via the proteasome proteolytic pathway as previously reported (16). No significant difference in PCGF protein abundance was observed between bortezomib- and DMSO-treated uninfected cells (Fig. 4C and D). There were no differences in ehrlichial morula morphology or count between bortezomib- and DMSO-treated cells 10 h after treatment, when PCGF analysis was performed.
E. chaffeensis infection results in altered Hox gene expression.
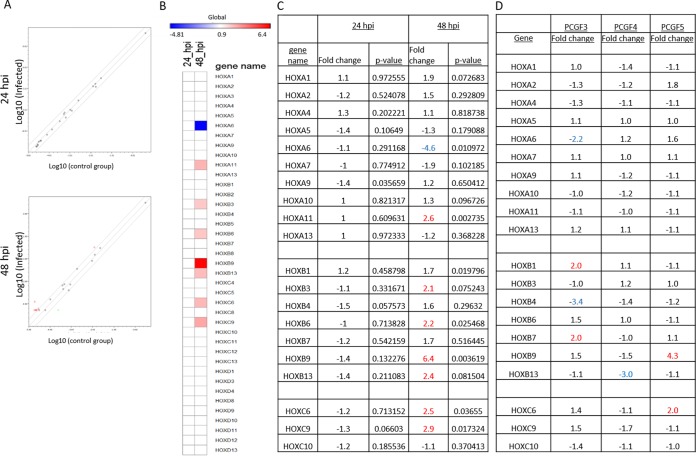
PRCs are the primary transcriptional regulators of Hox gene expression (27–29). As we observed a significant decrease in PCGF protein levels in E. chaffeensis-infected THP-1 cells, we investigated the changes in Hox gene expression in response to E. chaffeensis infection using a custom human Hox RT-qPCR array. The transcriptional activity of 39 human Hox genes at four different loci (HOXA, HOXB, HOXC, and HOXD) was measured in uninfected and E. chaffeensis-infected THP-1 cells at 24 and 48 hpi (Fig. 5). A basal level of expression was observed in 20 of 39 mammalian Hox genes in uninfected THP-1 cells (Fig. 5C). During infection, HOXA6 showed downregulation, but 7 of 20 Hox genes (35%) showed significant upregulation (>2-fold change) in expression at 48 hpi (Fig. 5B), suggesting greater accessibility of transcription machinery in these genomic regions.
FIG 5.
E. chaffeensis infection alters Hox gene expression. (A) Scatterplot analysis of PCR array results shows the differentially expressed genes between E. chaffeensis-infected and uninfected THP-1 cells at 24 (top panel) and 48 (bottom panel) hpi. Red dots represent upregulation, a black dot represents no significant change of expression, and green dots represent downregulation. The graph shows the logarithmic values of average gene expression under either condition. (B) Heat map showing relative expression levels of Hox genes at 24 (left column) and 48 (right column) hpi. Each position in the heat map represents an individual Hox gene (listed next to the heat map). The scale bar intensity demonstrates differential expression (red indicates induction, blue indicates repression, and white represents no significant change) from the average gene expression level in uninfected cells. The intensity of the color represents the amplitude of induction/repression. (C) List of Hox genes in which transcript levels were detected in at least any one of the conditions, i.e., uninfected and 24 and 48 hpi. Seven out of 20 transcriptionally active Hox genes showed a biologically significant (P ≤ 0.05) increase (≥2-fold increase; red) in their transcript levels compared to the control. (D) PCR array results demonstrating differential Hox gene expression in PCGF3 (left)-, PCGF4 (middle)-, and PCGF5 (right)-silenced THP-1 cells. Knockdowns were performed individually one at a time, and biologically significant changes (≥2-fold increase or decrease) in gene expression are color coded (red, increase; blue, decrease).
To confirm that the changes in Hox gene expression in THP-1 cells were associated with the decrease in PCGF isoforms, small interfering RNAs (siRNAs) were used to individually knock down isoform-specific PCGF expression in THP-1 cells and the transcriptional activity of the Hox genes was measured. Altered transcriptional activity in HOXA6, HOXB1, HOXB4, and HOXB7 was observed in PCGF3 knockdown, in HOXB13 in PCGF4 knockdown, and in HOXB9 and HOXC6 in PCGF5 knockdown (Fig. 5D). Consistent with other reports and our previous observation, we determined that the decrease in PCGF protein level affects the transcriptional activity of a subset of Hox genes.
E. chaffeensis infection results in an alteration in PRC1-mediated repressive histone marks.
Functionally, PRC1 is a ubiquitin ligase complex which facilitates monoubiquitination of H2AK119Ub. Conventionally, PRC1-mediated H2AK119Ub requires prior nucleation of PRC2 and placement of an H3K27me3 repressive mark. As our current and previous data strongly suggest that TRP120 recruits PCGF isoforms from the nucleus to the ehrlichial vacuole (14), the global levels of H2AK119Ub and H3K27me3 repressive histone marks were examined in E. chaffeensis-infected cells using Western blotting. Whole-cell lysates from uninfected and E. chaffeensis-infected (24 and 48 hpi) cells were separated on a sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred to nitrocellulose membranes, and probed with H2AK119Ub, H2A, H3K27me3, H3, and GAPDH antibodies. The band intensities were quantified using densitometry and normalized to GAPDH. The relative abundance of H2Ak119Ub to H2A was significantly decreased in E. chaffeensis-infected cells at 48 hpi compared to those cells at 24 hpi and uninfected cells (Fig. 6A and B). There was no significant difference in relative abundance of H3K27me3 in uninfected or E. chaffeensis-infected THP-1 cells (Fig. 6A and C). A decrease in PRC1-mediated repressive histone mark was probably due to the nuclear export of PCGF isoforms at 48 hpi disrupting the PRC1 complex (Fig. 2). Changes in PRC1- but not PRC2-mediated repressive histone marks in E. chaffeensis-infected cells suggest independent modulation of H2AK119Ub and H3K27me3 histone marks during E. chaffeensis infection.
FIG 6.
E. chaffeensis infection results in decreased PRC1-mediated repressive histone marks. (A) Western blot analysis of E. chaffeensis-infected and uninfected whole-cell lysates using H2AK119Ub- and H3K27me3-specific antibodies. GAPDH was used as the loading control, and H2A and H3 were used as nucleosome core particle controls. (B) GAPDH-normalized relative abundance of H2AK119 to H2A (n = 3; Student's two-tailed t test; *, P ≤ 0.05). (C) GAPDH-normalized relative abundance of H3K27me3 to H3 (n = 3). (D) Chromatin immunoprecipitation with H2AK119Ub from E. chaffeensis-infected or uninfected cells followed by promoter/gene enrichment analysis of HOXB9 with real-time qPCR demonstrating a decrease in PRC1-mediated repressive histone marks in the HOXB9 promoter. Error bars indicate standard deviations between experiments (n = 4; Student's two-tailed t test; **, P ≤ 0.001).
To demonstrate that the transcriptional upregulation of Hox genes is due to a decrease in PRC1-associated H2AK119Ub from the promoter region, we performed chromatin immunoprecipitation (ChIP) with ChIP-validated H2AK119Ub-specific antibody and performed promoter enrichment of HOXB9 using quantitative real-time qPCR with validated primers (30). The promoter region of HOXB9 was chosen for gene enrichment because it exhibited the highest transcriptional activity during E. chaffeensis infection (Fig. 5), was transcriptionally upregulated during siRNA-mediated silencing of PCGF5 (Fig. 5), and had a ChIP-validated primer pair for the promoter region (30). A significant decrease in H2AK119Ub was observed in the promoter region of HOXB9, suggesting a direct relationship between loss of these histone marks and the altered expression of HOXB9 in E. chaffeensis-infected cells (Fig. 6D).
Decreases in PCGF isoforms have significant effects on E. chaffeensis burden in the host cell.
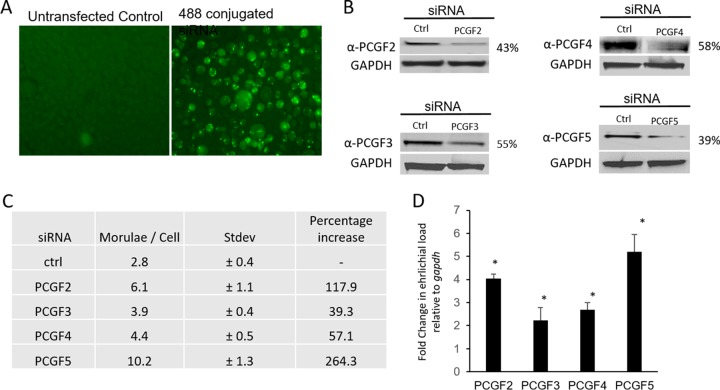
To understand the functional relevance of PCGF degradation during E. chaffeensis infection, uninfected THP-1 cells were first transfected with individual PCGF isoform-specific siRNA using Lipofectamine 3000 (Invitrogen). The efficiency of transfection was confirmed using Alexa Fluor 488-conjugated negative siRNA (Fig. 7A), and siRNA-mediated knockdown of PCGF isoforms was confirmed by Western immunoblotting (Fig. 7B). The relative abundance of PCGF isoforms in transfected cells was normalized to the GAPDH loading control and is represented as the percentage remaining after the knockdown compared to control siRNA. The PCGF siRNA-transfected cells demonstrate a protein knockdown of 42 to 61%. The PCGF knockdown cells were then infected with cell-free E. chaffeensis, and the infection status was determined at 48 hpi by bright-field microscopy after staining with Diff-Quick. The ehrlichial vacuole count per cell demonstrated an average increase in the number in PCGF knockdown cells, specifically PCGF2 (∼6 morulae/cell) and PCGF5 (∼10 morulae/cell), compared to the scrambled control (Fig. 7C; Fig. S4). The fold change in ehrlichial burden in PCGF knockdown cells relative to the control was quantified using real-time qPCR and normalized to host gapdh (Fig. 7D). The ehrlichial replication and the host cell division were normalized by comparing ehrlichial dsb copy numbers and human gapdh, in control and PcG knockdown cells, and the bacterial fold changes in PCGF knockdown cells were calculated using the 2−ΔΔCT method. There were 4-, 2.2-, 2.6-, and 5.2-fold increases in E. chaffeensis infection in cells with reduced protein expression of PCGF2, -3, -4, and -5, respectively.
FIG 7.
siRNA-mediated silencing of PCGF isoforms increases E. chaffeensis infection. THP-1 cells were transfected with isoform-specific siRNA and then infected with E. chaffeensis at 24 h posttransfection. (A) Alexa Fluor 488-conjugated siRNA-transfected cell, showing high efficiency of RNA transfection using Lipofectamine 3000. (B) Western blot analysis of the total cell lysate from control and siRNA-transfected THP-1 cells confirmed the decrease in PCGF2, PCGF3, PCGF4, and PCGF5 48 h posttransfection. GAPDH was used as the loading control. The relative abundance of PCGF isoforms in siRNA-transfected cells was determined after normalization to the loading control and then represented as the percentage remaining after the knockdown. (C) Table representing the percentage increase in ehrlichial morulae and the average number of morulae/cell for each PCGF isoform-specific knockdown. The average morula counts were determined by counting the number of morula present in each field of view and then dividing that by the number of cells counted. The experiment was repeated three times in duplicate, and the values shown are means ± standard deviations (Stdev). (D) The fold change in ehrlichial infection was determined by comparing the ehrlichial dsb to the host cell gapdh in individual PCGF knockdown using real-time qPCR at 48 hpi (n = 3; *, P ≤ 0.05).
DISCUSSION
Recently, there has been a growing appreciation of effector-mediated epigenetic regulation of host transcriptome by intracellular pathogens during infection (12, 31, 32). We have previously demonstrated that during E. chaffeensis infection, TRP effectors target genes encoding chromatin-modifying enzymes and interact with several chromatin-associated proteins, including PCGF5 (13, 26). The interaction of TRP120 with PCGF5 involves posttranslational modification of TRP120 in the carboxy-terminal domain, where it is conjugated to SUMO at a canonical SUMO motif (14). The study also demonstrated that the disruption of TRP120 sumoylation perturbed TRP120-PCGF5 interactions in the RING domain of PCGF5. Moreover, in a recent study, we have also demonstrated that Nedd4L, a HECT E3 ubiquitin ligase, is also involved in TRP120 ubiquitination and redistribution of PCGF5 to the ehrlichial vacuole (16).
PCGFs are the paralogous PcG members, assemble in multisubunit nuclear complexes, and regulate the transcriptional outcome of numerous mammalian genes, including Hox genes (33, 34). Though PcGs are well studied in developmental biology and oncology, this is the first description of an intracellular bacterium exploiting these epigenetic regulators to promote infection in the host cell. In this study, we have demonstrated that during early infection, TRP120 interacts with PCGFs in the nucleus, followed by redistribution of these proteins to the ehrlichial vacuole and subsequent degradation. Decreased host PCGF isoforms promote E. chaffeensis infection, and this coincides with altered Hox gene expression and associated repressive epigenetic marks.
A previous study has demonstrated that E. chaffeensis interacts with PCGF5, and TRP120 is also known to translocate to the nucleus, where it binds to host DNA (26). Thus, we investigated the possibility of a nuclear interaction of TRP120 with PCGF5 in E. chaffeensis-infected cells using confocal laser scanning microscopy. The micrographs revealed that during early infection, nuclear TRP120 strongly colocalizes with PCGF5. Based on a previous study, which demonstrated the importance of TRP120 sumoylation and interactions with PCGF5, it is highly likely that the nuclear interaction of TRP120 with PCGF5 is SUMO dependent (14). Moreover, SUMO modifications are often involved in diverse nuclear processes, including alteration in chromatin structure due to association with different repressor complexes, including PcGs (35).
PcG complexes are involved in heterochromatin formation and aggregated in a limited number of nuclear foci rather than dispersed throughout the genome (36–39). In this study, confocal laser microscopy revealed that PCGF isoforms are mainly present as punctate nuclear aggregates in uninfected cells, and during infection, these isoforms were dramatically redistributed from the nucleus to the ehrlichial vacuole by 48 hpi. The redistribution of PCGFs to the ehrlichial vacuole requires the prior interaction of TRP120 with PCGF isoforms, which involves posttranslational modification of TRP120 (14, 16). Recently, it was also demonstrated that TRP120 has a HECT E3 ligase domain with a functionally conserved catalytic site involved in polyubiquitination of PCGF5 (16). This finding further supports our observation that the PCGFs are subsequently degraded via the ubiquitin 26s proteasome proteolytic pathway during E. chaffeensis infection. Though we have seen a significant increase in the PCGF2 transcript level, there were no significant changes in PCGF2 levels, suggesting a rapid decay in mRNA during infection; however, further investigation is needed to define the basis of PCGF2 stability during E. chaffeensis infection.
The degradation of PCGF isoforms coincided with an alteration in the transcription of PRC-repressed Hox genes in E. chaffeensis-infected cells. A majority of Hox genes with altered transcription profiles in infected cells were upregulated, suggesting a direct relation between PCGF degradation and transcriptional activation of Hox genes. The data strongly suggest that during infection, certain Hox genes, like genes in the HOXB and HOXC clusters, are more actively transcribed due to degradation of PCGFs and displacement of PRC complexes. This observation agrees with a recent study demonstrating derepression of the HOXB cluster in embryonic stem cells (ESCs) in the absence of Ring1B, a PCGF-interacting core component of PRC1 (15, 40). Using siRNA-mediated PCGF knockdown, we also confirmed the role of PCGFs as one of the primary regulators of Hox genes in E. chaffeensis-infected cells. Differences in expression pattern in individual PCGF knockdowns can be attributed to the fact that Hox genes may be autoregulated by their own products, or controlled by other HOX proteins (41–43). The mechanism of these complex interactions remains mostly undefined.
Since PCGFs, the core component of a PRC1 complex, are degraded during E. chaffeensis infection, the levels of PRC1- and PRC2-mediated repressive histone marks in E. chaffeensis-infected cells were analyzed using Western blotting. A significant decrease in PRC1- but not PRC2-mediated repressive histone marks was observed at late stages of infection, suggesting that during infection, E. chaffeensis TRP effectors selectively target genes, mainly regulated by the PRC1 type of complexes by manipulating global levels of PcGs. This result is also consistent with another study, which recently reported the loss of H2AK119Ub in the absence of PCGF5 in hematopoietic stem and progenitor cells (44). Moreover, these data are in agreement with multiple recent studies that emphasize the independent role of PRC1 in gene repression (45–47).
PRC-mediated gene silencing can be due to either direct inhibition, where PRCs bind to a promoter region and interfere with the binding of the transcription machinery, or due to the formation of an inhibitory chromatin structure, nonconducive to the passage of transcription machinery (48). Genome-wide analysis of PRC1 and PRC2 targets in murine and human ESCs revealed that the PRCs mainly associate with the promoter regions of developmental regulators (49, 50). To determine whether the changes in Hox gene expression are related to the removal of PRC1-related repressive histone marks during E. chaffeensis infection, we performed chromatin immunoprecipitation using an antibody against H2Ak119Ub, followed by gene-specific enrichment using real-time PCR, with healthy and E. chaffeensis-infected cells. The result showed a Hox gene-specific decrease in the H2AK119Ub repressive mark in infected cells, demonstrating that the activation of Hox genes is due to the loss of PRC1-mediated H2AK119Ub marks from the Hox gene promoter.
siRNA-mediated silencing of PCGF isoforms significantly promotes E. chaffeensis infection, demonstrating the importance of these epigenetic regulators in maintaining infection. The PRC1 complexes serve as the epigenetic regulator of genes associated with several evolutionary conserved molecular pathways such as Wnt and Notch that are involved in cell fate determination, including regulation of self-renewal or proliferation, senescence or immortalization, and cell death. Moreover, recent studies have demonstrated that different PCGF isoforms play a significant role in conserved cell signaling pathways like Wnt and Notch, which are activated by E. chaffeensis during infection (51–53). Through this investigation, we conclude that E. chaffeensis TRP120 targets components of host epigenetic complexes to control diverse cellular and molecular pathways to promote E. chaffeensis infection.
MATERIALS AND METHODS
Cell culture and cultivation of E. chaffeensis.
Human monocytic leukemia cells (THP-1) were cultured in RPMI 1640 medium with l-glutamine and 25 mM HEPES buffer (Invitrogen), supplemented with 1 mM sodium pyruvate (Sigma, St. Louis, MO), 2.5 g/liter d-(+)-glucose (Sigma, St. Louis, MO), and 10% fetal bovine serum (HyClone, Logan, UT). E. chaffeensis (Arkansas strain) was cultivated in THP-1 cells as previously described (54).
siRNAs and antibodies.
siRNAs used in this study were PCGF2 (sc-38191), PCGF3 (sc-89157), PCGF4 (sc-29814), PCGF6 (sc-90663), and nontargeting siRNA (sc-37007) (Santa Cruz Biotechnology, Santa Cruz, CA). siRNA for PCGF5 (M-007089-01) was obtained from GE Dharmacon (Lafayette, CO). Alexa Fluor 488-labeled negative siRNA was obtained from Qiagen (Germantown, MD). Polyclonal mouse anti-TRP120 antibody used in this study was generated against a peptide (SKVEQEETNPEVLIKDLQDVAS) and was described previously (55). Polyclonal mouse anti-PCGF5 antibody used in this study was generated against a peptide (PKVDEEGDENEDDKDYHRS) and was described previously (56). Other antibodies used in this study were LaminB1 (sc-30264), GAPDH (sc-47724), PCGF2 (sc-10744) (Santa Cruz), PCGF3 (Invitrogen, Carlsbad, CA), PCGF4 (sc-10745), PCGF6 (sc-367209) (Santa Cruz), H3K27Me3 (Millipore, Billerica, MA), and H2AK119Ub (Cell Signaling, Beverly, MA) antibodies.
Immunofluorescence and confocal laser microscopy.
Uninfected and E. chaffeensis-infected THP-1 cells were collected at 3, 12, 24, 48, and 72 h postinfection (hpi) and adhered to glass slides by cytocentrifugation. The cells were fixed with 4% paraformaldehyde (PFA) for 20 min at room temperature, followed by washes in phosphate-buffered saline (PBS) twice. Fixed cells were blocked and permeabilized with 0.3% Triton X-100 in 2% bovine serum albumin (BSA) for 30 min and washed. The cells were then incubated with primary antibody (1:100) in PBS with 2% BSA for 1 h, washed, and incubated with Alexa Fluor 568 IgG (H+L) and/or Alexa Fluor 488 IgG (H+L) secondary antibodies (1:100) for 30 min. The slides were washed 3 times and mounted with ProLong Gold antifade reagent with DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen). Immunofluorescence images were captured with an Olympus BX61 epifluorescence microscope and analyzed using Slidebook software (v.5.0; Intelligent Imaging Innovations, Denver, CO). Confocal laser micrographs were obtained with an LSM 510 meta-confocal laser microscope and analyzed with LSM meta-software (v.4.0). Intensity correlation analysis (ICA) and corrected total cell fluorescence (CTCF) were measured using ImageJ (57, 58). Briefly, the region of interest in a micrograph was marked with a selection tool in ImageJ, and the ICA plugin was used to calculate Pearson's correlation coefficient. For total cell fluorescence after selection of the region of interest in a micrograph, the measurement tool was used to calculate CTCF.
Western immunoblot analysis.
Cells were washed twice with PBS, and lysates were prepared using CytoBuster protein extraction reagent (Novagen/EMD, Gibbstown, NJ) supplemented with cOmplete, mini, EDTA-free protease inhibitor (Roche, Basel, Switzerland), and 10 mM phenylmethenesulfonylfluoride (PMSF) (Sigma, St. Louis, MO). The cell lysates were centrifuged at 15,000 × g for 10 min at 4°C. The supernatants were collected, and protein concentration was then measured using a Pierce BCA protein assay kit (Thermo Fisher Scientific, Waltham, MA). Equal amounts of protein (15 to 30 μg/well) were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with primary antibodies. Horseradish peroxidase-conjugated mouse anti-rabbit or rabbit anti-goat IgG (H+L) secondary antibodies (Kirkegaard & Perry Laboratories, Gaithersburg, MD) were used and visualized by SuperSignal West Dura chemiluminescent substrate or ECL (Thermo Fisher Scientific).
Inhibition of proteasome activity.
E. chaffeensis-infected or uninfected THP-1 cells were treated with either 0.1% DMSO (control) or 100 nM bortezomib (59) for 10 h beginning at 38 h postinfection. After incubation, cell viability and morula morphology and number were determined by trypan blue staining and confirmed using a NucView488 and MitoView633 apoptotic assay kit (Biotium, Fremont, CA). The whole-cell lysates were prepared using CytoBuster protein extraction reagent (EMD Millipore) and subjected to SDS-PAGE separation and Western immunoblot analysis using PCGF isoform-specific antibodies. The band intensity of individual PCGF isoforms was measured and normalized with loading control (GAPDH).
RNA isolation and cDNA synthesis.
E. chaffeensis-infected or uninfected THP-1 cells were harvested at different time points (4, 10, 24, 48, and 72 h postinfection), and total RNA was isolated using an RNeasy minikit (Qiagen). On-column DNA digestion was performed using an RNase-free DNase kit (Qiagen). The concentration and purity of RNA were determined using a Nanodrop 100 spectrophotometer (Thermo Fisher Scientific, West Palm Beach, FL). cDNA was synthesized from total RNA (0.5 μg) using the RT2 first strand kit (Qiagen).
PCR array.
PCR arrays were performed using a 96-well custom profiler RT2 PCR array (Qiagen SA Biosciences, Germantown, MD) for 39 human Hox genes according to the manufacturer's instructions. First, amplified cDNA was diluted with RNase-free water and mixed with 2× RT2 SYBR green master mix (Bio-Rad, Hercules, CA). PCR master mix (25 μl) was dispensed into each well of the PCR array. Real-time PCR was performed on a Mastercycler EP Realplex2 S (Eppendorf, Hamburg, Germany). The cycling conditions used were as follows: 95°C for 10 min and 40 cycles of 95°C for 15 s and 60°C for 1 min. The real-time cycler software RealPlex 1.5 (Eppendorf) was used for PCR and data collection. The data were analyzed by using a PCR array data analysis Web portal (SA Bioscience). The beta-actin/ACTB gene was used for normalization of the sample data, and normalization to the housekeeping genes was performed by calculating the cycle threshold (ΔΔCT) for each gene of interest in the plate. Any CT value greater than 35 was considered to be a negative result. The RT2 Profiler PCR array data analysis software calculates the fold change based on the ΔΔCT method (60, 61). PCR array quality checks were performed using Web-based PCR array data analysis software (v.3.5; SABiosciences) for reproducibility. Reverse transcription control (RTC), positive PCR control (PPC), and human genomic DNA contamination control (HGDC) were included.
Quantitative chromatin immunoprecipitation.
E. chaffeensis-infected or uninfected THP-1 cells (1 × 107) were harvested at 48 hpi for chromatin immunoprecipitation (ChIP) using an EZ-Magna ChIP A/G kit (EMD Millipore). Briefly, cells were cross-linked with 1% formaldehyde (final concentration) for 10 min, followed by quenching with glycine and subsequent washing with PBS on ice. Cells were lysed using lysis buffer, and nuclei were released using a Dounce homogenizer. Sonication was performed on ice to obtain ≤1-kb nucleic acid fragments using a Sonics Vibra-Cell, and antibodies against H2AK119Ub and control IgG were used to perform ChIP. The cross-linked nucleic acids were eluted out of the solution by using the manufacturer's protocol and confirmed on a 2% agarose gel. The gene enrichment was performed using real-time PCR on a Mastercycler EP Realplex2 S (Eppendorf) using ChIP-validated primers and iQ SYBR green Supermix (Bio-Rad). Fold enrichment (2−ΔΔCT) was calculated after normalization of a signal over background.
RNA interference.
THP-1 cells (1.0 × 106) were transfected with 10 nM human siRNA using Lipofectamine 3000 (Invitrogen). Briefly, specific siRNA (3 μl) and Lipofectamine 3000 reagent (7.5 μl) were added to Opti-MEM medium (250 μl) (Invitrogen), incubated for 5 min at room temperature, and then added to the cell suspension in a 6-well plate. A control siRNA consisting of a scrambled sequence was used as a negative control, and a fluorescently labeled siRNA (Alexa Fluor 488-conjugated siRNA) was used as a control for transfection monitoring and optimization. At 1 day posttransfection, the cells were infected with cell-free E. chaffeensis (multiplicity of infection [MOI], ∼50). Infected cells were harvested at different time points for qPCR and the PCR array. The PCGF isoform knockdowns were confirmed by Western immunoblotting in siRNA-transfected THP-1 cells with isoform-specific antibody, and the knockdown efficiency was determined using densitometric analysis.
Quantification of E. chaffeensis by qPCR.
E. chaffeensis infection was evaluated by real-time qPCR as previously described (25). Briefly, E. chaffeensis-infected THP-1 cells (48 hpi) were pelleted, washed with PBS, and lysed in SideStep lysis and stabilization buffer (Agilent, Santa Clara, CA) at room temperature, and lysates were used in qPCRs. Amplicons for the ehrlichial dsb gene and the human GAPDH gene were obtained using dsb forward primer 5′-GCTGCTCCACCAATAAATGTATCCCT-3′, dsb reverse primer 5′-GTTTCATTAGCCAAGAATTCCGACACT-3′, human gapdh forward primer 5′-GGAGTCCACTGGCGTCTTCAC-3′, and human gapdh reverse primer 5′-GAGGCATTGCTGATGATCTTGAG-3′ in two separate qPCRs. The bacterial fold change was calculated using the 2−ΔΔCT method and CT values for the ehrlichial dsb and host GAPDH genes in PCGF knockdown and the transfected control cells.
Statistical analysis.
The statistical analysis of experimental groups was calculated using a two-tailed Student's t test.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by National Institute of Allergy and Infectious Disease grants AI115449 and AI106859. Shubhajit Mitra was supported by a James W. McLaughlin postdoctoral fellowship.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00845-17.
REFERENCES
- 1.Dawson JE, Anderson BE, Fishbein DB, Sanchez JL, Goldsmith CS, Wilson KH, Duntley CW. 1991. Isolation and characterization of an Ehrlichia sp. from a patient diagnosed with human ehrlichiosis. J Clin Microbiol 29:2741–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dunning Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen J, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet 2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rikihisa Y. 2010. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet Parasitol 167:155–166. doi: 10.1016/j.vetpar.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JZ, Sinha M, Luxon BA, Yu XJ. 2004. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect Immun 72:498–507. doi: 10.1128/IAI.72.1.498-507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rikihisa Y. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis: subversive manipulators of host cells. Nat Rev Microbiol 8:328–339. doi: 10.1038/nrmicro2318. [DOI] [PubMed] [Google Scholar]
- 6.Lina TT, Farris T, Luo T, Mitra S, Zhu B, McBride JW. 2016. Hacker within! Ehrlichia chaffeensis effector driven phagocyte reprogramming strategy. Front Cell Infect Microbiol 6:58. doi: 10.3389/fcimb.2016.00058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McBride JW, Walker DH. 2011. Molecular and cellular pathobiology of Ehrlichia infection: targets for new therapeutics and immunomodulation strategies. Expert Rev Mol Med 13:e3. doi: 10.1017/S1462399410001730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felsenfeld G. 2014. A brief history of epigenetics. Cold Spring Harb Perspect Biol 6:a018200. doi: 10.1101/cshperspect.a018200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Bannister AJ, Kouzarides T. 2011. Regulation of chromatin by histone modifications. Cell Res 21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rennoll-Bankert KE, Garcia-Garcia JC, Sinclair SH, Dumler JS. 2015. Chromatin-bound bacterial effector ankyrin A recruits histone deacetylase 1 and modifies host gene expression. Cell Microbiol 17:1640–1652. doi: 10.1111/cmi.12461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rolando M, Sanulli S, Rusniok C, Gomez-Valero L, Bertholet C, Sahr T, Margueron R, Buchrieser C. 2013. Legionella pneumophila effector RomA uniquely modifies host chromatin to repress gene expression and promote intracellular bacterial replication. Cell Host Microbe 13:395–405. doi: 10.1016/j.chom.2013.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Luo T, Kuriakose JA, Zhu B, Wakeel A, McBride JW. 2011. Ehrlichia chaffeensis TRP120 interacts with a diverse array of eukaryotic proteins involved in transcription, signaling, and cytoskeleton organization. Infect Immun 79:4382–4391. doi: 10.1128/IAI.05608-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dunphy PS, Luo T, McBride JW. 2014. Ehrlichia chaffeensis exploits host SUMOylation pathways to mediate effector-host interactions and promote intracellular survival. Infect Immun 82:4154–4168. doi: 10.1128/IAI.01984-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu B, Das S, Mitra S, Farris TR, McBride JW. 2017. Ehrlichia chaffeensis TRP120 moonlights as a HECT E3 ligase involved in self and host ubiquitination to influence protein interactions and stability for intracellular survival. Infect Immun 85:e00290-. doi: 10.1128/IAI.00290-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morey L, Helin K. 2010. Polycomb group protein-mediated repression of transcription. Trends Biochem Sci 35:323–332. doi: 10.1016/j.tibs.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 18.Grossniklaus U, Paro R. 2014. Transcriptional silencing by polycomb-group proteins. Cold Spring Harb Perspect Biol 6:a019331. doi: 10.1101/cshperspect.a019331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ciferri C, Lander GC, Maiolica A, Herzog F, Aebersold R, Nogales E. 2012. Molecular architecture of human polycomb repressive complex 2. Elife 1:e00005. doi: 10.7554/eLife.00005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mallo M, Alonso CR. 2013. The regulation of Hox gene expression during animal development. Development 140:3951–3963. doi: 10.1242/dev.068346. [DOI] [PubMed] [Google Scholar]
- 21.van Oostveen J, Bijl J, Raaphorst F, Walboomers J, Meijer C. 1999. The role of homeobox genes in normal hematopoiesis and hematological malignancies. Leukemia 13:1675–1690. doi: 10.1038/sj/leu/2401562. [DOI] [PubMed] [Google Scholar]
- 22.Magli MC, Largman C, Lawrence HJ. 1997. Effects of HOX homeobox genes in blood cell differentiation. J Cell Physiol 173:168–177. doi:. [DOI] [PubMed] [Google Scholar]
- 23.Argiropoulos B, Humphries RK. 2007. Hox genes in hematopoiesis and leukemogenesis. Oncogene 26:6766–6776. doi: 10.1038/sj.onc.1210760. [DOI] [PubMed] [Google Scholar]
- 24.Grier DG, Thompson A, Kwasniewska A, McGonigle GJ, Halliday HL, Lappin TR. 2005. The pathophysiology of HOX genes and their role in cancer. J Pathol 205:154–171. doi: 10.1002/path.1710. [DOI] [PubMed] [Google Scholar]
- 25.Luo T, Dunphy PS, McBride JW. 2017. Ehrlichia chaffeensis tandem repeat effector targets differentially influence infection. Front Cell Infect Microbiol 7:178. doi: 10.3389/fcimb.2017.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhu B, Kuriakose JA, Luo T, Ballesteros E, Gupta S, Fofanov Y, McBride JW. 2011. Ehrlichia chaffeensis TRP120 binds a G+C-rich motif in host cell DNA and exhibits eukaryotic transcriptional activator function. Infect Immun 79:4370–4381. doi: 10.1128/IAI.05422-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis EB. 1978. A gene complex controlling segmentation in Drosophila. Nature 276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- 28.Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol Cell 32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Cai L, Rothbart SB, Lu R, Xu B, Chen WY, Tripathy A, Rockowitz S, Zheng D, Patel DJ, Allis CD, Strahl BD, Song J, Wang GG. 2013. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol Cell 49:571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pennini ME, Perrinet S, Dautry-Varsat A, Subtil A. 2010. Histone methylation by NUE, a novel nuclear effector of the intracellular pathogen Chlamydia trachomatis. PLoS Pathog 6:e1000995. doi: 10.1371/journal.ppat.1000995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rennoll-Bankert KE, Dumler JS. 2012. Lessons from Anaplasma phagocytophilum: chromatin remodeling by bacterial effectors. Infect Disord Drug Targets 12:380–387. doi: 10.2174/187152612804142242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sparmann A, van Lohuizen M. 2006. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer 6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 34.Jaenisch R, Young R. 2008. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Dominguez M, Reyes JC. 2009. SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789:451–459. doi: 10.1016/j.bbagrm.2009.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. 1998. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol 142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ficz G, Heintzmann R, Arndt-Jovin DJ. 2005. Polycomb group protein complexes exchange rapidly in living Drosophila. Development 132:3963–3976. doi: 10.1242/dev.01950. [DOI] [PubMed] [Google Scholar]
- 38.Dietzel S, Niemann H, Bruckner B, Maurange C, Paro R. 1999. The nuclear distribution of Polycomb during Drosophila melanogaster development shown with a GFP fusion protein. Chromosoma 108:83–94. doi: 10.1007/s004120050355. [DOI] [PubMed] [Google Scholar]
- 39.Hernandez-Munoz I, Taghavi P, Kuijl C, Neefjes J, van Lohuizen M. 2005. Association of BMI1 with polycomb bodies is dynamic and requires PRC2/EZH2 and the maintenance DNA methyltransferase DNMT1. Mol Cell Biol 25:11047–11058. doi: 10.1128/MCB.25.24.11047-11058.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popperl H, Featherstone MS. 1992. An autoregulatory element of the murine Hox-4.2 gene. EMBO J 11:3673–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Popperl H, Bienz M, Studer M, Chan SK, Aparicio S, Brenner S, Mann RS, Krumlauf R. 1995. Segmental expression of Hoxb-1 is controlled by a highly conserved autoregulatory loop dependent upon exd/pbx. Cell 81:1031–1042. doi: 10.1016/S0092-8674(05)80008-X. [DOI] [PubMed] [Google Scholar]
- 43.Arcioni L, Simeone A, Guazzi S, Zappavigna V, Boncinelli E, Mavilio F. 1992. The upstream region of the human homeobox gene HOX3D is a target for regulation by retinoic acid and HOX homeoproteins. EMBO J 11:265–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Si S, Nakajima-Takagi Y, Aoyama K, Oshima M, Saraya A, Sugishita H, Nakayama M, Ishikura T, Koseki H, Iwama A. 2016. Loss of Pcgf5 affects global H2A monoubiquitination but not the function of hematopoietic stem and progenitor cells. PLoS One 11:e0154561. doi: 10.1371/journal.pone.0154561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LL, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cooper S, Dienstbier M, Hassan R, Schermelleh L, Sharif J, Blackledge NP, De Marco V, Elderkin S, Koseki H, Klose R, Heger A, Brockdorff N. 2014. Targeting polycomb to pericentric heterochromatin in embryonic stem cells reveals a role for H2AK119u1 in PRC2 recruitment. Cell Rep 7:1456–1470. doi: 10.1016/j.celrep.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalb R, Latwiel S, Baymaz HI, Jansen PW, Muller CW, Vermeulen M, Muller J. 2014. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat Struct Mol Biol 21:569–571. doi: 10.1038/nsmb.2833. [DOI] [PubMed] [Google Scholar]
- 48.Levine SS, King IF, Kingston RE. 2004. Division of labor in polycomb group repression. Trends Biochem Sci 29:478–485. doi: 10.1016/j.tibs.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 49.Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 50.Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo T, Dunphy PS, Lina TT, McBride JW. 2015. Ehrlichia chaffeensis exploits canonical and noncanonical host Wnt signaling pathways to stimulate phagocytosis and promote intracellular survival. Infect Immun 84:686–700. doi: 10.1128/IAI.01289-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lina TT, Dunphy PS, Luo T, McBride JW. 2016. Ehrlichia chaffeensis TRP120 activates canonical Notch signaling to downregulate TLR2/4 expression and promote intracellular survival. mBio 7:e00672-. doi: 10.1128/mBio.00672-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lina TT, Luo T, Velayutham TS, Das S, McBride JW. 2017. Ehrlichia activation of Wnt-PI3K-mTOR signaling inhibits autolysosome generation and autophagic destruction by the mononuclear phagocyte. Infect Immun 85:e00690-. doi: 10.1128/IAI.00690-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kuriakose JA, Miyashiro S, Luo T, Zhu B, McBride JW. 2011. Ehrlichia chaffeensis transcriptome in mammalian and arthropod hosts reveals differential gene expression and post transcriptional regulation. PLoS One 6:e24136. doi: 10.1371/journal.pone.0024136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luo T, Zhang X, McBride JW. 2009. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin Vaccine Immunol 16:982–990. doi: 10.1128/CVI.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wakeel A, Kuriakose JA, McBride JW. 2009. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect Immun 77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li Q, Lau A, Morris TJ, Guo L, Fordyce CB, Stanley EF. 2004. A syntaxin 1, Galpha (o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J Neurosci 24:4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burgess A, Vigneron S, Brioudes E, Labbe JC, Lorca T, Castro A. 2010. Loss of human Greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci U S A 107:12564–12569. doi: 10.1073/pnas.0914191107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oerlemans R, Franke NE, Assaraf YG, Cloos J, van Zantwijk I, Berkers CR, Scheffer GL, Debipersad K, Vojtekova K, Lemos C, van der Heijden JW, Ylstra B, Peters GJ, Kaspers GL, Dijkmans BA, Scheper RJ, Jansen G. 2008. Molecular basis of bortezomib resistance: proteasome subunit beta5 (PSMB5) gene mutation and overexpression of PSMB5 protein. Blood 112:2489–2499. doi: 10.1182/blood-2007-08-104950. [DOI] [PubMed] [Google Scholar]
- 60.Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 61.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.