Abstract
Enterococci, in particular vancomycin-resistant enterococci (VRE), are a leading cause of hospital-acquired infections. Promoting intestinal resistance against enterococci could reduce the risk of VRE infections. We investigated the effects of two Lactobacillus strains to prevent intestinal VRE. We used an intestinal colonisation mouse model based on an antibiotic-induced microbiota dysbiosis to mimic enterococci overgrowth and VRE persistence. Each Lactobacillus spp. was administered daily to mice starting one week before antibiotic treatment until two weeks after antibiotic and VRE inoculation. Of the two strains, Lactobacillus paracasei CNCM I-3689 decreased significantly VRE numbers in the feces demonstrating an improvement of the reduction of VRE. Longitudinal microbiota analysis showed that supplementation with L. paracasei CNCM I-3689 was associated with a better recovery of members of the phylum Bacteroidetes. Bile salt analysis and expression analysis of selected host genes revealed increased level of lithocholate and of ileal expression of camp (human LL-37) upon L. paracasei CNCM I-3689 supplementation. Although a direct effect of L. paracasei CNCM I-3689 on the VRE reduction was not ruled out, our data provide clues to possible anti-VRE mechanisms supporting an indirect anti-VRE effect through the gut microbiota. This work sustains non-antibiotic strategies against opportunistic enterococci after antibiotic-induced dysbiosis.
Introduction
The human gastrointestinal (GI) tract is colonised by a dense and diverse microbial community referred to as the gut microbiota. This highly complex microbial ecosystem is involved in many host physiological processes including improvement of the intestinal epithelial barrier, education of the immune system, and nutrient acquisition1. In adult mammals, the gut microbiota is dominated by two bacterial phyla—the Firmicutes and the Bacteroidetes—but the bacterial species present are highly diverse2,3. Mounting evidence demonstrates that some members of the sub-dominant fraction of the gut microbiota, referred to as pathobionts, can harbor potential pathogenic features. Pathobionts can expand under some circumstances resulting in the disruption of a well-balanced microbial ecosystem and suspected to be involved in opportunistic infections or chronic diseases4,5. In particular, pathobiont proliferation represents a threat in immunocompromised and frail elderly people causing infectious diseases6–9.
Enterococci are natural inhabitants of the subdominant human intestinal microbiota in adults2,3. They are considered pathobionts as many enterococci are harmless for healthy humans, but can be pathogenic under certain circumstances (e. g. prolonged antibiotic treatments, severe underlying diseases, and impaired immune system); causing urinary tract and intra-abdominal infections, bacteremia and infective endocarditis. Furthermore, Enterococcus spp. contribute to community-acquired intra-abdominal infections10,11 and count among the ten most frequently isolated micro-organisms in healthcare-associated infections12–14. As enterococci are intrinsically resistant to many antibiotics such as penicillin and cephalosporins, broad-spectrum antibiotic treatment is one of the conditions of overgrowth of enterococci at the expense of major members of the gut microbiota. Intestinal expansion of Enterococcus spp. is associated with an increased risk of developing bloodstream infection with the same bacterial species15,16. E. faecalis accounts for more than 60% of the hospital-acquired enterococcal infections17–19. Given the importance of gastrointestinal colonisation and proliferation as primary steps of E. faecalis infectious process and transmission between patients, prevention of E. faecalis overgrowth and persistence in the GI tract appears a good approach to limit the risks of infection following antibiotic treatment.
It has been previously demonstrated that ingested bacteria or probiotics can increase resistance mechanisms against intestinal pathogens20,21. This beneficial effect of probiotic strains can involve direct inhibitory effect by competition for nutrients or killing of the pathogen by an inhibitory molecule or indirect effect such as a positive impact on the gut microbiota composition or on the host defense mechanisms22. Despite recent clinical success of fecal microbiota transplantation in the reduction of VRE and identification of mouse commensal strains that restore colonisation resistance against VRE, relatively few studies evaluating the use of Lactobacillus spp. strains to prevent or limit VRE colonisation and overgrowth have been reported23–25. For instance, two studies reported a beneficial effect of Lactobacillus rhamnosus GG in human26,27. The use of strains with a qualified presumption of safety appears less problematic than fecal microbiota transfer25,28,29. As members of the lactic acid bacteria studied for health-promoting effects, several strains of lactobacilli were shown to reinforce the epithelial intestinal barrier and prevent development of pathogens30. Among them, Lactobacillus paracasei CNCM I-3689 decreases translocation and dissemination of Listeria monocytogenes in gnotobiotic and conventional mice31 and L. rhamnosus CNCM I-3690 improves intestinal barrier integrity in a murine model and exerts anti-inflammatory properties32,33.
We previously adapted an E. faecalis colonisation model in mice with conventional microbiota as developed by Donskey et al.34,35. In this model, mice are pre-treated with clindamycin that causes an imbalance of the gut microbiota including an increase of endogenous enterococci and allows transient colonisation of E. faecalis V583 strain, a representative of the leading hospital adapted lineage of E. faecalis in the United States and in several European countries36,37. The aim of this work was to evaluate the effect of the strains L. paracasei CNCM I-3689 and L. rhamnosus CNCM I-3690 on the colonisation and persistence of the E. faecalis V583 strain. We measured transient colonisation and persistence of VRE in the intestine of mice upon supplementation with Lactobacillus spp. strains. We observed an improvement of VRE reduction by L. paracasei CNCM I-3689. The effect of this strain on the gut microbiota and on the expression of a selection of host genes was analysed leading us to propose that part of the L. paracasei CNCM I-3689 anti-VRE effect may rely on a faster recovery of members of the phylum Bacteroidetes.
Results
Lactobacillus paracasei CNCM I-3689 reduces E. faecalis V583 persistence level
We previously showed that transient intestinal colonisation of clindamycin-treated mice by the vancomycin-resistant E. faecalis strain V583 paralleled the overgrowth of endogenous enterococci35,38. We used this model to examine the effect of the strains L. paracasei CNCM I-3689 and L. rhamnosus CNCM I-3690 on the colonisation and persistence of E. faecalis V583 according to the experimental protocol depicted in Supplementary Figure S1. After an adaptation period (D0), mice received a daily dose of 109 CFU of strain L. paracasei CNCM I-3689, L. rhamnosus CNCM I-3690 or control solution for the duration of the experiment (D21). After the first week of supplementation (D7), animals received clindamycin for 3 days (D7 to D9) and were then inoculated with strain E. faecalis V583 at D10. Numbers of strain E. faecalis V583, total enterococci, and lactobacilli were monitored. Three independent trials (trial 1, 2 and 3, Supplementary Figure S1) were carried out. In this model, the inoculated strain E. faecalis V583 transiently colonised the GI tract, reaching a maximum of 5 × 108 to 5 × 109 CFU/g, one day (D11) after inoculation in the control, L. paracasei CNCM I-3689 and L. rhamnosus CNCM I-3690 groups. In all trials the fecal counts of E. faecalis V583 decreased and stabilised at 105 CFU/g nine days (D18) after the end of antibiotic treatment in the control and L. rhamnosus groups. Conversely we observed a higher number of mice with E. faecalis V583 under the detection limit from D14 to D21 indicating a progressive reduction of carriage of E. faecalis V583 in the group receiving L. paracasei CNCM I-3689 (Fig. 1). At D21 E. faecalis V583 number was below the detection level (<102 CFU/g) in eight out of eighteen of the mice receiving L. paracasei CNCM I-3689. Comparison of the kinetics of VRE for L. paracasei and control groups in the different experiments (Fig. 2) established that numbers of E. faecalis V583 were significantly reduced in trials 1 (P = 0.0079) and 3 (P = 0.013). In contrast, strain L. paracasei CNCM I-3689 did not show significant reduction on numbers of strain E. faecalis V583 in trial 2. This result may be linked to a faster wash-out of E. faecalis V583 in the control group of this trial, indicative of a lower persistence of V583. Together these data showed that strain L. paracasei CNCM I-3689 contributes to intestinal reduction or clearance of vancomycin-resistant E. faecalis V583.
Figure 1.
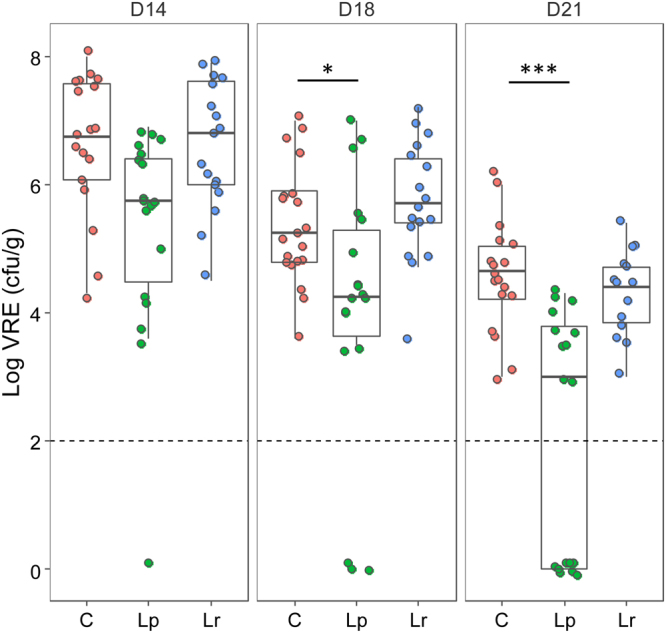
L. paracasei CNCM I‐3689 reduces the fecal levels of VRE E. faecalis V583 in the intestinal tract. E. faecalis V583 counts (CFU/g) at D14, D18 and D21 in mice receiving strain L. paracasei CNCM I-3689 (Lp), L. rhamnosus CNCM I-3690 (Lr) or NaCl (C). Each dot represents one mouse (n = 13 to 18). Horizontal bars represent the median for each condition and the dashed line indicates the detection limit. Statistical tests were performed using a Mann–Whitney test. Asterisks indicate a p-value considered statistically significant (*P = 0.057; ***P = 0.001).
Figure 2.
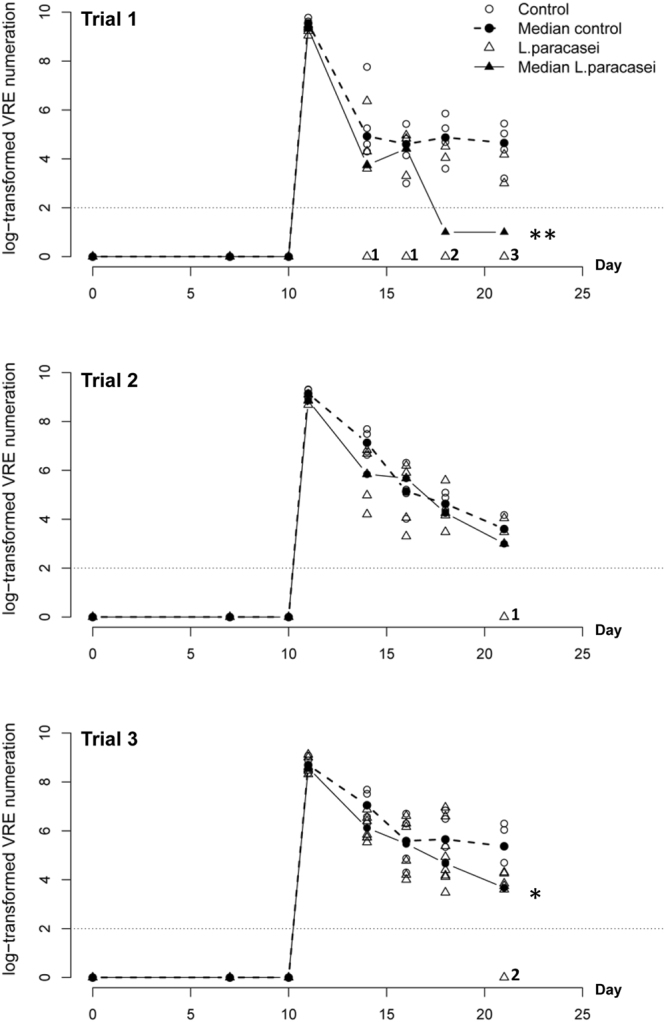
Kinetics of fecal levels of VRE in trials 1, 2 and 3. Trial 1 (n = 5 mice per group), trial 2 (n = 4 mice for control and n = 5 for L. paracasei group), trial 3 (n = 5 mice for control group and n = 8 for L. paracasei group). Values below the detection level at 102 were set to zero and numbers on the right of the empty triangles indicate the number of mice with VRE below the detection level. The continuous and dashed line curves show the median of log-transformed numeration from mice in control and L. paracasei groups computed at each time point. For time points with more than half of the values below 102 the mean of the two extreme values is represented, as the median was not uniquely defined and laid between 0 and the smallest non-zero value. Asterisks indicate a P-value considered statistically significant (*P < 0.05; **P < 0.01).
Clindamycin treatment was concomitant with transient increase of total enterococci, which reached the highest level (~5 × 109CFU/g) two days after the end of the antibiotic treatment (D11) and decreased to an average level (~107 cfu/g) within the next 5 days (Figure S2). Lactobacilli population also reached a maximum (>1010 CFU/g) at D11 (Figure S2). Similarly to total enterococci, no difference on total lactobacilli was detected between the three animal groups, indicating that administration of L. paracasei CNCM I-3689 or L. rhamnosus CNCM I-3690 had no major effect on total enterococci and lactobacilli when assessed using cultivation based approach.
Strain L. paracasei CNCM I-3689 improves resilience of Bacteroidetes members after clindamycin treatment
To examine whether strain L. paracasei CNCM I-3689 had an effect on the intestinal microbiota and to characterise changes in the microbiota that could be associated with a reduction of E. faecalis V583, we analysed the fecal microbiota of control and L. paracasei CNCM I-3689-supplemented mice by 16S rRNA gene sequencing. To circumvent potential interference of the presence of VRE on the microbiota, we performed two trials (Trials 4 and 5) by omitting inoculation of E. faecalis V583 at D11 as depicted in Supplementary Figure S1. Microbiota diversity and composition were compared for 3 to 8 mice for trials 1, 2, 4 and 5 at baseline (D0), 1 week after L. paracasei CNCM I-3689 supplementation (D7), and one (D11) and eleven (D21) days after the cessation of the clindamycin-treatment. In addition, microbiota was analysed the day before E. faecalis V583 inoculation (D10) for trials 1 and 2, and four days after the end of the clindamycin-treatment (D14) for trials 4 and 5. For trial 3, microbiota was analysed at baseline (D0) and 11 days after inoculation of E. faecalis V583 (D21) only. A total of 183 samples were analysed (Supplementary Table S1).
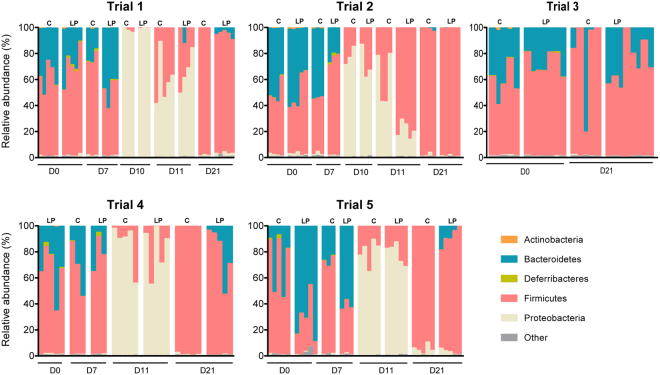
Because bacterial communities varied between mice and trials at baseline at genus level, phylum-based analysis is presented for each trial (Fig. 3 and Supplementary Table S2). At baseline, the mouse gut microbiota was dominated by Firmicutes and Bacteroidetes, accounting for >95% of total microbiota with presence of Proteobacteria and Actinobacteria at lower levels (accounting for less than 2%). Using a permutational analysis of variance (PERMANOVA), no major effect of L. paracasei CNCM I-3689 on microbiota composition was detected after daily supplementation for seven days (Supplementary Table S3). The effect of clindamycin treatment (D10 and D11) on gut microbiota was evaluated on both alpha and beta-diversity. A significant reduction of richness (as measured by number of Operational Taxonomic Units (OTUs)) and evenness (as measured by Shannon index) of the gut microbiota was observed (Supplementary Figure S3, P < 0.05). A significant shift in microbiota represented by a principal coordinate analysis (Supplementary Figure S4) was observed in each group of mice following clindamycin treatment (D11) for both weighted and unweighted UniFrac distances (PERMANOVA tests P = 0.001). At taxonomical level, the phylum Bacteroidetes was severely reduced to non detectable level, while a drastic bloom of Proteobacteria was observed (increase from less than 2% to up to 97%) following clindamycin administration. Eleven days (D21) after the end of the clindamycin treatment, despite a decrease of Proteobacteria close to baseline level (~2%) and an increase of the Firmicutes, the microbiota composition was still different from baseline, regardless of L. paracasei supplementation and E. faecalis V583 inoculation (Supplementary Table S4). In trials 4 and 5, Bacteroidetes were detected in all animals receiving L. paracasei CNCM I-3689 and absent in the control group (P = 10−5). A lower abundance of Firmicutes (P = 0.0039) and Proteobacteria (P = 0.075) following L. paracasei CNCM I-3689 supplementation was also observed. These results support that administration of L. paracasei CNCM I-3689 improved microbiota recovery after cessation of antibiotic-treatment. While all animals receiving L. paracasei CNCM I-3689 had detectable level of the Bacteroides genus, none had in the control group. Other genera from Bacteroidetes were affiliated mostly to the Bacteroidales order. They included an unknown genus from the S24-7 family and Parabacteroides (Porphyromonadaceae) in 70 and 60% of the mice, respectively (Supplementary Figure S5). When mice were inoculated with VRE, the phylum Bacteroidetes was more abundant upon L. paracasei CNCM I-3689-supplementation in trials 1 (with the exception of an outlier) and 3, but not in trial 2 (Fig. 3). The effect of L. paracasei CNCM I-3689 on the increase of Bacteroidetes (P = 0.0445) was statistically confirmed when data of trials 1 and 3 were pooled, supporting that Bacteroidetes recovery is associated with improved VRE reduction. Notably, the improved microbiota recovery in the absence of E. faecalis V583 suggests that E. faecalis V583 interferes on the effect of L. paracasei CNCM I-3689 supplementation. Furthermore, to examine whether a Lactobacillus strain with no anti-VRE effect promoted Bacteroidetes recovery, we performed a trial with L. rhamnosus CNCM I-3690 supplementation and without VRE inoculation. We analysed by 16S sequencing the fecal microbiota of control and L. rhamnosus CNCM I-3690 -supplemented mice collected at D0 and D21. Microbiota composition analysis at the phylum level revealed no difference between control and supplemented groups at D21 (Figure S6 and Supplementary Table S5). Notably, Bacteroidetes was not detected in any of the groups at D21. Together, our results show that of the two strains tested L. paracasei CNCM I-3689 specifically improves the microbiota recovery after antibiotic induced dysbiosis by promoting resilience of some genera from Bacteroidetes, especially Bacteroides and other genera of the Bacteroidales order. However, L. paracasei CNCM I-3689 has no major effect on the microbiota in non-antibiotic treated mice, neither during antibiotic treatment.
Figure 3.
L. paracasei CNCM I‐3689 supplementation improves Bacteroidetes recovery. Kinetics of the microbiota at phylum level at all days for control and L. paracasei supplemented groups in each trial. Trials 1 to 3 were performed in presence of E. faecalis V583 and trials 4 and 5 in absence of E. faecalis V583.
L. paracasei CNCM I-3689 does not inhibit growth of E. faecalis V583 in vitro
Our in vivo studies have demonstrated an impact of L. paracasei CNCM I-3689 strain on E. faecalis persistence in feces. To gain insights on the underlying mechanisms we investigated whether L. paracasei CNCM I-3689 strain exerts a direct effect on the growth of E. faecalis V583 in vitro through the production of a diffusible inhibitory molecule. We cultured E. faecalis V583 in the presence of culture supernatant of L. paracasei CNCM I-3689 neutralised at pH 7.0. No effect of L. paracasei CNCM I-3689 supernatant was observed on growth of E. faecalis V583 under the experimental conditions used (Supplementary Figure S7).
L. paracasei CNCM I-3689 modulates intestinal host responses in parallel to the reduction of fecal levels of VRE
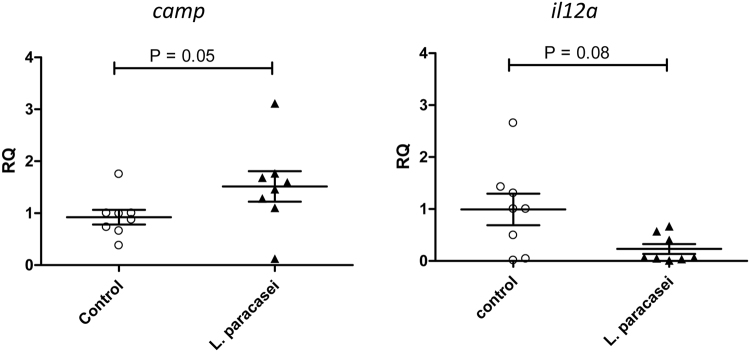
We further investigated whether the impact of L. paracasei CNCM I-3689 on the reduction of E. faecalis V583 was associated with modulation of the intestinal host responses in ileum, a major site of immune response. Expression of 42 genes involved in various intestinal defenses mechanisms such as cytokines, immune responses, regulation of cell proliferation and differentiation, the intestinal barrier, antimicrobial peptides was analysed using a custom-designed TaqMan Low Density Array (Supplementary Table S6). Analysis was carried out on the ileum of eight mice for control and L. paracasei groups of trials 1 and 3 at D21 where maximum effect of VRE reduction by L. paracasei CNCM I-3689 was observed. We identified two genes of interest when differences between control and L. paracasei groups were ranked by p-value: camp that encodes the anti-microbial peptide cathelicidin, and il12a that encodes the p35 subunit of the pro-inflammatory cytokine IL-12 (Supplementary Table S3). We further analysed the expression of these genes by single Taqman assays (Fig. 4). Expression of camp was increased in L. paracasei compared to control group (P = 0.05). We also observed a trend of decreased expression of il12a in L. paracasei compared to control group (P = 0.08). We also investigated the effect of L. paracasei administration on ileal and colonic epithelium structure through the analysis of tight junction and cell adhesion proteins ZO-1, claudin-1 and claudin-2 and of proliferation markers Ki67 and PCNA. We found no differences for ZO-1 and claudins at the ileal and colonic levels (data not shown). In contrast, our data revealed a significant increase both of Ki67- and PCNA-positive cells in the colonic epithelium after L. paracasei CNCM I-3689 supplementation (Supplementary Figure S8), suggesting L. paracasei CNCM I-3689 could contribute to the dynamic of the regeneration process of the small intestine.
Figure 4.
L. paracasei CNCM I‐3689 supplementation modulates the expression of camp and il12a expression in the ileum. Ileal camp and il12a gene expression were analysed by QPCR in Control or L. paracasei groups. RQ is the relative abundance of camp or Il12a mRNA normalised to those of gapdh and compared with values for control mice. 8 mice were analysed in both groups. Each point on the graph corresponds to an individual. *Significantly different from control mice values (P = 0.05).
Bile acids and SCFA analysis
We further investigated the impact of L. paracasei CNCM I-3689 on metabolites that were previously reported to have anti-pathogenic effects. SCFA were quantified in cecal contents at D21 from trials 1, 4 and 5, in which Bacteroidetes was increased following L. paracasei CNCM I3689 supplementation. Regarding, SCFA, there was no difference between controls and L. paracasei treated-mice in absolute amounts for acetate and butyrate (Supplementary Table S7). However L. paracasei CNCM I-3689 increased the absolute amount of propionate compared to control group (P < 0.05 in trials 1 and 5). Noticeably, very low amount of propionate detected in control groups may emphasise the difference.
In addition, bile acids were analysed from cecal contents at D21 from trial 3. Six primary and secondary bile acids were found to dominate the cecal content of control and L. paracasei mice: cholic and taurocholic acids (primary), and deoxycholic, lithocholic, ursodeoxycholic, and taurodeoxycholic acids (secondary). Although the variability was found to be high within groups, there was a trend towards higher level of the secondary bile acid lithocholic acid in L. paracasei treated mice (P = 0.057) (Supplementary Figure S9).
Discussion
Among intestinal pathobionts, vancomycin-resistant enterococci (VRE) are a leading cause of health-care associated and community-acquired infections associated with increased morbidity of patients and hospital costs. Promotion of the colonisation resistance provided by the gut microbiota is an attractive non-antibiotic alternative to minimise the colonisation and persistence of VRE. Few clinical studies have established that probiotics enhance intestinal barrier against intestinal pathogens20,21,39. The development of probiotics against multi-antibiotic resistant nosocomial enterococci is a significant area of unmet medical need with the goal of reducing pathogen transmission and dissemination. This study highlights that strain L. paracasei CNCM I-3689 reduces the fecal level of VRE and promotes the recovery of some dominant members of the gut microbiota, mostly Bacteroidetes, after antibiotic-induced dysbiosis. We found that Camp (LL-37) and IL-12a are candidate host factors modulated by the supplementation with L. paracasei CNCM I-3689 strain in the presence of E. faecalis. The candidate host factors are acting mainly at two different levels in terms of immune response: Camp as an antimicrobial peptide via innate mucosal immunity and IL-12a as a potential link with adaptive immunity via gut macrophages and DCs that could be modulated upon L. paracasei CNCM I-3689 supplementation. Although the expression of inhibitory molecules may differ in vivo and in vitro, the absence of in vitro inhibition of the strain L. paracasei CNCM I-3689 against V583 is not in favor of a direct anti-VRE mechanism such as bacteriocins as observed by Millete et al.40 or recently by Kommineni et al.41. Even if other direct mechanisms for the L. paracasei CNCM I-3689-mediated anti-VRE effect such as nutrient competition may occur, one possible mechanism could be indirect and involve Bacteroidetes and/or the host response.
Given complex interactions among microorganisms in the human microbiota, preventing carriage of antimicrobial-resistant enterococci may be challenging while decreasing the load may be sufficient to prevent infection and dissemination. We assessed the anti-VRE potential of two Lactobacillus strains showing interindividual differences combined with several independent trials. In this worst-case scenario, no preventive effect on the VRE overgrowth of the Lactobacillus strains supplementation was observed. Impact of probiotics on composition of human fecal microbiota in healthy adults has been reported to be minor42. Because of interindividual variability, larger scale trials and greater probing depth are needed to assess if these Lactobacillus strains have any effect on eubiotic microbiota. Alternatively, the response of disrupted microbiota to probiotics appears relevant43. Yet, we repeatedly detected a significant reduction of persisting VRE upon supplementation with L. paracasei CNCM I-3689 in trials 1 and 3, although not all mice responded. Interindividual variation of baseline microbiota has been shown to be a factor that participates to the response to a given intervention, either dietary, pharmaceutical or fecal transplantation42,44. Another confounding factor in our experimental conditions is that coprophagy of feces with high amount of VRE can fuel cross-contaminations by VRE. However, our results suggest that supplementation with L. paracasei CNCM I-3689 rather improves reduction of persisting E. faecalis V583 than eradicates E. faecalis from the gut microbiota. No effect of the strain L. rhamnosus CNCM I-3690 was observed although efficacy of Lactobacillus spp. strains from the same species against VRE was reported. The strain L. rhamnosus GG reduced at least transiently the number of VRE in adult or children VRE-positive patients within four and three weeks26,27, whereas supplementation of adult VRE-positive patients with L. rhamnosus Lcr35r had no effect on VRE carriage44. However, Vidal et al. reported that this strain was able to decrease VRE Enterococcus faecium in mice up to 34 days after inoculation. These studies highlight that probiotic effects are strain specific. Assessment of anti-VRE effect at the preclinical level in animal trials remains a way to increase the chance to identify good candidates against VRE in human.
Resilience of microbiota following an external challenge, including antibiotics is of high clinical relevance as a less resilient microbiota might predispose to diseases45. Administration of clindamycin has long lasting effect on members of Bacteroidetes phylum in human and mice, mainly Bacteroidaceae46–49. Here we showed that supplementation with L. paracasei CNCM I-3689 is correlated with partial recovery of the gut microbiota for instance Bacteroidetes phylum, mostly Bacteroidales order and with a higher reduction in VRE levels. Interestingly, significant enrichment of obligate anaerobes including Bacteroidetes has been inversely associated with E. faecalis levels50. Reintroduction of Bacteroidetes to antibiotic-treated mice would allow to assess if Bacteroidetes recovery and reduction in levels of fecal VRE are corelated or independent events. Ingestion of bacterial strains that can hasten the recovery of members of Bacteroidales order is therefore of high relevance as they include dominant members of the gut microbiota, in mice and human51 that have important roles on physiological and immunological functions52. Identification of the species and the underlying mechanisms that may mediate L. paracasei CNCM I-3689 effect on VRE reduction is challenging, especially because the presence of VRE seems to interfere with recovery of Bacteroidetes. Ubeda et al.53 demonstrated that reintroduction of a diverse intestinal microbiota containing Barnesiella intestihominis, a member of the Bacteroidales order, to densely VRE-colonised mice eliminates VRE from the intestinal tract. Of note, prospective analysis of patients’ gut microbiota undergoing allogeneic hematopoietic stem cell transplantation substantiated that high level of Barnesiella spp. correlates with resistance to intestinal VRE domination53. Recently, Caballero et al.28 demonstrated by fractionation of the colonic microbiota of ampicillin-treated mice that colonisation resistance against vancomycin-resistant Enterococcus faecium requires bacterial cooperation of four commensal species: two Bacteroidales ampicillin resistant (Bacteroides sartorii and Parabacteroides distasonis) that allow engraftment of two ampicillin sensitive Firmicutes (Clostridium bolteae and Blautia producta). B. producta was shown to directly inhibit VRE growth and to cooperate with B. sartorii, P. distasonis and C. bolteae for intestinal colonisation to mediate VRE clearance. Similarly, cooperative interaction between a β-lactamase producing strain of B. thetaiotaomicron and the microbiota prevented overgrowth of VRE54. On top of that, correlation of Bacteroides spp. preservation during Fidaxomicin treatment and reduced risk of acquisition and overgrowth of VRE further support Bacteroides as contributors of anti-VRE effect55,56. Thus, it is tempting to hypothesise a cooperative mechanism involving the L. paracasei CNCM I-3689 strain and members of the Bacteroidales order. In the case of mice, the transfer of a particular bacterium due to coprophagy may promote Bacteroidetes recovery. A reductionist approach in simplified ecosystems in gnotobiotic mice should help to identify the keystone species of anti-VRE effect. A limitation of the current study is that mice were co-housed in a single cage for all experiments in a ratio of 4–5 mice per cage. Although cages were changed every three days, this set up may raise the question of a cage effect and of the impact of coprophagy on our findings. One outlier mouse in a cage could influence the microbiota recovery (or the VRE levels as discussed previously). However, regarding the effect on the resilience at phyla level, previous work supports that the cage effect and coprophagy has more effect on lower taxonomic levels57. To overcome this confounding factor daily cage change and less mice housed per cage will be used for future experiments.
Mechanisms controlling intestinal expansion of VRE may involve microbiota-mediated inhibition through the production of inhibitory metabolites or nutrient competition, and/or host immune functions modulated by the microbiota22,58. Colonisation resistance against various pathogens associates with restoration of bacterial metabolites48,59,60. Short chain fatty acids are microbial metabolites that have been shown to exert various anti-pathogenic effects, such as direct growth inhibition or reduction of oxygen availability through the host response22,61. Secondary bile acids synthesised by the gut microbiota have a strong antibacterial activity62 and contribute to inhibit C. difficile intestinal colonisation60. The observed trend of higher level of lithocholate and possibly propionate in some trials upon L. paracasei CNCM I-3689 supplementation may contribute to anti-VRE effect and derive from improved recovery of the microbiota, including Bacteroidetes known to be propionate producers63. We could speculate a direct or indirect role of Bacteroidetes in the recovery of propionate and/or secondary bile salt metabolism by the microbiota. Host response analysis provides us clues on the late mechanism against VRE. In accordance with a previous study, we found that L. paracasei CNCM I-3689 induces colonic epithelial cell proliferation64. The rate of intestinal renewal has been shown to provide an important intrinsic defense system as it probably participates to a better reduction of pathogens65. Notably, the supplementation of L. paracasei CNCM I-3689 is associated with an increase of Camp (LL-37 in human), and a decrease of Il12. Cathelicidin LL-37 has potent antimicrobial effect against E. faecalis66 indicating a potential role for an antimicrobial activity against E. faecalis in the gut. No effect of L. paracasei CNCM I-3689 on camp nor il12a expression was observed in a previous study performed on a gnotobiotic model31, arguing for an indirect effect of L. paracasei CNCM I-3689 on the ileal expression of camp or il12a genes. It is tempting to postulate that L. paracasei CNCM I-3689 may have an indirect effect on intestinal host response through improved recovery of Bacteroidetes by potentially creating a more favourable niche. Interestingly, Koh and collaborators demonstrated that Bacteroidetes namely Bacteroides thetaiotaomicron stimulates LL-37/CRAMP intestinal expression that can promote colonisation resistance against Candida albicans, another opportunistic pathogen in immunocompromised patients67. Also, intestinal expansion of VRE is controlled by RegIIIg, which ileal expression is induced by gram-negative commensal bacteria outnumbered by Bacteroidetes68,69. Our study highlights that a better understanding of the role of the microbiota on the production of ileal antimicrobial peptides and bacterial metabolites is needed to better control intestinal expansion of VRE. Despite the complexity of the pathobiome interplay between the microbiota, the host and the pathobiont, this preclinical study reveals the strain L. paracasei CNCM I-3689 improves VRE-reduction and resilience of some members of the gut microbiota (mainly Bacteroidetes) in an antibiotic-induced dysbiosis. Preclinical and clinical studies are required to corroborate and elucidate the underlying anti-VRE mechanisms of L. paracasei CNCM I-3689 and to assess its efficacy in human.
Material and Methods
Bacterial strains and growth
E. faecalis V583 strain70 was grown in M17 supplemented with 0.5% glucose (M17G) at 37 °C under static conditions. Strains L. paracasei CNCM I-3689 and L. rhamnosus CNCM I-3690 were grown in MRS at 37 °C in static liquid medium or in anaerobic jars on plates. Bacterial inocula were prepared using bacteria collected by centrifugation 1 h after reaching stationary phase. Bacteria were washed twice with 0.9% saline solution and stored as dry frozen pellets at −80 °C35. Before inoculation, a frozen bacterial pellet was suspended in a saline solution and serial diluted before plating on M17G or MRS agar to determine the bacterial count and adjust concentration at 109 CFU of Lactobacillus strains in 0.1 ml for administration to mice.
In vivo experimental design
All animals were handled in strict accordance with good animal practice as defined by the local animal welfare bodies (Unité IERP, INRA Jouy-en-Josas, France). Animal work was carried out under the authority of license issued by the national Direction des Service Vétérinaires (accreditation number A78–187 to LR-G), and approved by COMETHEA, the appropriate local ethic committee (authorisation number 12/081). CF-1 mice originally purchased from Envigo (Indianapolis, USA) were raised under specific pathogen-free conditions at the CDTA-CNRS (Orléans, France). Mouse experiments were performed using male CF-1 mice aged 6–8-weeks and 4 to 8 mice per group. A maximum of 5 mice were housed in each cage and were fed with autoclaved food and water ad libitum. Mice received a daily dose of 109 CFU of L. paracasei CNCM I-3689 or L. rhamnosus CNCM I-3690 strain in 0.1 ml of 0.9% NaCl (saline solution) by orogastric inoculation using a feeding tube (Ecimed). Animals from the control group received 0.1 ml of saline solution. After one week of supplementation, a dose of 1.4 mg/day of clindamycin was administered subcutaneously daily for three days. In experiments referred as trials 1, 2 and 3 mice received 1010 colony-forming units (CFU) of E. faecalis VRE strain V583 by orogastric inoculation one day after stopping the clindamycin treatment (D10).
Stool samples were collected at D0, D7, D10 or D11, D14, D18 and D21. Fecal samples collected for 16S rRNA gene survey analysis of the whole microbiota were stored at −80 °C. Feces (from 50 to 100 mg/mice) kept at 4 °C were treated within 3 hours after sampling and processed at room temperature. From this stage, all steps were performed in sterile conditions. Samples were weighted and suspended at a dilution of 10−1. An adjusted volume of peptone water was added according to the weight (eg. 900 µl for 100 mg, 450 µl for 50 mg). A volume of 100 µl of the suspension (dilution -1) was used to perform decimal dilutions in peptone water until 10−8. Total enterococci count was monitored by plating onto BEA, and total lactobacilli on MRS medium at 37 °C under anaerobic condition (Gas pack) for 48 h. The number of E. faecalis V583 was followed by plating onto BEA supplemented with vancomycin at 6 µg/ ml. All mice were euthanized at the end of the experiment. Small intestine and colon tissues were recovered and immediately stored in liquid nitrogen or placed into paraformaldehyde solution 4% for further RNA extraction or histological and immunochemistry analysis, respectively.
Analysis of fecal microbiota by 454 pyrosequencing
DNA of fecal samples was extracted following Godon et al. protocol71. Control quality of DNA samples was assessed according to Life Sequencing instructions (Lifesequencing S.L., Valencia, Spain). The 16S rRNA genes were sequenced by Life Sequencing based on the analysis of the V3-V5 region72 using a 454 Life Sciences GS FLX + instrument (Roche).
Bioinformatic analyses were performed using QIIME v.1.973. Data from Life sequencing were assigned to the samples after filtering according to the following criteria: size between 200 and 1000 nt, quality above 25 over a 50 base pairs window, no mismatch authorised in primers and barcode sequences, and absence of polymers larger than 6 nt. Remaining reads were clustered into Operational Taxonomic Units (OTUs) defined at 97% identity using Usearch and representative sequences for each OTU were aligned and taxonomically assigned using Silva database (version 111). For alpha and beta diversity, samples were rarefied to 1500 sequences per sample. Alpha-diversity (that measures diversity within samples) was assessed using rarefaction curves for Shannon index, and numbers of observed OTUs. Beta diversity between samples was performed on weighted and unweighted Unifrac and Bray-Curtis distances using 1500 reads.
In vitro interaction in liquid culture
To assess possible effect of L. paracasei CNCM I-3689 strain on E. faecalis growth in vitro, the supernatant of an overnight culture of L. paracasei CNCM I-3689 was recovered by centrifugation at 15,000 g for 15 min. The pH of the supernatant was adjusted to reach approximately 7 (6.8–7.2) with NaOH. Then, the supernatant was filtrated on 0.2 µm. Five ml of filtrated supernatant were added to 5 ml of M17G. The resulting conditioned medium was inoculated with 100 µL of an overnight culture of E. faecalis V583. E. faecalis growth was compared to growth in a control tube of equal volumes of MRS and M17G. E. faecalis V583 growth was monitored by measuring OD at 600 nm and plating.
Quantification of expression of selected mRNAs in the small intestine
A custom-designed TaqMan Low Density Array (TLDA) card was configured into 8 identical 48-gene sets. The list of genes analysed is given in Supplementary Table S3.
Total RNAs were extracted from small intestine fragments using the mirVana miRNA isolation Kit (Life Technologies) according to the recommendations of the manufacturer. RNA quality was assessed with the Agilent 2100 Bioanalyzer (Agilent Technologies). RNA integrity number (RIN) was equivalent among groups and was of 8.1 ± 0.3 (n = 16). Complementary DNA (cDNA) was generated by reverse transcription (RT) with random hexamers and the High-Capacity cDNA Reverse Transcription d’Applied. cDNA, corresponding to 50 ng of starting RNA, was mixed with TaqMan Universal PCR Master Mix (Applied Biosystems, Inc.) and loaded into one of the eight fill ports on the TLDA microfluidic card. The cards were briefly centrifuged for 1 min at 1300 × g to distribute the reaction mix to each of the reaction wells and were then sealed to prevent well-to-well contamination. PCR amplifications were performed on a QuantStudio Real Time PCR Detection System (Applied Biosystems) under the following thermal-cycling conditions: 95 °C for 10 minutes followed by 40 cycles of 95 °C for 15 seconds and 60 °C for 1 minute. Cycle threshold (Cq) values were extracted using Real-Time qPCR Analysis software (Applied Biosystems) [using Symphony™ Suite analysis software (www.lifetechnologies.com)]. The fold-change (Rq or relative quantification) in the expression of target genes between Lactobacilli-supplemented and control mice were calculated using the comparative 2−ΔΔCq method74,75. Results obtained were normalised to those for the Ubiquitin gene (Mm01201237_m1) as internal control and compared with the mean target gene expression in NaCl treated mice. Further validation of data obtained from TLDA was carried out using single Taqman assays. camp and Il-12a mRNA were analysed using Applied Biosytems designed Taqman assays Mm00438285_m1 and Mm00434165_m1 respectively. The assay Mm99999915_g1 corresponding to gapdh (glyceraldehyde-3-phosphate dehydrogenase) was used as reference gene in single assay measurement.
Histology and immunochemistry
Samples were fixed in 4% paraformaldehyde (24 h, 4 °C), dehydrated, and embedded in paraffin, according to standard histological protocols. Sections (5 μm) of tissues were mounted on SuperFrost Plus slides (Thermo Fisher, Waltham, MA, USA). After dewaxing and rehydratation, sections were heated at 97 °C in 10 mM citrate buffer (pH 6.0) for 40 min. Nonspecific binding was blocked using protein block serum-free (X0909; DakoCytomation) for 1 h at room temperature. Sections were incubated with primary antibodies diluted in antibody diluent (S3022; DakoCytomation) overnight at 4 °C. The primary antibodies used were anti-Z0–1 (1:500; LifeTech), anti-claudin-1 (1:500; InVitrogen), anti-claudin-2 (1:500; InVitrogen), anti-PCNA (1:1000; GeneTex) and anti-Ki67 (1:50, DakoCytomation). Nuclei were stained with Hoechst before mounting the slides using Fluorescent Mounting Media (Dako). Sections were scanned using a Pannoramic Scan digital slide scanner (3DHistech) and analysed using digital slide scanner Pannoramic scan (3Dhistech). For Ki67 and PCNA 10 crypts were measured per mice and per cut.
Short Chain Fatty Acids (SCFA) analysis
SCFA (acetic, propionic and butyric acid) were analysed and concentrations were determined with gas chromatography (Nelson 1020, Perkin‐Elmer, St Quentin en Yvelines, France) as described previously76. Results are expressed in relative percentage of each SCFA.
Quantification of cecal bile acids
Cecal contents (5 controls and 8 from L. paracasei treated mice) from trial 3 were used to quantify primary and secondary bile acids. Bile acids analysis was performed at Bioaster (Lyon, France). Samples were prepared from 50 to 60 mg of −80 °C frozen cecal content using aqueous methanol extraction process. Bile acids extracts were analysed by LC-MS/MS on a triple quadrupole Thermo Quantum Ultra (SN: TQU00665) combined to a Dionex Ultimate 3000 HPLC system (SN: 8074045 & 8087183). Samples were separated on a C18 column (2,7 μM, 150 × 2,1 mm) from Ascentis Express using a methanol/water gradient containing 5 mM ammonium acetate and 0,012% formic acid. All bile acid standards: Lithocholic acid, Chenodeoxycholic acid, Deoxycholic acid, Ursodeoxycholic acid, Cholic acid, Glycodeoxycholic acid, Glycoursodeoxycholic acid, Glychenodeoxycholic acid, Glychocolic acid, Taurolithocholic acid, Taurodeoxycholic acid, Taurocholic acid, were bought from Sigma Aldrich. Results of bile salt quantification were expressed in peak area, corrected for sample weight.
Statistical analysis
Comparison of kinetics of VRE in control and L. paracasei groups was performed separately on each trial. The difference at each time point was tested by Mann-Whitney test. The P-values p11, p14, p16, p18, p21 corresponding to days 11, 14, 16, 18, 21 were gathered using Fisher’s combined probability method: the statistic S = −Σ log(pt) equal to the sum of the inverse of the log-transformed gets larger as soon as a significant difference between groups is observed at one time point. A permutation p-value was then computed by random reallocation of group labels (control and L. paracasei).
The PERMANOVA (Permutational multivariate analysis of variance) test implemented in the function adonis2 of the R-package vegan was used to test global differences in microbiota composition (a) between D0 and D7 separately on control and L. paracasei groups grouping mice from the five trials, (b) between D0 and D21 separately on each trial, (c) between D0 and D21 separately for control and L. paracasei groups and for trials with and without VRE inoculation (trials 1 and 3 and trials 4 and 5, respectively). In order to erase individuals effect, in each comparison between two groups, only mice with measurements in both groups were considered. PERMANOVA test is based on prior calculation of a matrix of two-by-two distance between all pairs of samples. In our analysis, we considered the following distances: weighted- and unweighted-Unifrac distance, and Bray-Curtis distance based on OTU. These three distances led to similar conclusions.
The difference of abundances between control and L. paracasei groups for each phylum was assessed using two distinct statistical tests: Mann-Whitney test for phyla with less than 10% of zeros and a Fisher presence/absence test for phyla with more than 50% of zeros. For phyla, which proportion of zeros ranged between 10% and 50%, the minimum of the two p-values was considered.
Short chain fatty acid, bile salts and host response data were analysed by the Mann-Whitney test (GraphPad 4.03).
Electronic supplementary material
Acknowledgements
We thank the staff of the IERP and CDTA Orléans animal units for skilful animal care. We also thank C. Bevilacqua of the ICE and J. Rivière and A. Boukadiri of the histology plateform for methodological support. We are thankful B. Goustard-Langelier for TLDA advice and B. Laroche and S. Labarthe for discussion on data analysis. This work was supported by INRA and Danone Research.
Author Contributions
Conceived and designed the experiments: L.C., M.D., L.R.G., C.C., J.v.H.V., G.G., P.S. Performed the experiments: L.C., B.C., M.F., C.C., L.R.G. Analysed the data: L.C., M.D., S.P., C.C., L.R.G., P.S. Wrote the paper: M.D., C.C., S.P., L.R.G., P.S. All authors reviewed the manuscript.
Competing Interests
L.R.G., C.C., S.P. and P.S. are employees of I.N.R.A., a national research institute. M.D., J.v.H.V. and G.G. are employees of Danone Nutricia Research. This work was co-funded by I.N.R.A. and Danone Nutricia Research and covered supplies, subcontracting and salaries of L.C. and B.C. who were paid by I.N.R.A. M.F. was a trainee subsidized by I.N.R.A. Seven co-authors, L.C., M.D., C.C., G.G., L.R.G., J.v.H.V. and P.S. have a joined patent application related to this work (International Application No. PCT/IB2015/052752). It covers the effect of use of L. paracasei CNCM I-3689 on the recovery of the intestinal microbiota diversity after dysbiosis and on the decrease of Enterococcus faecalis in the intestinal microbiota.
Footnotes
Laureen Crouzet and Muriel Derrien contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23437-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Sommer F, Backhed F. The gut microbiota-masters of host development and physiology. Nat Rev Microbiol. 2013;11:227–238. doi: 10.1038/nrmicro2974. [DOI] [PubMed] [Google Scholar]
- 2.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li J, et al. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 4.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol. 2011;23:473–480. doi: 10.1016/j.coi.2011.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weinstein JW, et al. Resistant enterococci: a prospective study of prevalence, incidence, and factors associated with colonization in a university hospital. Infect Control Hosp Epidemiol. 1996;17:36–41. doi: 10.2307/30142363. [DOI] [PubMed] [Google Scholar]
- 7.Alverdy J, et al. Gut-derived sepsis occurs when the right pathogen with the right virulence genes meets the right host: evidence for in vivo virulence expression in Pseudomonas aeruginosa. Ann Surg. 2000;232:480–489. doi: 10.1097/00000658-200010000-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liss BJ, et al. Intestinal colonisation and blood stream infections due to vancomycin-resistant enterococci (VRE) and extended-spectrum beta-lactamase-producing Enterobacteriaceae (ESBLE) in patients with haematological and oncological malignancies. Infection. 2012;40:613–619. doi: 10.1007/s15010-012-0269-y. [DOI] [PubMed] [Google Scholar]
- 9.Montassier E, Batard E, Gastinne T, Potel G, de La Cochetiere MF. Recent changes in bacteremia in patients with cancer: a systematic review of epidemiology and antibiotic resistance. Eur J Clin Microbiol Infect Dis. 2013;32:841–850. doi: 10.1007/s10096-013-1819-7. [DOI] [PubMed] [Google Scholar]
- 10.Sartelli M. A focus on intra-abdominal infections. World J Emerg Surg. 2010;5:9. doi: 10.1186/1749-7922-5-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montravers P, et al. Clinical and microbiological profiles of community-acquired and nosocomial intra-abdominal infections: results of the French prospective, observational EBIIA study. J Antimicrob Chemother. 2009;63:785–794. doi: 10.1093/jac/dkp005. [DOI] [PubMed] [Google Scholar]
- 12.Magill SS, et al. Multistate point-prevalence survey of health care-associated infections. N Engl J Med. 2014;370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mendes RE, et al. Longitudinal (2001–14) analysis of enterococci and VRE causing invasive infections in European and US hospitals, including a contemporary (2010–13) analysis of oritavancin in vitro potency. J Antimicrob Chemother. 2016;71:3453–3458. doi: 10.1093/jac/dkw319. [DOI] [PubMed] [Google Scholar]
- 14.Jones RN, et al. Resistance surveillance program report for selected European nations (2011) Diagn Microbiol Infect Dis. 2014;78:429–436. doi: 10.1016/j.diagmicrobio.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 15.Ubeda C, et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J Clin Invest. 2010;120:4332–4341. doi: 10.1172/JCI43918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taur Y, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin Infect Dis. 2012;55:905–914. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Remschmidt C, et al. Surveillance of Antibiotic Use and Resistance in Intensive Care Units (SARI) Dtsch Arztebl Int. 2017;114:858–865. doi: 10.3238/arztebl.2017.0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sievert DM, et al. Antimicrobial-resistant pathogens associated with healthcare-associated infections: summary of data reported to the national healthcare safety network at the centers for disease control and prevention, 2009–2010. Infect Control Hosp Epidemiol. 2013;34:1–14. doi: 10.1086/668770. [DOI] [PubMed] [Google Scholar]
- 19.Lake JG, et al. Pathogen Distribution and Antimicrobial Resistance Among Pediatric Healthcare-Associated Infections Reported to the National Healthcare Safety Network, 2011–2014. Infect Control Hosp Epidemiol. 2018;39:1–11. doi: 10.1017/ice.2017.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. 2013;13:790–801. doi: 10.1038/nri3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Derrien M, Vlieg vanH. J. E. Fate, activity, and impact of ingested bacteria within the human gut microbiota. Trends Microbiol. 2015;23:354–366. doi: 10.1016/j.tim.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 22.Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. 2013;138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sohn KM, Cheon S, Kim YS. Can Fecal Microbiota Transplantation (FMT) Eradicate Fecal Colonization With Vancomycin-Resistant Enterococci (VRE)? Infect Control Hosp Epidemiol. 2016;37:1519–1521. doi: 10.1017/ice.2016.229. [DOI] [PubMed] [Google Scholar]
- 24.Stripling J, et al. Loss of Vancomycin-Resistant Enterococcus Fecal Dominance in an Organ Transplant Patient With Clostridium difficile Colitis After Fecal Microbiota Transplant. Open Forum Infect Dis. 2015;2:ofv078. doi: 10.1093/ofid/ofv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crouzet L, Rigottier-Gois L, Serror P. Potential use of probiotic and commensal bacteria as non-antibiotic strategies against vancomycin-resistant enterococci. FEMS Microbiol Lett. 2015;362:fnv012. doi: 10.1093/femsle/fnv012. [DOI] [PubMed] [Google Scholar]
- 26.Manley KJ, Fraenkel MB, Mayall BC, Power DA. Probiotic treatment of vancomycin-resistant enterococci: a randomised controlled trial. Med J Aust. 2007;186:454–457. doi: 10.5694/j.1326-5377.2007.tb00995.x. [DOI] [PubMed] [Google Scholar]
- 27.Szachta P, Ignys I, Cichy W. An evaluation of the ability of the probiotic strain Lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J Clin Gastroenterol. 2011;45:872–877. doi: 10.1097/MCG.0b013e318227439f. [DOI] [PubMed] [Google Scholar]
- 28.Caballero S, et al. Cooperating Commensals Restore Colonization Resistance to Vancomycin-Resistant Enterococcus faecium. Cell Host Microbe. 2017;21:592–602. doi: 10.1016/j.chom.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dubberke ER, et al. Clearance of Vancomycin-Resistant Enterococcus Concomitant With Administration of a Microbiota-Based Drug Targeted at Recurrent Clostridium difficile Infection. Open Forum Infect Dis. 2016;3:ofw133. doi: 10.1093/ofid/ofw133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turpin W, Humblot C, Thomas M, Guyot JP. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int J Food Microbiol. 2010;143:87–102. doi: 10.1016/j.ijfoodmicro.2010.07.032. [DOI] [PubMed] [Google Scholar]
- 31.Archambaud C, et al. Impact of lactobacilli on orally acquired listeriosis. Proc Natl Acad Sci USA. 2012;109:16684–16689. doi: 10.1073/pnas.1212809109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Laval L, et al. Lactobacillus rhamnosus CNCM I-3690 and the commensal bacterium Faecalibacterium prausnitzii A2-165 exhibit similar protective effects to induced barrier hyper-permeability in mice. Gut Microbes. 2015;6:1–9. doi: 10.4161/19490976.2014.990784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grompone G, et al. Anti-inflammatory Lactobacillus rhamnosus CNCM I-3690 strain protects against oxidative stress and increases lifespan in Caenorhabditis elegans. PLoS One. 2012;7:e52493. doi: 10.1371/journal.pone.0052493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Donskey CJ, Hanrahan JA, Hutton RA, Rice LB. Effect of parenteral antibiotic administration on persistence of vancomycin-resistant Enterococcus faecium in the mouse gastrointestinal tract. J Infect Dis. 1999;180:384–390. doi: 10.1086/314874. [DOI] [PubMed] [Google Scholar]
- 35.Rigottier-Gois L, et al. The Surface Rhamnopolysaccharide Epa of Enterococcus faecalis Is a Key Determinant of Intestinal Colonization. J Infect Dis. 2015;211:62–71. doi: 10.1093/infdis/jiu402. [DOI] [PubMed] [Google Scholar]
- 36.Kuch A, et al. Insight into antimicrobial susceptibility and population structure of contemporary human Enterococcus faecalis isolates from Europe. Journal of Antimicrobial Chemotherapy. 2012;67:551–558. doi: 10.1093/jac/dkr544. [DOI] [PubMed] [Google Scholar]
- 37.Nallapareddy SR, Wenxiang H, Weinstock GM, Murray BE. Molecular characterization of a widespread, pathogenic, and antibiotic resistance-receptive Enterococcus faecalis lineage and dissemination of its putative pathogenicity island. J. Bacteriol. 2005;187:5709–5718. doi: 10.1128/JB.187.16.5709-5718.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matos RC, et al. Enterococcus faecalis prophage dynamics and contributions to pathogenic traits. PLoS Genet. 2013;9:e1003539. doi: 10.1371/journal.pgen.1003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butel MJ. Probiotics, gut microbiota and health. Med Mal Infect. 2014;44:1–8. doi: 10.1016/j.medmal.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 40.Millette M, et al. Capacity of human nisin- and pediocin-producing lactic acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl Environ Microbiol. 2008;74:1997–2003. doi: 10.1128/AEM.02150-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kommineni S, et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature. 2015;526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kristensen NB, et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med. 2016;8:52. doi: 10.1186/s13073-016-0300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McFarland LV. Use of probiotics to correct dysbiosis of normal microbiota following disease or disruptive events: a systematic review. BMJ Open. 2014;4:e005047. doi: 10.1136/bmjopen-2014-005047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Derrien M, Veiga P. Rethinking Diet to Aid Human-Microbe Symbiosis. Trends Microbiol. 2017;25:100–112. doi: 10.1016/j.tim.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 45.Sommer F, Anderson JM, Bharti R, Raes J, Rosenstiel P. The resilience of the intestinal microbiota influences health and disease. Nat Rev Microbiol. 2017;15:630–638. doi: 10.1038/nrmicro.2017.58. [DOI] [PubMed] [Google Scholar]
- 46.Lawley TD, et al. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect Immun. 2009;77:3661–3669. doi: 10.1128/IAI.00558-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buffie CG, et al. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to Clostridium difficile-induced colitis. Infect Immun. 2012;80:62–73. doi: 10.1128/IAI.05496-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jump RL, et al. Metabolomics analysis identifies intestinal microbiota-derived biomarkers of colonization resistance in clindamycin-treated mice. PLoS One. 2014;9:e101267. doi: 10.1371/journal.pone.0101267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 2007;1:56–66. doi: 10.1038/ismej.2007.3. [DOI] [PubMed] [Google Scholar]
- 50.Pham TA, et al. Epithelial IL-22RA1-mediated fucosylation promotes intestinal colonization resistance to an opportunistic pathogen. Cell Host Microbe. 2014;16:504–516. doi: 10.1016/j.chom.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao L, et al. A catalog of the mouse gut metagenome. Nat Biotechnol. 2015;33:1103–1108. doi: 10.1038/nbt.3353. [DOI] [PubMed] [Google Scholar]
- 52.Mazmanian SK, Liu CH, Tzianabos AO, Kasper DL. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–118. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 53.Ubeda C, et al. Intestinal microbiota containing Barnesiella species cures vancomycin-resistant Enterococcus faecium colonization. Infect Immun. 2013;81:965–973. doi: 10.1128/IAI.01197-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stiefel U, Nerandzic MM, Pultz MJ, Donskey CJ. Gastrointestinal colonization with a cephalosporinase-producing bacteroides species preserves colonization resistance against vancomycin-resistant Enterococcus and Clostridium difficile in cephalosporin-treated mice. Antimicrob Agents Chemother. 2014;58:4535–4542. doi: 10.1128/AAC.02782-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deshpande A, et al. Effect of Fidaxomicin versus Vancomycin on Susceptibility to Intestinal Colonization with Vancomycin-Resistant Enterococci and Klebsiella pneumoniae in Mice. Antimicrob Agents Chemother. 2016;60:3988–3993. doi: 10.1128/AAC.02590-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nerandzic MM, Mullane K, Miller MA, Babakhani F, Donskey CJ. Reduced acquisition and overgrowth of vancomycin-resistant enterococci and Candida species in patients treated with fidaxomicin versus vancomycin for Clostridium difficile infection. Clin Infect Dis. 2012;55(Suppl 2):S121–126. doi: 10.1093/cid/cis440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hildebrand F, et al. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14:R4. doi: 10.1186/gb-2013-14-1-r4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vogt SL, Pena-Diaz J, Finlay BB. Chemical communication in the gut: Effects of microbiota-generated metabolites on gastrointestinal bacterial pathogens. Anaerobe. 2015;34:106–115. doi: 10.1016/j.anaerobe.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 59.Lewis BB, Pamer EG. Microbiota-Based Therapies for Clostridium difficile and Antibiotic-Resistant Enteric Infections. Annu Rev Microbiol. 2017;71:157–178. doi: 10.1146/annurev-micro-090816-093549. [DOI] [PubMed] [Google Scholar]
- 60.Buffie CG, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2015;517:205–208. doi: 10.1038/nature13828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rivera-Chavez F, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell Host Microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Begley M, Gahan CG, Hill C. The interaction between bacteria and bile. FEMS Microbiol Rev. 2005;29:625–651. doi: 10.1016/j.femsre.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 63.Reichardt N, et al. Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J. 2014;8:1323–1335. doi: 10.1038/ismej.2014.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jones RM, et al. Symbiotic lactobacilli stimulate gut epithelial proliferation via Nox-mediated generation of reactive oxygen species. EMBO J. 2013;32:3017–3028. doi: 10.1038/emboj.2013.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cliffe LJ, et al. Accelerated intestinal epithelial cell turnover: a new mechanism of parasite expulsion. Science. 2005;308:1463–1465. doi: 10.1126/science.1108661. [DOI] [PubMed] [Google Scholar]
- 66.Scheb-Wetzel M, Rohde M, Bravo A, Goldmann O. New insights into the antimicrobial effect of mast cells against Enterococcus faecalis. Infect Immun. 2014;82:4496–4507. doi: 10.1128/IAI.02114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan D, et al. Activation of HIF-1alpha and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21:808–814. doi: 10.1038/nm.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Brandl K, et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature. 2008;455:804–807. doi: 10.1038/nature07250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kinnebrew MA, et al. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534–543. doi: 10.1086/650203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahm DF, et al. In vitro susceptibility studies of vancomycin-resistant Enterococcus faecalis. Antimicrob. Agents Chemother. 1989;33:1588–1591. doi: 10.1128/AAC.33.9.1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Godon JJ, Zumstein E, Dabert P, Habouzit F, Moletta R. Molecular microbial diversity of an anaerobic digestor as determined by small-subunit rDNA sequence analysis. Appl Environ Microbiol. 1997;63:2802–2813. doi: 10.1128/aem.63.7.2802-2813.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sim K, et al. Improved detection of bifidobacteria with optimised 16S rRNA-gene based pyrosequencing. PLoS One. 2012;7:e32543. doi: 10.1371/journal.pone.0032543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 75.Rajkumar T, et al. A 7 gene expression score predicts for radiation response in cancer cervix. BMC Cancer. 2009;9:365. doi: 10.1186/1471-2407-9-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lan A, et al. Increased induction of apoptosis by Propionibacterium freudenreichii TL133 in colonic mucosal crypts of human microbiota-associated rats treated with 1,2-dimethylhydrazine. Br J Nutr. 2008;100:1251–1259. doi: 10.1017/S0007114508978284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.