ABSTRACT
Accurate diagnosis and early treatment of tuberculosis (TB) and latent TB infection (LTBI) are vital to prevent and control TB. The lack of specific biomarkers hinders these efforts. This study's purpose was to screen immunological markers that discriminate Mycobacterium tuberculosis infection outcomes in a setting where it is endemic, Ethiopia. Whole blood from 90 participants was stimulated using the ESAT-6/CFP-10 antigen cocktail. The interferon gamma (IFN-γ)-based QuantiFERON diagnostic test was used to distinguish between LTBI and uninfected control cases. Forty cytokines/chemokines were detected from antigen-stimulated plasma supernatants (SPSs) and unstimulated plasma samples (UPSs) using human cytokine/chemokine antibody microarrays. Statistical tests allowed us to identify potential biomarkers that distinguish the TB, LTBI, and healthy control groups. As expected, the levels of IFN-γ in SPSs returned a high area under the receiver operating characteristic curve (AUC) value comparing healthy controls and LTBI cases (Z = 0.911; P < 0.001). The SPS data also indicated that interleukin 17 (IL-17) abundance discriminates LTBI from healthy controls (Z = 0.763; P = 0.001). RANTES and MIP-1β were significantly elevated in SPSs of TB-infected compared to healthy controls (P < 0.05), while IL-12p40 and soluble tumor necrosis factor receptor II (sTNF-RII) were significantly increased in active TB cases compared to the combined LTBI and control groups (P < 0.05). Interestingly, quantitative changes for RANTES were observed using both SPSs and UPSs, with P values of 0.013 and 0.012, respectively, in active TB versus LTBI cases and 0.001 and 0.002, respectively, in active TB versus healthy controls. These results encourage biomarker verification studies for IL-17 and RANTES. Combinations of these cytokines may complement IFN-γ measurements to diagnose LTBI and distinguish active TB from LTBI cases.
KEYWORDS: immunological marker, cytokine array, tuberculosis, RANTES, IL-17, Mycobacterium, Ethiopia
INTRODUCTION
Tuberculosis (TB) is an infectious disease transmitted through inhalation of droplet nuclei into the lung alveoli. Disease outcome is dependent on the ability of the host to elicit a potent cell-mediated immune response which develops 3 to 8 weeks after the initial infection and results in macrophage activation (1). In more than 90% of all cases, cell-mediated immune responses effectively control disease progression and result in latent TB infection (LTBI) (2). Less than 10% of these, equivalent to 8 to 12 million new cases, advance to active TB and cause 3 million deaths globally each year (3). TB remains a formidable threat to human health and necessitates steps toward accelerated development of new vaccines, drugs, and diagnostics. As shown for interferon gamma (IFN-γ), assays to detect LTBI with relatively high specificity and sensitivity can be developed. Novel biomarkers targeting host immune responses against Mycobacterium tuberculosis may result in improved clinical tests (4).
Rapid M. tuberculosis detection and the identification of TB symptoms are critical to effective control of this disease. However, many people with active TB do not experience symptoms in early stages of the disease and may not seek care or are not correctly diagnosed when seeking care, thus serving as reservoirs for M. tuberculosis transmission. Early and specific diagnosis of active TB is important to slow the interhuman transmission. The standard diagnostic test, acid-fast staining followed by microscopic detection of the bacterium, has poor sensitivity and is unsuitable to monitor therapeutic effects, because it is restricted to viable M. tuberculosis cells in the sample (5, 6). The time frame of sputum cultures to observe the pathogen's growth in vitro, more than 6 weeks, is too long. The recently developed GeneXpert MTB/RIF test has a few disadvantages, including low sensitivity in smear-negative samples (68%), a short shelf life, high cost, requirement for a stable electric supply, and a critical temperature ceiling (7). Human immune response-associated tests are an alternative to fill the current void of rapid TB diagnostics for use at the point of care.
Treatment of LTBI can prevent the development and reemergence of active TB. The WHO has described the diagnosis and treatment of LTBI as a cornerstone of successful reduction of the TB epidemic (8). A commonly used test to screen for LTBI is the IFN-γ release assay (IGRA). The assay quantifies T cells that recognize and prevent the bacterial spread in peripheral blood. It is an enzyme-linked immunosorbent assay (ELISA) that determines if an individual has been infected with M. tuberculosis. A positive test with elevated IFN-γ levels suggests a persistent immune response caused by active TB or LTBI. While specific and sensitive, this LTBI test does not discern latency from active TB. This is a problem in high-TB-burden settings (9). Recent data suggest that LTBI is a more complex phenomenon. Household and community contacts with negative results in the IFN-γ test may progress to active TB in a time frame of a few months to a few years (10). The tests may also be indeterminate or negative for children and immunocompromised patients infected with M. tuberculosis (11, 12). Furthermore, imaging data pointed to individual granulomas that are not synchronized in their metabolic activities and highlight the difficulties in discerning LTBI from an active TB state within a single patient (2, 13). Quantitative surveys of other cytokines or chemokines may bring more diagnostic specificity to cases of indeterminate IFN-γ test results.
Quantitative blood plasma-based cytokine and chemokine studies for improved TB diagnostics have yielded inconsistent results to date. They include cytokine (micro)arrays using unstimulated plasma samples comparing patients with active TB, those with LTBI, and healthy controls (14, 15) and samples derived from plasma stimulated with TB-specific antigens (16–21). Rivera-Ordaz et al. used peripheral blood mononuclear cells from patients with active TB and LTBI to stimulate secretion of cytokines with TB antigens and measured their abundances before and after stimulation (22). Luminex or Milliplex technology-based multiplexed bead arrays were used in some of these studies (14–16). Differences in the assay platforms used may contribute to the overall inconclusive results on the association of cytokine abundance changes with TB outcomes. Most studies did not include chemokines regulating T-cell activation, such as RANTES (the full name is “Regulated on Activation, Normal T-Cell Expressed and Secreted”). This signaling molecule is involved in the chemoattraction of memory T cells. Except for IFN-γ, biomarker verification studies able to distinguish active-TB and LTBI outcomes, requiring hundreds of clinical samples, have not been conducted to date. We sought to assess whether Human Inflammation Antibody Array 3 (HIAA), which measures 40 inflammatory cytokines and chemokines (classified by function in Table 1) and is available from Ray Biotech, Inc., Norcross, GA, provides new insights into biomarker candidates comparing blood plasma levels from TB patients prior to directly observed treatment, short course (DOTS), or another antibiotic treatment with those from LTBI individuals and healthy controls. This would set the stage for biomarker verification studies and, potentially, development of clinically useful tests for TB diagnosis.
TABLE 1.
Classification of 40 cytokines/chemokines based on their function
| Type | Cytokines/chemokines |
|---|---|
| Proinflammatory cytokines | IFN-γ, IL-1α, IL-1β, IL-6, IL-11, IL-12p40, IL-12p70, IL-16, IL-17, ICAM-1, TNF-α, TNF-β |
| Anti-inflammatory cytokines | IL-4, IL-10, IL-13, TGF-β |
| Chemokines | IL-8, eotaxin/CCL11, eotaxin-2/CCL24, CSF1, CSF2, CSF3, I-309/CCL1, IP-10, MCP1, MCP2, MIG, MIP-1α, MIP-1β, MIP-1δ, RANTES/CCL5 |
| Soluble receptors | sIL-6R, sTNF-RI, sTNF-RII |
| Growth factors | IL-3, Il-7, IL-2, IL-15, PGDF-BB, TIMP-2 |
RESULTS
Sociodemographic characteristics of study participants.
Of the 90 study participants, 16 (53.3%), 16 (53.3%), and 14 (46.7%) were males for active TB cases, LTBI subjects, and healthy community controls, respectively. There was no significant gender difference in the three groups (χ2 = 0.220; P = 0.896). The mean age of individuals with active TB was 34.52 years, with a standard deviation (SD) of 17.48; the mean age of individuals with LTBI was 37.15 years, with an SD of 11.69; and the mean age of healthy control subjects was 35.71 years, with an SD of 12.52. There was no significant age difference in the three groups (P = 0.685). One healthy subject was excluded from the analysis due to outlier values in normality testing. Four smear- and culture-positive cases were cases of infection with nontuberculous mycobacteria (NTM) based on genotyping data; the involved subjects likewise were excluded from the analysis.
Diagnostic ability of 40 plasma supernatant cytokines/chemokines to discriminate between subjects with LTBI and healthy controls.
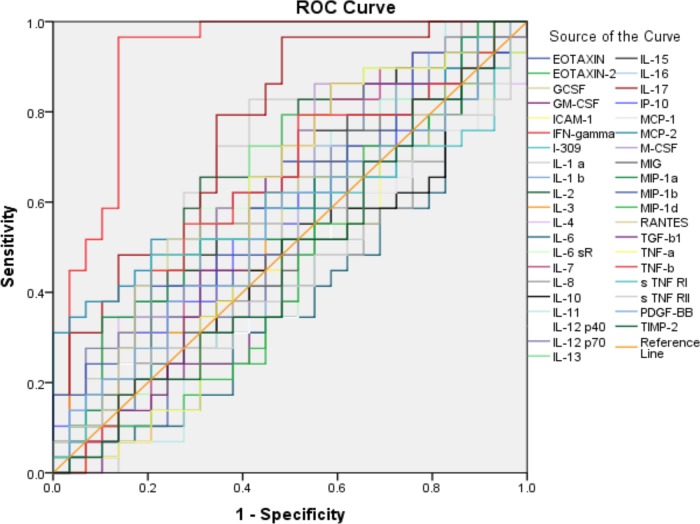
In this study, we used antigen-stimulated blood plasma supernatants (SPSs) and unstimulated plasma samples (UPSs). Given that the definition of the LTBI group was based on IGRA results, we expected IFN-γ abundance levels derived from Human Inflammation Antibody Array 3 (HIAA) assays to return high P and area under the receiver operating characteristic (ROC) curve (AUC) values in the comparison of the healthy control and LTBI groups. Indeed, this was observed (Z = 0.91; P < 0.001). This finding is consistent with good accuracy for quantification of IFN-γ using the array. We generated ROC curves for all 40 cytokines and chemokines measured in SPSs to determine their discriminatory power comparing the LTBI and healthy control cases. Single cytokines or chemokines were predictors, with AUC values ranging from 0.39 for IL-6 to 0.76 for interleukin 17 (IL-17) (see the supplemental material). Median abundances of IFN-γ (as expected) and IL-1α, IL-2, and IL-17 were significantly higher in the LTBI group than in healthy controls (P < 0.05). The low P value for IL-17 (<0.01) suggests that this cytokine, measured in SPSs, is a promising biomarker candidate, as it pertains to the detection of TB latency (AUC, Z = 0.763 and P = 0.001) (Fig. 1). Given the IGRA-based definition of the LTBI cohort and no additional clinical information to substantiate this diagnosis, a study with a larger number of participants and more clinical information to support the distinction of latent and active TB cases would be useful to determine if IL-17 is a diagnostic biomarker of equal value to IFN-γ.
FIG 1.
ROC curve for 40 cytokines/chemokines measured in stimulated plasma supernatants (SPSs) via the HIAA method comparing LTBI and healthy community control groups. The 40 AUC values were analyzed by generating ROC curves for all 40 analytes. LTBI subjects were positive for IFN-γ release in the QuantiFERON-TB ELISA (n = 30); healthy controls were negative (n = 26). Discriminatory values were excellent for IFN-γ (AUC = 0.911; P < 0.001) and also good for IL-17 (AUC = 0.763; P = 0.001).
Cytokine/chemokine abundance surveys comparing TB-infected subjects with healthy controls and active TB cases with LTBI/healthy subjects.
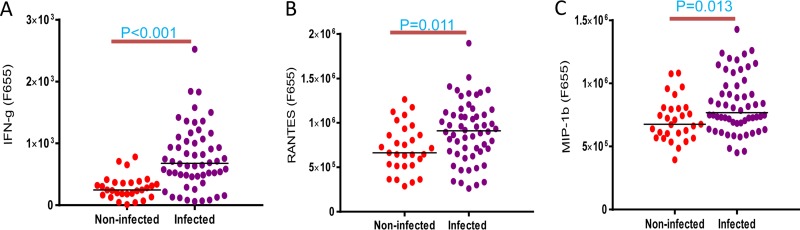
HIAA experiments were performed comparing 29 data sets from healthy controls and 56 data sets from TB-infected subjects (that is, 30 LTBI and 26 active TB cases) using SPSs as the sample source. Independent-sample t tests were used to identify cytokines/chemokines with statistically significant abundance differences. The abundances of IFN-γ (2.2-fold) and the chemokines RANTES (1.8-fold) and MIP-1β (1.75-fold) were elevated in TB-infected subjects versus the healthy control group (P < 0.05) (Fig. 2). The abundances of IL-12p40 (3.5-fold) and soluble tumor necrosis factor receptor II (sTNF-RII) (1.8-fold) were increased in active-TB patients compared to joint LTBI and healthy groups (P < 0.05) (data not shown). Statistically significant quantitative differences for other cytokines/chemokines were not determined in these two-cohort comparisons.
FIG 2.
Differential abundance values of cytokines/chemokines comparing all TB-infected cases (LTBI and active TB cases) with the healthy control group measured at a fluorescence emission wavelength of 655 nm. As described in the legend to Fig. 1, the analytes were measured in SPSs from the TB-infected group (n = 56) and healthy control group (n = 29). IFN-γ, RANTES, and MIP-1β abundances were moderately elevated in the TB-infected cohort, with statistical significance. Independent-sample t testing was performed, and P values of <0.05 were considered statistically significant.
Differentially abundant cytokines/chemokines based on comparisons of all three groups.
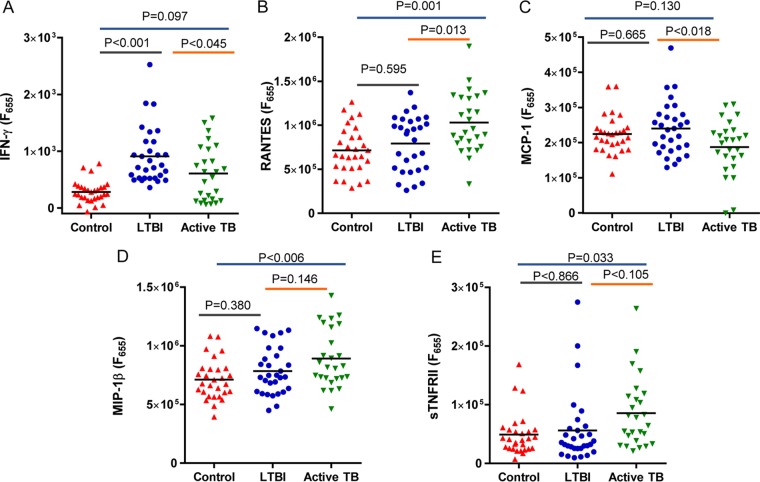
As determined by the HIAA method, the abundance of RANTES was significantly higher in subjects with active TB than in the LTBI group (P = 0.013) and in those with active TB than in the healthy controls (P = 0.001) as measured in SPSs. For active TB patients, the analysis yielded a significantly higher averaged abundance of MIP-1β than in the control group (P = 0.006), but statistical significance was not observed for the comparison of active TB versus LTBI. The abundance of sTNF-RII was also significantly increased in subjects with active TB versus healthy controls (P = 0.033). In contrast, MCP-1 was decreased in abundance in active TB compared to LTBI cases (P = 0.018). Interestingly, IFN-γ did not reveal abundance differences in the comparison of active TB cases and the LTBI cohort (Fig. 3).
FIG 3.
Cytokine/chemokine abundances showing significant differences between the active-TB (n = 26), LTBI (n = 30), and control (n = 29) groups in SPS measurements using one-way ANOVA. RANTES abundance differences were observed for active TB cases compared to both LTBI (P = 0.013) and healthy controls (P = 0.001). MIP-1β (P = 0.006) and sTNF-RII (P = 0.033) had different abundances for the active-TB versus healthy control cases. The relative abundance of MCP-1 was significantly lower in active-TB cases than in LTBI cases (P = 0.018).
Cytokine/chemokine abundance profiles in unstimulated plasma samples.
To assess whether the stimulation of immune cells with M. tuberculosis antigens in blood was essential to measure quantitative differences in cytokines/chemokines, analogous experiments were performed using UPS aliquots. No IFN-γ abundance differences were observed comparing the healthy control and LTBI groups (P = 1.00), healthy control and active TB groups (P = 0.706), and LTBI and active-TB groups (P = 0.691). In contrast, consistent with the results observed in SPS analyses, abundance levels of RANTES were also significantly elevated in active TB versus LTBI cases (P = 0.012) and in active-TB versus healthy control cases (P = 0.002) using UPS aliquots in the HIAAs (Fig. 4). No elevated abundances for TNF-RII, MCP-1, or MIP-1β in UPSs were detected in comparisons of the above-mentioned groups (P > 0.05). Accordingly, no other cytokines/chemokines showed significant abundance differences in the disease group comparisons using UPSs (P > 0.05).
FIG 4.
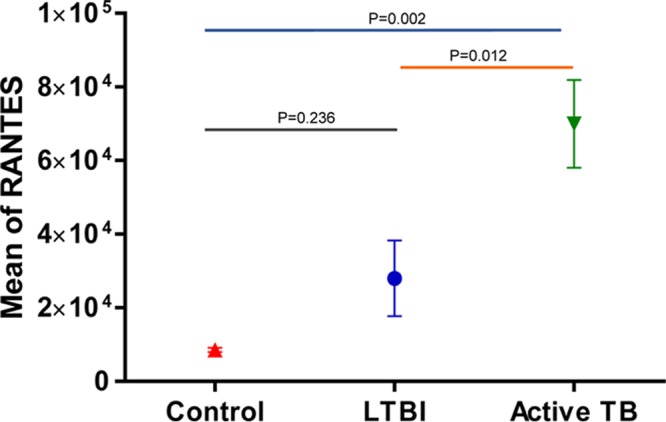
Mean abundances of RANTES in TB patients, LTBI subjects, and negative controls in HIAA analysis with unstimulated plasma samples measured at a fluorescence emission wavelength of 655 nm. RANTES differentiated active TB cases from LTBI and negative controls in unstimulated plasma samples using a one-way ANOVA test.
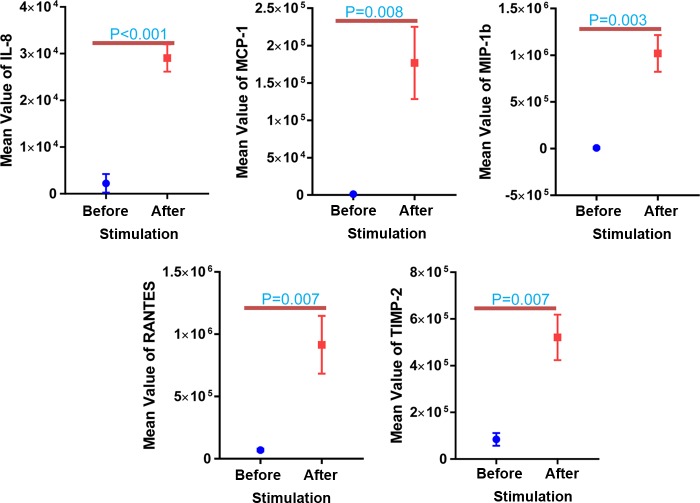
IL-8, MCP-1, MIP-1β, RANTES, and TIMP-2 showed highly significant differences (P < 0.01) in abundance following M. tuberculosis antigen stimulation (in SPSs) in active-TB and healthy control cases (Fig. 5). Only macrophage colony-stimulating factor (M-CSF) (P = 0.036) and MIP-1β (P = 0.012) showed significant changes in the LTBI versus healthy control group (data not shown).
FIG 5.
Plasma cytokine/chemokine abundance differences comparing SPSs (after stimulation) and UPSs (before stimulation) as sample sources. Normalized relative abundance values were measured at a fluorescence emission wavelength of 655 nm. The paired t test was used for statistical analyses. A P value of <0.01 was considered highly significant. IL-8, MCP-1, MIP-1β, RANTES, and TIMP-2 are analytes quantified compared to the negative control.
DISCUSSION
The currently used diagnostic tests for active TB (microscopy and culture) are not sensitive enough and too slow for point-of-care applications, respectively (5, 6). Though transcriptional signatures can distinguish active TB from LTBI (23, 24) and further predict progression of LTBI to active TB (25), translating these signatures into tests for diagnosis is challenging anywhere, but especially in resource-poor settings. The currently used diagnostic test for LTBI, an IGRA, has poor specificity for active TB in high-burden settings. The QuantiFERON-TB Gold in-tube test (QFT-IT) is a third-generation ELISA based on IFN release and was approved by the FDA for LTBI diagnosis. Compared to previous tests, it has a clearer definition for positive versus negative results and lower interference associated with Mycobacterium bovis BCG vaccination and NTM infections (26–28). IFN-γ is a powerful proinflammatory cytokine that plays a major role in maintaining TB latency through macrophage activation and in overcoming phagosome maturation blocking, nitrogen and oxygen radical production, and promotion of antigen recognition by enhancing major histocompatibility complex class II (MHC-II) expression on antigen-presenting cells (29, 30). Pai et al. reviewed 20 studies and concluded that the specificity of IGRAs for the detection of LTBI is high (31).
We screened 40 cytokines/chemokines, including IFN-γ, via antibody arrays to identify biomarker candidates in a first step (the discovery stage) toward the development of a diagnostic test able to distinguish active TB from LTBI and LTBI from healthy controls with improved specificity and sensitivity. Using SPSs, measurements of IFN-γ were largely consistent with QFT-IT data (the LTBI group was defined based on the QFT-IT). IFN-γ levels had the highest P and AUC values for healthy control and LTBI samples. In our HIAA-based experiments, the cytokine with the 2nd highest AUC value was IL-17, an immunological marker also potentially useful to identify latently TB-infected human subjects.
IL-17 is a proinflammatory cytokine produced by Th17 and gamma delta lymphocytes and plays a major role in TB protection via recruitment of neutrophils, macrophages, and Th1 lymphocytes to the site of infection. This mechanism appears to contribute to inhibiting M. tuberculosis dissemination (32). While some studies reported increased release of IL-17 in patients with active TB (33, 34), other studies described increased release of IL-17 in LTBI patients (35) and a decreased release of IL-17 in patients with active TB (36). Our findings suggest that IL-17 levels are increased in LTBI cases compared to those of negative controls and active TB. Our data agree with other results (35) from which it was concluded that following Mycobacterium bovis BCG vaccination and infection with M. tuberculosis, IL-17 acts as an effector similar to IFN-γ and, together with IFN-γ, contributes to TB protection.
IFN-γ and IL-2 were also reported to inhibit Th17 differentiation and IL-17 production, data that are not consistent with the average abundance increases of IFN-γ, IL-1α, and IL-2 in parallel with IL-17 in LTBI patients (as observed in our survey). This may be explained by the absence of significant changes in abundance of transforming growth factor β1 (TGF-β1), a cytokine with a key role in Th17 cell differentiation through its ability to inhibit the differentiation of Th1 cells (37–39). Elevated abundance of IL-17 in LTBI patients may point to the involvement of neutrophils, which play a role in protection from acute TB and the manifestation of latency. The exact role of altered IL-17 expression in the context of inflammatory versus protective effects at the tissue site of TB infection remains to be determined.
Combining the active TB and LTBI groups into one group, the chemokines RANTES and MIP-1β showed statistically significant differences, in addition to IFN-γ, compared to the healthy group. Combining the LTBI and healthy control groups, the cytokines IL-12p40 (3.5-fold) and sTNF-RII were significantly decreased compared to the levels in active-TB patients. Although preliminary, the data suggest additional opportunities to explore cytokines and chemokines for the rapid diagnosis of active TB from M. tuberculosis antigen-stimulated blood samples to replace or complement acid-fast staining and the diagnosis-delaying M. tuberculosis culture tests.
The most interesting findings in our study were those on two proinflammatory chemokines, MIP-1β and especially RANTES. In infections with M. tuberculosis, RANTES and MIP-1β potentially recruit and activate different types of leukocytes (40, 41). The main cellular expression source of RANTES is T cells. RANTES is a chemoattractant for CD4+ T cells, CD45RO memory T cells, and mononuclear monocytes (42). RANTES is thought to be primarily released by mononuclear phagocytes in response to M. tuberculosis infection. The high levels of RANTES that we observed in SPSs of TB-infected compared to noninfected individuals strengthen the notion that T lymphocytes can produce RANTES. Alveolar T lymphocytes, which are increased during active TB, could be a source of RANTES once they are released into peripheral blood (43). Interestingly, the ability of RANTES to differentiate active TB from negative controls as well as active TB from LTBI has no complement to IFN-γ quantification: IGRAs are unable to distinguish LTBI from active TB using SPSs. Equally important, quantitative differences for RANTES in these group comparisons were significant not only when we used SPSs but also when we used unstimulated plasma samples (UPSs). RANTES quantification may ultimately result in simpler, less expensive clinical tests that do not require M. tuberculosis antigen stimulation. Elevated levels of a chemokine without antigen stimulation are the basis of more rapid tests to identify M. tuberculosis infections (44). MIP-1β abundance was significantly higher in TB-infected subjects than in healthy controls, but not different in the active-TB and LTBI cohorts. We speculate that MIP-1β expression is not induced in early and asymptomatic stages of M. tuberculosis infection. Our findings are consistent with functional roles of RANTES and MIP-1β in the pathogenesis of other infectious and noninfectious diseases (45, 46).
We show that IL-17 may be a complementary or alternative biomarker for LTBI diagnosis. Since our LTBI group definition was based on IGRA data, we cannot directly compare the specificity results for IFN-γ and IL-17. Our cytokine/chemokine array studies were conducted with a cohort of multiethnic pastoralist communities in a specific geographic area. To generalize the importance of IL-17, RANTES, and MIP-1β as candidate biomarkers differentiating TB disease states and/or LTBI from healthy individuals, the cohorts will need to be expanded and diversified. If such studies confirm some of our preliminary results, biomarker validation studies with thousands of clinical samples can be conducted. The ultimate goals are specific, less expensive, point-of-care immunological tests for diagnosis of TB in symptomatic and asymptomatic stages.
MATERIALS AND METHODS
Study subjects.
Ninety human subjects 18 to 80 years of age were enrolled under the study name “Systems Biology for Molecular Analysis of Tuberculosis in Ethiopia” in the Southern Ethiopian South Omo zone during 2014 and 2015. The zone is characterized by numerous ethnically distinct pastoralist groups with different sociocultural practices and lifestyles and limited access to regular health care. Human subject protocols and enrollment forms were carefully designed and approved by the Ethics Committee of Addis Ababa University (AAU) and the Internal Review Board of the J. Craig Venter Institute (JCVI) in January 2014 (see “Ethics statement” below). Patients with active pulmonary TB were recruited from regional and local clinics before antibiotic treatment began and from communities in selected districts in the South Omo zone. Approximately 3,300 apparently healthy control subjects, some of whom were later diagnosed with LTBI, were recruited from the same districts. The QFT-IT was used to screen healthy controls for evidence of LTBI, as recommended by the Food and Drug Administration and Centers for Disease Control and Prevention in the United States and by the WHO (26–28), following the manufacturer's instructions. Medical data were collected to rule out the possibility that the control/LTBI group had clinical signs and symptoms of other respiratory diseases (26). Due to the remote setting and difficulties in recontacting and communicating with the enrolled subjects, medical data were not collected after enrollment. The active-TB group was defined via a positive test for acid-fast bacilli in microscopic examinations and M. tuberculosis growth using Löwenstein-Jensen (LJ) medium cultures. Additional medical data were also collected. Subjects suffering from allergies, serious malnutrition, malignancies, and immunodeficiencies (e.g., HIV patients, patients receiving immunosuppressive therapies, and patients with congenital immunodeficiency) were excluded from the study.
Plasma samples.
Three milliliters of whole blood was drawn from most of the enrolled subjects via venipuncture and collected into heparin test tubes, followed by centrifugation for 15 min at 1,000 × g to recover the plasma fraction. The UPSs were stored at −80°C and shipped on dry ice until further use.
Preparation of stimulated plasma samples for cytokine/chemokine assays.
Whole-blood samples were collected from 90 enrolled participants (30 healthy controls, 30 individuals with LTBI, and 30 patients with active TB) and stimulated with the ESAT-6/CFP-10 antigen cocktail. Briefly, 1 ml of heparinized blood was diluted in RPMI 1640 medium containing l-glutamine supplemented with penicillin (100 U/ml) and streptomycin (100 μg/ml) to a final dilution of 1:10. The ESAT-6/CFP-10 cocktail was used to stimulate cytokine release from blood samples at a final 10-μg/ml concentration. As positive and negative controls, phytohemagglutinin antigen (10 μg/ml) in RPMI 1640 medium and medium only were used, respectively. After 48 h of incubation at 37°C with 5% CO2, these stimulated plasma supernatants (SPSs) were harvested and stored at −80°C and shipped on dry ice until further use.
Measurement of cytokines and chemokines using an antibody microarray-based method.
A commercial product to measure inflammation-associated cytokines and chemokines in a microarray format was used, the G-series HIAA 3 (Ray Biotech, Inc., Norcross, GA). Procedures recommended by the manufacturer were employed to quantify the analytes from UPSs and SPSs, including the Ray Biotech Q Analyzer program for data analysis. Briefly, frozen samples were thawed, vortexed, and centrifuged for 10 min at 10,000 × g to remove particulate matter. After 30 min of incubation with a sample diluent, HIAA glass chips were washed, and each well was overlaid with 70 μl of UPS or SPS, covered with adhesive film, and incubated at room temperature (RT) for 2 h. Following a wash step with wash buffers I and II, each subarray was covered with an equal volume (70 μl) of biotin-conjugated anti-cytokine antibodies, incubated for 2 h at RT, washed with wash buffers I and II, and incubated at 4°C overnight, followed by another wash step with wash buffer II. Alexa Fluor 647-conjugated streptavidin was added, followed by incubation for 2 h at RT. The fluorescence signals (Cy5 range; 655-nm emission wavelength) were scanned and extracted using a Genepix 4000B laser scanner (Axon Instruments, Foster City, CA). Normalization steps included (i) subtracting background signals, (ii) normalizing to positive controls, and (iii) comparing signal intensities for antigen-specific array spots among different array images. Relative normalized abundance values for all cytokine/chemokine analytes in all samples were computed. We chose fold change cutoff values recommended by the manufacturer, 1.5 and 0.65 for increase and decrease in abundance, respectively, for the analyte in group-based comparisons.
Statistical tests.
Differences in cytokine/chemokine abundances among the active TB, LTBI, and healthy subject cohorts were compared by one-way analysis of variance (ANOVA) using IBM SPSS software version 20. Multiple comparisons were performed using the least significant difference (LSD) post hoc test when the variance between samples was equal or Dunnett's T3 test when the variances were not equal. The independent-sample t test was used to identify differences in relative analyte abundance levels comparing active TB with the combination of the other two groups. The median analyte differences between groups were evaluated by a nonparametric analysis (Mann-Whitney U test). Comparisons of data pertaining to SPSs (stimulated) and UPSs (unstimulated) in the assays were performed using a paired-sample t test. P values of <0.05 were considered statistically significant. GraphPad Prism 7 software was used for graphing the dot plots. Diagnostic values for 40 analytes measured in SPSs using the HIAA and of 1 analyte (IFN-γ) using the IGRA were determined by AUCs.
Ethics statement.
Ethical approval for the study was obtained from Addis Ababa University, Aklilu Lemma Institute of Pathobiology Research and Ethics Committee, as well as from the National Research Ethics Committee of Ethiopia (reference no. 3.10/785/07). Written consent was obtained from each study participant after the objective of the study was clearly explained. Blood sample collection was undertaken under aseptic conditions by licensed medical laboratory professionals. Volunteers with signs and symptoms of active TB or any other disease during the enrollment period were treated in nearby health facilities at the expense of this study.
Supplementary Material
ACKNOWLEDGMENTS
This study was financially supported by the National Institutes of Health (NIH) through its H3Africa consortium program (grant reference no. U01HG007472-01).
We thank the study participants, DAAD In-country scholarship, and JCVI, USA, staffs. We also thank Harinder Singh and Mahlet Chanyalew for technical assistance.
We declare that there are no competing interests.
T.T. was involved in the design, data collection, laboratory work, statistical analysis and interpretation, and manuscript drafting; K.K. was involved in the laboratory work, interpretation, and critical revision; B.W., M.H., and A.Z. were involved in data collection and critical revision; M.L. and G.M. were involved in data analysis and interpretation and critical revision; and R.P. and G.A. were involved in design, interpretation, and critical revision. All authors have read and approved the paper for submission.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/IAI.00759-17.
REFERENCES
- 1.Smith I. 2003. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin Microbiol Rev 16:463–496. doi: 10.1128/CMR.16.3.463-496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry CE III, Boshoff H, Dartois V, Dick T, Ehrt S, Flynn J, Schnappinger D, Wilkinson RJ, Young D. 2009. The spectrum of latent tuberculosis: rethinking the goals of prophylaxis. Nat Rev Microbiol 7:845–855. doi: 10.1038/nrmicro2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Raviglione MC, Snider DE, Kochi A. 1995. Global epidemiology of tuberculosis: morbidity and mortality of a worldwide epidemic. JAMA 273:220–226. [PubMed] [Google Scholar]
- 4.WHO. 2008. Joint TDR/EC expert consultation on biomarkers in Tuberculosis. Report of the joint TDR/EC expert consultation to evaluate the potential roles of biomarkers in the management of HIV-infected and HIV-uninfected patients with tuberculosis, Geneva, Switzerland, 2 to 3 July 2008 WHO, Geneva, Switzerland. [Google Scholar]
- 5.Steingart KR, Henry M, Ng V, Hopewell PC, Ramsay A, Cunningham J, Urbanczik R, Perkins M, AbdelAziz M, Pai M. 2006. Fluorescence versus conventional sputum smear microscopy for tuberculosis: a systematic review. Lancet Infect Dis 6:570–581. doi: 10.1016/S1473-3099(06)70578-3. [DOI] [PubMed] [Google Scholar]
- 6.Steingart KR, Ramsay A, Pai M. 2007. Optimizing sputum smear microscopy for the diagnosis of pulmonary tuberculosis. Expert Rev Anti Infect Ther 4:327–331. doi: 10.1586/14787210.5.3.327. [DOI] [PubMed] [Google Scholar]
- 7.WHO. 2014. Xpert MTB/RIF implementation manual. Technical and operational ‘how-to’: practical considerations. WHO/HTM/TB/2014.1 WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 8.WHO. 2015. The End TB strategy: guidelines on the managements of latent tuberculosis. WHO/HTM/TB 2015.01 WHO, Geneva, Switzerland. [Google Scholar]
- 9.Denkinger CM, Pai M, Patel M, Menzies D. 2013. Gamma interferon release assay for monitoring of treatment response for active tuberculosis: an explosion in the spaghetti factory. J Clin Microbiol 51:607–610. doi: 10.1128/JCM.02278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pai M, Denkinger CM, Kik SV, Rangaka MX, Zwerling A, Oxlade O, Metcalfe JZ, Cattamanchi A, Dowdy DW, Dheda K, NiazBanaei N. 2014. Gamma interferon release assays for detection of Mycobacterium tuberculosis infection. Clin Microbiol Rev 27:3–20. doi: 10.1128/CMR.00034-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Santin M, Muñoz L, Rigau D. 2012. Interferon-γ release assays for the diagnosis of tuberculosis and tuberculosis infection in HIV-infected adults: a systematic review and meta-analysis. PLoS One 7:e32482. doi: 10.1371/journal.pone.0032482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mandalakas AM, Detjen AK, Hesseling AC, Benedetti A, Menzies D. 2011. Interferon-gamma release assays and childhood tuberculosis: systematic review and meta-analysis. Int J Tuberc Lung Dis 15:1018–1032. doi: 10.5588/ijtld.10.0631. [DOI] [PubMed] [Google Scholar]
- 13.Lenaerts A, Barry CE, Dartois V. 2015. Heterogeneity in tuberculosis pathology, microenvironments and therapeutic responses. Immunol Rev 264:288–307. doi: 10.1111/imr.12252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner D, Hörster R, Lange B, Lange C. 2008. Evaluation of T-cell interferon-gamma-release assays for the diagnosis of latent and active tuberculosis. Dtsch Med Wochenschr 133:354–357. doi: 10.1055/s-2008-1046718. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, Jiang T, Wei L, Yang X, Wang C, Zhang Z, Xu D, Chen Z, Yang F, Li JC. 2013. The discovery and identification of a candidate proteomic biomarker of active tuberculosis. BMC Infect Dis 13:506. doi: 10.1186/1471-2334-13-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Frahm M, Goswami ND, Owzar K, Hecker E, Mosher A, Cadogan E, Nahid P, Ferrari G, Stout JE. 2011. Discriminating between latent and active tuberculosis with multiple biomarker responses. Tuberculosis 91:250–256. doi: 10.1016/j.tube.2011.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kellar KL, Gehrke J, Weis SE, Mahmutovic-Mayhew A, Davila B, Zajdowicz MJ, Scarborough R, LoBue PA, Lardizabal AA, Daley CL, Reves RR, Bernardo J, Campbell BH, Whitworth WC, Mazurek GH. 2011. Multiple cytokines are released when blood from patients with tuberculosis is stimulated with Mycobacterium tuberculosis antigens. PLoS One 6:e26545. doi: 10.1371/journal.pone.0026545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murthy MK, Kaliappan T, Raja A. 2011. Cytokine and chemokine responses to selected early secreted antigenic target-6 and culture filtrate protein-10 peptides in tuberculosis. J Interferon Cytokine Res 31:299–307. doi: 10.1089/jir.2010.0048. [DOI] [PubMed] [Google Scholar]
- 19.Borgström E, Andersen P, Atterfelt F, Julander I, Källenius G, Maeurer M, Rosenkrands I, Widfeldt M, Bruchfeld J, Gaines H. 2012. Immune responses to ESAT-6 and CFP-10 by FASCIA and multiplex technology for diagnosis of M. tuberculosis infection; IP-10 is a promising marker. PLoS One 7:e43438. doi: 10.1371/journal.pone.0043438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anbarasu D, Raja CP, Raja A. 2013. Multiplex analysis of cytokines/chemokines as biomarkers that differentiate healthy contacts from tuberculosis patients in high endemic settings. Cytokine 61:747–754. doi: 10.1016/j.cyto.2012.12.031. [DOI] [PubMed] [Google Scholar]
- 21.Armand M, Chhor V, de Lauzanne A, Guérin-El Khourouj V, Pédron B, Jeljeli M, Gressens P, Faye A, Sterkers G. 2014. Cytokine responses to quantiferon peptides in pediatric tuberculosis: a pilot study. J Infect 68:62–70. doi: 10.1016/j.jinf.2013.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Rivera-Ordaz A, Gonzaga-Bernachi J, Serafín-López J, Hernández-Pando R, Van Soolingen D, Estrada-Parra S, Estrada-García I, Chacón-Salinas R. 2012. Mycobacterium tuberculosis Beijing genotype induces differential cytokine production by peripheral blood mononuclear cells of healthy BCG vaccinated individuals. Immunol Invest 41:144–156. doi: 10.3109/08820139.2011.596604. [DOI] [PubMed] [Google Scholar]
- 23.Kaforou M, Wright VJ, Oni T, French N, Anderson ST, Bangani N, Banwell CM, Brent AJ, Crampin AC, Dockrell HM, Eley B, Heyderman RS, Hibberd ML, Kern F, Langford PR, Ling L, Mendelson M, Ottenhoff TH, Zgambo F, Wilkinson RJ, Coin LJ, Levin M. 2013. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case-control study. PLoS Med 10:e1001538. doi: 10.1371/journal.pmed.1001538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Anderson ST, Kaforou M, Brent AJ, Wright VJ, Banwell CM, Chagaluka G, Crampin AC, Dockrell HM, French N, Hamilton MS, Martin L, Hibberd ML, Kern F, Langford PR, Ling L, Mlotha R, Ottenhoff THM, Pienaar S, Pillay V, Anthony G, Twahir H, Wilkinson RJ, Coin LJ, Heyderman RS, Levin M, Eley B, the ILULU Consortium and KIDS TB Study Group. 2014. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med 370:1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zak DE, Penn-Nicholson A, Scriba TJ, Thompson E, Suliman S, Amon LM, Mahomed H, Erasmus M, Whatney W, Hussey GD, Abrahams D, Kafaar F, Hawkridge T, Verver S, Hughes EJ, Ota M, Sutherland J, Howe R, Dockrell HM, Boom WH, Thiel B, Ottenhoff T, Mayanja-Kizza H, Crampin AC, Downing K, Hatherill M, Valvo J. 2016. A prospective blood RNA signature for tuberculosis disease risk. Lancet 387:2312–2322. doi: 10.1016/S0140-6736(15)01316-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazurek GH, Jereb J, Vernon A, LoBue P, Goldberg S, Castro K. 2010. Updated guidelines for using interferon gamma release assays to detect Mycobacterium tuberculosis infection—United States, 2010. MMWR Recomm Rep 59(RR-5):1–25. [PubMed] [Google Scholar]
- 27.Food and Drug Administration. QuantiFERON-TB Gold In-Tube P010033/S011. Pre-market approval. https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?start_search=1&pmanumber=P010033&sortcolumn=an_desc Accessed 20 September 2017.
- 28.WHO. 2011. Tuberculosis IGRA test policy statement 2011; the use of TB interferon-gamma release assays (IGRAs) in low- and middle-income countries. WHO, Geneva, Switzerland. [PubMed] [Google Scholar]
- 29.Shen ZT, Nguyen TT, Daniels KA, Welsh RM, Stern LJ. 2013. Disparate epitopes mediating protective heterologous immunity to unrelated viruses share peptide-MHC structural features recognized by cross-reactive T cells. J Immunol 191:5139–5152. doi: 10.4049/jimmunol.1300852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clute SC, Naumov YN, Watkin LB, Aslan N, Sullivan JL, Thorley-Lawson DA, Luzuriaga K, Welsh RM, Puzone R, Celada F, Selin LK. 2010. Broad cross-reactive TCR repertoires recognizing dissimilar Epstein-Barr and influenza A virus epitopes. J Immunol 185:6753–6764. doi: 10.4049/jimmunol.1000812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pai M, Zwerling A, Menzies D. 2008. Systematic review: T-cell based assays for the diagnosis of latent tuberculosis infection: an update. Ann Intern Med 149:177–184. doi: 10.7326/0003-4819-149-3-200808050-00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shah K, Lee WW, Lee SH, Kim SH, Kang SW, Craft J, Kang I. 2010. Dysregulated balance of Th17 and Th1 cells in systemic lupus erythematosus. Arthritis Res Ther 12:R53. doi: 10.1186/ar2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marín ND, París SC, Rojas M, Garcí LF. 2012. Reduced frequency of memory T cells and increased Th17 responses in patients with active tuberculosis. Clin Vaccine Immunol 19:1667–1676. doi: 10.1128/CVI.00390-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu L, Cui G, Jia H, Zhu Y, Ding Y, Chen J, Lu C, Ye P, Gao H, Li L, Ma W, Lyu J, Diao H. 2016. Decreased IL-17 during treatment of sputum smear-positive pulmonary tuberculosis due to increased regulatory T cells and IL-10. J Transl Med 14:179. doi: 10.1186/s12967-016-0909-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li Q, Li J, Tian J, Zhu B, Zhang Y, Yang K, Ling Y, Hu Y. 2012. IL-17 and IFN-γ production in peripheral blood following BCG vaccination and Mycobacterium tuberculosis infection in human. Eur Rev Med Pharmacol Sci 16:2029–2036. [PubMed] [Google Scholar]
- 36.Shu CC, Wu MF, Wang JY, Lai HC, Lee LN, Chiang BL, Yu CJ. 2017. Decreased T helper 17 cells in tuberculosis is associated with increased percentages of programmed death ligand 1, T helper 2 and regulatory T cells. Respir Res 18:128. doi: 10.1186/s12931-017-0580-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo KV. 2006. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature 441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 38.Santarlasci V, Maggi L, Capone M, Frosali F, Querci V, Palma RD, Liotta F, Cosmi L, Maggi E, Romagnani S, Annunziato F. 2009. TGF-beta indirectly favors the development of human Th17 cells by inhibiting Th1 cells. Eur J Immunol 39:207–215. doi: 10.1002/eji.200838748. [DOI] [PubMed] [Google Scholar]
- 39.Oukka M. 2008. Th17 cells in immunity and autoimmunity. Ann Rheum Dis 67(Suppl 3):iii26–iii29. [DOI] [PubMed] [Google Scholar]
- 40.Hasan Z, Cliff JM, Dockrell HM, Jamil B, Irfan M, Ashraf M, Hussain R. 2009. CCL2 responses to Mycobacterium tuberculosis are associated with disease severity in tuberculosis. PLoS One 4:e8459. doi: 10.1371/journal.pone.0008459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monin L, Khader SA. 2014. Chemokines in tuberculosis: the good, the bad and the ugly. Semin Immunol 26:552–558. doi: 10.1016/j.smim.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schall TJ. 1991. Biology of the RANTES/SIS cytokine family. Cytokine 3:165–183. doi: 10.1016/1043-4666(91)90013-4. [DOI] [PubMed] [Google Scholar]
- 43.Sadek M, Sada E, Toossi Z, Schwander SK, Rich EA. 1998. Chemokines induced by infection of mononuclear phagocytes with mycobacteria and present in lung alveoli during active pulmonary tuberculosis. Am J Respir Cell Mol Biol 19:513–521. doi: 10.1165/ajrcmb.19.3.2815. [DOI] [PubMed] [Google Scholar]
- 44.Cannas A, Calvo L, Chiacchio T, Cuzzi G, Vanini V, Lauria FN, Pucci L, Girardi E, Goletti D. 2010. IP-10 detection in urine is associated with lung diseases. BMC Infect Dis 10:333. doi: 10.1186/1471-2334-10-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Deshmane SL, Kremlev S, Amini S, Sawaya BE. 2009. Monocyte chemoattractant protein-1 (MCP-1): an overview. J Interferon Cytokine Res 29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Murdoch C, Finn A. 2000. Chemokine receptors and their role in inflammation and infectious diseases. Blood 95:3032–3043. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.