Abstract
Current knowledge on the genetic basis of nonalcoholic fatty liver disease (NAFLD) suggests that variants contributing not only to the disease predisposition but histological severity as well are located in genes that regulate lipid metabolism. We explored the role of rs641738 C/T located in TMC4 (transmembrane channel-like 4) exon 1 (p.Gly17Glu) and 500 bases- downstream of MBOAT7 gene (TMC4/MBOAT7), in the genetic risk for developing NAFLD in a case-control study. Our sample included 634 individuals (372 patients with NAFLD diagnosed by liver biopsy and 262 control subjects); genotyping was performed by a Taqman assay. Genotype frequencies in controls (CC: 84, CT: 137, TT: 41) and patients (CC: 134, CT: 178, TT: 60) were in Hardy-Weinberg equilibrium; minor allele frequency 40.8%. Our sample had 84–99% power if an additive genetic model is assumed for estimated odds ratios of 1.3–1.5, respectively. We found no evidence of association between rs641738 and either NAFLD (Cochran-Armitage test for trend, p = 0.529) or the disease severity (p = 0.61). Low levels of MBOAT7 protein expression were found in the liver of patients with NAFLD, which were unrelated to the rs641738 genotypes. In conclusion, the role of rs641738 in the pathogenesis of NAFLD is inconclusive.
Introduction
Current understanding of the genetic basis of nonalcoholic fatty liver disease (NAFLD) suggests that variants contributing not only to the disease susceptibility but also histological severity are located in genes that regulate lipid metabolism. Specifically, the missense p.Ile148Met (rs738409) variant located in PNPLA3 (patatin-like phospholipase domain-containing 3) has been consistently associated with increased liver fat content and NAFLD severity, including fibrosis, across different populations around the world1–3. The risk effect of rs738409 on developing NAFLD is the strongest ever reported for a common variant modifying the genetic susceptibility of the disease, representing ~5.3% of the total variance2,3 and a moderate odds ratio (OR) of NAFLD and NASH of ~ 3.33. Likewise, the missense p.Glu167Lys (rs58542926) variant located in TM6SF2 (transmembrane 6 superfamily member 2) gene, while protecting against cardiovascular disease (CVD)4,5, has been associated with a modest risk of liver fat accumulation (OR ~2.13)5, NAFLD, and the NAFLD severity6–11.
Interestingly, a missense (p.Gly17Glu, rs641738 C/T) variant located in exon 1 of TMC4 (transmembrane channel-like 4) gene and intergenic downstream of MBOAT7 gene has been associated with a modest risk of developing NAFLD (OR ~1.37), NASH, and fibrosis12. However, these findings were based on a large report involving patients of European descent12. Nonetheless, the authors observed that the effect of rs641738 was restricted to European-Caucasian individuals, while not being significant in African American and Hispanic population12. Unfortunately, the association of rs641738 and NAFLD could not be replicated in other populations around the world, including Europeans from different cohorts, except for a small study that included cases-only (n = 125)13. For instance, a recent study including a large sample (n = 515) of patients with NAFLD recruited from several centers across Germany showed that rs641738 was associated with a marginal effect on liver fibrosis (p = 0.046) without any effect on NAFLD or liver function test14. Similarly, results yielded by analyzing the data pertaining to a small cohort of patients that underwent bariatric surgery in two European centers failed to confirm any association of rs641738 and NAFLD15. Likewise, studies from Asia failed to find an association of the variant with NAFLD or NASH16–18.
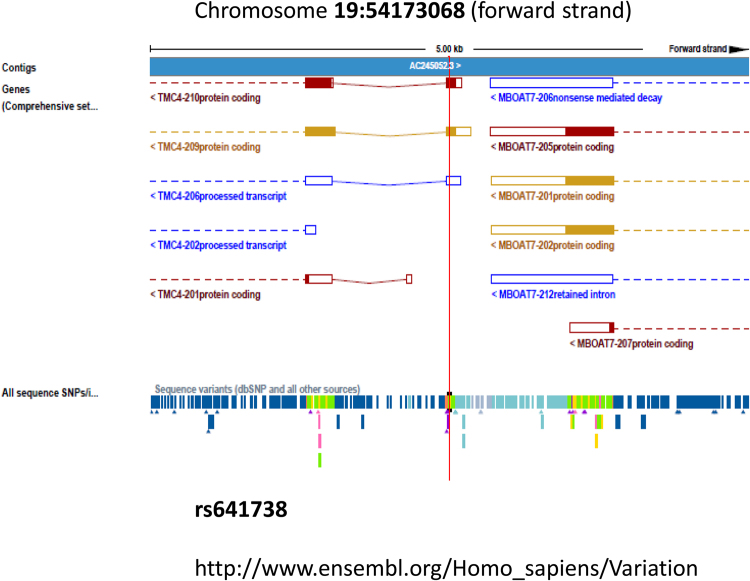
The rs641738 variant is mapped 500 bases downstream of MBOAT7 (membrane-bound O-acyltransferase domain-containing 7) locus (https://www.ncbi.nlm.nih.gov/snp/rs641738). Likewise, data from the genome assembly shows this variant located in exon 1 of the TMC4 genomic region (19:54173068, GRCh38.p7 assembly) (Fig. 1). Annotation details provided by The Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org/), shows that rs641738 as a transcript variant of TMC4 locus. For that reason, previous reports refer to rs641738 as MBOAT7 variant or TMC4/MBOAT7 (https://www.ncbi.nlm.nih.gov/snp/rs641738).
Figure 1.
Genomic location of rs641738 in the forward strand of chromosome 19: 54,173,068. Figure shows the genomic assembly as a blue bar composed of individual contigs; rs641738 (outlined by a vertical red line) is shown in a 5 kb region along with surrounding variations. TMC4 locus is located in chromosome 19: 54,160,168–54,173,171; MBOAT7 locus is located in chromosome 19: 54,173,412–54,189,443.
In addition, speculations on the putative biological role of MBOAT7 in the pathogenesis of NAFLD still persist because the protein encoded by this gene is a lysophosphatidylinositol acyltransferase, which has specificity for arachidonoyl-CoA as an acyl donor.
Combined, available evidence suggests that associations of rs641738 with NAFLD and NASH remain to be either confirmed or refuted. Hence, we performed a hospital-based case-control study to explore the association between rs641738 and NAFLD, including adult patients in whom the histological disease severity was confirmed by liver biopsy.
In addition, we explored the protein expression pattern of MBOAT7 in the liver of patients with NAFLD to provide evidence of whether the protein encoded by this locus might be involved in the biology of the disease.
Results
The rs641738 is not associated with NAFLD or the histological disease severity
Clinical and biochemical features of patients and controls are disclosed in Tables 1 and 2.
Table 1.
Clinical and biochemical features of patients and controls in the cross-sectional study of patients with NAFLD and Metabolic Syndrome (MetS).
| Variables | Control subjects | NAFL | NASH |
|---|---|---|---|
| Number of subjects | 241 | 113 | 153 |
| Female, % | 46 | 56 | 67 |
| Age, years | 47 ± 14 | 54 ± 0.8 | 50 ± 0.9 |
| BMI, kg/m2 | 25 ± 4.2 | 31.5 ± 5.4# | 34 ± 6.0+,* |
| Fasting plasma glucose, mg/dL | 84 ± 15 | 97 ± 22# | 129 ± 119+,* |
| Fasting plasma insulin, μU/ml | 6.8 ± 4.7 | 13 ± 9# | 16 ± 11+,* |
| HOMA-IR index | 1.4 ± 1.0 | 3 ± 2.1# | 5.1 ± 6.6+,* |
| Total cholesterol, mg/dL | 205 ± 42 | 207 ± 53 | 211 ± 43 |
| HDL-cholesterol, mg/dL | 57 ± 16 | 53 ± 25 | 50 ± 14 |
| LDL-cholesterol, mg/dL | 123 ± 38 | 126 ± 48 | 125 ± 42 |
| Triglycerides, mg/dL | 120 ± 77 | 154 ± 76# | 192 ± 119+ |
| ALT, U/L | 20 ± 6.0 | 56 ± 60# | 73 ± 54+* |
| AST, U/L | 17.5 ± 6.6 | 35 ± 17# | 51 ± 33+* |
| Histological Features | |||
| Degree of steatosis, % | — | 47 ± 25 | 61 ± 21* |
| Lobular inflammation (0–3) | — | 0.6 ± 0.6 | 1.2 ± 0.6* |
| Hepatocellular ballooning (0–2) | — | 0.03 ± 0.19 | 0.9 ± 0.6* |
| Fibrosis Stage | — | 0.03 ± 0.3 | 1.4 ± 1.24* |
| NAFLD activity score (NAS) | — | 2.6 ± 1.1 | 4.5 ± 1.4* |
NAFL: nonalcoholic fatty liver, NASH: nonalcoholic steatohepatitis BMI: body mass index; HOMA: homeostatic model assessment; ALT and AST: Serum alanine and aspartate aminotransferase. Results are expressed as mean ± SD.
#p < 0.001 Indicates NAFL vs. controls, *p < 0.001 indicates comparisons between NAFL and NASH, and +p < 0.001 denotes comparisons between NASH and control subjects.
P value stands for statistical significance using Mann-Whitney U test, except for female/male proportion that p value stands for statistical significance using Chi-square test.
Table 2.
Clinical and biochemical features of morbid obese patients recruited from the bariatric surgery cohort.
| Variables | Control patients (morbid obese-no NAFLD) | NAFL | NASH |
|---|---|---|---|
| Number of subjects | 21 | 54 | 52 |
| Female, % | 75 | 67 | 59 |
| Age, years | 38.2 ± 10 | 39 ± 9 | 46 ± 12 |
| BMI, kg/m2 | 53 ± 11 | 53 ± 13 | 48 ± 12 |
| Fasting plasma glucose, mg/dL | 95.5 ± 13.4 | 105 ± 30# | 146 ± 76+,* |
| Fasting plasma insulin, μU/ml | 12 ± 5.5 | 13 ± 6.5# | 33 ± 56+,* |
| HOMA-IR index | 2.64 ± 1.4 | 3.2 ± 1.6# | 19.6 ± 52+,* |
| Total cholesterol, mg/dL | 187 ± 37 | 185 ± 35 | 197 ± 54 |
| HDL-cholesterol, mg/dL | 40 ± 9 | 45 ± 10 | 41 ± 6.7 |
| LDL-cholesterol, mg/dL | 122 ± 32 | 124 ± 27 | 135 ± 47 |
| Triglycerides, mg/dL | 130 ± 57 | 149 ± 54 | 200 ± 119+ |
| ALT, U/L | 21 ± 6 | 34 ± 25# | 40 ± 18+ |
| AST, U/L | 15.7 ± 5 | 26 ± 18# | 30 ± 13+ |
| Histological Features | |||
| Degree of steatosis, % | — | 36.5 ± 26 | 51 ± 27* |
| Lobular inflammation (0–3) | — | 0.3 ± 0.54 | 1.5 ± 0.96* |
| Hepatocellular ballooning (0–2) | — | 0.20 ± 0.4 | 1.1 ± 0.6* |
| Fibrosis Stage | — | 0.3 ± 1.67 | 1.5 ± 1.4* |
| NAFLD activity score (NAS) | — | 2.2 ± 1.37 | 4.8 ± 1* |
NAFL: nonalcoholic fatty liver, NASH: nonalcoholic steatohepatitis BMI: body mass index; HOMA: homeostatic model assessment; ALT and AST: Serum alanine and aspartate aminotransferase. Results are expressed as mean ± SD.
#p < 0.001 Indicates NAFL vs. controls, *p < 0.001 indicates comparisons between NAFL and NASH, and +p < 0.001 denotes comparisons between NASH and control subjects.
P value stands for statistical significance using Mann-Whitney U test, except for female/male proportion that p value stands for statistical significance using Chi-square test.
Genotype frequencies in controls (n = CC: 84, CT: 137, TT: 41, p = 0.22) and patients (CC: 134, CT: 178, TT: 60, p = 0.94) were in Hardy-Weinberg equilibrium (HWE). The minor allele frequency (MAF) in our sample was 40.8%, in line with that reported in the 1000 Genomes Project for the T allele in all populations (37%) and Europeans (44%) (1000 Genomes Project, Phase 3, http://www.ensembl.org).
The association analysis of rs641738 and NAFLD showed no effect of the variant on the susceptibility of NAFLD (Cochran-Armitage test for trend χ2 = 0.397, p = 0.529). The variant was associated with neither NASH nor the disease severity (p = 0.61). No association was found with fibrosis status (fibrosis yes/no) (p = 0.95), lobular inflammation (p = 0.46), or NAFLD- NAS score (p = 0.25). However, in univariate analysis we observed a significant association with circulating triglycerides (TG) (p = 0.004). The rs641738 was not associated with glucose metabolism, HOMA-index, total, HDL, LDL-cholesterol or other MetS components.
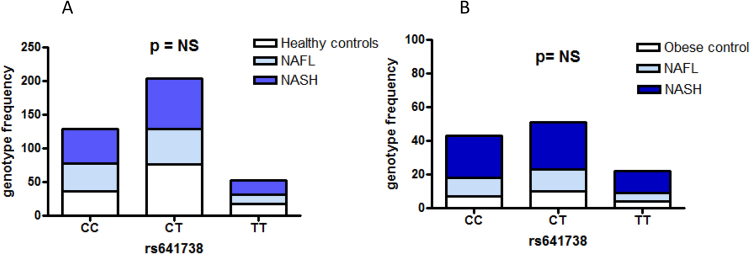
Genotype frequencies of rs641738 according to the disease status (control subjects, patients with simple steatosis-NAFL and NASH) in the two studied groups are shown in Fig. 2A,B.
Figure 2.
Genotype frequencies of rs641738 according the disease status (control subjects, patients with simple steatosis-NAFL and NASH). (A) Results from the cross-sectional study of patients with NAFLD and Metabolic Syndrome. (B) Results from a cohort-study of morbid-obese patients that underwent bariatric surgery.
MBOAT7 is expressed in the liver of patients with NAFLD at low levels
In order to provide evidence supporting a putative role of MBOAT7 in the biology of NAFLD, we further explored whether the protein encoded by this gene is expressed in the liver.
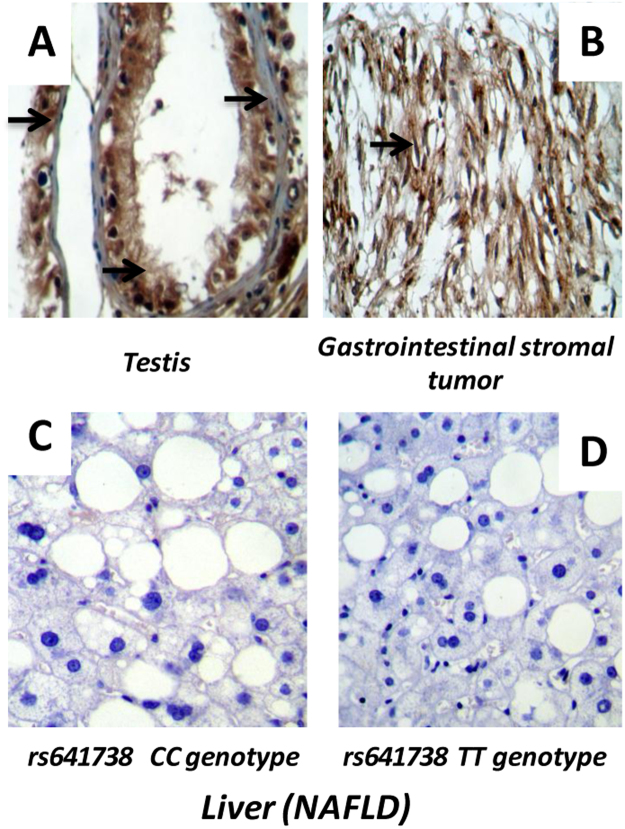
As positive control tissues, we included a sample of testis and a sample of gastrointestinal stromal tumor retrieved from the collection of our Pathology Department in which we observed a strong immunoreactivity of MBOAT7 (Fig. 3A,B). In contrast, in the liver of patients with NAFLD, we found very low expression levels of the protein assessed by immunohistochemistry (Fig. 3C,D). Thus, our results are comparable to the information displayed in the Human Protein Atlas (http://www.proteinatlas.org/ENSG00000125505-MBOAT7/tissue). Furthermore, we found no differences in the liver MBOAT7 expression pattern between rs641738 genotypes (CC 0.8 ± 0.27 vs. TT 0.9 ± 0.22, p = 0.69) (Fig. 3C,D).
Figure 3.
Expression profile of MBOAT7 in the liver tissue of patients with NAFLD. (A) A representative sample of testis and (B) A representative sample of gastrointestinal stromal tumor, which were used as positive control tissues; arrows denote strong immunoreactivity. (C) and (D) A representative sample of a patient with NAFLD carrying the rs641738 CC and TT genotype, respectively. Protein expression was assessed by imunohistochemistry in ten patients with NAFLD (CC n = 5 vs. TT n = 5) by two independent Pathologists and a semiquantitative score (0–4). As the samples presented very low levels of staining no sample was classified as having an score higher than 1. Mann-Whitney U test was used to analyse statistical significance.
Discussion
In this study, we explored the role of the missense rs641738 variant in the susceptibility of NAFLD and the disease severity. We did not find statistically significant differences in genotypic or allelic frequencies for the variant in either the predisposition of NAFLD or NASH, or other related histological features. Genotype frequencies in controls and cases were in HWE, and sample size estimation showed at least 84% power for the additive genetic model even if a very modest effect (OR: 1.3) is considered. Power calculation based on a OR of 1.3 is justified by previous evidence of association of the variant and NAFLD (OR 1.37)12 or liver fibrosis (OR 1.41)12 in European American population; in fact, our entire sample is composed of individuals of self-reported European ancestry. In addition, for polymorphisms with minor allele frequencies >0.2 (like the one observed for rs641738 MAF 0.40), the ORs are expected be in the range of 1.1–1.519. However, our study is underpowered for ORs 1.2–1.25 and for all histological features analyses.
In contrast to some reports in the literature indicating a significant association of the variant with NASH, liver damage and fibrosis in individuals of European descent but not other ethnicities12, our study suggests that it is highly unlikely that rs641738 plays a role in the genetic susceptibility of NAFLD, at least in our population. A note of caution regarding the lack of association of the variant and liver fibrosis in our sample should be added because it could be explained by insufficient power.
Likewise, Krawczyk and coworkers failed to detect an association of the rs641738 and NAFLD or liver function test14, while a marginal but positive effect of the variant on liver fibrosis (OR 1.41 95% CI 1.003–1.982, p = 0.048) was observed.
Meanwhile this manuscript was under the peer review process, several reports on the role of rs641738 were published16–18,20; specifically, there were large studies that included well characterized patients diagnosed by liver biopsy16,17. Interestingly, these studies showed a negative association of the variant with NAFLD16–18,20 and NASH or liver fibrosis16,17.
A detailed summary of the available evidence is shown in Table 3.
Table 3.
A summary of studies that explored the association between the rs641738 and NAFLD, liver histology, and related traits.
| First author, Year, Reference | Country, Ancestry | Study design | Sample size and features | Association with NAFLD as disease trait | Effect on liver histology and liver enzymes |
|---|---|---|---|---|---|
| Studies that reported lack of association of rs641738 and NAFLD | |||||
| Current study (Sookoian S, 2018 | Argentina, Caucasian descent | Case-control hospital based | 634 individuals (372 patients with NAFLD diagnosed by liver biopsy and 262 control subjects) | No evidence of association | No evidence of association with NASH, fibrosis, or liver enzymes |
| Lin YC, 201818 | Taiwan, Han Chinese population | Population based; 189 (22.7%) had hepatic steatosis | 831 obese children aged 7–15 years. NAFLD diagnosed by liver ultrasonographic examination | No evidence of association | No evidence of association with liver enzymes |
| Koo BK, 201717 | Korea, Asian descent. | Case-control hospital based | 525 individuals (416 patients with NAFLD diagnosed by liver biopsy and 109 healthy controls) | No evidence of association | No evidence of association with NASH, fibrosis, or liver enzymes |
| Dold L, 201720 | Germany, Caucasian descent | Case-control hospital based | 291 individuals (142 HIV-infected patients and 149 healthy blood donors). Liver stiffness assessed by Fibroscan and NAFLD by liver ultrasound. | No evidence of association | No evidence of association with liver enzymes or liver stiffness |
| Krawczyk M, 201714 | Germany, Caucasian descent | Cases only | 515 patients with NAFLD. Liver biopsy was performed in a sub-group of 320 patients. | No evidence of association | Association with liver fibrosis (P = 0.046) |
| Krawczyk M, 201615 | Germany, Caucasian descent | Cases only | 84 obese individuals scheduled for bariatric surgery | No evidence of association | No evidence of association with NASH, fibrosis, or liver enzymes |
| Kawaguchi T, 201816 | Japan, Asian descent | Case-control hospital based | 8608 individuals (936 histologically proven NAFLD patients and 7,672 general-population controls) | No evidence of association with hepatic steatosis | No evidence of association with NASH, fibrosis, or liver enzymes |
| Studies that reported association of rs641738 and NAFLD and NASH | |||||
| Mancina R, 201612 | Multi-ethnic | Two stages. First stage: population based. Second stage: cases-only, hospital based | 3854 participants from the Dallas Heart Study (a multi-ethnic population-based sample of Dallas County residents; hepatic triglyceride content evaluated by liver spectroscopy n = 2736) and 1149 European individuals (NAFLD evaluated by liver biopsy). | First stage: association with NAFLD in the global analysis. Stratification by ethnic groups: association only significant in African Americans but not European) | Association with hepatic steatosis (p = 0.015), NASH (p = 0.05), and fibrosis stage F2-4 (p = 0.012). |
| Luukkonen P, 201613 | Finland | Cases only | 125 patients with NAFLD assessed by liver biopsy | Not assessed | Association with hepatic steatosis degree (p = 0.03), inflammation (p = 0.04) and fibrosis stage F2-4 (p = 0.01) |
| Viitasalo A, 201631 | Finland | Population based | 467 Caucasian children aged 6–9 years, *no assessment of NAFLD by any imaging method. | Not assessed | Association with plasma ALT levels (additional adjustment for body fat percentage show not association with liver enzymes (P = 0.063) |
While the reasons behind these discrepancies are unclear, several potential explanations should be considered.
The first explanation relates to putative discrepancies at the population level and the design of the extant studies on the effect of rs641738 on either hepatic steatosis or hepatic triglyceride content (HTGC)—as measured by liver spectroscopy—both of which contribute to inconsistencies among different datasets. For example, in their analyses, Mancina et al. stratified the data by ethnic groups of the population-based Dallas Heart Study (DHS), and observed a positive significant effect (p = 0.019) of rs641738 on HTGC content (continuous variable) that was restricted to African Americans. In contrast, association with hepatic steatosis (NAFLD as a disease trait) remained significant in European Americans (OR: 1.37; 95% CI: 1.09–1.72; p = 0.007) but not in African Americans12. The biological reasons behind such discrepancies, while interesting, are certainly hard to explain.
A second explanation could be a false positive association between the variant and NAFLD ascribed to deviations from HWE or insufficient genotyping accuracy. Mancina et al. showed that rs641738 was associated with NASH and the disease severity in European population; however, genotype frequencies deviated from HWE (p = 0.017)12. Nevertheless, HWE is statistically a null hypothesis as it assumes there is no evolution in the population. In fact, disease-associated allele can be deviated from HWE in a disease population (cases) but not in controls.
A third but yet unexplored explanation relates to a putative gene × environment interaction, the occurrence of which seems to be limited to the European cohort of Mancina et al.’s study12. Nevertheless, this possibility is hard to conceive, as the variant was not associated with NAFLD in patients from other European countries, including Germany14,15; although the results of the German study could be a remote example of this situation.
In our sample we observed that rs641738 was associated with triglyceride levels and so, it might indirectly regulate intermediate steps of fatty acid (polyunsaturated fatty acids-PUFA and PUFA-containing TG) biosynthesis. Still, the association of the variant with TG was not observed among individuals included in previous reports12, except for one study from Germany15; hence, the biological meaning of this observation remains unknown.
Moreover, a distant but noteworthy explanation could pertain to disparities in the minor allele frequency (MAF) among different populations around the world, including our sample. However, the frequency of the risk allele in our population is comparable to that reported in the 1000 Genomes Project for Caucasians (44%); hence, it seems unlikely that discrepancies among studies arise from racial differences. It is still possible, however, that this variant may bare a population-specific association with NAFLD. It is also likely that rs641738 may work via an interaction with other injuries or unknown environmental factors, especially when these factors may be distributed differently among different populations.
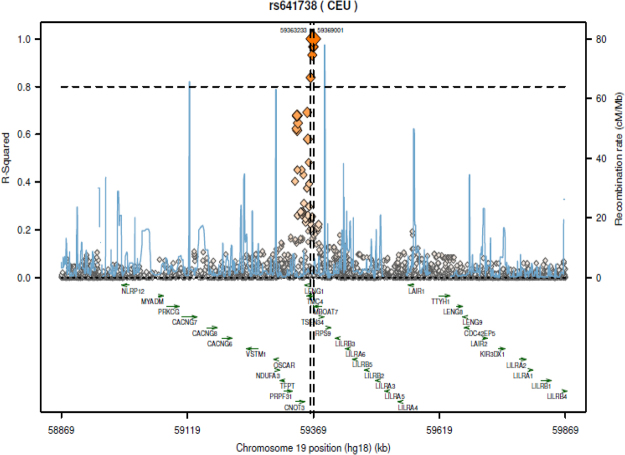
A final plausible explanation is that rs641738 is not necessarily the causal variant; thus, other SNPs in strong linkage disequilibrium (LD) could explain an impact on the phenotype. Exploration of variants in high LD with rs641738 shows at least five SNPs, (Fig. 4 and Table 4), including rs8736 a 3′ UTR variant of MBOAT7/2 kb upstream variant of TMC4.
Figure 4.
Exploration of potential regulatory variants in linkage disequilibrium with rs641738. Plot was retrieved from SNAP, a web-based tool for identification and annotation of proxy SNPs (https://personal.broadinstitute.org/plin/snap/index.php). SNAP finds proxy SNPs based on linkage disequilibrium and physical distance. Pair-wise linkage disequilibrium is pre-calculated based on phased genotype data from the International HapMap Project. The plot shows the associated region (chromosome 19, rs641738), defined by the contiguous region that contains all proxy SNPs with r2 > 0.8.
Table 4.
A description of variants in strong linkage disequilibrium (LD) with rs641738, including potential functionality and location in locus (variant feature).
| Proxy | Distance | R 2 | D Prime | Chr | Locus* | Variant feature* |
|---|---|---|---|---|---|---|
| rs641738 | — | — | — | chr19 | MBOAT7/TMC4 | TMC4: Missense Variant/MBOAT7: 500B Downstream Variant |
| rs626283 | 238 | 0.967 | 1 | chr19 | MBOAT7/TMC4 | TMC4: 2KB Upstream Variant/MBOAT7: 500B Downstream Variant |
| rs2576452 | 330 | 0.967 | 1 | chr19 | TMC4 | Intron Variant |
| rs8736 | 426 | 0.967 | 1 | chr19 | MBOAT7/TMC4 | TMC4: Upstream Variant/MBOAT7: 3 Prime UTR Variant2KB |
| rs4806498 | 2021 | 0.932 | 0.966 | chr19 | TMC4 | Intron variant |
| rs60204587 | 5342 | 0.838 | 0.963 | chr19 | TMC4 | Intron variant |
Chr: chromosome, UTR: untranslated region.
*https://www.ncbi.nlm.nih.gov/snp (GRCh38.p7).
As a final point, we would like to comment on the significant dissimilarities that indeed exist between the biological function of MBOAT7 and TMC4. In fact, while MBOAT7 is a protein involved in the pathway of phospholipid metabolism, TMC4 is involved in the transport of ions. More specifically, MBOAT7 encodes a member of the membrane-bound O-acyltransferases family of integral membrane proteins that exhibit acyltransferase activity. The encoded protein is a lysophosphatidylinositol acyltransferase that has specificity for arachidonoyl-CoA as an acyl donor; this protein is involved in the reacylation of phospholipids as a part of the phospholipid remodeling pathway known as the Land cycle.
Analysis of eQTLs (expression quantitative trait loci), which denote correlations between genotype and tissue-specific gene expression levels, shows that rs641738 is associated with eQTLs in the liver (Tables 5 and 6) and other tissues and cell types as well (Table 6). Likewise, other MBOAT7-variants, including some that are in strong LD with rs641738, are associated with many eQTLs in the liver tissue (Table 5). Complete details of significant Single-Tissue eQTLs for MBOAT7 (ENSG00000125505.12) and TMC4 (ENSG00000167608.7) in the liver tissue are shown in Table 5.
Table 5.
Significant Single-Tissue eQTLs for MBOAT7 (ENSG00000125505.12) and TMC4 (ENSG00000167608.7) in the liver tissue.
| Query Gencode Id | SNP Id | P-Value | Effect Size |
|---|---|---|---|
| MBOAT7 (ENSG00000125505.12) | |||
| ENSG00000125505.12 | rs11668882 | 8.80E-10 | −0.43 |
| ENSG00000125505.12 | rs8736 | 1.00E-09 | −0.43 |
| ENSG00000125505.12 | rs2576452 | 1.10E-09 | −0.43 |
| ENSG00000125505.12 | rs372932354 | 1.40E-09 | −0.42 |
| ENSG00000125505.12 | rs641738 | 2.80E-09 | −0.42 |
| ENSG00000125505.12 | rs626283 | 2.80E-09 | −0.42 |
| ENSG00000125505.12 | rs4806498 | 3.60E-09 | −0.42 |
| ENSG00000125505.12 | rs60204587 | 4.10E-09 | −0.43 |
| ENSG00000125505.12 | rs10416555 | 5.90E-08 | −0.61 |
| ENSG00000125505.12 | rs36656 | 0.0000011 | −0.34 |
| ENSG00000125505.12 | rs77215230 | 0.0000012 | −0.6 |
| ENSG00000125505.12 | rs1050527 | 0.0000012 | −0.6 |
| ENSG00000125505.12 | rs11084313 | 0.0000018 | −0.58 |
| ENSG00000125505.12 | rs8100978 | 0.0000043 | −0.36 |
| TMC4 (ENSG00000167608.7) | |||
| ENSG00000167608.7 | rs776138589 | 6.80E-20 | 1.6 |
| ENSG00000167608.7 | rs43211 | 2.80E-08 | −0.67 |
| ENSG00000167608.7 | rs117643023 | 3.60E-08 | 1.4 |
| ENSG00000167608.7 | rs4806716 | 6.20E-08 | −0.67 |
| ENSG00000167608.7 | rs8100978 | 1.30E-07 | −0.63 |
| ENSG00000167608.7 | rs36663 | 1.40E-07 | −0.65 |
| ENSG00000167608.7 | rs36659 | 1.40E-07 | −0.65 |
| ENSG00000167608.7 | rs42319 | 2.10E-07 | −0.64 |
| ENSG00000167608.7 | rs36642 | 2.20E-07 | −0.65 |
| ENSG00000167608.7 | rs36641 | 2.30E-07 | −0.64 |
| ENSG00000167608.7 | rs593073 | 3.80E-07 | −0.64 |
| ENSG00000167608.7 | rs3816129 | 3.90E-07 | −0.63 |
| ENSG00000167608.7 | rs36658 | 4.10E-07 | −0.64 |
| ENSG00000167608.7 | rs8101186 | 4.40E-07 | −0.6 |
| ENSG00000167608.7 | rs653560 | 4.60E-07 | −0.64 |
| ENSG00000167608.7 | rs40168 | 6.90E-07 | −0.61 |
| ENSG00000167608.7 | rs36624 | 8.10E-07 | −0.59 |
| ENSG00000167608.7 | rs40167 | 0.000001 | −0.6 |
| ENSG00000167608.7 | rs635608 | 0.0000012 | −0.6 |
| ENSG00000167608.7 | rs7595 | 0.0000012 | −0.6 |
| ENSG00000167608.7 | rs36621 | 0.0000012 | −0.6 |
| ENSG00000167608.7 | rs12975696 | 0.0000013 | −0.59 |
| ENSG00000167608.7 | rs40357 | 0.000002 | −0.59 |
| ENSG00000167608.7 | rs183716 | 0.000002 | −0.59 |
| ENSG00000167608.7 | rs39714 | 0.000002 | −0.59 |
| ENSG00000167608.7 | rs36623 | 0.000002 | −0.59 |
| ENSG00000167608.7 | rs36622 | 0.0000027 | −0.6 |
eQTL: expression quantitative trait loci. Data Source: The Genotype-Tissue Expression (GTEx) project (Data Source: GTEx Analysis Release V7 (dbGaP Accession phs000424.v7.p2). The eQTL effect allele is the alternative allele relative to the reference allele in the human genome reference, not the minor allele. Query was specifically done on MBOAT7 locus (ENSG00000125505.12).
Table 6.
Analysis of eQTLs (expression quantitative trait loci) denoting correlations between rs641738 and cell tissue-specific gene expression levels.
| Query SNP | LD-eQTL | exGene | Association | LD | Study |
|---|---|---|---|---|---|
| rs641738 | rs641738 | TMC4 | 6.00E-11 | 1 | LV: Caucasian liver donors |
| rs641738 | rs641738 | MBOAT7 | 3.65E-12 | 1 | LV: Caucasian liver donors |
| rs641738 | rs641738 | TFPT | 3.88E-03 | 1 | LV: Caucasian liver donors |
| rs641738 | rs641738 | MBOAT7 | 9.364 | 1 | LV2: Liver donors |
| rs641738 | rs641738 | TMC4 | 2.001e-08 | 1 | EGEUV_EUR: 1000 Genome-EUR |
| rs641738 | rs2576452 | TMC4 | 7.283e-09 | 0.923 | MuTHER_Fat |
| rs641738 | rs626283 | TMC4 | 6.631e-09 | 1 | MuTHER_Fat |
| rs641738 | rs641738 | TMC4 | 7.858e-09 | 1 | MuTHER_Fat |
| rs641738 | rs8736 | TMC4 | 5.979e-09 | 0.92 | MuTHER_Fat |
| rs641738 | rs2576452 | TMC4 | 1.234e-19 | 0.923 | MuTHER_Skin |
| rs641738 | rs626283 | TMC4 | 3.031e-20 | 1 | MuTHER_Skin |
| rs641738 | rs641738 | TMC4 | 5.080e-20 | 1 | MuTHER_Skin |
| rs641738 | rs8736 | TMC4 | 1.052e-19 | 0.92 | MuTHER_Skin |
Table shows tissue-specific eQTL associations were identified by comparing eQTL data from six cell types: LCLs, B cells, Monocytes, Brain, Liver, and Skin.
Data was extracted from the integrated eQTL database, which is available at: http://www.exsnp.org/LDeQTL. Query was specifically done on rs641638.
All eQTL association data in this database were collected from 16 publicly available studies that had been performed on various human tissues and populations.
MuTHER: Multiple Tissue Human Expression Resource.
The observation that rs641738 is associated with eQTLs in non-liver tissues, including fat, might reinforce the possibility of unexplored associations between the variant and the disease. For example, a recent study showed that associations between common gene variants and NAFLD are uncovered by adiposity degree21; this point could explain some of the above mentioned observations.
Information regarding protein expression is much limited. We have specifically assessed the protein expression pattern of MBOAT7 in the liver of patients with NAFLD and we observed no evidence of robust immunoreactivity. Similarly, we failed to observe any association between liver-MBOAT7 expression and rs641738 genotypes (CC, TC and TT) (Fig. 3). Contrasting evidence was published elsewhere suggesting that rs641738 T allele was associated with reduced mRNA and hepatic protein MBOAT7 expression in patients with advanced fibrosis12,22. Then, a putative yet uncovered in cis effect of rs641738 (i.e., pertaining to mRNA stability or translation) might explain participation of the variant in lipid metabolism by regulating MBOAT7 expression.
On the other hand, TMC4 encodes for a membrane protein involved in the transport channel expressed in the peripheral nervous system; TMC4 belongs to the calcium-dependent chloride channel (ca-clc) family highly expressed among epithelia (kidney, small intestine, colon)23. Interestingly, there is evidence supporting the presence of chloride channels not only in the plasma membrane of hepatocytes but in multiple intracellular compartments as well24. The involvement of ion channels in the pathogenesis of NAFLD and/or in pathways associated with hepatic fibrogenesis remains elusive; nevertheless, it certainly represents an interesting path for future research. In fact, this observation could explain previously reported associations of rs641738 and fibrosis in patients with chronic hepatitis C and B25,26, and NAFLD12–14. Still, the exact effect/mechanisms by which the variant could regulate liver fibrogenesis remains uncertain. We reinforce the importance of precision in identifying the genomic location and the biological function of a given variant, as this would increase not only the understanding of the genetic component of NAFLD but also its relationship with the disease pathogenesis.
In conclusion, the rs641738 is not associated with NAFLD in our population. The association of the variant and NAFLD as disease trait could not be replicated in population-based or hospital based studies from Asia16–18,20 or Germany14. Nevertheless, the association with liver histology, including fibrosis was only observed in patients with NAFLD of European ancestry12–14; this finding could not be replicated in studies that included Asian population16,17. Hence, larger studies are required before any definitive conclusion can be reached.
Patients and Methods
Patients and control subjects: selection criteria
The study included a sample of 634 unrelated individuals, of which 262 were controls subjects and 372 were patients who have histopathologic-proven features of NAFLD. Patients and controls were selected from two different hospital-based settings, including a cross-sectional study of patients diagnosed with NAFLD and Metabolic Syndrome (MetS) in the Liver Unit, Hospital Abel Zubizarreta, Buenos Aires, Argentina, and an independent cohort of morbid-obese patients that underwent bariatric surgery in the Surgery Department, Hospital de Alta Complejidad en Red El Cruce, Buenos Aires, Argentina.
All investigations performed were conducted in accordance with the guidelines of the 1975 Declaration of Helsinki. Informed and written consent for study participation from all individuals was obtained in accordance with the procedures approved by the ethical committee of our institution (protocol number: 104/HGAZ/09, 89/100 and 1204/2012).
Exclusion criteria: Secondary causes of steatosis, including alcohol abuse (≥30 g alcohol daily for men and ≥20 g for women), total parenteral nutrition, hepatitis B and hepatitis C virus infection, and the use of drugs known to precipitate steatosis were excluded. In addition, patients with any of the following diseases were excluded from participation: autoimmune liver disease, metabolic liver disease, Wilson’s disease, and α-1-antitrypsin deficiency.
Control subjects that matched patients with NAFLD-MetS were selected from subjects attending our hospital for check-up purposes whose age and sex matched the NAFLD patients. In addition to the standard heath examination, all non-obese control individuals were subjected to a liver ultrasonographic (US) examination. They were included in the study if they did not have evidence of fatty change or biochemical abnormalities. Furthermore, control subjects were confirmed not to have any of the features of the metabolic syndrome as defined by the National Cholesterol Education Program Adult Treatment Panel III and did not abuse alcohol.
In the population of morbid obese patients, control subjects were obese patients who also underwent bariatric surgery and had not features of NAFLD demonstrated in the liver biopsy.
The case participants and the controls were selected during the same study period from the same population of patients attending the above mentioned institution, and all of them share the same demographic characteristics (occupation, educational level, place of residence, and ethnicity).
Physical, anthropometric, biochemical evaluation and histological
Health examinations included anthropometric measurements, a questionnaire on health-related behaviours and biochemical determinations.
The disease severity was assessed by liver biopsy that was performed before any intervention with ultrasound guidance and a modified 1.4-mm-diameter Menghini needle (Hepafix, Braun, Germany) under local anesthesia on an outpatient basis or during bariatric surgery. All liver biopsies were evaluated by the same pathologist.
A portion of each liver biopsy specimen was routinely fixed in 40 g/l formaldehyde (pH 7.4), embedded in paraffin, and stained with hematoxylin and eosin, Masson trichrome, and silver impregnation for reticular fibers. All the biopsies were at least 3 cm in length and contained a minimum of 8 portal tracts. The degree of steatosis was assessed according to the system developed by Kleiner et al., based on the percentage of hepatocytes containing macrovesicular fat droplets27. NASH and NAFLD Activity Score (NAS)27,28 were defined as reported previously; a NAS threshold of 5 was used for further comparisons with variables of interest, NASH was defined as steatosis plus mixed inflammatory-cell infiltration, hepatocyte ballooning and necrosis, Mallory’s hyaline, and any stage of fibrosis, including absent fibrosis27,28.
Genotype and association analysis, and power and sample size calculation
The genetic analyses were done on genomic DNA extracted from white blood cells. Genotyping of rs641738 was performed using a TaqMan genotyping assay (dbSNP rs641738 assay C___8716820_10, # 4351379; Applied Biosystems, California 92008, USA) according to manufacturer’s instructions. To ensure genotyping quality, we included DNA samples as internal controls, hidden samples of known genotype, and negative controls (water). The overall genotype completion rate was 100%.
To account for possible population stratification, we used a collection of 13 SNPs at different loci (located in chromosomes 4, 15, 17, 13, 1, and 3) and then analyzed the data with the Structure program Version 229 as we explained elsewhere2. We found no evidence of stratification in our sample because the cases and the controls showed similar Q values and the Structure program assigned a similar distance to clusters with no further improvement in the fitting model by adding up to four clusters (the ln of likelihood was maximum for K = 1). Moreover, all the participants in this study self-reported a Caucasian ethnicity as a surrogate of ancestry, which is consistent with the observed MAF.
Using the CaTS power calculator for genetic association studies30 and assuming a prevalence of NAFLD of 0.30, minor allele frequency (MAF) T = 0.40 and an odds ratio (OR) of 1.3–1.5, our sample had 84–99% power, respectively, for the additive genetic model.
Liver Immunohistochemistry
Four-micrometer sections were mounted onto silane coated glass slides to ensure section adhesion through subsequent staining procedures. Briefly, sections were deparaffinized, rehydrated, washed in phosphate buffer solution (PBS), and treated with 3% H2O2 in PBS for 20 min at room temperature to block endogenous peroxidase. Following microwave heat-induced epitope retrieval in 0.1 M citrate buffer at pH 6.0 for 20 min, the slides were incubated with a dilution of 1:100 of rabbit polyclonal antibody for Human Anti-MBOAT7 (ARP49811_T100, Aviva Systems Biology, San Diego, CA 92121 USA). Immunostaining was performed using the VECTASTAIN Elite ABC Kit (Vector Lab. CA, USA) detection system. Subsequently, slides were immersed in a 0.05% 3,3′-diaminobenzidine solution in 0.1 M Tris buffer, pH 7.2, containing 0.01% H2O2. After a brown color developed, slides were removed and the reaction was stopped by immersion in PBS. Negative controls were carried out with rabbit serum diluted to the same concentration as the primary antibody. MBOAT7 immunostaining was evaluated in a blinded fashion regarding any of the histological and clinical characteristics of the patients. The extent of staining was scored according to its amount and intensity by a 4-point scoring system as follows: 0 = no staining, 1 = positive staining in less than 20% of cells, 2 = 21–50% of positive cells, and 3 = positive staining in more than 50% of cells. The sections were observed in bright field microscopy with a microscope Axiostar plus (Carl Zeiss, Germany) at a magnification of X400. As control tissue we used a sample of testis retrieved from the collection of tissues of the Pathology Department.
Statistical analysis
Quantitative data were expressed as mean ± SD unless otherwise indicated. As a significant difference in SD was observed between the groups in most of the variables and the distribution was significantly skewed in most cases, we chose to be conservative and assessed the differences between the groups using nonparametric Mann–Whitney U or Kruskal-Wallis tests. The Cochran–Armitage test for trend was used in the categorical data analysis to assess the presence of association between the variant and disease severity and a regression analysis for an ordinal multinomial distribution (Probit as the Link function) with disease severity as the dependent (response) variable coding controls; NAFL and NASH subjects as 0, 1, and 2, respectively; age, HOMA, and BMI as continuous predictor variables; and sex and rs641738 genotypes (0, 1, 2) as grouping variables. Moreover, logistic regression analysis was included for the evaluation of the association between genotypes and histological disease severity (NAS, ballooning, fibrosis, and inflammation: present coded as 1 or absent coded as 0). To assess the association between genotypes with NAFLD or quantitative traits, we used a chi-square test and logistic regression or ANCOVA and multiple regression, adjusting for co-variables, such as age, HOMA, BMI, and rs738409. For ordinal multinomial analysis, logistic analysis, or ANCOVA, we adjusted for co-variables that were not normally distributed through log-transformation. Correlation between two variables was done using the Spearman’s rank correlation test. The CSS/Statistica program package version 6.0 (StatSoft, Tulsa, OK, USA) was used in these analyses.
Data availability
All data generated or analyzed during this study are included in this published article.
Acknowledgements
This study was partially supported by grants PICT 2014-0432, PICT 2014-1816 and PICT 2015-0551 (Agencia Nacional de Promoción Científica y Tecnológica, FONCyT). SS, DF and CJP belong to Consejo Nacional de Investigaciones Científicas (CONICET).
Author Contributions
S.S.: study concept and design; data acquisition; performed liver biopsies and collected biological material; data analysis and interpretation; general study supervision; drafting of the manuscript; securing funding. D.F.: genotyping. M.G. and G.O.C.: performed liver biopsies and collected biological samples. J.S.M.: imunohistochemistry; C.G.: histological diagnosis. C.J.P.: study concept and design; data acquisition; data analysis and interpretation; statistical analysis; drafting of the manuscript; general study and supervision and securing funding.
Competing Interests
The authors declare no competing interests.
Footnotes
Silvia Sookoian and Carlos J. Pirola jointly supervised this work.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Silvia Sookoian, Email: sookoian.silvia@lanari.fmed.uba.ar.
Carlos J. Pirola, Email: pirola.carlos@lanari.fmed.uba.ar
References
- 1.Romeo S, et al. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2008;40:1461–1465. doi: 10.1038/ng.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sookoian S, et al. A nonsynonymous gene variant in the adiponutrin gene is associated with nonalcoholic fatty liver disease severity. J. Lipid Res. 2009;50:2111–2116. doi: 10.1194/jlr.P900013-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology. 2011;53:1883–1894. doi: 10.1002/hep.24283. [DOI] [PubMed] [Google Scholar]
- 4.Mahdessian H, et al. TM6SF2 is a regulator of liver fat metabolism influencing triglyceride secretion and hepatic lipid droplet content. Proc. Natl. Acad. Sci. USA. 2014;111:8913–8918. doi: 10.1073/pnas.1323785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pirola CJ, Sookoian S. The dual and opposite role of the TM6SF2-rs58542926 variant in protecting against cardiovascular disease and conferring risk for nonalcoholic fatty liver: A meta-analysis. Hepatology. 2015;62:1742–1756. doi: 10.1002/hep.28142. [DOI] [PubMed] [Google Scholar]
- 6.Dongiovanni P, et al. Transmembrane 6 superfamily member 2 gene variant disentangles nonalcoholic steatohepatitis from cardiovascular disease. Hepatology. 2015;61:506–514. doi: 10.1002/hep.27490. [DOI] [PubMed] [Google Scholar]
- 7.Kozlitina J, et al. Exome-wide association study identifies a TM6SF2 variant that confers susceptibility to nonalcoholic fatty liver disease. Nat. Genet. 2014;46:352–356. doi: 10.1038/ng.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu YL, et al. TM6SF2 rs58542926 influences hepatic fibrosis progression in patients with non-alcoholic fatty liver disease. Nat. Commun. 2014;5:4309. doi: 10.1038/ncomms5309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sookoian S, et al. Genetic variation in transmembrane 6 superfamily member 2 and the risk of nonalcoholic fatty liver disease and histological disease severity. Hepatology. 2015;61:515–525. doi: 10.1002/hep.27556. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, Liu Z, Peng Z, Liu W. The TM6SF2 rs58542926 T Allele Is Significantly Associated with Nonalcoholic Fatty Liver Disease in Chinese. J. Hepatol. 2015;62:1438–1439. doi: 10.1016/j.jhep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 11.Wong VW, Wong GL, Tse CH, Chan HL. Prevalence of the TM6SF2 variant and non-alcoholic fatty liver disease in Chinese. J. Hepatol. 2014;61:708–709. doi: 10.1016/j.jhep.2014.04.047. [DOI] [PubMed] [Google Scholar]
- 12.Mancina RM, et al. The MBOAT7-TMC4 Variant rs641738 Increases Risk of Nonalcoholic Fatty Liver Disease in Individuals of European Descent. Gastroenterology. 2016;150:1219–1230. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luukkonen PK, et al. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J. Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 14.Krawczyk M, et al. Combined effects of the TM6SF2rs58542926, PNPLA3 rs738409 and MBOAT7 rs641738 variants on NAFLD severity: multicentre biopsy-based study. J. Lipid Res. 2017;58:247–255. doi: 10.1194/jlr.P067454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krawczyk M, et al. PNPLA3p.I148M variant is associated with greater reduction of liver fat content after bariatric surgery. Surg. Obes. Relat Dis. 2016;12:1838–1846. doi: 10.1016/j.soard.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 16.Kawaguchi T, et al. Risk estimation model for nonalcoholic fatty liver disease in the Japanese using multiple genetic markers. PLoS. One. 2018;13:e0185490. doi: 10.1371/journal.pone.0185490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koo, B. K. et al. Additive effects of PNPLA3 and TM6SF2 on the histological severity of non-alcoholic fatty liver disease. J. Gastroenterol. Hepatol. [Epub ahead of print] (2017). [DOI] [PubMed]
- 18.Lin, Y. C., Chang, P. F., Chang, M. H., & Ni, Y. H. Genetic determinants of hepatic steatosis and serum cytokeratin-18 fragment levels in Taiwanese children. Liver Int. [Epub ahead of print] (2018). [DOI] [PubMed]
- 19.Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat. Protoc. 2007;2:2492–2501. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dold L, et al. Genetic polymorphisms associated with fatty liver disease and fibrosis in HIV positive patients receiving combined antiretroviral therapy (cART) PLoS. One. 2017;12:e0178685. doi: 10.1371/journal.pone.0178685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stender S, et al. Adiposity amplifies the genetic risk of fatty liver disease conferred by multiple loci. Nat. Genet. 2017;49:842–847. doi: 10.1038/ng.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donati B, et al. MBOAT7 rs641738 variant and hepatocellular carcinoma in non-cirrhotic individuals. Sci. Rep. 2017;7:4492. doi: 10.1038/s41598-017-04991-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348, 648–660 (2015). [DOI] [PMC free article] [PubMed]
- 24.Li X, Weinman SA. Chloride channels and hepatocellular function: prospects for molecular identification. Annu. Rev. Physiol. 2002;64:609–633. doi: 10.1146/annurev.physiol.64.090501.145429. [DOI] [PubMed] [Google Scholar]
- 25.Thabet K, et al. MBOAT7 rs641738 increases risk of liver inflammation and transition to fibrosis in chronic hepatitis C. Nat. Commun. 2016;7:12757. doi: 10.1038/ncomms12757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thabet K, et al. The MBOAT7 variant rs641738 increases inflammation and fibrosis in chronic hepatitis B. Hepatology. 2017;65:1840–1850. doi: 10.1002/hep.29064. [DOI] [PubMed] [Google Scholar]
- 27.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 28.Brunt EM, Kleiner DE, Wilson LA, Belt P, Neuschwander-Tetri BA. Nonalcoholic fatty liver disease (NAFLD) activity score and the histopathologic diagnosis in NAFLD: distinct clinicopathologic meanings. Hepatology. 2011;53:810–820. doi: 10.1002/hep.24127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian C, Gregersen PK, Seldin MF. Accounting for ancestry: population substructure and genome-wide association studies. Hum. Mol. Genet. 2008;17:R143–R150. doi: 10.1093/hmg/ddn268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat. Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 31.Viitasalo A, et al. Association of MBOAT7 gene variant with plasma ALT levels in children: the PANIC study. Pediatr. Res. 2016;80:651–655. doi: 10.1038/pr.2016.139. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.