ABSTRACT
Toxoplasmosis is caused by infection with the protozoan parasite Toxoplasma gondii, which has the capacity to infect all warm-blooded animals worldwide. Toxoplasmosis is a major cause of visual defects in the Colombian population; however, the association between genetic polymorphisms in cytokine genes and susceptibility to ocular toxoplasmosis has not been studied in this population. This work evaluates the associations between polymorphisms in genes coding for the cytokines tumor necrosis factor alpha (TNF-α) (rs1799964, rs1800629, rs1799724, rs1800630, and rs361525), interleukin 1β (IL-1β) (rs16944, rs1143634, and rs1143627), IL-1α (rs1800587), gamma interferon (IFN-γ) (rs2430561), and IL-10 (rs1800896 and rs1800871) and the presence of ocular toxoplasmosis (OT) in a sample of a Colombian population (61 patients with OT and 116 healthy controls). Genotyping was performed with the “dideoxynucleotide (ddNTP) primer extension” technique. Functional-effect predictions of single nucleotide polymorphisms (SNPs) were done by using FuncPred. A polymorphism in the IL-10 gene promoter (−1082G/A) was significantly more prevalent in OT patients than in controls (P = 1.93e−08; odds ratio [OR] = 5.27e+03; 95% confidence interval [CI] = 3.18 to 8.739; Bonferroni correction [BONF] = 3.48e−07). In contrast, haplotype “AG” of the IL-10 gene promoter polymorphisms (rs1800896 and rs1800871) was present at a lower frequency in OT patients (P = 7e−04; OR = 0.10; 95% CI = 0.03 to 0.35). The +874A/T polymorphism of IFN-γ was associated with OT (P = 3.37e−05; OR = 4.2; 95% CI = 2.478 to 7.12; BONF = 6.07e−04). Haplotype “GAG” of the IL-1β gene promoter polymorphisms (rs1143634, rs1143627, and rs16944) appeared to be significantly associated with OT (P = 0.0494). The IL-10, IFN-γ, and IL-1β polymorphisms influence the development of OT in the Colombian population.
KEYWORDS: ocular toxoplasmosis, single nucleotide polymorphisms, cytokines, susceptibility
INTRODUCTION
Toxoplasmosis is caused by infection with the protozoan parasite Toxoplasma gondii, which has the capacity to infect all warm-blooded animals worldwide (1). It is estimated that 30 to 70% of the human population is infected with this parasite, and essentially the entire human population is at risk of infection (2). A limited number of people develop symptoms, suggesting that host susceptibility and strain disparity can play a role in the variability of clinical symptoms (3). For instance, in Colombia, where seroprevalence studies show that almost half of the population is infected (47% according a national study) (4), the incidence of symptomatic ocular toxoplasmosis (OT) is 3 new episodes per 100,000 inhabitants per year (5). It is estimated that around 10% of newly infected people develop the ocular form of toxoplasmosis (6). OT is characterized by intraocular inflammation and is the most common clinical manifestation of toxoplasmosis (7). Particularly, OT causes severe pathology in the eye in South America that is more severe than in other parts of the world, and it is characterized by deviated T helper 2 (Th2) immune responses (8, 9). Lesions can originate both from congenital infection and from infections acquired after birth (10, 11). The lesions can affect the macula and other layers of the retina and the choroid, resulting in retinochoroiditis, the most frequent cause of posterior uveitis in immunocompetent patients (12). Ocular manifestations can have an early or late onset, with primary or recurrent clinical manifestations, and present different degrees of ocular involvement that vary according to the immune status of the individual and by infection with different T. gondii strains (13–15). The relative contribution of the host inflammatory response elicited, versus parasite proliferation, to the development of retinal destruction has not yet been completely defined (16). Cytokines, chemokines, and their receptors play a key role in the regulation of the type and magnitude of the immune response. Whether the ocular manifestations resulting from infection by T. gondii are attributable to host or parasite genetic factors, differences in the exposure rate, or all these factors remains uncertain (17).
One important issue to study is the polymorphisms in genes coding for cytokines involved in the immune response to Toxoplasma. Infection in humans is characterized by the presence of high levels of cytokines, such as interleukin 12 (IL-12), tumor necrosis factor alpha (TNF-α), and gamma interferon (IFN-γ), all of which have been associated with ocular pathology (18). Besides the involvement of both CD8+ T lymphocytes and natural killer (NK) cells, the immune response to T. gondii infection induces a strong Th1 response orchestrated by CD4+ T cells and is dominated by the production of proinflammatory mediators. However, while the Th1 response prevents parasite replication, a strong Th1 response may also cause immune-mediated tissue damage, contributing to the severity of ocular toxoplasmosis. More recently, Th17 cells, characterized by the production of IL-17, a potent inducer of inflammation, were identified as key contributors to immunopathological responses in OT (19, 20). Taken together, a variety of gene polymorphisms might be involved in OT and may create an individual risk profile for a given patient (21).
Functional genetic polymorphisms in cytokine genes may interfere with or enhance the expression of cytokines and play a role in the genetic regulation of inflammatory responses and resistance or susceptibility to infectious diseases. Single nucleotide polymorphisms (SNPs) are useful to identify polymorphisms associated with susceptibility to a particular disease. Natural selection has favored the introduction of biallelic SNPs into cytokine genes, which results in variation in the production and level of protein rather than variation in its quality (22). Previous studies indicated that polymorphisms in the IL-1, IL-10, IFN-γ, and TNF-α genes or promoters are associated with a higher frequency of OT in people from Brazil (21, 23–25). Not unexpectedly, host cytokine gene polymorphisms have been a focus of interest in toxoplasmic chorioretinitis. Cytokines, in particular IFN-γ and TNF-α, play an essential role in resistance to T. gondii infections (26, 27). These cytokines activate innate immunity and macrophages, a major first line of defense.
Polymorphisms in genes encoding various cytokines have been shown to be connected with susceptibility to parasitic diseases. Indeed, individuals in Brazil who were homozygous for the A allele (+874T/A) of the IFN-γ gene had a high risk of OT if they possessed the A/A genotype, compared to a negative-control group (23). In addition, experimental data demonstrated a relevant role for the anti-inflammatory cytokine IL-10 in modulating acute OT; thus, the IL-10 gene polymorphism (IL-10 −1082A allele; AA and AG genotypes) was associated with the occurrence of OT (24). More recently, a study conducted by Cordeiro et al. (28) similarly associated an IL-6 polymorphism (−174G/C) with the occurrence, but not recurrence, of OT in Brazilian patients. Those authors also showed that the recurrence of toxoplasmic retinochoroiditis was associated with an IL-1α (−889C/T) polymorphism, related to an increase in IL-1α expression (24). Another study in Poland demonstrated that the major C allele at the IL-1β +3954C>T SNP was significantly more frequently detected among fetuses and neonates with congenital T. gondii infection than among uninfected persons. The outcomes reported in that study suggested that the presence of a mutated T allele in or marking the gene with this IL-1β SNP has a protective function against the development of congenital toxoplasmosis (infection acquired in utero during gestation). However, the mechanism of the role of the IL-1β +3954C>T SNP has yet to be investigated in a detailed molecular study (29). A study of a TNF-α gene polymorphism (−308G/A) in Brazil also demonstrated that the occurrence or recurrence of toxoplasmic retinochoroiditis was not associated with this polymorphism (21). Together, data from those studies suggest that the genetic control of the immune response is relevant for the pathogenesis of toxoplasmic chorioretinitis.
However, given the complexity of both parasite biology and the host immune system, it is unlikely that genetic variation at a single locus, as shown by SNP analyses, would provide an adequate explanation for the interindividual differences in host immune responses that result in diverse clinical manifestations. To this end, the identification of gene-gene interactions could enhance the power and accuracy of predicting disease outcomes of a complex disorder (30). For a better description of the genetic architecture of disease susceptibility and unambiguous identification of factors responsible for both causality and predisposition to a disease, functional appraisal of disease-associated polymorphisms is essential. There is widespread recognition that differences in gene expression may be an important source of phenotypic diversity in complex diseases (31, 32) and that noncoding polymorphisms contribute to the variance and etiology of a trait by regulating the expression of nearby genes.
To explore the plausible regulatory mechanisms exerted by cytokine SNPs, we have characterized allele-specific events by studying their transcriptional differences in terms of reporter gene activities and allelic expression imbalance (AEI). Basu et al. provided detailed insights into the molecular effects of cis-regulatory variants in controlling cytokine gene expression in Plasmodium falciparum-mediated malaria. However, that study underscored the possibility that this complex trait involves even more complex regulatory intricacies than previously anticipated (33). To date, few studies have reported an association between IL-10, TNF-α, IL-1, and IFN-γ gene polymorphisms and the development of OT, and none of those studies was conducted with a Colombian population. In this paper, we conducted a case-control study to investigate the association between TNF-α (rs1799964, rs1800629, rs1799724, rs1800630, and rs361525), IL-1β (rs16944, rs1143634, and rs1143627), IL-1α (rs1800587), IFN-γ (rs2430561), and IL-10 (rs1800896 and rs1800871) gene polymorphisms and the risk of OT in a Colombian population.
RESULTS
Genetic ancestries of cases and controls are similar.
The apportionment of genetic ancestral contributions in cases and controls was estimated as the mean of each ancestry across individuals, using Admixture v1.3 software. Considering the historical formation of the Colombian population, we assumed an essentially trihybrid contribution from Native Americans, Europeans, and Africans (i.e., K = 3). Ancestry analyses were conducted by using the Human Genome Diversity Project and Centre d'Etude du Polymorphisme Humain (HGDP-CEPH) populations as a reference (for Africa n = 43 cases [P = 0.07] and n = 10 controls [P = 0.05]; for Europe, n = 43 cases [P = 0.68] and n = 10 controls [P = 0.70]; and for America, n = 43 cases [P = 0.25] and n = 10 controls [P = 0.25]). No differences in the genetic ancestries in cases and controls were detected (P = 0.91). The ancestry information for cases and controls was similar to that previously reported by Ossa et al. (34) for the Central West Andean region of Colombia (where the present study was carried out), with the European contribution to the genetic background being higher for this population.
Case-control association analysis indicates that the IL-10 −1082A/G and the IFN-γ +874A/T polymorphisms are associated with ocular toxoplasmosis.
We investigated the distribution of 12 SNPs in 61 Colombian OT patients (cases) and 116 healthy controls. The genotype distribution of all polymorphisms did not deviate significantly from the Hardy-Weinberg equilibrium (HWE). The frequencies of the genotypes of polymorphisms in proinflammatory and anti-inflammatory cytokine genes in cases and controls are presented in Table 1. Among the cytokine genes, significantly higher frequencies of IL-1, IFN-γ, and IL-10 polymorphisms were observed for the OT group. We found that the IFN-γ polymorphism at this position (+874) was most frequently found in the OT group. Notably, the GG genotype with the IL-10 −1082A/G polymorphism was highly associated with the OT phenotype (P = 1.47e−05). Among variants of IL-10, the strongest association was found for the SNP −1082A/G (rs1800896), where the GG genotype was highly prevalent in patients in comparison to healthy subjects. The IL-10 −819G/A polymorphism was not associated with OT (P = 0.756).
TABLE 1.
Genotypic frequencies for each SNP evaluated
| Official gene symbol | Alternative gene names | Gene ID | Polymorphism |
Genotype, no. (%) of individuals |
P value | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rs IDa | Position in gene | Chromosome location | Cases | Healthy controls | ||||||||
| TNF | TNF-α, DIF, TNFA, TNFSF2, TNLG1F | 7124 | rs1799964 | −1031 | 6:31574531 | TT, 39 (67.2) | TC, 18 (31.0) | CC, 1 (1.7) | TT, 76 (65.5) | TC, 38 (32.8) | CC, 2 (1.7) | |
| TNF | TNF-α, DIF, TNFA, TNFSF2, TNLG1F | 7124 | rs1800630 | −863 | 6:31574699 | CC, 39 (76.5) | CA, 10 (19.6) | AA, 2 (3.9) | CC, 88 (77.2) | CA, 26 (22.8) | AA, 0 (0.0) | |
| TNF | TNF-α, DIF, TNFA, TNFSF2, TNLG1F | 7124 | rs1799724 | −857 | 6:31574705 | GG, 35 (61.4) | GA, 21 (36.8) | AA, 1 (1.8) | GG, 78 (67.2) | GA, 34 (29.3) | AA, 4 (3.5) | |
| TNF | TNF-α, DIF, TNFA, TNFSF2, TNLG1F | 7124 | rs361525 | −238 | 6:31575324 | CC, 53 (89.8) | CT, 5 (8.5) | TT, 1 (1.7) | CC, 103 (88.8) | CT, 12 (10.3) | TT, 1 (0.9) | |
| TNF | TNF-α, DIF, TNFA, TNFSF2, TNLG1F | 7124 | rs1800629 | −308 | 6:31575254 | GG, 51 (85.0) | GA, 9 (15.0) | AA, 0 (0.0) | GG, 100 (86.2) | GA, 16 (13.8) | AA, 0 (0.0) | |
| IL1A | IL-1, IL-1A, IL1F1, IL-1-ALPHA | 3552 | rs1800587 | −889 | 2:112785383 | GG, 33 (55.0) | GA, 23 (38.3) | AA, 4 (6.7) | GG, 59 (50.9) | GA, 46 (39.7) | AA, 11 (9.5) | |
| IL1B | IL-1, IL1F2, IL-1-BETA | 3553 | rs16944 | −511 | 2:112837290 | GG, 15 (24.6) | GA, 29 (47.5) | AA, 17 (27.9) | GG, 37 (31.9) | GA, 56 (48.3) | AA, 23 (19.8) | 0.391 |
| IL1B | IL-1, IL1F2, IL-1-BETA | 3553 | rs1143634 | 3954 | 2:112832813 | GG, 41 (68.3) | GA, 17 (28.3) | AA, 2 (3.3) | GG, 83 (71.5) | GA, 29 (25.0) | AA, 4 (3.5) | |
| IL1B | IL-1, IL1F2, IL-1-BETA | 3553 | rs1143627 | −31 | 2:112836810 | GG, 14 (22.9) | GA, 27 (44.3) | AA, 20 (32.8) | GG, 37 (31.9) | GA, 55 (47.4) | AA, 24 (20.7) | 0.169 |
| IFNG | IFG, IFI | 3458 | rs2430561 | 874 | 12:68158742 | TT, 2 (4.3) | TA, 21 (44.7) | AA, 24 (51.1) | TT, 47 (40.5) | TA, 46 (39.7) | AA, 23 (19.8) | |
| IL-10 | CSIF, TGIF, GVHDS, IL-10, IL10A | 3586 | rs1800871 | −819 | 1:206773289 | GG, 34 (57.6) | GA, 19 (32.2) | AA, 6 (10.2) | GG, 61 (52.6) | GA, 44 (37.9) | AA, 11 (9.5) | 0.756 |
| IL-10 | CSIF, TGIF, GVHDS, IL-10, IL10A | 3586 | rs1800896 | −1082 | 1:206773552 | AA, 8 (15.1) | AG, 14 (26.4) | GG, 31 (58.5) | AA, 57 (50.0) | AG, 40 (35.1) | GG, 17 (14.9) | 1.47e−05 |
rs ID, reference SNP ID number.
In Table 2, we present the frequencies of the TNF-α −238G/A, TNF-α −308G/A, TNF-α −1031T/C, TNF-α −857C/T, and TNF-α −863C/A polymorphisms. No differences between the observed and expected distributions of genotypes were found between cases and controls. Our study is the first to investigate the association between TNF-α gene polymorphisms and the occurrence of OT in Colombian populations. Our finding was unexpected, as the role of TNF-α in OT seems to be relevant. In addition, for the IL-1 SNPs, we did not find significant differences in the frequencies of the G/A genotype.
TABLE 2.
Allelic distribution of cytokine gene polymorphisms in Colombian patients with ocular toxoplasmosis and healthy controlsa
| Chr | dbSNP ID | A1 | A2 | MAF |
GMAF | |
|---|---|---|---|---|---|---|
| Cases | Controls | |||||
| 1 | rs1800871 | G | A | 0.2627 | 0.2845 | 0.43 |
| 1 | rs1800896 | A | G | 0.283 | 0.3246 | 0.27 |
| 2 | rs1800587 | G | A | 0.2583 | 0.2931 | 0.27 |
| 2 | rs1143634 | G | A | 0.175 | 0.1595 | 0.13 |
| 2 | rs1143627 | G | A | 0.4508 | 0.444 | 0.47 |
| 2 | rs16944 | G | A | 0.4836 | 0.4397 | 0.49 |
| 6 | rs1799964 | T | C | 0.1724 | 0.181 | 0.22 |
| 6 | rs1800630 | C | A | 0.1373 | 0.114 | 0.15 |
| 6 | rs1799724 | G | A | 0.2018 | 0.181 | 0.10 |
| 6 | rs1800629 | G | A | 0.075 | 0.06897 | 0.09 |
| 6 | rs361525 | C | T | 0.05932 | 0.6034 | 0.06 |
| 12 | rs2430561 | T | A | 0.266 | 0.3966 | 0.28 |
Chr, chromosome; A1, alelle 1; A2, allele 2; MAF, minor allele frequency; GMAF, global minor allele frequency.
The allele distributions (minor allele frequency [MAF]) of the polymorphisms in cytokines genes for each group are summarized in Table 2. In order to determine whether the less-represented alleles of SNPs in candidate genes (IL-1, IFN-γ, TNF-α, and IL-10 genes) were an independent risk factor for toxoplasmosis, we performed an association test (Table 3).
TABLE 3.
Association analysis of MAF alleles in cytokine polymorphisms in patients with ocular toxoplasmosis and healthy controlsa
| Chr | Gene | Position | CHISQ | P | OR | L95 | U95 | BONF |
|---|---|---|---|---|---|---|---|---|
| 1 | IL-10 −819G/A | 206773288 | 0.1851 | 0.6671 | 0.8962 | 0.5439 | 1.477 | 1 |
| 1 | IL-10 −1082A/G | 206773551 | 45.04 | 1.93e−08 | 5.27 | 3.18 | 8.739 | 3.48e−07 |
| 2 | IL-1α −889G/A | 112785382 | 0.473 | 0.4916 | 0.8401 | 0.5111 | 1.381 | 1 |
| 2 | IL-1β +3954G/A | 112832812 | 0.1384 | 0.7099 | 1.118 | 0.6212 | 2.012 | 1 |
| 2 | IL-1β −31G/A | 112836809 | 3.546 | 0.05969 | 1.526 | 0.9819 | 2.371 | 1 |
| 2 | IL-1β −511G/A | 112837289 | 1.892 | 0.169 | 1.361 | 0.8768 | 2.112 | 1 |
| 6 | TNF-α −1031T/C | 31574530 | 0.03925 | 0.843 | 0.9425 | 0.5244 | 1.694 | 1 |
| 6 | TNF-α −863C/A | 31574698 | 0.3567 | 0.5503 | 1.236 | 0.6161 | 2.48 | 1 |
| 6 | TNF-α −857G/A | 31574704 | 0.2151 | 0.6428 | 1.143 | 0.6489 | 2.015 | 1 |
| 6 | TNF-α −308G/A | 31575253 | 0.04365 | 0.8345 | 1.095 | 0.4687 | 2.556 | 1 |
| 6 | TNF-α −238C/T | 31575323 | 0.001451 | 0.9696 | 0.982 | 0.3853 | 2.503 | 1 |
| 9 | IFN-γ +874T/A | 68158741 | 30.48 | 3.37e−05 | 4.2 | 2.478 | 7.12 | 6.07e−04 |
Chr, chromosome; CHISQ, chi-square value; OR, odds ratio; L95 and U95, lower and upper bounds of the 95% confidence intervals for the odds ratio, respectively; BONF, Bonferroni correction. Boldface indicates that statistical differences are significant.
Most of the SNPs analyzed had no significant effect, while the differences proved to be significant for the rs1800896 and rs2430561 SNPs after the application of the Bonferroni correction (BONF). In our study, the −1082G allele (P = 1.93e−08; odds ratio [OR] = 5.27; 95% confidence interval [CI] = 3.18 to 8.73; BONF = 3.48e−07) and the +874A allele (P = 3.37e−05; OR = 4.2; 95% CI = 2.478 to 7.12; BONF = 6.07e−04) were present at high frequencies and were significantly represented in OT patients compared with controls. In the case of the rs1143627 polymorphism, it initially showed a significant effect (P = 0.05969; OR = 1.53; 95% CI = 0.98 to 2.37) that disappeared when the Bonferroni correction was applied (BONF = 1.00).
So far, several studies have shown some involvement of the proinflammatory cytokine IL-1β in the immune response after T. gondii infection. No associations between the observed and expected distributions of the IL-1α −889T/C, IL-1β +3954G/A, IL-1β −511C/T, TNF-α −308G/A, TNF-α −238C/T, TNF-α −863C/A, TNF-α −857G/A, and TNF-α −1031T/C alleles were found. There were no significant differences in the allelic frequencies of each of these polymorphisms in patients with OT compared to control subjects (P = 0.4916, P = 0.7099, P = 0.169, P = 0.8345, P = 0.9696, P = 0.5503, P = 0.6428, and P = 0.843, respectively).
Haplotypes of the IL-1β and IL-10 gene promoter polymorphisms are associated with susceptibility to ocular toxoplasmosis.
In order to analyze the haplotype effect of cytokine genes, we considered those genes with enough SNPs analyzed (at least two in each gene). The IFN-γ gene was not analyzed by the haplotype approach. We also estimated the TNF-α haplotype frequencies (rs1799964, rs1800629, rs1799724, rs1800630, and rs361525) and evaluated the association of these variants with OT. We observed five haplotype combinations (TGGCC, TGACC, CGGAC, TAGCC, and CGGCT). However, a significant association of the distributions of the haplotype frequencies between cases and healthy controls was not found.
Haplotype “GAA” of the IL-1β gene promoter polymorphisms (rs16944, rs1143634, and rs1143627) appeared to be significantly associated with susceptibility to ocular toxoplasmosis (P = 0.038). Haplotypes “GG” and “GA” of the IL-10 gene promoter polymorphism (rs1800896 and rs1800871) are also significantly associated with OT (P = 4.0132e−07 and P = 1.1072e−06, respectively). Frequencies are shown in Table 4.
TABLE 4.
Haplotype frequencies of IL-10, TNF-α, and IL-1β gene polymorphisms in patients with ocular toxoplasmosisb
| SNP | Haplotype | Frequency | P valuea |
|---|---|---|---|
| IL-10 gene region (rs1800896, rs1800871) | GG | 0.405 | 4.01e−03 |
| GA | 0.319 | 1.11e−02 | |
| AA | 0.231 | 0.0321 | |
| AG | 0.045 | 0.001 | |
| TNF-α gene region (rs1799964, rs11800629, rs1799724, rs1800630, rs361525) | TGGCC | 0.569 | NA |
| TGACC | 0.162 | 0.6 | |
| CGGAC | 0.111 | 0.72 | |
| TAGCC | 0.07 | 0.6 | |
| CGGCT | 0.04 | 0.73 | |
| IL-1β gene region (rs16944, rs1143634, rs1143627) | AGG | 0.458 | NA |
| GGA | 0.357 | O.61 | |
| GAA | 0.159 | 0.038 | |
| GGG | 0.017 | 0.71 |
NA, not applicable.
Boldface indicates that statistical differences are significant.
Coexpression analysis identifies connectivity of the IL-1β and IL-1α genes mediated by Ribonuclease k6 (RNASE6).
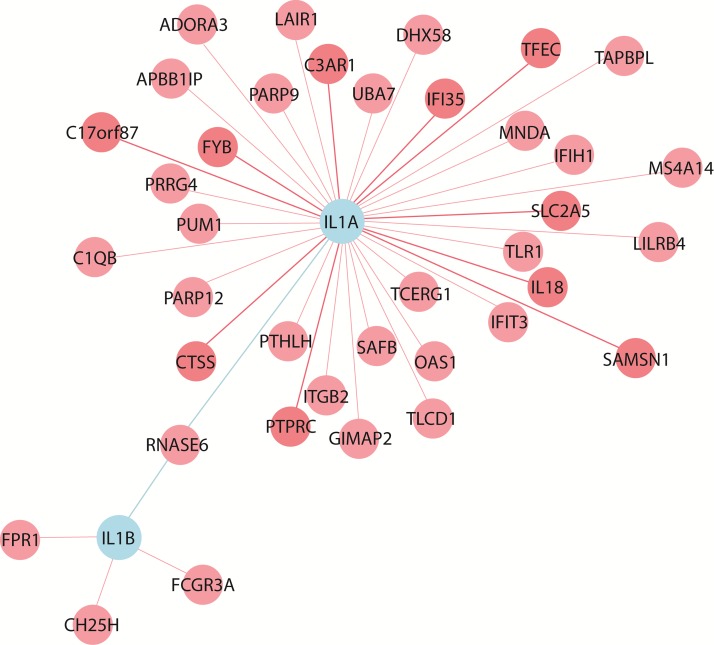
We simultaneously analyzed the 12 SNPs for the most significant gene-gene interactions associated with OT. Most of the polymorphisms involved in multifactor dimensionality reduction (MDR) combinations were not associated with disease in the single-site analysis. We further evaluated a possible epistatic effect among the less-represented alleles of SNPs in candidate genes. To this end, we applied a statistical approach to perform an analysis of the combined epistatic effect of SNPs. We did not find a significant risk for ocular toxoplasmosis in the presence of any MAF alleles from the rs1800896 and rs2430561 SNPs (Table 5). Coexpression analyses identified a connectivity of the IL-1β and IL-1α genes, mediated by RNASE6. No coexpression with IL-10, IFN-γ, and TNF-α (Fig. 1) was observed. Additional work is needed to assess the still unknown gene-gene interactions. Raj et al. performed an expression quantitative trait locus (eQTL) study on purified CD4+ T cells and monocytes from 461 healthy individuals and found that susceptibility alleles for certain diseases had an overrepresentation of either T cell-specific or monocyte-specific eQTLs (35). We used the SCAN database to check for coexpression between the genes identified in this paper and the genes identified by Raj et al., whose SNPs had P values of <10−8 (36). We found that there was coexpression between these two groups of genes (Table 6).
TABLE 5.
Gene-gene interactions between the IL-10 −1082A/G and IFN-γ +874T/A gene polymorphisms
| rs1800896 haplotype | No. of individuals with rs2430561 haplotype (frequency) |
||
|---|---|---|---|
| A/A | A/T | T/T | |
| G/G | 20 (0.112) | 19 (0.107) | 5 (0.028) |
| G/A | 9 (0.050) | 22 (0.124) | 20 (0.112) |
| A/A | 17 (0.096) | 23 (0.129) | 24 (0.135) |
FIG 1.
Coexpression analysis of IL-1α, IL-1β, TNF-α, IFN-γ, and IL-10, using NP de novo coexpression analysis (http://www.wzgenomics.cn/NPdenovo/index.php). The reference data set was HBT (The Human Brain Transcriptome), the minimum Pearson correlation coefficient was 0.8, and the maximum number of nodes of the network was 80.
TABLE 6.
Coexpression between genes identified in this paper and those identified by Raj et al.,a as determined by using the SCAN databaseb
| SNP | Gene | Coexpressed gene | P value |
|---|---|---|---|
| rs5016378 | ABCC4 | TNF | 0.00003 |
| rs6496603 | ANPEP | TNF | 0.00003 |
| rs3857405 | ARSB | TNF | 0.00003 |
| rs10882987 | AVPI1 | IL1A | 0.00003 |
| rs11571700 | BRCA2 | IL1A | 0.00001 |
| rs7335538 | CARS2 | IL-10 | 0.0001 |
| rs10823760 | CDH23 | IL1A | 0.00003 |
| rs2185415 | CDH23 | IL1A | 0.00003 |
| rs12782689 | CDH23 | IL1A | 0.00006 |
| rs12763836 | CDH23 | IL1A | 0.0001 |
| rs12772205 | CDH23 | IL1A | 0.0001 |
| rs10873263 | COQ6 | TNF | 0.00006 |
| rs10503215 | CSMD1 | IL-10 | 0.00009 |
| rs3849831 | CSMD1 | TNF | 0.0001 |
| rs260709 | EDAR | TNF | 0.000002 |
| rs12090415 | EPHB2 | TNF | 0.00003 |
| rs16947233 | FBXW8 | TNF | 0.00007 |
| rs7133609 | FBXW8 | TNF | 0.0001 |
| rs6549191 | FRMD4B | IL-10 | 0.0001 |
| rs488532 | GCNT2 | IL-10 | 0.00003 |
| rs4853066 | HK2 | IL1A | 0.0001 |
| rs2069727 | IFNG | C1orf112 | 0.00009 |
| rs2069727 | IFNG | CUL7 | 0.0001 |
| rs2069722 | IFNG | DPYSL2 | 0.00008 |
| rs2069722 | IFNG | FOXJ2 | 0.00004 |
| rs2069727 | IFNG | LRRCC1 | 0.00004 |
| rs2069727 | IFNG | MSH5 | 0.0001 |
| rs2069727 | IFNG | RAB23 | 0.00006 |
| rs2069727 | IFNG | SLC4A7 | 0.0001 |
| rs2069718 | IFNG | SPIRE1 | 0.0001 |
| rs2069727 | IFNG | TIFA | 0.0001 |
| rs3024491 | IL-10 | CD52 | 0.0001 |
| rs1304037 | IL1A | IER3 | 0.0001 |
| rs1304037 | IL1A | MRPS7 | 0.000001 |
| rs1800587 | IL1A | MRPS7 | 0.00004 |
| rs9407340 | KANK1 | IL-10 | 0.0001 |
| rs6853658 | KCNIP4 | TNF | 0.00002 |
| rs655487 | KCNMA1 | TNF | 0.0001 |
| rs2219172 | L3MBTL4 | IL-10 | 0.0001 |
| rs11710266 | LIMD1 | TNF | 0.0001 |
| rs2667975 | LYN | IL-10 | 0.00003 |
| rs373696 | LYRM4 | IL-10 | 0.00007 |
| rs195063 | MAN1A1 | IL1A | 0.0001 |
| rs3791328 | MGAT5 | TNF | 0.00006 |
| rs7174277 | MYO1E | TNF | 0.00001 |
| rs6692267 | NEGR1 | IL1A | 0.00005 |
| rs11908460 | PLCB1 | TNF | 0.0001 |
| rs13325518 | PPM1L | TNF | 0.000002 |
| rs16831830 | PPM1L | TNF | 0.0001 |
| rs10771415 | PZP | TNF | 0.0001 |
| rs4714758 | TMEM63B | IL-10 | 0.0001 |
| rs228587 | TPK1 | IL1A | 0.0001 |
| rs10840100 | TRIM66 | TNF | 0.00003 |
| rs1286422 | TTC7B | IL-10 | 0.00007 |
| rs1286439 | TTC7B | IL-10 | 0.00007 |
| rs1290434 | TTC7B | IL-10 | 0.00007 |
| rs2343 | TTC7B | TNF | 0.00005 |
| rs1396049 | TXK | IL1A | 0.00007 |
| rs16954363 | VPS53 | TNF | 0.00009 |
| rs12450330 | VPS53 | TNF | 0.0001 |
| rs16954271 | VPS53 | TNF | 0.0001 |
| rs7207469 | VPS53 | TNF | 0.0001 |
| rs7220509 | VPS53 | TNF | 0.0001 |
| rs292553 | WDR91 | IL1A | 0.0001 |
| rs292557 | WDR91 | IL1A | 0.0001 |
| rs4504197 | ZMAT3 | TNF | 0.00006 |
| rs6769215 | ZMAT3 | TNF | 0.00006 |
See reference 35.
P values for coexpression according to the SCAN database are included.
DISCUSSION
Ocular toxoplasmosis can cause visual alterations, retinochoroiditis, and, in some cases, loss of sight. T. gondii infection induces a response by T helper 1 cells, which produce IFN-γ, a cytokine that is involved in resistance to toxoplasmosis (37). Chorioretinitis during active OT involves all chorioretinal layers. This inflammatory process is characterized by the appearance of exudative, white-appearing lesions with soft borders; during the active phase, these lesions evolve with necrosis and the deposition of pigment, resulting in chronic inactive and irreversible atrophic and hyperpigmented retinal scars, which are sequelae of the acute process. The effector cells that attack and destroy infected retinal neurons include CD4+ and CD8+ T cells, B cells, and macrophages. Macrophages have also been occasionally detected in the choroid underlying retinal lesions (38). In patients with active retinochoroidal lesions due to congenital infection, an expansion of monocytes and NK cells in blood was found (39). The expansion, migration, and activation of these cells might be associated with chemokine/cytokine cross talk (40). Information concerning intraocular cytokine levels in OT has been obtained from aqueous humor (AH) samples, but further studies are needed in order to determine the precise source of these mediators and their contributions to pathogenesis.
Similar to results in the mouse model of ocular toxoplasmosis, resistance to OT is associated with the ability to produce IL-12 and IFN-γ in response to parasite antigens (41). Infection by Toxoplasma induces high levels of IFN-γ in humans, as can be deduced from reports of both asymptomatic and symptomatic T. gondii-seropositive individuals, compared to negative controls (42). However, by comparing the levels of this cytokine in individuals with disease to those in individuals who are infected but without symptoms, only a slight increase in the level of this cytokine was reported for T. gondii-seropositive asymptomatic individuals compared to individuals with OT (43). During our studies concerning intraocular cytokine levels in OT, we analyzed only a very few patients in order to establish differences in the levels of cytokines between those with vertically transmitted OT and those with OT acquired after birth. However, an analysis of the IFN-γ/IL-10 ratio in ex vivo-stimulated peripheral mononuclear cells suggests that patients who acquired infection in utero have higher levels of IL-10 production (9).
To explain why infected people can become sick although they produce IFN-γ, an imbalance in the cytokine network should be explored. Polymorphisms in the cytokine-encoding genes, including proinflammatory cytokines such as TNF-α, IL-6, IL-12, and IFN-γ and anti-inflammatory cytokines such as IL-10 and transforming growth factor β (TGF-β), were previously shown to be associated with several diseases (44, 45).
It is well recognized that differences in gene expression may be an important source of phenotypic diversity in complex diseases and that noncoding polymorphisms contribute to the variance and etiology of a trait by regulating the expression of nearby genes (31, 46). The present study is the first to demonstrate the association between polymorphisms in the IL-10 gene promoter (−819G/A and −1082A/G) and OT in a Colombian population. The link between OT and the GG genotype at the IL-10 promoter SNPs provides evidence that abnormalities in the genetic control of cytokine levels may be relevant in influencing the human immune response in OT. IL-10 is an anti-inflammatory and immune-regulatory cytokine, which induces T cell anergy by downregulating the expression of the genes coding for the costimulatory molecule B7-1/B7-2, major histocompatibility complex (MHC) class II, proinflammatory cytokines (IFN-γ, TNF-α, and IL-12), and chemokines secreted by activated macrophages (47, 48). One of the most important SNPs in the promoter region of the IL-10 gene is the −1082A/G (rs1800896) polymorphism, which may have an effect on the transcriptional binding site; thus, this SNP may alter the rate of gene expression. It has been reported that 50 to 75% of the variation in IL-10 production is genetically controlled (49).
The human IL-10 gene is located on chromosome 1 and has been mapped to the junction between 1q31 and 1q32 (50). Several polymorphic sites in the IL-10 gene have been identified. Three polymorphic sites (−1082A/G [rs1800896] −819C/T [rs1800871], and −592C/A [rs1800872]) are located in the promoter region of the gene. Individuals homozygous for the −1082G allele have higher circulating IL-10 levels, higher expression levels of IL-10 mRNA, and higher levels of production of IL-10 following in vitro stimulation (51, 52). Indeed, the presence of the G allele at position −1082 correlates with a higher level of IL-10 protein production in vitro and in the pleural fluid of patients with active tuberculosis (50, 53, 54). Data from those studies suggest that carriers of the −1082G allele are likely to have a high risk for the progression of OT because the −1082G allele may suppress the immune response by increasing the expression level of IL-10.
IFN-γ is an important Th1 cytokine that is secreted by NK cells and T cells, and its production plays a critical role in macrophage activation in order to control Toxoplasma infection. The IFN-γ-encoding gene is located on chromosome 12q24 and consists of four exons with three intervening regions (55). A polymorphism in the first intron of the IFN-γ gene at position 874 (rs2430561) directly influences IFN-γ production levels (56). The IFN-γ +874T/A SNP is located within a putative nuclear factor κB (NF-κB) binding site, and NF-κb specifically binds to the DNA sequence containing the T allele (57). Thus, the T allele might be responsible for the induction of IFN-γ production at a higher level. Indeed, it has been shown that the T and A alleles most likely correlate with high and low expression levels of IFN-γ, respectively (58, 59). The +874T allele is absolutely linked to the 12-CA-repeat microsatellite, while the +874A allele is adjacent to a non-12-CA repeat (56). Our results indicate that the A allele was significantly associated with the development of OT. However, for revealing the biological significance of these SNPs in susceptibility to ocular toxoplasmosis, further studies are needed in different populations.
IL-1β, a proinflammatory pleiotropic cytokine, is a member of the IL-1 family that possesses the ability to stimulate the expression of genes associated with inflammation and the immune response, including cyclooxygenase type 2, phospholipase A2, and inducible nitric oxide synthase (60). Additionally, another important proinflammatory property of IL-1β is its capacity to increase the expression of adhesion molecules such as intercellular adhesion molecule 1 (ICAM-1) on endothelial and other cell surfaces. IL-1α and IL-1β bind to IL-1 receptor type I, eliciting signal transduction and the corresponding biological effects. At least three SNPs in the IL-1β gene have been reported, all of which represent a C/T base transition at positions −511 and −31 in the promoter and at position +3954 in the exon (61–64). Despite the importance of IL-1β during the immune response to T. gondii, no associations of the allelic frequencies of these polymorphisms between patients and controls were found.
Data presented in this work refer to specific SNPs in some of the cytokine genes involved in the immune response to T. gondii; however, additional studies are required to explain how these SNPs can affect the immune response in OT. SNPs may influence protein conformation (topology), promoter activity, pre-mRNA and RNA splicing, or transcriptional regulation affecting cytokine production (65). Some SNPs in the IL-4 and TNF-α genes create a new binding site for the OCT-1 transcription factor affecting gene expression (66, 67). In addition, the variability in IL-10 production associated with the presence of the IL-10 −819G/A polymorphism is related to the activation of poly(ADP-ribose) polymerase 1 (PPAR-1) and the suppression of IL-10 transcription (68). Further studies to examine the relationship between SNPs in cytokine genes and the presence of recurrences, a major concern in OT, would be useful. We have not yet analyzed the relationship of SNPs with recurrences in this cohort.
Our data suggest that genetic susceptibility to ocular involvement during Toxoplasma infection exists, which provides some possible clues to understanding why some Toxoplasma-infected people have ocular involvement. Although the eye has immune privilege, the ocular immune response is connected with the peripheral response. We previously demonstrated that patients with ocular toxoplasmosis have different peripheral immune responses compared to those of patients without ocular involvement (9). Both retina and brain are damaged by Toxoplasma infection in humans. The presence of particular receptors in neural cells (including the retina) and the absence of mechanisms in the eye to compensate for the effects of IL-10 and IFN-γ gene polymorphisms are possible explanations for this tissue tropism.
Functional studies are needed to understand how our SNPs affect the cytokine balance in OT. The clinical presentation of OT is heterogenous in terms of the size and number of lesions and the presence and location of recurrences (macula, optic nerve, and peripheral retina). Different cytokine profiles might contribute to this clinical heterogeneity.
A key objective in biological research is to identify all molecules within a living cell and how they interact. However, the functions of many genes are still not understood. Gene coexpression networks can be used for various purposes, including candidate disease gene prioritization, functional gene annotation, and the identification of regulatory genes (69). Coexpression analysis identified the connectivity of the IL-1β and IL-1α genes mediated by RNASE6.
Our study provides evidence that common genetic variants in Th1 (IL-1, IFN-γ, and TNF-α) and Th2 (IL-10) genes are associated with the risk of developing OT in patients from Colombia. These results are similar to those of previous studies of patients with OT in Brazil (70) and congenital toxoplasmosis in Poland (29) and are consonant in their broad conclusions and generalizations about the proinflammatory and downmodulatory cytokine genes that are associated with our findings in a cohort of patients with congenital toxoplasmosis in the United States. These genes include P2X7R (70, 71), NALP1 (72–74), ALOX12 (75), HLA classes I and II (76–79), and ERAP1 (80), all of which influence the immune response. Our findings are in agreement with current knowledge about immunity to T. gondii.
In future studies, it may be useful for devising diagnostic and therapeutic approaches in OT to determine whether these polymorphisms alter cytokine production in patients with ocular compromise and if they influence the clinical presentation of disease. To this end, the identification of gene-gene interactions could enhance the power and accuracy of predicting disease outcomes of a complex disorder. For a better description of the genetic architecture of disease susceptibility and unambiguous identification of factors responsible for both causality and predisposition to a disease, functional appraisal of disease-associated polymorphisms is essential (30, 32, 81).
MATERIALS AND METHODS
Subjects.
The Ethics Committee of the Universidad Tecnológica de Pereira approved this study's protocol. The group of patients was composed of 61 cases (mean age ± standard deviation [SD], 37.37 ± 17.27 years), with a male/female ratio of 1.44. Patients from the Bogotá and Quindío regions in Colombia with OT were diagnosed as previously described (5, 8). These patients had diagnostic confirmation by sampling of aqueous humor. The control group was divided in two subgroups. Subgroup 1 was composed of 22 Colombian patients with symptoms of uveitis for whom ocular toxoplasmosis was excluded as the cause of disease, and subgroup 2 was composed of 94 healthy individuals aged 36.29 ± 13.81 years (male/female ratio, 0.84).
Biological samples and diagnostic assays for ocular toxoplasmosis.
Both patients (n = 83) and healthy controls (n = 94) included in the study consented to provide blood samples. In addition, patients with ocular diseases (ocular toxoplasmosis [n = 61] or other causes of uveitis [n = 22]) provided ocular fluid samples. Seven patients did not have aqueous humor sampling because they had only a single functional eye, for which the sampling risk was considered too high. Blood samples were used to obtain serum and peripheral blood mononuclear cells (PBMCs) for further DNA extraction and PCR tests. AH samples (0.1 to 0.2 ml) were obtained at the Ophthalmological Center, Clínica Barraquer, under sterile conditions after topical anesthesia and sent to a laboratory for analysis. All serum samples were analyzed for anti-Toxoplasma IgG and IgM antibody titers by using commercial enzyme-linked immunosorbent assays (ELISAs) according to the manufacturer's recommendations (Vidas Toxo IgG II [reference number 30210] and Toxo IgM [reference number 30202]; bioMérieux, France). Those with positive IgG results underwent local ocular antibody production testing. In order to detect intraocular anti-Toxoplasma antibodies, the Goldmann-Witmer coefficient (GWC) was calculated as follows: (anti-Toxoplasma IgG in aqueous humor/total IgG in aqueous humor)/(anti-Toxoplasma IgG in serum/total IgG in serum) (8, 82). The specific anti-Toxoplasma IgG antibody titers in aqueous humor samples were determined by an ELISA as described previously (83). An index of <2 was considered a positive result for ocular Toxoplasma infection.
DNA extraction from blood samples.
A tube with heparin was used to collect blood samples, which were then processed for DNA extraction. The Wizard Genomic DNA purification kit (Promega, Madison, WI, USA) was used, and procedures were performed as recommended by the manufacturer. Briefly, DNA from white blood cells was obtained by incubating samples with a cell lysis solution for 10 min at room temperature. After centrifugation at 13,000 × g for 20 s at room temperature, the supernatant was discarded, and the pellet was recovered. The cellular proteins were then removed by a salt precipitation step, and genomic DNA was concentrated and desalted by isopropanol precipitation.
Genotyping. (i) Primer design and multiplex PCR amplification.
The polymorphisms to be studied were selected based on data from previous studies carried out with patients with ocular toxoplasmosis or other human diseases (21, 23–25). Genes and SNPs were selected based on their functional significance and previous reports on an association with any disease condition. A list of cytokine candidate genes and their selected polymorphisms indicating the SNPs, reference SNP ID numbers (rs IDs), chromosomal positions, locations, and predicted functional effects is shown in Table 7. Functional-effect predictions of SNPs were done by using the FuncPred (Functional SNP Prediction) tool (https://snpinfo.niehs.nih.gov/snpinfo/snpfunc.html). Functional prediction of the deleterious effect, if any, of the associated SNPs with respect to functional categories such as protein coding, splicing regulation, transcriptional regulation, and posttranslation was assessed by using the F-SNP program (http://compbio.cs.queensu.ca/F-SNP/). We selected polymorphisms in the following genes: TNF-α (rs1799964, rs1800629, rs1799724, rs1800630, and rs361525), IL-1β (rs16944, rs1143634, and rs1143627), IL-1α (rs1800587), IFN-γ (rs2430561), and IL-10 (rs1800896 and rs1800871). Each primer set was designed by using Primer3 software (http://frodo.wi.mit.edu/primer3/) to generate amplicons (including each SNP) of <150 bp by setting each primer binding site closer to the SNP. Each primer was checked for potential structures of the self-dimer by using AutoDimer software (http://www.cstl.nist.gov/strbase/AutoDimerHomepage/AutoDimerProgramHomepage.htm). PCR was carried out by using a Veriti thermal cycler (Applied Biosystems, Foster City, CA, USA) for a final volume of 10 μl containing 1 to 10 ng genomic DNA, 1× Qiagen Multiplex PCR master mix (Qiagen, Hilden, Germany), and 0.2 to 0.6 μM each primer (Table 8) under following conditions: an initial denaturation step at 95°C for 10 min and 35 cycles of 94°C for 1 min, 60°C for 90 s, and 72°C for 50 s, followed by a final extension step at 72°C for 7 min.
TABLE 7.
Cytokine candidate genes and gene polymorphisms evaluated in a Colombian populationa
| Gene | Locus | SNP | rs ID | Location | Predicted functional effect(s) |
|---|---|---|---|---|---|
| IL-1α | 2q13 | −889G>A | rs1800587 | Promoter | TFBS, splicing (ESE or ESS) |
| IL-1β | 2q13 | +3954G>A | rs1143634 | Exon 4 | sSNP, splicing (ESE or ESS) |
| −31G>A | rs1143627 | Promoter | TFBS | ||
| −511G>A | rs16944 | Promoter | TFBS | ||
| IL-10 | 1q31 | −819G>A | rs1800871 | Promoter | TFBS |
| −1082A>G | rs1800896 | Promoter | TFBS | ||
| IFN-γ | 12q24.1 | +874T>A | rs2430561 | Intron 1 | NF-κB binding site |
| TNF-α | 6p21.3 | −308G>A | rs1800629 | Promoter | TFBS |
| −238C>T | rs361525 | Promoter | TFBS | ||
| −863C>A | rs1800630 | Promoter | TFBS | ||
| −857G>A | rs1799724 | Promoter | TFBS | ||
| −1031T>C | rs1799964 | Promoter | TFBS |
rs ID, reference SNP ID number; TFBS, transcription factor binding site; ESE, exonic splicing enhancer; ESS, exonic splicing silencer; sSNP synonymous SNP.
TABLE 8.
Primer sequences used for multiplex PCR amplification and extension probes used for multiplex primer extension
| Gene | dbSNP ID | Variation | Forward primer sequence (5′–3′) | Reverse primer sequence (5′–3′) | Extension probe sequence (5′–3′) |
|---|---|---|---|---|---|
| TNF-α −308 | rs1800629 | G/A | GCCCCTCCCAGTTCTAGTTC | AAAGTTGGGGACACACAAGC | AATAGGTTTTGAGGGGCATG |
| TNF-α −1031 | rs1799964 | T/C | CTGTGGGGAGAACAAAAGGA | CTCCTACCCATTGCTGTGGT | GCAAAGGAGAAGCTGAGAAGA |
| TNF-α −863 | rs1800630 | C/A | ACCACAGCAATGGGTAGGAG | CTCTGGGGTCCCTGATTTTT | CTCTCTCTCGAGTATGGGGACCCCC |
| TNF-α −857 | rs1799724 | G/A | ACCACAGCAATGGGTAGGAG | ACTCTGGGGTCCCTGATTTT | CTCTCTCTCTCTCTACATGGCCCTGTCTTC |
| TNF-α −238 | rs361525 | C/T | GCCCCTCCCAGTTCTAGTTC | GCACCTTCTGTCTCGGTTTC | CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTACTCCCCATCCTCCCTGCTG |
| IL-1β −511 | rs16944 | G/A | GCCCTCCCTGTCTGTATTGA | GTCTTGCAGGGTTGTGTGAG | CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTTTGGGTGCTGTTCTCTGCCTC |
| IL-1β +3954 | rs1143634 | G/A | TCCATATCCTGTCCCTGGAG | TGTTCTTAGCCACCCCACTC | CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTAAGCCTCGTTATCCCA |
| IL-1β −31 | rs1143627 | G/A | CTTGTGCCTCGAAGAGGTTT | GCTACTCCTTGCCCTTCCAT | CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCAGTTTCTCCCTCGCTGTTTTTAT |
| IL-1α −889 | rs1800587 | G/A | GGGTGACAAGCTCCTGGTTA | TAGGCTGGCCACAGGAATTA | CTCTCTCTCTCTCTCTCTCTCTCTCTCTGGATTTTTACATATGAGCCTTCAATG |
| IFN-γ +874 | rs2430561 | T/A | GCAAAGCCACCCCACTATAA | CTTCGTTGCTCACTGGGATT | CTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCTCGAT |
| IL-10 −1082 | rs1800896 | A/G | ATGGAGGCTGGATAGGAGGT | TTCCCCAGGTAGAGCAACAC | CTCTCTCTCTCTCTCTACTAAGGCTTCTTTGGGA |
| IL-10 −819 | rs1800871 | G/A | GTGCTCACCATGACCCCTAC | TCAACTTCTTCCACCCCATC | CTCTCTCTCTCTCTCTCTGCAAACTGAGGCACAGAGAT |
(ii) Multiplex SBE reaction and electrophoresis.
The PCR product was cleaned with 1 μl of ExoSAP-IT (Affymetrix, Santa Clara, CA, USA). The product was incubated at 37°C for 45 min and then heated at 85°C for 15 min to inactivate the enzyme. By using this preamplification product as the template, reactions were carried out to detect SNP variants using a minisequencing method (SBE [single-base extension]). SBE reactions were performed with a total volume of 6 μl containing 2 μl of the preamplification product, 2.5 μl of SNaPshot reaction mix (Applied Biosystems), and 1.5 μl of the SBE primer mixture at a concentration of 0.2 μM (Table 8), with amplification for 25 cycles at 96°C for 10 s, 50°C for 5 s, and 60°C for 30 s. Excess nucleotides were removed by the addition of 1 U of SAP (shrimp alkaline phosphatase; Amersham Pharmacia, Fairfield, CT, USA) according to the manufacturer's instructions. One microliter of the SBE reaction product was analyzed by capillary electrophoresis (CE) using an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems) with run module SNP36-POP-4. The data were analyzed by using GeneMapper v3.2 software (Applied Biosystems).
(iii) Ancestry-informative marker (AIM) genotyping.
Ancestry analysis was carried out on a subgroup of 43 cases and 10 controls in which DNA samples were available for the evaluation of 46 ancestry-informative insertion deletion markers (AIM-INDELs) distributed across the autosomal genome. PCRs were carried out in a single multiplex reaction mixture containing all 46 primer pairs according to a protocol described previously by Pereira et al. (84). The following PCR cycling conditions were used: an initial step for 15 min at 95°C followed by 27 cycles at 94°C for 30 s, 60°C for 1.5 min, and 72°C for 45 s and a final extension step at 72°C for 60 min. Dye-labeled PCR-amplified fragments were separated by capillary electrophoresis and detected by using an ABI Prism 3100-Avant genetic analyzer (Applied Biosystems). Automated allele calls were obtained with GeneMapper v.3.2 software (Applied Biosystems).
(iv) Gene-gene interaction analysis.
Gene-gene interactions among cytokine genes associated with ocular toxoplasmosis were detected by using the open-source MDR software package, version 3.0.1 (available at http://www.epistasis.org/software.html), in order to explore potential gene-gene interactions by combining the genotype data from this study (85, 86).
Statistical analysis.
Allelic, genotypic, and haplotypic frequencies were calculated and compared by using PLINK software (87). Association was also analyzed by using dominant, recessive, and additive genetic models. A subgroup analysis based on gender was also carried out. Deviations from the Hardy-Weinberg equilibrium (HWE) were tested for all polymorphisms in controls by comparing observed and expected genotype frequencies using the Haploview v4.1 program (www.broad.mit.edu/mpg/haploview). The linkage disequilibrium (LD) structure among SNPs and haplotype blocks was visualized by generating LD plots using Haploview. ORs and CIs were calculated. Bonferroni correction was used, and significance was set at a P value of <0.05. The genetic etiology of complex diseases is considered to involve interactions among multiple genetic variants and environmental conditions. Gene-gene interactions among cytokine genes associated with ocular toxoplasmosis were detected by using the open-source MDR software package, version 3.0.1 (available at http://www.epistasis.org/software.html) (88). Given the complexity of the statistical analysis, only combinations with P values of <0.005 (Fisher test) were taken into account.
ACKNOWLEDGMENTS
We gratefully acknowledge financial support from the Universidad Tecnológica de Pereira, Colombia (projects 5-14-1, and 5-11-3); Colciencias, Colombia (project 111056934589, contract 469-2013); the Universidad del Quindío, Colombia; and the Escuela Superior de Oftalmología-Instituto Barraquer de America, Colombia.
Finally, we thank the Toxoplasmosis Center at the University of Chicago and the Universidad Autónoma de Manizales for supporting the Ph.D. fellowship to C.A.N.-G. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
REFERENCES
- 1.Gómez-Marín JE. 2010. Protozoología médica: protozoos parásitos en el contexto latinoamericano. Editorial El Manual Moderno, Bogotá, Colombia. [Google Scholar]
- 2.Hill DE, Chirukandoth S, Dubey JP. 2005. Biology and epidemiology of Toxoplasma gondii in man and animals. Anim Health Res Rev 6:41–61. doi: 10.1079/AHR2005100. [DOI] [PubMed] [Google Scholar]
- 3.Pfaff AW, de-la-Torre A, Rochet E, Brunet J, Sabou M, Sauer A, Bourcier T, Gomez-Marin JE, Candolfi E. 2014. New clinical and experimental insights into Old World and neotropical ocular toxoplasmosis. Int J Parasitol 44:99–107. doi: 10.1016/j.ijpara.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Canon-Franco WA, Lopez-Orozco N, Gomez-Marin JE, Dubey JP. 2014. An overview of seventy years of research (1944–2014) on toxoplasmosis in Colombia, South America. Parasit Vectors 7:427. doi: 10.1186/1756-3305-7-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de-la-Torre A, Lopez-Castillo CA, Gomez-Marin JE. 2009. Incidence and clinical characteristics in a Colombian cohort of ocular toxoplasmosis. Eye (Lond) 23:1090–1093. doi: 10.1038/eye.2008.219. [DOI] [PubMed] [Google Scholar]
- 6.de-la-Torre A, Gonzalez G, Diaz-Ramirez J, Gomez-Marin JE. 2007. Screening by ophthalmoscopy for Toxoplasma retinochoroiditis in Colombia. Am J Ophthalmol 143:354–356. doi: 10.1016/j.ajo.2006.09.048. [DOI] [PubMed] [Google Scholar]
- 7.Maenz M, Schluter D, Liesenfeld O, Schares G, Gross U, Pleyer U. 2014. Ocular toxoplasmosis past, present and new aspects of an old disease. Prog Retin Eye Res 39:77–106. doi: 10.1016/j.preteyeres.2013.12.005. [DOI] [PubMed] [Google Scholar]
- 8.de-la-Torre A, Sauer A, Pfaff AW, Bourcier T, Brunet J, Speeg-Schatz C, Ballonzoli L, Villard O, Ajzenberg D, Sundar N, Grigg ME, Gomez-Marin JE, Candolfi E. 2013. Severe South American ocular toxoplasmosis is associated with decreased Ifn-γ/Il-17a and increased Il-6/Il-13 intraocular levels. PLoS Negl Trop Dis 7:e2541. doi: 10.1371/journal.pntd.0002541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torres-Morales E, Taborda L, Cardona N, De-la-Torre A, Sepulveda-Arias JC, Patarroyo MA, Gomez-Marin JE. 2014. Th1 and Th2 immune response to P30 and ROP18 peptides in human toxoplasmosis. Med Microbiol Immunol 203:315–322. doi: 10.1007/s00430-014-0339-0. [DOI] [PubMed] [Google Scholar]
- 10.Boothroyd JC, Grigg ME. 2002. Population biology of Toxoplasma gondii and its relevance to human infection: do different strains cause different disease? Curr Opin Microbiol 5:438–442. doi: 10.1016/S1369-5274(02)00349-1. [DOI] [PubMed] [Google Scholar]
- 11.Vasconcelos-Santos DV. 2012. Ocular manifestations of systemic disease: toxoplasmosis. Curr Opin Ophthalmol 23:543–550. doi: 10.1097/ICU.0b013e328358bae5. [DOI] [PubMed] [Google Scholar]
- 12.Furtado JM, Winthrop KL, Butler NJ, Smith JR. 2013. Ocular toxoplasmosis I: parasitology, epidemiology and public health. Clin Exp Ophthalmol 41:82–94. doi: 10.1111/j.1442-9071.2012.02821.x. [DOI] [PubMed] [Google Scholar]
- 13.Carneiro ACAV, Andrade GM, Costa JGL, Pinheiro BV, Vasconcelos-Santos DV, Ferreira AM, Su C, Januário JN, Vitor RWA. 2013. Genetic characterization of Toxoplasma gondii revealed highly diverse genotypes for isolates from newborns with congenital toxoplasmosis in southeastern Brazil. J Clin Microbiol 51:901–907. doi: 10.1128/JCM.02502-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilbert RE, Freeman K, Lago EG, Bahia-Oliveira LM, Tan HK, Wallon M, Buffolano W, Stanford MR, Petersen E, European Multicentre Study on Congenital Toxoplasmosis. 2008. Ocular sequelae of congenital toxoplasmosis in Brazil compared with Europe. PLoS Negl Trop Dis 2:e277. doi: 10.1371/journal.pntd.0000277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pleyer U, Schluter D, Manz M. 2014. Ocular toxoplasmosis: recent aspects of pathophysiology and clinical implications. Ophthalmic Res 52:116–123. doi: 10.1159/000363141. [DOI] [PubMed] [Google Scholar]
- 16.Roberts F, McLeod R. 1999. Pathogenesis of toxoplasmic retinochoroiditis. Parasitol Today 15:51–57. doi: 10.1016/S0169-4758(98)01377-5. [DOI] [PubMed] [Google Scholar]
- 17.Khan A, Jordan C, Muccioli C, Vallochi AL, Rizzo LV, Belfort R Jr, Vitor RW, Silveira C, Sibley LD. 2006. Genetic divergence of Toxoplasma gondii strains associated with ocular toxoplasmosis, Brazil. Emerg Infect Dis 12:942–949. doi: 10.3201/eid1206.060025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Garweg JG, Candolfi E. 2009. Immunopathology in ocular toxoplasmosis: facts and clues. Mem Inst Oswaldo Cruz 104:211–220. doi: 10.1590/S0074-02762009000200014. [DOI] [PubMed] [Google Scholar]
- 19.Dutra MS, Bela SR, Peixoto-Rangel AL, Fakiola M, Cruz AG, Gazzinelli A, Quites HF, Bahia-Oliveira LM, Peixe RG, Campos WR, Higino-Rocha AC, Miller NE, Blackwell JM, Antonelli LR, Gazzinelli RT. 2013. Association of a NOD2 gene polymorphism and T-helper 17 cells with presumed ocular toxoplasmosis. J Infect Dis 207:152–163. doi: 10.1093/infdis/jis640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauer A, Pfaff AW, Villard O, Creuzot-Garcher C, Dalle F, Chiquet C, Pelloux H, Speeg-Schatz C, Gaucher D, Prevost G, Bourcier T, Candolfi E. 2012. Interleukin 17A as an effective target for anti-inflammatory and antiparasitic treatment of toxoplasmic uveitis. J Infect Dis 206:1319–1329. doi: 10.1093/infdis/jis486. [DOI] [PubMed] [Google Scholar]
- 21.Cordeiro CA, Moreira PR, Costa GC, Dutra WO, Campos WR, Orefice F, Teixeira AL. 2008. TNF-alpha gene polymorphism (−308G/A) and toxoplasmic retinochoroiditis. Br J Ophthalmol 92:986–988. doi: 10.1136/bjo.2008.140590. [DOI] [PubMed] [Google Scholar]
- 22.Vasconcelos RH, Montenegro SM, Azevedo EA, Gomes YM, Morais CN. 2012. Genetic susceptibility to chronic Chagas disease: an overview of single nucleotide polymorphisms of cytokine genes. Cytokine 59:203–208. doi: 10.1016/j.cyto.2012.04.035. [DOI] [PubMed] [Google Scholar]
- 23.de Albuquerque MC, Aleixo ALQDC, Benchimol EI, Leandro ACCS, das Neves LB, Vicente RT, Bonecini-Almeida MDG, Amendoeira MRR. 2009. The IFN-γ+874T/A gene polymorphism is associated with retinochoroiditis toxoplasmosis susceptibility. Mem Inst Oswaldo Cruz 104:451–455. doi: 10.1590/S0074-02762009000300009. [DOI] [PubMed] [Google Scholar]
- 24.Cordeiro CA, Moreira PR, Costa GC, Dutra WO, Campos WR, Orefice F, Teixeira AL. 2008. Interleukin-1 gene polymorphisms and toxoplasmic retinochoroiditis. Mol Vis 14:1845–1849. [PMC free article] [PubMed] [Google Scholar]
- 25.Cordeiro CA, Moreira PR, Andrade MS, Dutra WO, Campos WR, Orefice F, Teixeira AL. 2008. Interleukin-10 gene polymorphism (−1082G/A) is associated with toxoplasmic retinochoroiditis. Invest Ophthalmol Vis Sci 49:1979–1982. doi: 10.1167/iovs.07-1393. [DOI] [PubMed] [Google Scholar]
- 26.Masek KS, Hunter CA. 2013. Pro-inflammatory responses in macrophages during Toxoplasma gondii infection. Madame Curie Bioscience Database. [Google Scholar]
- 27.Suzuki Y, Sa Q, Gehman M, Ochiai E. 2011. Interferon-gamma- and perforin-mediated immune responses for resistance against Toxoplasma gondii in the brain. Expert Rev Mol Med 13:e31. doi: 10.1017/S1462399411002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cordeiro CA, Moreira PR, Bessa TF, Costa GC, Dutra WO, Campos WR, Oréfice F, Young LH, Teixeira AL. 2013. Interleukin-6 gene polymorphism (−174 G/C) is associated with toxoplasmic retinochoroiditis. Acta Ophthalmol (Copenh) 91:e311–e314. doi: 10.1111/aos.12046. [DOI] [PubMed] [Google Scholar]
- 29.Wujcicka W, Gaj Z, Wilczynski J, Nowakowska D. 2015. Contribution of IL6-174 G>C and IL1B +3954 C>T polymorphisms to congenital infection with Toxoplasma gondii. Eur J Clin Microbiol Infect Dis 34:2287–2294. doi: 10.1007/s10096-015-2481-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fehrmann RS, Jansen RC, Veldink JH, Westra HJ, Arends D, Bonder MJ, Fu J, Deelen P, Groen HJ, Smolonska A, Weersma RK, Hofstra RM, Buurman WA, Rensen S, Wolfs MG, Platteel M, Zhernakova A, Elbers CC, Festen EM, Trynka G, Hofker MH, Saris CG, Ophoff RA, van den Berg LH, van Heel DA, Wijmenga C, Te Meerman GJ, Franke L. 2011. Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 7:e1002197. doi: 10.1371/journal.pgen.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. 2009. Mapping complex disease traits with global gene expression. Nat Rev Genet 10:184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jais PH. 2005. How frequent is altered gene expression among susceptibility genes to human complex disorders? Genet Med 7:83–96. doi: 10.1097/01.GIM.0000153665.55420.C3. [DOI] [PubMed] [Google Scholar]
- 33.Basu M, Das T, Ghosh A, Majumder S, Maji AK, Kanjilal SD, Mukhopadhyay I, Roychowdhury S, Banerjee S, Sengupta S. 2012. Gene-gene interaction and functional impact of polymorphisms on innate immune genes in controlling Plasmodium falciparum blood infection level. PLoS One 7:e46441. doi: 10.1371/journal.pone.0046441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ossa H, Aquino J, Pereira R, Ibarra A, Ossa RH, Perez LA, Granda JD, Lattig MC, Groot H, Fagundes de Carvalho E, Gusmao L. 2016. Outlining the ancestry landscape of Colombian admixed populations. PLoS One 11:e0164414. doi: 10.1371/journal.pone.0164414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, Imboywa S, Von Korff A, Okada Y, Patsopoulos NA, Davis S, McCabe C, Paik HI, Srivastava GP, Raychaudhuri S, Hafler DA, Koller D, Regev A, Hacohen N, Mathis D, Benoist C, Stranger BE, De Jager PL. 2014. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344:519–523. doi: 10.1126/science.1249547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gamazon ER, Zhang W, Konkashbaev A, Duan S, Kistner EO, Nicolae DL, Dolan ME, Cox NJ. 2010. SCAN: SNP and copy number annotation. Bioinformatics 26:259–262. doi: 10.1093/bioinformatics/btp644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki Y, Conley FK, Remington JS. 1989. Importance of endogenous IFN-gamma for prevention of toxoplasmic encephalitis in mice. J Immunol 143:2045–2050. [PubMed] [Google Scholar]
- 38.Roberts F, Mets MB, Ferguson DJ, O'Grady R, O'Grady C, Thulliez P, Brezin AP, McLeod R. 2001. Histopathological features of ocular toxoplasmosis in the fetus and infant. Arch Ophthalmol 119:51–58. [PubMed] [Google Scholar]
- 39.Machado AS, Carneiro AC, Bela SR, Andrade GM, Vasconcelos-Santos DV, Januario JN, Coelho-dos-Reis JG, Ferro EA, Teixeira-Carvalho A, Vitor RW, Martins-Filho OA, Ufmg Congenital Toxoplasmosis Brazilian Group. 2014. Biomarker analysis revealed distinct profiles of innate and adaptive immunity in infants with ocular lesions of congenital toxoplasmosis. Mediators Inflamm 2014:910621. doi: 10.1155/2014/910621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Araujo TE, Coelho-Dos-Reis JG, Bela SR, Carneiro A, Machado AS, Cardoso LM, Ribeiro AL, Dias MHF, Queiroz Andrade GM, Vasconcelos-Santos DV, Januario JN, Teixeira-Carvalho A, Vitor RWA, Ferro EAV, Martins-Filho OA, UFMG Congenital Toxoplasmosis Brazilian Group. 2017. Early serum biomarker networks in infants with distinct retinochoroidal lesion status of congenital toxoplasmosis. Cytokine 95:102–112. doi: 10.1016/j.cyto.2017.02.018. [DOI] [PubMed] [Google Scholar]
- 41.Vallochi AL, Nakamura MV, Schlesinger D, Martins MC, Silveira C, Belfort R Jr, Rizzo LV. 2002. Ocular toxoplasmosis: more than just what meets the eye. Scand J Immunol 55:324–328. doi: 10.1046/j.1365-3083.2002.01052.x. [DOI] [PubMed] [Google Scholar]
- 42.Fatoohi F, Cozon GJ, Wallon M, Kodjikian L, Peyron F. 2006. Systemic T cell response to Toxoplasma gondii antigen in patients with ocular toxoplasmosis. Jpn J Ophthalmol 50:103–110. doi: 10.1007/s10384-005-0295-8. [DOI] [PubMed] [Google Scholar]
- 43.Yamamoto JH, Vallochi AL, Silveira C, Filho JK, Nussenblatt RB, Cunha-Neto E, Gazzinelli RT, Belfort R Jr, Rizzo LV. 2000. Discrimination between patients with acquired toxoplasmosis and congenital toxoplasmosis on the basis of the immune response to parasite antigens. J Infect Dis 181:2018–2022. doi: 10.1086/315494. [DOI] [PubMed] [Google Scholar]
- 44.Barrett S, Collins M, Kenny C, Ryan E, Keane CO, Crowe J. 2003. Polymorphisms in tumour necrosis factor-alpha, transforming growth factor-beta, interleukin-10, interleukin-6, interferon-gamma, and outcome of hepatitis C virus infection. J Med Virol 71:212–218. doi: 10.1002/jmv.10472. [DOI] [PubMed] [Google Scholar]
- 45.MacLean L, Chisi JE, Odiit M, Gibson WC, Ferris V, Picozzi K, Sternberg JM. 2004. Severity of human African trypanosomiasis in East Africa is associated with geographic location, parasite genotype, and host inflammatory cytokine response profile. Infect Immun 72:7040–7044. doi: 10.1128/IAI.72.12.7040-7044.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nicolae DL, Gamazon E, Zhang W, Duan S, Dolan ME, Cox NJ. 2010. Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet 6:e1000888. doi: 10.1371/journal.pgen.1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Couper KN, Blount DG, Riley EM. 2008. IL-10: the master regulator of immunity to infection. J Immunol 180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 48.Fulton SA, Cross JV, Toossi ZT, Boom WH. 1998. Regulation of interleukin-12 by interleukin-10, transforming growth factor-beta, tumor necrosis factor-alpha, and interferon-gamma in human monocytes infected with Mycobacterium tuberculosis H37Ra. J Infect Dis 178:1105–1114. doi: 10.1086/515698. [DOI] [PubMed] [Google Scholar]
- 49.Reuss E, Fimmers R, Kruger A, Becker C, Rittner C, Hohler T. 2002. Differential regulation of interleukin-10 production by genetic and environmental factors—a twin study. Genes Immun 3:407–413. doi: 10.1038/sj.gene.6363920. [DOI] [PubMed] [Google Scholar]
- 50.Turner DM, Williams DM, Sankaran D, Lazarus M, Sinnott PJ, Hutchinson IV. 1997. An investigation of polymorphism in the interleukin-10 gene promoter. Eur J Immunogenet 24:1–8. doi: 10.1111/j.1365-2370.1997.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 51.Galley HF, Lowe PR, Carmichael RL, Webster NR. 2003. Genotype and interleukin-10 responses after cardiopulmonary bypass. Br J Anaesth 91:424–426. doi: 10.1093/bja/aeg174. [DOI] [PubMed] [Google Scholar]
- 52.Suarez A, Castro P, Alonso R, Mozo L, Gutierrez C. 2003. Interindividual variations in constitutive interleukin-10 messenger RNA and protein levels and their association with genetic polymorphisms. Transplantation 75:711–717. doi: 10.1097/01.TP.0000055216.19866.9A. [DOI] [PubMed] [Google Scholar]
- 53.Liang L, Zhao YL, Yue J, Liu JF, Han M, Wang H, Xiao H. 2011. Interleukin-10 gene promoter polymorphisms and their protein production in pleural fluid in patients with tuberculosis. FEMS Immunol Med Microbiol 62:84–90. doi: 10.1111/j.1574-695X.2011.00791.x. [DOI] [PubMed] [Google Scholar]
- 54.Meenakshi P, Ramya S, Shruthi T, Lavanya J, Mohammed HH, Mohammed SA, Vijayalakshmi V, Sumanlatha G. 2013. Association of IL-1beta +3954 C/T and IL-10-1082 G/A cytokine gene polymorphisms with susceptibility to tuberculosis. Scand J Immunol 78:92–97. doi: 10.1111/sji.12055. [DOI] [PubMed] [Google Scholar]
- 55.Calvo J, Martínez N, Etxagibel A, Calleja S, Sáez-Torres C, Sedeño M, Julià R, Muncunill J, Matamoros N, Gayà A. 2002. Allelic frequencies of polymorphic variants of cytokine genes (IL1A, IL1B, IL1RN, IL6, IL10, IL12p40, and IFNG) in a Spanish population. Inmunologia 21:76–86. [Google Scholar]
- 56.Pravica V, Perrey C, Stevens A, Lee JH, Hutchinson IV. 2000. A single nucleotide polymorphism in the first intron of the human IFN-gamma gene: absolute correlation with a polymorphic CA microsatellite marker of high IFN-gamma production. Hum Immunol 61:863–866. doi: 10.1016/S0198-8859(00)00167-1. [DOI] [PubMed] [Google Scholar]
- 57.Rossouw M, Nel HJ, Cooke GS, van Helden PD, Hoal EG. 2003. Association between tuberculosis and a polymorphic NFkappaB binding site in the interferon gamma gene. Lancet 361:1871–1872. doi: 10.1016/S0140-6736(03)13491-5. [DOI] [PubMed] [Google Scholar]
- 58.Cavalcanti YV, Brelaz MC, Neves JK, Ferraz JC, Pereira VR. 2012. Role of TNF-alpha, IFN-gamma, and IL-10 in the development of pulmonary tuberculosis. Pulm Med 2012:745483. doi: 10.1155/2012/745483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tso HW, Ip WK, Chong WP, Tam CM, Chiang AK, Lau YL. 2005. Association of interferon gamma and interleukin 10 genes with tuberculosis in Hong Kong Chinese. Genes Immun 6:358–363. doi: 10.1038/sj.gene.6364189. [DOI] [PubMed] [Google Scholar]
- 60.Dinarello CA. 2002. The IL-1 family and inflammatory diseases. Clin Exp Rheumatol 20:S1–S13. [PubMed] [Google Scholar]
- 61.di Giovine FS, Takhsh E, Blakemore AI, Duff GW. 1992. Single base polymorphism at −511 in the human interleukin-1 beta gene (IL1 beta). Hum Mol Genet 1:450. doi: 10.1093/hmg/1.6.450. [DOI] [PubMed] [Google Scholar]
- 62.Guasch JF, Bertina RM, Reitsma PH. 1996. Five novel intragenic dimorphisms in the human interleukin-1 genes combine to high informativity. Cytokine 8:598–602. doi: 10.1006/cyto.1996.0080. [DOI] [PubMed] [Google Scholar]
- 63.Nicklin MJ, Weith A, Duff GW. 1994. A physical map of the region encompassing the human interleukin-1 alpha, interleukin-1 beta, and interleukin-1 receptor antagonist genes. Genomics 19:382–384. doi: 10.1006/geno.1994.1076. [DOI] [PubMed] [Google Scholar]
- 64.Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. 1992. A TaqI polymorphism in the human interleukin-1 beta (IL-1 beta) gene correlates with IL-1 beta secretion in vitro. Eur J Clin Invest 22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 65.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, Wang HH, Yao A, Chen YT, Hsu CN. 2006. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res 34:W635–W641. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gervaziev YV, Olenina LV, Krasotkina JV, Lupatov AY, Mazurina SA, Gervazieva VB. 2010. Oct-1 is responsible for the C-33T polymorphism effect in the IL-4 promoter. Int J Immunogenet 37:13–20. doi: 10.1111/j.1744-313X.2009.00883.x. [DOI] [PubMed] [Google Scholar]
- 67.Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, Kwiatkowski D. 1999. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet 22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 68.Kang X, Kim HJ, Ramirez M, Salameh S, Ma X. 2010. The septic shock-associated IL-10-1082 A > G polymorphism mediates allele-specific transcription via poly(ADP-ribose) polymerase 1 in macrophages engulfing apoptotic cells. J Immunol 184:3718–3724. doi: 10.4049/jimmunol.0903613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amar D, Safer H, Shamir R. 2013. Dissection of regulatory networks that are altered in disease via differential co-expression. PLoS Comput Biol 9:e1002955. doi: 10.1371/journal.pcbi.1002955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jamieson SE, Peixoto-Rangel AL, Hargrave AC, Roubaix LA, Mui EJ, Boulter NR, Miller EN, Fuller SJ, Wiley JS, Castellucci L, Boyer K, Peixe RG, Kirisits MJ, Elias LDS, Coyne JJ, Correa-Oliveira R, Sautter M, Smith NC, Lees MP, Swisher CN, Heydemann P, Noble AG, Patel D, Bardo D, Burrowes D, McLone D, Roizen N, Withers S, Bahia-Oliveira LM, McLeod R, Blackwell JM. 2010. Evidence for associations between the purinergic receptor P2X(7) (P2RX7) and toxoplasmosis. Genes Immun 11:374–383. doi: 10.1038/gene.2010.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lees MP, Fuller SJ, McLeod R, Boulter NR, Miller CM, Zakrzewski AM, Mui EJ, Witola WH, Coyne JJ, Hargrave AC, Jamieson SE, Blackwell JM, Wiley JS, Smith NC. 2010. P2X7 receptor-mediated killing of an intracellular parasite, Toxoplasma gondii, by human and murine macrophages. J Immunol 184:7040–7046. doi: 10.4049/jimmunol.1000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ewald SE, Chavarria-Smith J, Boothroyd JC. 2014. NLRP1 is an inflammasome sensor for Toxoplasma gondii. Infect Immun 82:460–468. doi: 10.1128/IAI.01170-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gorfu G, Cirelli KM, Melo MB, Mayer-Barber K, Crown D, Koller BH, Masters S, Sher A, Leppla SH, Moayeri M, Saeij JP, Grigg ME. 2014. Dual role for inflammasome sensors NLRP1 and NLRP3 in murine resistance to Toxoplasma gondii. mBio 5:e01117-13. doi: 10.1128/mBio.01117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Witola WH, Mui E, Hargrave A, Liu S, Hypolite M, Montpetit A, Cavailles P, Bisanz C, Cesbron-Delauw MF, Fournie GJ, McLeod R. 2011. NALP1 influences susceptibility to human congenital toxoplasmosis, proinflammatory cytokine response, and fate of Toxoplasma gondii-infected monocytic cells. Infect Immun 79:756–766. doi: 10.1128/IAI.00898-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Witola WH, Liu SR, Montpetit A, Welti R, Hypolite M, Roth M, Zhou Y, Mui E, Cesbron-Delauw MF, Fournie GJ, Cavailles P, Bisanz C, Boyer K, Withers S, Noble AG, Swisher CN, Heydemann PT, Rabiah P, Muench SP, McLeod R. 2014. ALOX12 in human toxoplasmosis. Infect Immun 82:2670–2679. doi: 10.1128/IAI.01505-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Brown CR, McLeod R. 1990. Class I MHC genes and CD8+ T cells determine cyst number in Toxoplasma gondii infection. J Immunol 145:3438–3441. [PubMed] [Google Scholar]
- 77.Cong H, Mui EJ, Witola WH, Sidney J, Alexander J, Sette A, Maewal A, McLeod R. 2010. Human immunome, bioinformatic analyses using HLA supermotifs and the parasite genome, binding assays, studies of human T cell responses, and immunization of HLA-A*1101 transgenic mice including novel adjuvants provide a foundation for HLA-A03 restricted CD8+T cell epitope based, adjuvanted vaccine protective against Toxoplasma gondii. Immunome Res 6:12. doi: 10.1186/1745-7580-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cong H, Mui EJ, Witola WH, Sidney J, Alexander J, Sette A, Maewal A, McLeod R. 2011. Towards an immunosense vaccine to prevent toxoplasmosis: protective Toxoplasma gondii epitopes restricted by HLA-A*0201. Vaccine 29:754–762. doi: 10.1016/j.vaccine.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.El Bissati K, Zhou Y, Dasgupta D, Cobb D, Dubey JP, Burkhard P, Lanar DE, McLeod R. 2014. Effectiveness of a novel immunogenic nanoparticle platform for Toxoplasma peptide vaccine in HLA transgenic mice. Vaccine 32:3243–3248. doi: 10.1016/j.vaccine.2014.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tan TG, Mui E, Cong H, Witola WH, Montpetit A, Muench SP, Sidney J, Alexander J, Sette A, Grigg ME, Maewal A, McLeod R. 2010. Identification of T. gondii epitopes, adjuvants, and host genetic factors that influence protection of mice and humans. Vaccine 28:3977–3989. doi: 10.1016/j.vaccine.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Frazer KA, Murray SS, Schork NJ, Topol EJ. 2009. Human genetic variation and its contribution to complex traits. Nat Rev Genet 10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 82.de-la-Torre A, Pfaff AW, Grigg ME, Villard O, Candolfi E, Gomez-Marin JE. 2014. Ocular cytokinome is linked to clinical characteristics in ocular toxoplasmosis. Cytokine 68:23–31. doi: 10.1016/j.cyto.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Villard O, Filisetti D, Roch-Deries F, Garweg J, Flament J, Candolfi E. 2003. Comparison of enzyme-linked immunosorbent assay, immunoblotting, and PCR for diagnosis of toxoplasmic chorioretinitis. J Clin Microbiol 41:3537–3541. doi: 10.1128/JCM.41.8.3537-3541.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Pereira R, Phillips C, Pinto N, Santos C, dos Santos SE, Amorim A, Carracedo A, Gusmao L. 2012. Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS One 7:e29684. doi: 10.1371/journal.pone.0029684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Moore JH. 2004. Computational analysis of gene-gene interactions using multifactor dimensionality reduction. Expert Rev Mol Diagn 4:795–803. doi: 10.1586/14737159.4.6.795. [DOI] [PubMed] [Google Scholar]
- 86.Moore JH, Williams SM. 2009. Epistasis and its implications for personal genetics. Am J Hum Genet 85:309–320. doi: 10.1016/j.ajhg.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Moore JH, Gilbert JC, Tsai CT, Chiang FT, Holden T, Barney N, White BC. 2006. A flexible computational framework for detecting, characterizing, and interpreting statistical patterns of epistasis in genetic studies of human disease susceptibility. J Theor Biol 241:252–261. doi: 10.1016/j.jtbi.2005.11.036. [DOI] [PubMed] [Google Scholar]