Abstract
Iron is essential for growth and proliferation of mammalian cells. The maintenance of cellular iron homeostasis is regulated by iron regulatory proteins (IRPs) through binding to the cognate iron-responsive elements in target mRNAs and thereby regulating the expression of target genes. Irp1 or Irp2-null mutation is known to reduce the cellular iron level by decreasing transferrin receptor 1 and increasing ferritin. Here, we report that Irp1 or Irp2-null mutation also causes downregulation of frataxin and IscU, two of the core components in the iron-sulfur cluster biogenesis machinery. Interestingly, while the activities of some of iron-sulfur cluster-containing enzymes including mitochondrial aconitase and cytosolic xanthine oxidase were not affected by the mutations, the activities of respiratory chain complexes were drastically diminished resulting in mitochondrial dysfunction. Overexpression of human ISCU and frataxin in Irp1 or Irp2-null cells was able to rescue the defects in iron-sulfur cluster biogenesis and mitochondrial quality. Our results strongly suggest that iron regulatory proteins regulate the part of iron sulfur cluster biogenesis tailored specifically for mitochondrial electron transport chain complexes.
Introduction
Iron is essential for growth and proliferation of mammalian cells mainly because iron is a crucial component of heme and iron-sulfur clusters (Fe-S) therefore indispensable for DNA synthesis, oxygen transport and ATP production. Iron deficiency is known to cause anaemia and permanent neurocognitive and motor impairments1,2. Yet, excess cellular iron could lead to the generation of reactive oxygen species that damage macromolecules such as DNA, lipid, and proteins. Excess iron in brain has been found to be associated with common neurodegenerative disorders, including Alzheimer’s, Parkinson’s, and Friedreich’s ataxia1,3. Therefore, organisms and cells must precisely regulate iron metabolism.
In mammals, cellular iron metabolism is regulated by iron regulatory protein (IRP) 1 and 24,5. IRPs post-transcriptionally regulate the expression of target protein by binding to iron responsive elements (IREs) located within the 5′- or 3′- untranslated region (UTR) of the target gene transcripts. These transcripts mostly encode iron metabolism proteins, including the iron storage protein, ferritin, and iron import protein, transferrin receptor 1 (TfR1)5,6. When cells are iron-deficient, IRPs bind IREs located in the 5′-UTR of ferritin to inhibit the translation and in the 3′-UTR of TfR1 to stabilize the mRNA to facilitate iron import. When cells are iron-sufficient, IRP1 is converted to a [4Fe-4S]-containing aconitase and IRP2 is degraded through iron mediated proteasomal degradation7–9, which increases ferritin translation and promotes TfR1 mRNA degradation to prevent more iron absorption and avoid the excess iron-induced injury. Thus, IRPs play critical roles in cellular iron homeostasis.
The mouse models of Irp1 and Irp2 deficiency have been generated10–12. Irp1−/− mice display polycythemia due to derepression of the Irp1-specific target mRNA of hypoxia-inducible factor 2α13–15. Two mouse models of global Irp2 deficiency display dysregulation of ferritin and TfR1 and abnormal iron content in several tissues, and develop microcytic anaemia and erythropoietic protoporphyria16,17. Mice lacking Irp2 also show symptoms of neurological disorders with motor neuron death11,18,19. The motor neuron death observed in the Irp2-null mice is likely caused in part by the diminished size of the ‘functional’ iron pool (compared to ‘total’) due to the reduced TfR1 expression and increased ferritin expression20. Importantly, the role of IRPs in mitochondrial iron homeostasis has also been uncovered21. Irp1 activation is thought to be critical for sustaining the mitochondrial iron supply therefore maintaining normal mitochondrial function22. Irp2-null mice showed tissue-specific iron insufficiency and compromised motor neurons and their mitochondrial function20. However, the exact mechanism by which IRPs sustain normal mitochondrial function has yet to be determined.
Both Fxn and IscU are essential proteins, with Fxn acting as an iron chaperone for iron delivery or an allosteric factor that modulates the cysteine desulfurase activity (see review23), and IscU as a scaffold protein for Fe-S cluster biogenesis. Deficiency of human FXN causes Friedreich ataxia, a neurodegenerative mitochondrial disease24. A muscle-specific alternative mis-splicing of human ISCU renders ISCU myopathy, also known as “myopathy with deficiency of succinate dehydrogenase and aconitase”25. It is widely believed that deficiency of Fe-S cluster biogenesis is causative of mitochondrial dysfunction in both above mentioned diseases and that iron subcellular mislocalization is a common phenomenon in the disorders of Fe-S biogenesis3. Thus, we wonder whether the biological manifestation of IRPs deficiency shares the same or part of the mechanism of Fxn or IscU deficiency.
In this study, we confirmed that murine embryonic fibroblasts (MEFs), derived from global Irp1 or Irp2 deficient mice, showed cellular iron starvation due to the low expression of TfR1 and the increased expression of ferritin. More importantly, we found that depletion of IRPs led to deficiencies of Fxn and IscU. Furthermore, the IRP depletion-induced deficiencies in Fxn and IscU impaired specifically Fe-S cluster-dependent mitochondrial respiratory chain activities, but not the activity of either mitochondrial aconitase or cytosolic xanthine oxidase (Xod). Our results suggest that Irp affects the Fe-S cluster biogenesis tailored specifically for mitochondrial electron transport chain (ETC) complexes.
Results
Irp1 or Irp2 ablation induces downregulation of the components of Fe-S cluster biogenesis machinery
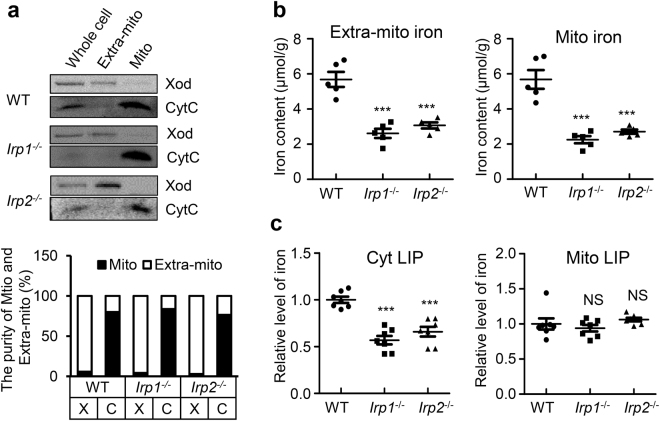
It has been reported that mice lacking Irp1 show systemic iron deficiency in the later age13 and lacking Irp2 show neurodegenerative symptoms11,26, which is thought due to the reduction of “functional iron pool”20. To gain further understanding of this phenotype, we used MEFs derived from global Irp1 or Irp2 deficient mice and measured iron content in mitochondrial and extra-mitochondrial (mito and extra-mito) fractions. First, fractionation was performed to separate mito from extra-mito fraction. Xod and cytochrome C (CytC) were used as markers to define the purity of extra-mito and mito fractions, respectively (Fig. 1a, upper panel). We quantified the purity of extra-mito and mito fractions by calculating the relative percentage of Xod or CytC in extra-mito and mito fractions after normalisation with total proteins as shown in Fig. 1a (bottom panel). Using ferrozine assays, we verified that in comparison with wild type (WT), MEFs from systemic knockout of Irp1 or Irp2 were iron deficient in both extra-mitochondrial and mitochondrial compartments (Fig. 1b). No significant difference between Irp1−/− and in Irp2−/− cells was observed. To further detect the cytosolic and mito labile iron pool (LIP, chelatable iron), we used both Calcein-AM and RPA, respectively, two well accepted iron probes, by monitoring the decay of these two fluorescent dyes. As shown in Fig. 1c, the cytosolic iron decreased significantly in both Irp1−/− and Irp2−/− cells comparing with WT cells, while the mito available iron content exhibited little if any changes (Fig. 1c).
Figure 1.
Irp1/Irp2 ablation induces cellular iron deficient, but not deficient in mitochondrial labile iron pool (LIP). (a) The purity of extra-mitochondrial (Extra-mito) and mitochondrial (Mito) fractions was analysed by Western blot. Xod and CytC were used as extra-mito and mito markers, respectively. A representative image set is presented. The purity of Extra-mito and Mito fractions was quantified by calculating the relative percentage of Xod or CytC in Extra-mito and Mito fractions after normalisation with total proteins (bottom panel). (b) Extra-mito and Mito total iron levels in mouse embryonic fibroblasts (MEFs) of Irp1−/−, Irp2−/−, and wild type (WT) measured by Ferrozine assays. (c) Levels of cytosolic (Cyt) and Mito LIP in MEFs of Irp1−/−, Irp2−/−, and WT measured by Calcein-AM and Rhodamine B-[(1, 10-phenanthroline-5-yl)-aminocarbonyl] benzyl ester (RPA), respectively. Values represent mean ± SEM (n = 8). A one-way ANOVA was performed. ***p < 0.001 compared with WT.
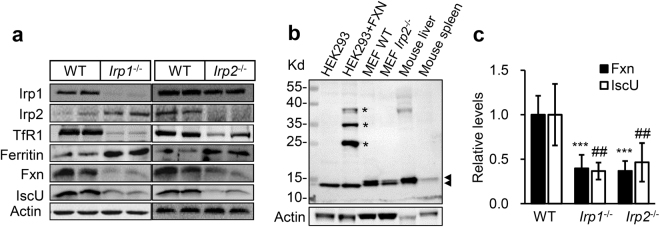
Since cellular iron homeostasis is mainly controlled by the IRP/IRE machinery in mammalian cells, we verified Irps-posttranscriptionally regulated target proteins ferritin and TfR1. In comparison with WT cells, the protein level of TfR1 drastically decreased and of ferritin increased in both Irp1−/− and Irp2−/− cells (Fig. 2a), which is consistent with the previous studies13,20. These results confirm that Irp1 or 2 deficiency reduces iron uptake and increases intracellular iron chelation, which both cause cellular iron deprivation.
Figure 2.
Irp1/Irp2-ablation drastically downregulates the components, Fxn and IscU, of Fe-S cluster biogenesis machinery in MEFs. (a) Western blot analysis of iron-related proteins including Irp1, Irp2, TfR1, Fxn, IscU, and ferritin in MEFs of Irp1−/−, Irp2−/−, and WT. Actin was used as a loading control. A representative image set is presented. (b) Verification of self-made FXN antibody. HEK293 + FXN: overexpressed 6×myc-tagged human full-length FXN in human cell line HEK293; Three asterisks indicate the precursor, intermediate, and mature forms of FXN; two arrows indicate the mouse and human endogenous mature forms of FXN. (c) Quantification of Fxn and IscU levels in MEFs of Irp1−/−, Irp2−/−, and WT. Values represent mean ± SEM (n = 3, each duplicates). A one-way ANOVA was performed. ##p < 0.01 for IscU, ***p < 0.001 for Fxn compared with WT.
Iron is revealed as a key regulator of mitochondrial biogenesis27 and expression of Fxn and IscU is regulated by iron28,29, we therefore examined whether Fxn and IscU levels were altered in Irp1−/− and Irp2−/− cells. We first confirmed our self-made anti-Fxn antibody30,31, then compared Fxn and IscU protein levels among WT, Irp1−/−, and Irp2−/− cells by immunoblotting. As shown in Fig. 2b, the antibody against Fxn was able to specifically detect the endogenous Fxn in both human and mouse samples as well as the overexpressed human FXN. When overexpressed, FXN (precursor, intermediate, and mature forms32,33 were clearly detected as marked with asterisks (Fig. 2b lane 2). Importantly, the levels of Fxn and IscU were markedly lower in both Irp1−/− and Irp2−/− cells than in WT cells (Fig. 2a), with quantification shown a more than 50% reduction (Fig. 2c). The data revealed that Irp deficiency resulted in a significant reduction in both Fxn and IscU levels.
Downregulation of the components of Fe-S biogenesis machinery in Irp1 and 2 deficient fibroblasts is specifically associated with impaired mitochondrial respiratory chain
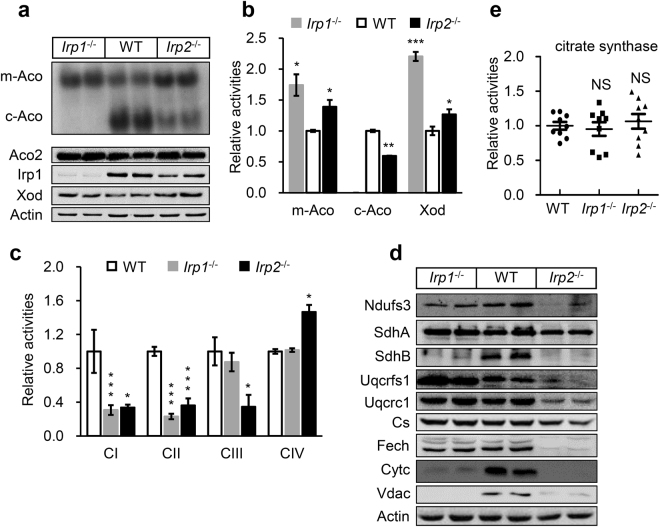
Both Fxn and IscU are known as two of core components for Fe-S cluster biogenesis. Reduction of both Fxn and IscU suggests that Fe-S cluster dependent pathways were likely defective in Irp1−/− and Irp2−/− cells. To ascertain whether this is the case, we first measured the activities of Fe-S cluster-dependent enzyme aconitase of WT, Irp1−/−, and Irp2−/− cells using an in-gel assay. As shown in Fig. 3a, the cytosolic aconitase activity (c-Aco) in Irp2−/− cells was indeed decreased compared with WT (Fig. 3a), and its protein level was lower in this mutant than in WT cells. Surprisingly, no reduction was detected for mitochondrial aconitase (m-Aco) in both Irp1−/− and Irp2−/− cells. Instead, both activities and protein levels were clearly higher in the mutants than in WT cells. Additionally, we examined the cytosolic Fe-S cluster-containing enzyme Xod in the Irp1−/− or Irp2−/− cells. Similar to m-Aco described above, the protein level and activity of Xod were also higher in Irp1−/− or Irp2−/− cells than in WT (Fig. 3a and b). Based on these data, it appears that Irp1 deficiency has similar effects to Irp2 deficiency on some of Fe-S enzymes with increased both protein level and activity.
Figure 3.
Downregulation of the components of Fe-S cluster biogenesis machinery in Irp1 and 2 deficient fibroblasts is specifically associated with impaired electron transport chain (ETC). (a) A representative graph of in-gel assays of mitochondrial (m-Aco, encoded by Aco2) and cytosolic (c-Aco, encoded by Irp1) aconitases in Irp deficient MEF cells (upper panel). The protein levels of Aco2, Irp1, and cytosolic Fe-S containing enzyme xanthine oxidative (Xod) were detected with western blotting. (b) The activities of Xod, m-Aco, and c-Aco were quantified. (c) Activities of ETC complexes were determined in Irp deficient MEFs. (d) Western blot analysis of mitochondrial proteins including Ndufs3 (a subunit of CI), SdhA and SdhB (subunits of CII), Uqcrc1 and Uqcrfs1 (subunits of CIII), Fech (matrix protein), CytC (intermembrane space protein), Vdac (outer membrane protein), and citrate synthase (Cs, matrix non-Fe-S protein). A representative image set is presented. (e) Activities of citrate synthase, which is a mitochondrial non-Fe-S enzyme, were determined. CI/CII/CIII/CIV: Complex I/II/III/IV. Values represent mean ± SEM (n = 10 for (e), n = 3, each duplicates for (b) and (c)). A one-way ANOVA was performed. *p < 0.05, **p < 0.01, ***p < 0.001 compared with WT.
However, iron deficiency or downregulation of Fe-S cluster biogenesis generally brings about the low activities of Fe-S cluster dependent enzymes25,34,35. We wonder if the phenotype we observed above is universal for Irp deficiency. Next, Fe-S cluster-richest and dependent ETC were analysed in the Irp1−/− and Irp2−/− cells. We measured the activities of complex I, II, III and IV. As shown in Fig. 3c, the activities of both complex I and II in the mutant cells were only about 40% of the WT level. The activity of complex III was reduced only in Irp2−/− cells, not in Irp1−/− cells. Complex IV activity was at the WT level in Irp1−/− cells and 30% higher in the Irp2−/− cells than in WT (Fig. 3c). The negative effects of Irp deficiency are specific on ETC complexes, which are known to be functionally dependent on Fe-S clusters except complex IV.
To further analyse the effects of Irp deficiency on complex I, II, and III, we also detected the levels of complex subunits by immunoblotting. As shown in Fig. 3d, the levels of Fe-S cluster-containing subunit Ndufs3 (Complex I) and SdhB (complex II) were lower in both Irp1−/− and Irp2−/− cells than in the WT cells. The level of SdhA, a non-Fe-S subunit that complexes with SdhB, was higher in Irp1−/− cells and lower in Irp2−/− cells than in WT cells. The expression of two subunits of complex III, Uqcrc1 (not containing Fe-S cluster) and Uqcrfs1 (Fe-S cluster-containing), was markedly decreased in Irp2−/− cells and mildly increased in Irp1−/− cells. To see a broader effect of Irp deficiency on mitochondrial biogenesis, we detected more other mitochondrial proteins, such as ferrochelatase (Fech, a matrix protein), CytC (an intermembrane protein), and Vdac (an outer membrane protein). The protein levels of CytC and Vdac were significantly decreased in both Irp1−/− and Irp2−/− cells compared with WT cells (Fig. 3d). However, the level of Fech was not changed in Irp1−/− cells and decreased in Irp2−/− cells (Fig. 3d). For comparison, we measured the enzymatic activity and protein level for citrate synthase, a critical and non-Fe-S cluster dependent enzyme in Krebs cycle. As shown in Fig. 3d and e, the activity and protein level in both Irp1−/− and Irp2−/− cells were similar to those in the WT. Taken together, these results reveal that Irp deficiency, particularly Irp2−/−, mainly affects Fe-S cluster dependent protein levels and activities of respiratory chain complexes.
The increased expression of FXN and ISCU in Irp1 and 2 deficient fibroblasts reverses the cellular iron content and restores the activities of complexes
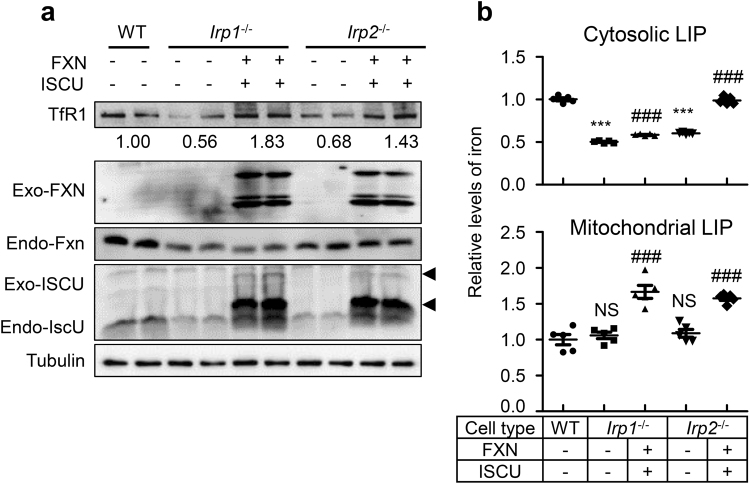
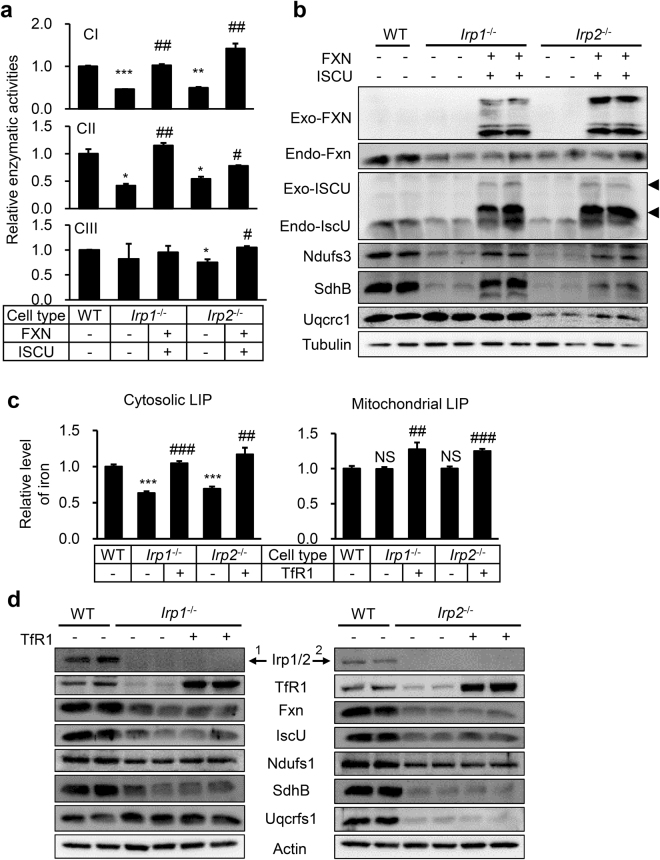
Because most of mitochondrial Fe-S cluster dependent ETC proteins are affected, we examined the role of IscU and Fxn downregulation in Irp depletion-induced malfunction of mitochondria. Human ISCU and FXN were expressed in Irp depleted cells to assess the effect on ETC basing on the conserved function of these proteins in human and rodents. Western blot analysis confirmed the successful expression of exogenous FXN and ISCU (Fig. 4a). Then, we measured the relative levels of cytosolic and mitochondrial available iron level after co-expression of FXN and ISCU. Very interestingly, both cytosolic and mitochondrial iron content increased (Fig. 4b), which was consistent with upregulated TfR1 (Fig. 4a). To determine whether overexpressed FXN and ISCU could improve the Fe-S cluster supply to mitochondrial ETC complexes in Irp deficient cells, we first measured the activities of complexes. The results showed that complex I and II activities remarkably increased in Irp1−/− and Irp2−/− cells (Fig. 5a, top and middle panels) and that complex III activity also increased in Irp2−/− cells after co-expression of FXN and ISCU (Fig. 5a, bottom panel). Next, we checked whether the increased activities of ETC complexes were coordinated with the expression levels of their subunits. The levels of all tested subunits were found to significantly increase in Irp1−/− and Irp2−/− cells after expression of exogenous FXN and ISCU (Fig. 5b). To further proof that the decrease of Fxn and IscU was not simply due to iron deficiency, we examined whether overexpression of TfR1 would improve the mitochondrial function in Irp1−/− and Irp2−/− cells. Although the cytosolic and mitochondrial labile iron pool increased after overexpression of TfR1 (Fig. 5c), the protein levels of Fxn and IscU were not changed and of mitochondrial complex subunits also kept constant (Fig. 5d). These results indicate that IRPs regulate respiratory chain function likely through modulating the expression of IscU and Fxn for Fe-S biogenesis and its targeted delivery.
Figure 4.
Co-expression of human FXN and ISCU in Irp deficient MEFs elevates the cellular iron content. (a) Western blot analysis of endogenous (Endo-) and exogenous (Exo-) FXN, ISCU, and TfR1 in MEF cells after co-transfection with pcDNA3.1-FXN-myc (encoding 6×myc-tagged human full-length of FXN) and pXS-ISCU-myc (encoding 1×myc-tagged human mitochondrial ISCU). A representative image set is presented. The arrows indicate the precursor and mature forms of exogenous ISCU. (b) The relative levels of cytosolic and mitochondrial LIP were measured with Calcein-AM and RPA, respectively (detail see Materials and Methods). Values represent mean ± SEM (n = 3, each duplicates for (a), n = 5 for (b)). A one-way ANOVA was performed. ***p < 0.001 compared with WT. ###p < 0.001 compared the expression-plasmid transfected cells with empty plasmid transfected cells (+FXN + ISCU vs. -FXN-ISCU).
Figure 5.
Co-expression of FXN and ISCU in Irp1 or 2 deficient MEFs rescues the mitochondrial function. (a) Activities of ETC CI, CII, and CIII were determined in Irp1 or Irp2 deficient MEFs after co-transfection with pcDNA3.1-FXN-myc and pXS-ISCU-myc (same plasmids as used in Fig. 4). Values represent mean ± SEM, n = 3, each duplicates. *p < 0.05, **p < 0.01, ***p < 0.001 compared with WT. #p < 0.05, ##p < 0.01 compared + FXN + ISCU with -FXN-ISCU. (b) Western blot analysis of endogenous and exogenous FXN and ISCU, and mitochondrial respiratory chain proteins including Ndufs3 (a subunit of CI), SdhB (a subunit of CII), Uqcrc1 (a subunit of CIII) in MEFs after co-transfection with pcDNA3.1-FXN-myc and pXS-ISCU-myc. The arrows indicate the precursor and mature forms of exogenous ISCU. A representative dataset is presented. (c) The relative levels of cytosolic and mitochondrial LIP were measured with Calcein-AM and RPA, respectively. Values represent mean ± SEM (n = 3, each duplicates). ***p < 0.001 vs. WT. ##p < 0.01, ###p < 0.001, +TfR1 vs. −TfR1. (d) Western blot analysis of iron related proteins including TfR1, Irp1 or Irp2, Fxn, IscU, and mitochondrial respiratory chain proteins including Ndufs1 (a subunit of CI), SdhB (a subunit of CII), Uqcrfs1 (a subunit of CIII) in MEFs after transfection with plasmid pcMV3-TfR1. A representative dataset is presented.
The co-expression of FXN and ISCU in Irp1 or Irp2-ablation cells improves mitochondrial integrity
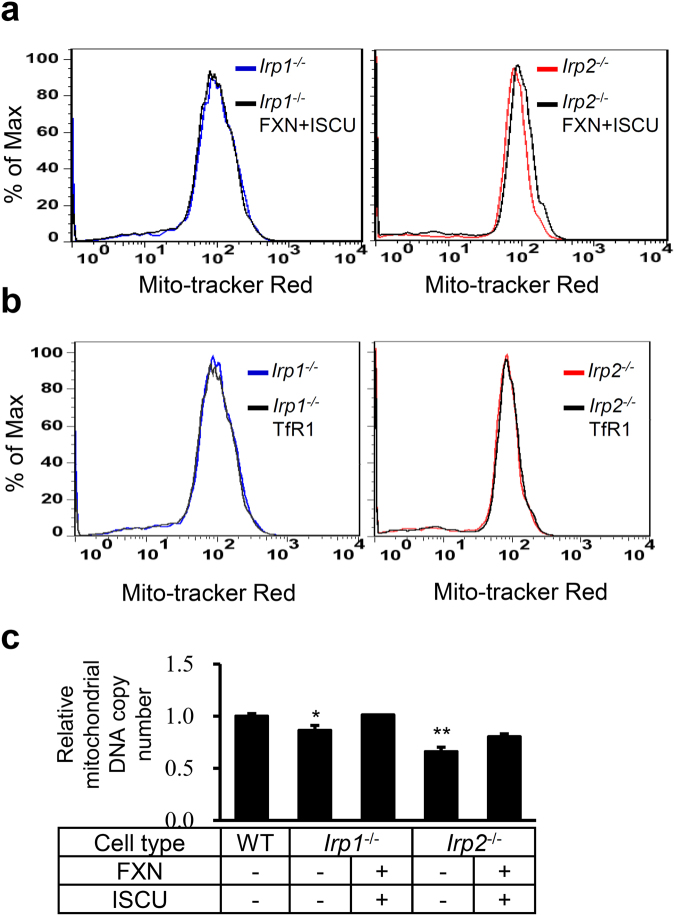
To further confirm the effect of co-expression of FXN and ISCU in Irp depletion cells on mitochondria, we examined the integrity and quantity of mitochondria by measuring the intensity of mito-tracker staining and the copy number of mitochondrial DNA (mtDNA), respectively. Mito-tracker stains mitochondria in living cells and its accumulation is dependent upon mitochondrial membrane potential (MMP), a subcellular marker for mitochondrial integrity. The intensity of mito-tracker staining represents the level of MMP, revealed by Flow cytometry. The data repeatedly showed a clear right shift of the peak representing increased MMP in Irp2−/− cells (Fig. 6a, right panel) after FXN and ISCU co-expression, but not in Irp1−/− cells (Fig. 6a, left panel). However, this shift was not observed when TfR was overexpressed (Fig. 6b). To estimate the mitochondrial quantity, we measured the relative copy number of mtDNA presented by the ratio of mtDNA to nuclear DNA (nDNA). Irp1 or Irp2 deficiency significantly reduced the relative mitochondrial DNA copy number. We did observe the alteration of the ratio in mutant cells between before and after FXN and ISCU co-expression, but without significance (Fig. 6c). Our data indicate that enhanced mitochondrial Fe-S cluster biogenesis by increasing expression of the core components improves the mitochondrial quality and quantity in Irp depletion cells.
Figure 6.
Overexpression of FXN and ISCU, but not TfR1, in Irp1 or 2 deficient MEFs improves the mitochondrial quality. MEFs were stained with Mito-tracker and analysed by Flow cytometry to represent the mitochondrial membrane potential (MMP) 24 h post transfection with plasmids pcDNA3.1-FXN-myc and pXS-ISCU-myc (a) or pcMV3-TfR1 (b). A representative graph is presented. (c) Relative mitochondrial DNA copy number after co-overexpression of FXN and ISCU in Irp ablation cells. n = 3, each duplicates. *p < 0.05, **p < 0.01 compared with WT.
Discussion
In this study, we report that Irp1 or Irp2-null mutation causes the decreased expression of Fxn and IscU, two important components of Fe-S cluster biogenesis machinery. Though Irp ablation down-regulates the expression of Fxn and IscU, the enzymatic activities of Fe-S cluster-containing aconitase and Xod were not diminished. Surprisingly, mitochondrial respiratory chain is severely impaired. Moreover, increasing the expression of FXN and ISCU reversed the Irp depletion-induced deficits including cellular iron content and activities of complex I, II, and III. Our results indicate that compromised function of the respiratory chain in response to Irp-depletion could be due to the downregulation of Fxn and IscU, which specifically reduces the acquisition of Fe-S cluster by respiratory complexes in MEFs.
Irp1 and Irp2 regulate the expression of target proteins posttranscriptionally by binding to IREs in transcripts that mostly encode iron metabolism proteins for iron uptake, storage, or export5,6. Their targeted deletion causes changes in the levels of their direct targets including reduced levels of TfR and increased levels of ferritin in tissues12,19,20. In current in vitro study, the decreased TfR1 and increased ferritin were verified in each Irp deficient MEFs, to a severe extent in Irp1−/− cells. Our results support the notion that both Irps are important contributors as iron regulatory proteins, giving priority to different cell types or tissues12–15.
Iron deprivation could strongly impair the mitochondrial respiratory chain and mitochondrial biogenesis27,36–38. Irp ensures adequate cellular iron supply that includes supply for proper mitochondrial function21, in which work both Irp1 and Irp2 were silenced in hepatocytes. The individual crucial roles of Irp122 and Irp220,39 in mitochondrial function have also been raised. Here we provide more evidence to verify the important roles of Irp1 and Irp2 for functional mitochondria. We speculate that the impaired mitochondrial function in Irp deficient cells may be caused by insufficient Fe-S cluster biogenesis due to the reduced expression of components, namely Fxn and IscU, of Fe-S synthesis machinery. Unexpectedly, the activities of mitochondrial aconitase and Xod, two Fe-S cluster-containing enzymes encoded by Aco2 and Xod, respectively, increased under normal culture condition. This could be, only partially, explained by the modestly increased protein levels of both Aco2 and Xod. Indeed, Aco2 has an IRE localized in the 5′-UTR of its mRNA40. Under the condition of Irp depletion-induced iron deprivation, the increased expression of Aco2 suggested that both Irps contribute to the regulation of Aco2. The increased activities of m-Aco and Xod could be also due to the increase of HIF41,42, one of which subunits, HIF1/2a, might be stabilized in IRP deficient cells due to the low level of iron43,44. However, the Fe-S cluster cofactor is essential for these enzymes to gain the enzymatic activities. Alternatively, Fe-S cluster might be stabilized under the iron deficient condition. The increased activities suggest that mitochondrial iron supply be not significantly limited in Irp depletion cells though severe cellular iron deficiency was observed. This result was confirmed by that WT and Irp mutants exhibited comparable available iron access in mitochondria.
To further clarify whether the decreased Fxn and IscU in Irp1 or Irp2 deficiency cells impaired other Fe-S enzymes, we detected the activities of ETC complexes, which contain several Fe-S clusters as important prosthetic groups for electron transport function. Our in vitro data showed significantly decreased complex I and II activities in both Irp-ablation cells. Similar results were previously observed in vivo in Irp2−/− mice20. This impairment might involve HSC20, an Fe-S cluster co-chaperone protein. FXN interacts with mitochondrial HSC20, and this interaction is iron-dependent45. HSC20 also interacts with multiple proteins involved in Fe-S biogenesis including the ISCU/Nfs1 complex and the chaperone GRP7545. Recent studies have shown that HSC20 guides selection of Fe-S cluster delivery by binding to a conserved leucine- tyrosine- arginine (LYR) motif presented in specific recipient Fe-S proteins or in accessory factors that likely assist Fe-S cluster insertion into target apoproteins46. Majority of HSC20 interactants are the components of complex I and II, including NDUFA6 and NDUFB9, DNUFS1/7/8, NDUFV1/2, SDHAF1, SDHB and more47,48. These suggest that IRP-deficiency may not only impair Fe-S biogenesis but also the HSC20-mediated delivery.
In this study we also found decreased complex III activity in addition to the decreased complex I and II activities in Irp2−/− MEFs. Different from Irp2−/− cells, Irp1−/− cells only showed defects of complex I and II activities. Expression of Uqcrc1 and Uqcrfs1, two subunits of complex III (non-Fe-S cluster and Fe-S cluster-containing proteins, respectively), dramatically decreased in Irp2−/− cells, while Irp1−/− cells even expressed slightly more Uqcrc1 and Uqcrfs1 than WT cells to maintain the complex III activity. Given the instability of proteins devoid of their Fe-S49,50 as for SdhB and Ndufs3 in this study due to the iron starvation, a compensatory mechanism would raise the expression of non-Fe-S cluster-containing proteins as for SdhA and Uqcrc1. This was only observed in Irp1−/− cells, not in Irp2−/− cells. We did not check the expression of SDHAF1, a LYR complex-II specific assembly factor, important for SDH activity by interaction with SDHB51. The pathogenic mutations of SDHAF1 abrogate binding to SDHB, which impairs biogenesis of holo-SDHB and results in LONP1-mediated degradation of SDHB52. The decreased expression of SdhA in Irp2−/− cells might also induce downregulation of SdhB protein expression53. Alternatively, Irp ablation-induced iron deprivation might induce a decrease at mRNA levels of the components of complexes through dynamic alterations of histone acetylation and methylation27,37. Thus, Irp ablation not only affects cellular iron content, but also affects the expression of components of Fe-S biogenesis machinery and subunits of ETC complexes to further trigger the deficiency of complex activities. This selective effect of Irp2 depletion on mitochondrial respiratory chain might partially explain the symptoms of neurological disorders in Irp2−/− mice11,16,20,26. However, overexpression of human FXN and ISCU in Irp1−/− and Irp2−/− cells significantly improves the mitochondrial function and recovers the deficits of ETC complex subunits. Curiously, genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension54, the same preclinical phenotype raised from Irp1 depletion13. Irp1 activation sustains mitochondrial iron supply and function rather than driving detrimental iron overload in Fxn deficient mice22. These results support that the reduced expression of Fxn and IscU is involved in the effects of Irp1 and Irp2 deficiency on impaired mitochondrial function to further cause the phenotypes in Irp1−/−and Irp2−/− mice.
Collectively, our data provide in vitro evidence that depletion of Irp1 or Irp2 downregulates the expression of Fxn and IscU and specifically compromises the activities of Fe-S cluster-containing ETC in MEFs. The current results reveal the role of Irp in securing mitochondrial function through regulating the expression of the core components, Fxn and IscU, of Fe-S cluster biogenesis machinery. Our data also imply that Irps specifically tailor Fe-S delivery for mitochondrial ETC complexes.
Materials and Methods
Cell culture and transfection
All media and reagents for cell culture were purchased from Invitrogen (Shanghai, China). MEFs (generously given by Dr. Tracey Rouault) derived from wild type (WT) and global Irp1 and Irp2 deficient mice were cultured in DMEM medium with 10% heat inactivated fetal bovine serum, 4 mM glutamine, penicillin, and streptomycin. All cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. For transfection, HG-Trans293TM transfection reagent (Genomeditech, Shanghai, China) was used according to the supplier’s manuals. The transfected plasmids included pXS-ISCU-myc, pcDNA3.1-FXN-myc, and pcMV3-TfR1 for overexpression of ISCU, FXN, and TfR, respectively. Cells were harvested 24 h post transfection for further analysis.
Subcellular fractionation
Mitochondria were isolated from cultured WT, Irp1−/−, and Irp2−/− cells, respectively, with the specialized cell mitochondrial isolation kit (Beyotime, Jiangsu, China) following the manufacturer’s instructions. Briefly, cells were harvested in mitochondrial isolation buffer with PMSF and transferred to a grinder for crushing until the isolation became homogeneous. The mitochondria were isolated by differential centrifugation at 600 g and 3500 g for 10 min at 4 °C, respectively. The latter pellet was washed with PBS one time, then stored in mitochondrial lysis buffer as mitochondrial fraction. The latter supernatant was transferred into new EP tubes and centrifuged at 12000 g for 10 min at 4 °C. This supernatant was considered as extra-mitochondrial fraction.
Ferrozine iron assays and labile iron pool (LIP) measurement
Iron content was measured using a colorimetric ferrozine-based assay with some modifications55. Briefly, 22 μl concentrated HCl (11.6 mol/L) was added to 100 μl cell lysate (~500 μg total protein). The mixed sample was heated at 95 °C for 20 min, then centrifuged at 12,000 g for 10 min. Supernatant was transferred very gently into fresh tubes. Ascorbate was added to reduce the Fe (III) into Fe (II). After 2 min of incubation at room temperature, ferrozine and saturate ammonium acetate (NH4Ac) were sequentially added to each tube and the absorbance was measured at 570 nm (BioTek EL x 800, Shanghai, China) within 30 min.
Labile iron was measured using the iron-sensitive probes Calcein-AM (Aladdin, Shanghai, China) and Rhodamine B-[(1, 10-phenanthroline-5-yl)-aminocarbonyl] benzyl ester (RPA, Squarix GmbH, Elbestr, Germany). Briefly, 2 × 105 WT, Irp1−/−, and Irp2−/− cells were incubated with 10 μM Calcein-AM for 10 min at 37 °C in PBS and then washed one time with PBS buffer. Cells were obtained in 100 μl PBS. Cytosolic LIP was measured using fluorescent microplate reader at 495 nm (excitation) and 530 nm (emission). For mitochondrial LIP, Cells were incubated with 10 μM RPA for 15 min at 37 °C in HBSS, then incubated with 100 μl HBSS buffer for 15 min following washing once with HBSS. Mitochondrial iron was measured at 543 nm (excitation) and 601 nm (emission) using fluorescent microplate reader.
Western blot analysis
Proteins from lysates were prepared and resolved by 12% SDS-PAGE at 100 V, transferred for 1.5 h at 250 mA onto Nitrocellulose membranes, and analysed by immunoblotting as described previously56. The information for primary antibodies is as follows: anti-ferritin (cat# 69090), SdhA (cat# 137040), and SdhB (cat# 178423) from Abcam (Cambridge, MA, USA), anti-Xod (cat# 55156-1-AP), citrate synthase (cat# 16131-1-AP), aconitase 2 (Aco2) (cat# 11134-1-AP), Ndufs1 (cat# 12444-1-AP), Ndufs3 (cat# 15066-1-AP), Uqcrc1 (cat# 21705-1-AP), and Uqcrfs1 (cat# 1843-1-AP) from Proteintech Group Inc. (Chicago, IN, USA), anti-Actin (cat# BM0627) from Boster (Wuhan, China), anti-Tubulin (cat# T0198) from Sigma-Aldrich (St. Louis, MO, USA), anti-TfR1 antibody (cat# 136800) from Zymed (San Francisco, CA, USA), anti-IscU, Fech, Irp1, and Irp2, anti-Fxn (self-made)31, anti-CytC (cat# 1896-1) from Epitmics (Burlingame, CA, USA), anti-Vdac (cat# 4661S) from Cell Signaling Technology Inc (Shanghai, China).
Enzymatic activities
In-gel aconitase activity assays were performed as described previously28. Related chemicals used were purchased from Sigma-Aldrich. The activities of complex I, II, III, and IV, Xod, and citrate synthase were measured following the manufacturer’s protocols, respectively. Purchase information is as follows: Complex I from Abcam, Complex II, Complex IV, and Citrate synthase from Comin Biotechnology Co. (Suzhou, Jiangsu, China), Complex III from Biovision Inc. (Milpitas, CA, USA), Xod from Nanjing Jiancheng Bioengineering Institute (Nanjing, China).
Flow cytometric analysis
WT, Irp1−/−, and Irp2−/− cells were incubated with Mito-tracker Red for 5 min at room temperature in PBS and then washed twice with PBS. Cells were harvested, pelleted by centrifugation and resuspended in PBS. The fluorescence intensity was measured by flow cytometry (BD, Franklin, NJ) and data were analysed using Flowjo software.
Quantitative real-time PCR (qRT-PCR)
Total DNA was prepared with QiaAmp DNA mini kit (Qiagen, Germantown, MD) according to the manufacturer’s instructions. qRT-PCR experiments were performed with SYBR Green PCR master mixture (Thermo Fisher Scientific). The relative copy number of mtDNA was determined by comparing the copy number of mtDNA-locating CytB to that of nDNA-locating Act. The used primer sequences (5′–3′) were: TTCATGTCGGACGAGGCTTA and CTGTGGCACCTCAGAATGAT for mouse CytB; ATAAGTGGCCTTGGAGTGTG and GTACGACCAGAGGCATACAG for mouse Act.
Statistical analysis
The values were expressed as mean ± SEM from, at least, three independent experiments. A one-way analysis of variance (ANOVA) was carried out using SPSS ver. 22.0 software (IBM Corporation, Armonk, NY, USA). Significance was considered at p < 0.05.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Acknowledgements
This work was supported by the National Basic Research Program of China (grant number: 2015CB856300) and by the National Natural Science foundation of China (grant numbers: 31571218, 31371060).
Author Contributions
H.L. and K.L. conceived and designed experiments. H.L., H.Z., S.H., L.S., C.S., and J.W. performed experiments. K.L., Q.T., and E.G.M. contributed to data analysis. H.L. and K.L. wrote the manuscript. All authors read and approved the final manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Youdim MB. Brain iron deficiency and excess; cognitive impairment and neurodegeneration with involvement of striatum and hippocampus. Neurotox. Res. 2008;14:45–56. doi: 10.1007/BF03033574. [DOI] [PubMed] [Google Scholar]
- 2.Rao R, Georgieff MK. Iron in fetal and neonatal nutrition. Semin. Fetal. Neonatal. Med. 2007;12:54–63. doi: 10.1016/j.siny.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rouault TA. Iron metabolism in the CNS: implications for neurodegenerative diseases. Nat. Rev. Neurosci. 2013;14:551–564. doi: 10.1038/nrn3453. [DOI] [PubMed] [Google Scholar]
- 4.Anderson CP, Shen M, Eisenstein RS, Leibold EA. Mammalian iron metabolism and its control by iron regulatory proteins. Biochim. Biophys. Acta. 2012;1823:1468–1483. doi: 10.1016/j.bbamcr.2012.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142:24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 6.Rouault TA. The role of iron regulatory proteins in mammalian iron homeostasis and disease. Nat. Chem. Biol. 2006;2:406–414. doi: 10.1038/nchembio807. [DOI] [PubMed] [Google Scholar]
- 7.Moroishi T, Nishiyama M, Takeda Y, Iwai K, Nakayama KI. The FBXL5-IRP2 Axis Is Integral to Control of Iron Metabolism In Vivo. Cell. Metab. 2011;14:339–351. doi: 10.1016/j.cmet.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 8.Salahudeen AA, et al. An E3 Ligase Possessing an Iron-Responsive Hemerythrin Domain Is a Regulator of Iron Homeostasis. Science. 2009;326:722–726. doi: 10.1126/science.1176326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vashisht AA, et al. Control of iron homeostasis by an iron-regulated ubiquitin ligase. Science. 2009;326:718–721. doi: 10.1126/science.1176333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galy B, Ferring D, Hentze MW. Generation of conditional alleles of the murine Iron Regulatory Protein (IRP)-1 and -2 genes. Genesis. 2005;43:181–188. doi: 10.1002/gene.20169. [DOI] [PubMed] [Google Scholar]
- 11.LaVaute T, et al. Targeted deletion of the gene encoding iron regulatory protein-2 causes misregulation of iron metabolism and neurodegenerative disease in mice. Nat. Genet. 2001;27:209–214. doi: 10.1038/84859. [DOI] [PubMed] [Google Scholar]
- 12.Meyron-Holtz EG, et al. Genetic ablations of iron regulatory proteins 1 and 2 reveal why iron regulatory protein 2 dominates iron homeostasis. Embo. J. 2004;23:386–395. doi: 10.1038/sj.emboj.7600041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghosh MC, et al. Deletion of iron regulatory protein 1 causes polycythemia and pulmonary hypertension in mice through translational derepression of HIF2alpha. Cell. Metab. 2013;17:271–281. doi: 10.1016/j.cmet.2012.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkinson N, Pantopoulos K. IRP1 regulates erythropoiesis and systemic iron homeostasis by controlling HIF2alpha mRNA translation. Blood. 2013;122:1658–1668. doi: 10.1182/blood-2013-03-492454. [DOI] [PubMed] [Google Scholar]
- 15.Anderson SA, et al. The IRP1-HIF-2alpha axis coordinates iron and oxygen sensing with erythropoiesis and iron absorption. Cell. Metab. 2013;17:282–290. doi: 10.1016/j.cmet.2013.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooperman SS, et al. Microcytic anemia, erythropoietic protoporphyria, and neurodegeneration in mice with targeted deletion of iron-regulatory protein 2. Blood. 2005;106:1084–1091. doi: 10.1182/blood-2004-12-4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galy B, et al. Altered body iron distribution and microcytosis in mice deficient in iron regulatory protein 2 (IRP2) Blood. 2005;106:2580–2589. doi: 10.1182/blood-2005-04-1365. [DOI] [PubMed] [Google Scholar]
- 18.Galy B, et al. Iron homeostasis in the brain: complete iron regulatory protein 2 deficiency without symptomatic neurodegeneration in the mouse. Nat. Genet. 2006;38:967–969. doi: 10.1038/ng0906-967. [DOI] [PubMed] [Google Scholar]
- 19.Zumbrennen-Bullough KB, et al. Abnormal Brain Iron Metabolism in Irp2 Deficient Mice Is Associated with Mild Neurological and Behavioral Impairments. PloS one. 2014;9:e98072. doi: 10.1371/journal.pone.0098072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong SY, et al. Iron Insufficiency Compromises Motor Neurons and Their Mitochondrial Function in Irp2-Null Mice. PloS one. 2011;6:e25404. doi: 10.1371/journal.pone.0025404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galy B, et al. Iron regulatory proteins secure mitochondrial iron sufficiency and function. Cell. Metab. 2010;12:194–201. doi: 10.1016/j.cmet.2010.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Martelli A, et al. Iron regulatory protein 1 sustains mitochondrial iron loading and function in frataxin deficiency. Cell. Metab. 2015;21:311–322. doi: 10.1016/j.cmet.2015.01.010. [DOI] [PubMed] [Google Scholar]
- 23.Chiang S, et al. Frataxin and the molecular mechanism of mitochondrial iron-loading in Friedreich’s ataxia. Clin. Sci. 2016;130:853–870. doi: 10.1042/CS20160072. [DOI] [PubMed] [Google Scholar]
- 24.Campuzano V, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271:1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 25.Mochel F, et al. Splice mutation in the iron-sulfur cluster scaffold protein ISCU causes myopathy with exercise intolerance. Am. J. Hum. Genet. 2008;82:652–660. doi: 10.1016/j.ajhg.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith SR, et al. Severity of neurodegeneration correlates with compromise of iron metabolism in mice with iron regulatory protein deficiencies. Ann. N. Y. Acad. Sci. 2004;1012:65–83. doi: 10.1196/annals.1306.006. [DOI] [PubMed] [Google Scholar]
- 27.Rensvold JW, et al. Complementary RNA and protein profiling identifies iron as a key regulator of mitochondrial biogenesis. Cell. Rep. 2013;3:237–245. doi: 10.1016/j.celrep.2012.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tong WH, Rouault TA. Functions of mitochondrial ISCU and cytosolic ISCU in mammalian iron-sulfur cluster biogenesis and iron homeostasis. Cell. Metab. 2006;3:199–210. doi: 10.1016/j.cmet.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 29.Li K, Besse EK, Ha D, Kovtunovych G, Rouault TA. Iron-dependent regulation of frataxin expression: implications for treatment of Friedreich ataxia. Hum. Mol. Genet. 2008;17:2265–2273. doi: 10.1093/hmg/ddn127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hao S, et al. Phosphorylation of Akt by SC79 Prevents Iron Accumulation and Ameliorates Early Brain Injury in a Model of Experimental Subarachnoid Hemorrhage. Molecules. 2016;21:325. doi: 10.3390/molecules21030325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hao S, Xu F, Li K. [Production and application of polyclonal antibody against mouse frataxin] Sheng wu gong cheng xue bao = Chinese journal of biotechnology. 2013;29:1313–1322. [PubMed] [Google Scholar]
- 32.Xia HY, et al. Novel Frataxin Isoforms May Contribute to the Pathological Mechanism of Friedreich Ataxia. PloS one. 2012;7(ARTN):e47847. doi: 10.1371/journal.pone.0047847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schmucker S, Argentini M, Carelle-Calmels N, Martelli A, Puccio H. The in vivo mitochondrial two-step maturation of human frataxin. Hum.Mol.Genet. 2008;17:3521–3531. doi: 10.1093/hmg/ddn244. [DOI] [PubMed] [Google Scholar]
- 34.Pain D, Dancis A. Roles of Fe-Sproteins: from cofactor synthesis to iron homeostasis to protein synthesis. Curr. Opin. Genet. Dev. 2016;38:45–51. doi: 10.1016/j.gde.2016.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rotig A, et al. Aconitase and mitochondrial iron-sulphur protein deficiency in Friedreich ataxia. Nat. Genet. 1997;17:215–217. doi: 10.1038/ng1097-215. [DOI] [PubMed] [Google Scholar]
- 36.Cartier LJ, Ohira Y, Chen M, Cuddihee RW, Holloszy JO. Perturbation Of Mitochondrial Composition In Muscle by Iron-Deficiency - Implications Regarding Regulation Of Mitochondrial Assembly. J. Biol. Chem. 1986;261:3827–3832. [PubMed] [Google Scholar]
- 37.Rensvold JW, Krautkramer KA, Dowell JA, Denu JM, Pagliarini DJ. Iron Deprivation Induces Transcriptional Regulation of Mitochondrial Biogenesis. J. Biol. Chem. 2016;291:20827–20837. doi: 10.1074/jbc.M116.727701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willis WT, Dallman PR. Impaired control of respiration in iron-deficient muscle mitochondria. Am. J.Physiol. 1989;257:C1080–1085. doi: 10.1152/ajpcell.1989.257.6.C1080. [DOI] [PubMed] [Google Scholar]
- 39.Cloonan SM, et al. Mitochondrial iron chelation ameliorates cigarette smoke-induced bronchitis and emphysema in mice. Nat. Med. 2016;22:163–174. doi: 10.1038/nm.4021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HY, LaVaute T, Iwai K, Klausner RD, Rouault TA. Identification of a conserved and functional iron-responsive element in the 5’-untranslated region of mammalian mitochondrial aconitase. J. Biol. Chem. 1996;271:24226–24230. doi: 10.1074/jbc.271.39.24226. [DOI] [PubMed] [Google Scholar]
- 41.Tsui KH, et al. Hypoxia upregulates the gene expression of mitochondrial aconitase in prostate carcinoma cells. J. Mol. Endocrinol. 2013;51:131–141. doi: 10.1530/JME-13-0090. [DOI] [PubMed] [Google Scholar]
- 42.Abooali M, et al. Crucial involvement of xanthine oxidase in the intracellular signalling networks associated with human myeloid cell function. Sci. Rep. 2014;4:6307. doi: 10.1038/srep06307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ribas V, Garcia-Ruiz C, Fernandez-Checa JC. Mitochondria, cholesterol and cancer cell metabolism. Clin. Transl. Med. 2016;5:22. doi: 10.1186/s40169-016-0106-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kaelin WG, Jr., Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Shan YX, Cortopassi G. HSC20 interacts with frataxin and is involved in ironsulfur cluster biogenesis and iron homeostasis. Hum. Mol. Genet. 2012;21:1457–1469. doi: 10.1093/hmg/ddr582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maio N, et al. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell. Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: New insights into the molecular mechanisms of cluster delivery. Bba-Mol. Cell. Res. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maio N, Kim KS, Singh A, Rouault TA. A Single Adaptable Cochaperone-Scaffold Complex Delivers Nascent Iron-Sulfur Clusters to Mammalian Respiratory Chain Complexes I-III. Cell metabolism. 2017;25(945–953):e946. doi: 10.1016/j.cmet.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guillon B, et al. Frataxin deficiency causes upregulation of mitochondrial Lon and ClpP proteases and severe loss of mitochondrial Fe-S proteins. Febs J. 2009;276:1036–1047. doi: 10.1111/j.1742-4658.2008.06847.x. [DOI] [PubMed] [Google Scholar]
- 50.Sheftel AD, et al. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell. 2012;23:1157–1166. doi: 10.1091/mbc.E11-09-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ghezzi D, et al. SDHAF1, encoding a LYR complex-II specific assembly factor, is mutated in SDH-defective infantile leukoencephalopathy. Nat. Genet. 2009;41:654–656. doi: 10.1038/ng.378. [DOI] [PubMed] [Google Scholar]
- 52.Maio N, et al. Disease-Causing SDHAF1 Mutations Impair Transfer of Fe-S Clusters to SDHB. Cell. Metab. 2016;23:292–302. doi: 10.1016/j.cmet.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pantaleo MA, et al. Analysis of all subunits, SDHA, SDHB, SDHC, SDHD, of the succinate dehydrogenase complex in KIT/PDGFRA wild-type GIST. Eur. J. Hum. Genet. 2014;22:32–39. doi: 10.1038/ejhg.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.White K, et al. Genetic and hypoxic alterations of the microRNA-210-ISCU1/2 axis promote iron-sulfur deficiency and pulmonary hypertension. EMBO. Mol. Med. 2015;7:695–713. doi: 10.15252/emmm.201404511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Riemer J, Hoepken HH, Czerwinska H, Robinson SR, Dringen R. Colorimetric ferrozine-based assay for the quantitation of iron in cultured cells. Anal. Biochem. 2004;331:370–375. doi: 10.1016/j.ab.2004.03.049. [DOI] [PubMed] [Google Scholar]
- 56.Yan H, Zhang D, Hao S, Li K, Hang CH. Role of Mitochondrial Calcium Uniporter in Early Brain Injury After Experimental Subarachnoid Hemorrhage. Mol. Neurobiol. 2015;52:1637–1647. doi: 10.1007/s12035-014-8942-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.