Abstract
In clinical genetics, detection of single nucleotide polymorphisms (SNVs) as well as copy number variations (CNVs) is essential for patient genotyping. Obtaining both CNV and SNV information from WES data would significantly simplify clinical workflow. Unfortunately, the sequence reads obtained with WES vary between samples, complicating accurate CNV detection with WES. To avoid being dependent on other samples, we developed a within-sample comparison approach (WISExome). For every (WES) target region on the genome, we identified a set of reference target regions elsewhere on the genome with similar read frequency behavior. For a new sample, aberrations are detected by comparing the read frequency of a target region with the distribution of read frequencies in the reference set. WISExome correctly identifies known pathogenic CNVs (range 4 Kb–5.2 Mb). Moreover, WISExome prioritizes pathogenic CNVs by sorting them on quality and annotations of overlapping genes in OMIM. When comparing WISExome to four existing CNV detection tools, we found that CoNIFER detects much fewer CNVs and XHMM breaks calls made by other tools into smaller calls (fragmentation). CODEX and CLAMMS seem to perform more similar to WISExome. CODEX finds all known pathogenic CNVs, but detects much more calls than all other methods. CLAMMS and WISExome agree the most. CLAMMS does, however, miss one of the known CNVs and shows slightly more fragmentation. Taken together, WISExome is a promising tool for genome diagnostics laboratories as the workflow can be solely based on WES data.
Introduction
Recent technological breakthroughs in DNA analysis methods have not only had a huge impact on genetic research, but also on genome diagnostics. Finding a genetic diagnosis is important as it helps patients and family members in understanding the disease, supports the search for possible treatments, and determines reproductive options in subsequent pregnancies [1]. Currently, most patients with genetic disorders are tested through standard practices such as array-based techniques for detecting copy number variations (either by array comparative genomic hybridization, array-CGH, or by a single nucleotide polymorphism array, SNP-array) [2], or Sanger sequencing of single genes [3, 4], but these methods do not always provide a diagnosis. This has recently changed through the upswing of next-generation sequencing (NGS), which allows for the parallel sequencing of gene panels, whole exomes (WES, whole-exome sequencing), or whole genomes (WGS, whole-genome sequencing). Although WGS obtains nearly the whole-genomic sequence of a patient, its costs are currently still too high for routine testing. As an affordable alternative, WES captures exon-specific regions, called targets, and uses target-specific probe sets to read out these targets (Fig. 1a). While providing a highly accurate way to obtain single nucleotide variation (SNV) information [5], WES data does not allow for straightforward copy number variation (CNV) analysis. The main reason being the non-uniform distribution of reads because: (1) target regions cover only 2% of the genome [6, 7], and (2) the varying amplification efficiency of target regions [8–10]. Moreover, this effect is not consistent over different samples as quality of DNA and environmental differences during sample preparation directly influence probe effectiveness [11]. Consequently, additional array analysis is still used to obtain CNV information for a patient. WES diagnostics would thus greatly benefit from a reliable tool to obtain CNV information from WES data only, as it would eliminate the need for additional separate analyses.
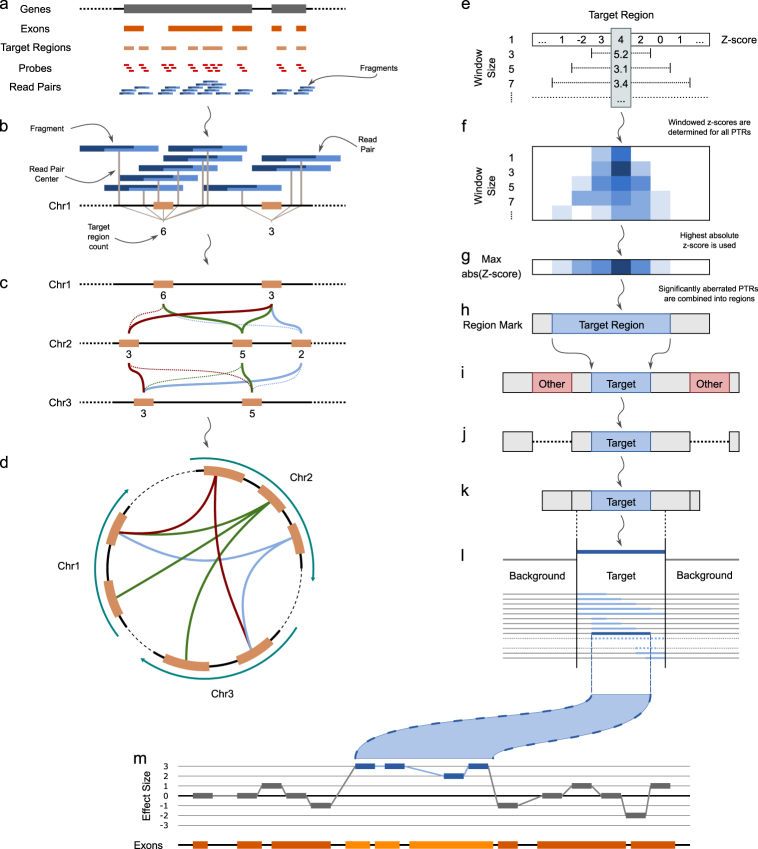
Fig. 1.
a Overview of all regions of importance for WES data. Genes (gray boxes) consist of exons (orange boxes), which are covered by target regions (light orange boxes), for which probes are designed (red boxes) that target a unique sub-sequence of the fragment. Paired-end reads (blue boxes) cover fragments. b Determination of target region read count: a detected fragment is mapped on the reference genome, and consequently assigned to the nearest target region (orange box) based on the center of the fragment (gray lines). c Selection of reference target regions based on the difference in read counts over a set of training samples. For illustrative purposes, we show only one sample and a few target regions on three different chromosomes. The selected reference target regions for the target regions on chromosome 2 are indicated by the straight colored lines. In this example, only reference target regions are considered when the read count differs at most 1 read. Dotted lines are tested but showed a larger difference. d Schematic overview of the selected reference target regions for the target regions on chromosome 2 as shown in c. e Based on its reference set, a z-score for every target region can be calculated (level window size 1). Z-scores of neighboring target probes are aggregated using a combined z-score (here Stouffer’s z-score) for different numbers of neighbors, using odd window sizes from 3 to 15. f Target regions with a significant z-score in any of the windows are marked and kept for further analysis (blue shaded boxes). g For every target region, a final z-score is determined by taking the maximal (positive or negative) z-score across all window sizes (at the target region position). h Stretches of significantly aberrated target regions (blue shaded boxes) are marked as a putative CNV segment. i, j, k To fine-tune the borders of the CNV, the putative CNV segment is considered with all non-putative CNV regions on the same chromosome (i shows the extension, j prunes the putatively aberrated CNVs, k is the result). l To find the exact borders of the CNV, the location of borders of the putative CNV are changed over all possible positions within and directly neighboring the putative CNV region (shaded blue lines in l). The segment that shows the largest difference in mean effect size within the segment compared to the mean effect size outside the segment is selected as the aberrated CNV segment (dark blue line in l). m For visualization purposes, the z-scores (vertical axis) for each target region are plotted across their genomic position (x-axis). Designated aberrated target regions (regions within a detected CNV segment) are colored blue, others gray. The bottom line shows the position of the exons (orange boxes).
Basically, CNVs in WES data can be detected by comparing the read count for a region to the expected read count distribution for that region, representing possible variations in read counts when assuming a diploid genome for that region. When the observed read count for a region differs significantly from the expected read count, this region is designated to be aberrant, either amplified (when having more reads than expected) or deleted (when having less). Generally, the expected read count distribution is derived from a training set of normal (diploid) samples. Several methods for CNV detection in WES data have been developed [12–14], all based on this same principle. For clinical genetic diagnostics of rare diseases, it is important to also detect few and small CNVs, as opposed to mostly large and highly abundant CNVs in cancer [13]. As tools for cancer diagnostics aim to find the distribution of large CNVs as well as their ploidy, they lack the precision required to find intra-genic CNVs. CoNIFER [15] and XHMM [16] are generally believed to perform well for identifying CNVs in a genetic diagnosis setting, based on several comparison studies [10, 17, 18]. Recently, CLAMMS [19] and CODEX [20] have been introduced that claim to outperform CoNIFER and XHMM.
CNVs are generally detected using a training set of normal (diploid) samples to capture the expected read count distribution. Consequently, next to experimental variation, the expected read count distribution also captures the between-sample (biological) variations, which principally would not be necessary. Avoiding the incorporation of the between-sample variation in the expected read distribution potentially increases the sensitivity of CNV detection. We previously developed WISECONDOR for trisomy detection on cell-free fetal DNA based on a within-sample comparison to deal with the fluctuations in read distribution [21]. This within-sample comparison approach assumes that all target region amplifications within a sample undergo the same experimental variations. Hence, if we know which target regions, elsewhere on the genome, respond similarly to the experimental variation, these regions can serve as a reference set and the expected read count distribution can be derived from read counts across this set of reference target regions as measured within the same sample. Here, we present WISExome: a CNV detection method for WES data based on this within-sample comparison principle. Figure 1 gives an overview of the complete procedure and a more elaborate description can be found in the “Methods” section.
We show that WISExome successfully replicates array analysis and compares favorably to other CNV detection tools. Together, the results suggest that WISExome can be used in diagnostics as replacement of array, removing the requirement for separate array analysis for WES diagnostics.
Materials and methods
Sample preparation
Whole-exome sequencing was performed as previously described [22]: Genomic DNA was isolated from blood for 336 samples and prepared using the SeqCap EZ Human Exome Library v3 kit (Roche, Basel, Switzerland), then sequenced using an Illumina HiSeq 2500. Reads were mapped with BWA (0.7.10) to Hg19 [23]. We removed duplicate reads (as marked by Picard Tools 1.111), reads with a mapping quality below 30, and reads that were not part of a read pair. Samples were split into a training set of 319 samples and a test set of 17 samples.
Array analysis
Array analysis was carried out on the high-resolution CytoScan HD array platform (Affymetrix, a part of Thermo Fisher Scientific, Santa Clara, CA, USA) according to the manufacturer’s protocols. This array consists of over 2.6 million copy number markers. Analysis was done using Nexus software (BioDiscovery, El Segundo, CA, USA), using SNPRank segmentation with a minimum of 20 probes per segment and the significance threshold set at 1e-5.
WISExome
WISExome determines excess of target read counts (enriched or depleted) as compared to the expected target read count based on a reference set of target regions within the genome. In the following, we explain the different steps of WISExome, shown in Fig. 1: (1) determining the target region counts from WES read data; (2) identification of the set of reference target regions for a target region; (3) CNV detection; and (4) fine-tuning identified CNVs using a segmentation algorithm. Finally, we explain WISExome’s scoring metric for so-called CNVs and its annotation of CNVs.
Target region read count
In WES data, DNA is fragmented, specific target regions within exons are captured, and subsequently enriched before sequencing (Fig. 1a, target level).
For every target region, there are several probes that are designed to recognize unique subsequences (Fig. 1a, probe level). Paired-end sequencing of the probe-selected fragments covers fragments (Fig. 1b), providing mappable reads. As experimental variation mostly influences the target region enrichment, we are interested in read counts for target regions instead of the probe, fragment, or exon level (the latter is commonly used in other CNV detection tools). Mapped fragments were linked to the closest target region based on the distance between the center of the mapped fragment and the center of a target region (Fig. 1b). To ensure specificity, we require an overlap of at least 20 bp of the mapped fragments with the linked target region. Target region counts, being the sum of mapped fragments linked to that target region, are normalized by the total target region count over the whole genome in the sample, resulting in target region count frequencies.
Creating a reference target region set
The basic idea behind the within-comparison approach is that for every target region, we find target regions on other parts of the genome that behave similarly to experimental variations; the reference target region set. By assuming a sparse number of CNVs, the target region read counts of the reference set represent the within-sample variation of a diploid read count for the associated target region. To find similarly varying target regions, we make use of a training set of samples with no known notable phenotypes. Note that this training set is only used once, to identify reference target regions. Because of lab-specific changes in read depth behavior, this set of reference samples is best obtained from the same lab as the test samples. During testing, we only use information of the test sample, and not the observed read counts in the training samples as other CNV detection tools do. For a target region of consideration, we correlate the observed read counts over all training samples to the read counts of all other target regions. Here, we used the squared Euclidean distance on the target read count frequencies across the 319 training samples (Fig. 1c), and selected the 100 target regions with the lowest distance to build up the reference set for the target region of consideration. We do this for all target regions, so every target region has its own 100 reference target regions. For target regions on the X chromosome, we find their reference target regions considering read count variations across female training samples only. To avoid reference target regions overlapping the CNV, we require that reference target regions lie on other chromosomes than the target region for which the reference set is being build. To avoid that the reference target regions are not similar enough to the target region of consideration, we prune the list of 100 reference target regions. First, the mean and variance of the squared Euclidean distances of the closest reference target for every target region (i.e., the distances of the top-1 target regions in each reference set) is calculated. Then, for every reference set, we remove target regions from the reference set that have a distance larger than the mean plus three times the standard deviation of the top-1 distances (z-score >3). As a result, the number of reference target regions will differ for every target region, see Fig. 1d. Those that have less than 10 references are ignored in further analysis, and are denoted as unreliable target regions (Supplementary Figs. S1, S2, and S3a show that results do not change much when varying this setting).
Finding CNVs
For each target region, its own reference set specifies an expected read count distribution. Hence, we can statistically test whether the observed read count of the target region points toward the region being aberrant or not. For this purpose, we use a z-score with the target region read count frequency as input, and with the mean and variance estimated from the read count frequencies of the reference target regions, measured in the same sample. A target region with a z-score larger than 5.64 (family-wise error rate (FWER) corrected significance level of 0.05, see Supplementary Section SM1) is considered to be amplified, and a target region with a z-score smaller than −5.64 is considered to be deleted. Applying this procedure to the (normal) training samples showed that some target regions are frequently being called, probably due to either large variations in target amplification or because they are part of a common CNV. We decided to exclude target regions being called in more than four of the training samples from further analyses (also denoted as unreliable target regions). This removed 4226 (1.15%) out of 366,795 target regions (Supplementary Figs. S1, S2, and S3b show that WISExome is not sensitive to increasing this threshold). As CNVs generally will be larger than target regions, we improved sensitivity by aggregating z-scores of neighboring target regions. For every target region, we calculated a combined z-score (here, Stouffer’s Z) for differently sized windows (odd window sizes up to 15; larger windows did not change the results, data not shown), see Fig. 1e, f. The z-score for the target region is then set equal to the largest (positive or negative) z-score across all windows, see Fig. 1g. A target region is called aberrant if the aggregated z-score is significant (absolute value larger than 5.64), see Fig. 1h. Consecutive called target regions then make up a CNV. Finally, we take the effect size of the aberration across the CNV into account. Hereto, first, the effect size of a target region is defined by the target region read count divided by the expected target region read count (based on the reference set). The effect size of the CNV is subsequently defined as the median effect size across the target regions it covers. Only CNVs that deviate more than (the arbitrarily chosen) cutoff of 35% from its expectation (i.e., CNV effect size smaller than 0.65 or larger than 1.35) are considered true CNVs.
Fine-tuning detected CNVs
Due to the aggregation of neighboring target regions into the z-score of a target region (the windowing), the borders of the CNVs will not be precise. For example, a strongly aberrant target region might cause that the aggregated z-score of neighboring target regions also becomes significant. In other words, the aggregation improved sensitivity at the cost of precision. Hence, we wanted to fine-tune the borders of every detected CNV to improve the precision of the CNV calls. For that, we devised a segmentation algorithm. Every detected CNV is first extended with eight target regions on each side, Fig. 1i. For all possible segmentations in this region (i.e., all possible start and end positions of the putative CNV), the mean effect size of target regions within the segmentation are compared to the effect sizes of all target regions on the same chromosome that are called unaberrated, using a Student’s t test with a pooled variance (Supplementary Section SM2). The segmentation that maximizes this Student’s t test is chosen as the fine-tuned CNV, see also Fig. 1j–l. Finally, we require that the mean effect size of the fine-tuned CNV, again to be at least 35%.
Quality score
Every call is annotated with a quality score reflecting the percentage of reliable target regions. For this score, we consider whether the neighboring target regions are unreliable. This is done because these unreliable neighboring target regions influence the fine-tuning of the CNV borders. The quality score for the CNV is then the number of reliable target regions minus the sum of the number of unreliable target regions covered by the CNV and the number of all unreliable neighboring target regions. Hence, a detected CNV with many unreliable neighboring target regions, or a detected CNV with many scattered unreliable target regions within the CNV will get low scores. The default setting for the CNV quality score we used was 6 (six more reliable target calls than unreliable calls).
CNV annotation
Additionally, calls are annotated with OMIM phenotype key scores of all (partially) overlapping genes using the OMIM API [24], i.e., the OMIM score for the CNV equals the maximum OMIM phenotype scores of the genes that overlap with the CNV. This score describes which method is used to link the gene to the disorder and reflects the certainty of a gene causing a specific disorder. For example, if the molecular basis of the disorder is known, this key is 3. If the gene is linked to a disorder through statistical methods only, the key is 2.
Other tools
We ran XHMM (downloaded from GitHub @ 18 June 2015), CoNIFER (version 0.2.2; released 17 September 2012), CODEX (GitHub, commit 3d40ac9 @ 7 April 2017), and CLAMMS (GitHub, commit 3e19892 @ 10 April 2017) according to their default settings. All tools were run on the same samples as WISExome, as described in “Sample preparation” section. XHMM and CoNIFER do not distinguish between training and test samples, CODEX and CLAMMS used the same division in training and test samples as WISExome. Additional information on decisions and settings can be found in the Supplementary Section SM3.
Results
Replication array analysis
To test for compliance with array and MLPA analysis, we tested WISExome on 17 test samples with at least one known pathogenic CNV each (20 CNVs total) as identified by array analysis (18 CNVs) or MLPA (2 CNVs, kit P170-B2 for the APP gene, MRC-Holland, Amsterdam, the Netherlands). We were able to correctly identify all known pathogenic CNVs as shown in Supplementary Table S1 and Supplementary Fig. S4. There are differences in start and end positions, but these are mostly because the array platform has probes in inter-exonic regions, whereas the WES probes lie in exons only. On average, WISExome finds 33 calls per sample without size filters, and 15 calls per sample when filtering at a minimum of 15 Kb (standard array resolution in current clinical practice), which is comparable to the array analysis. As several tools have been developed to call CNVs from exome data, we compared WISExome to XHMM, CoNIFER, CODEX, and CLAMMS (using default settings, see “Methods” section). An overview of how their calls overlap with known CNVs, as validated by array analysis, is shown in Fig. 2, Supplementary Fig. S4 and Supplementary Table S1.
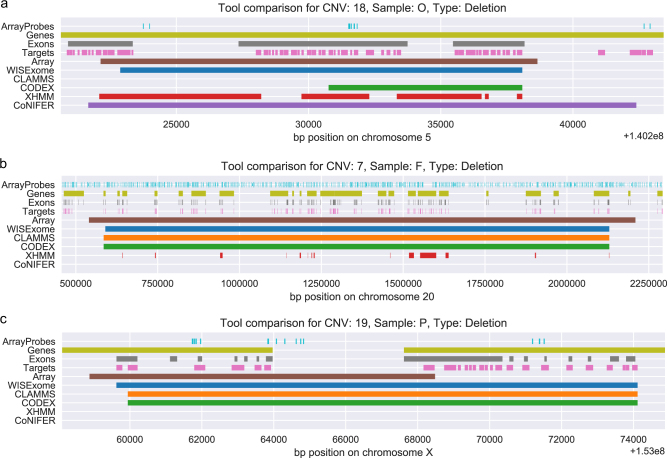
Fig. 2.
Detected CNV segments by WISExome (blue), CLAMMS (orange), CODEX (green), XHMM (red), and CoNIFER (purple) for three known pathogenic CNVs according to the array analysis (brown). The region is annotated by the array probes (cyan), genes (citron), exons (gray), and target regions (pink). CNVs shown here are marked in Supplementary Table S1 as numbers 18 (a), 7 (b), and 19 (c). a CLAMMS failed to identify this region as aberrated. WISExome, XHMM, and CoNIFER mark roughly the same area as part of a CNV, while CODEX made a relatively small call. b WISExome, CLAMMS, and CODEX are in near-perfect agreement on the CNV. XHMM shows a very fragmented call, failing to identify several affected genes in between its calls. CoNIFER does not make a call. c WISExome, CLAMMS, and CODEX all identify the region to the right as aberrated, while the CNV should have been shorter according to the array. Both XHMM and CoNIFER fail to make a call.
CoNIFER made few calls and only five overlapped the known CNVs in our test data. Adjusting the SVD argument yielded slight variations in results, but no setting was satisfactory. One of the few calls CoNIFER did make is shown in Fig. 2a. While other tools, except for WISExome, found a smaller area to be affected by the known CNV, CoNIFER appears to overestimate the CNV length.
XHMM identified most known CNVs partially (Fig. 2a, b) and failed to identify one known deletion and three duplications (numbered 13, 15, 17, and 19 in Supplementary Table S1, respectively), one missed CNV is shown in Fig. 2c, all results are shown in Supplementary Fig. S4. Most notably, XHMM tends to break up single CNV calls of the array in multiple small regions.
CODEX showed strong similarities to WISExome in results; it calls all CNVs known from the array analysis (Supplementary Table S1 and Supplementary Fig. S4). While generally extremely sensitive, Fig. 2a shows that CODEX only found the rightmost half of the CNV, while WISExome, XHMM, and CoNIFER agree on the upstream start position for this CNV.
CLAMMS also showed very similar behavior to WISExome (Supplementary Table S1 and Supplementary Fig. S4). However, CLAMMS was unable to identify one known CNV in our data set, shown in Fig. 2a. This CNV was not missed by any of the other tools, including CoNIFER.
Differences in detected regions
As it is unlikely that the array caught all CNVs in our data, we extended our comparison to include all calls made by the selected tools. Figure 3a shows an overview of the size of the detected regions by the different tools, whereas Fig. 3b shows the number of detected regions. Most tools detect roughly the same number of regions, except CODEX, which makes considerably more calls. WISExome and CoNIFER show relatively few and large calls. CLAMMS and XHMMs show fragmented calling behavior, which is reflected by their small median call size (Fig. 3a and Supplementary Fig. S5).
Fig. 3.
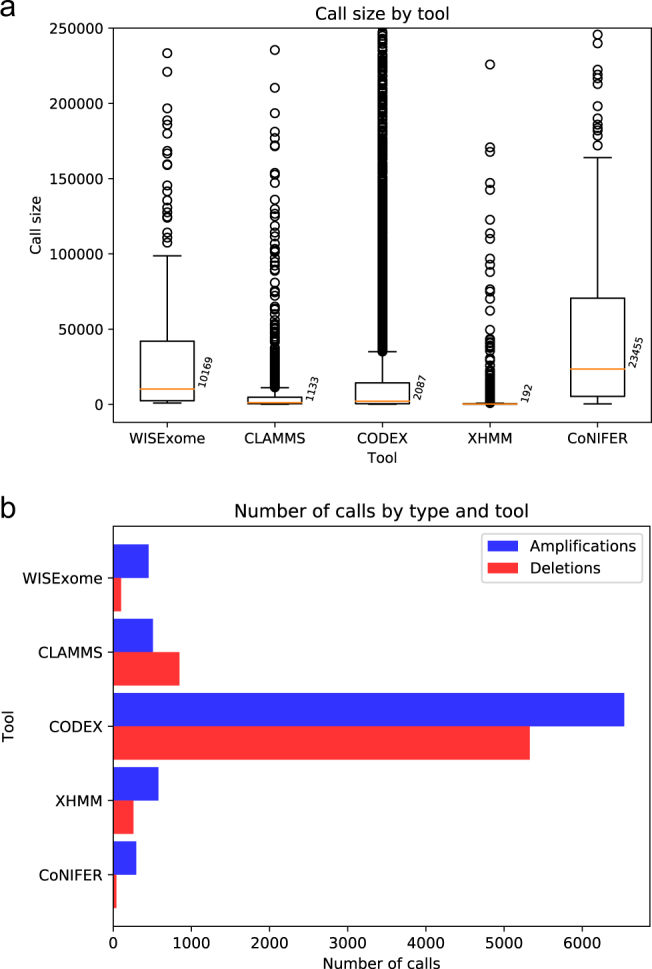
a Boxplot showing the size distribution (vertical axis) of calls made by the five CNV detection tools (horizontal axis). The annotated numbers next to boxplots show the median size for that tool. CoNIFER has the least fragmentation in its calls, and XHMM the most. CLAMMS calls are also fragmented and have a median size of only 1133 bp. To improve visibility of the box plots, outliers above 250,000 were cropped. Supplementary Fig. S5a shows this boxplot without the crop. b Number of amplifications and deletions per tool after thresholding, clearly showing CODEX’s huge number of calls compared to other tools.
To find out how the detected regions differ between tools, we compared the size of the union of the exons detected by a set of tools with the size of the intersection of the detected exons by these tools. When tools agree, their intersection should equal their union. The results for every possible combination of tools are shown in Fig. 4b. The figure is dominated by the high amount of calls by CODEX (consistently resulting in a large union), splitting the plot in combinations that either include or exclude CODEX. WISExome and CLAMMS show a large agreement in the detected regions, i.e., their intersection is 1/4th of the union (dotted line), whereas other tools have intersections lower than 1/10th of the union (dashed line).
Fig. 4.
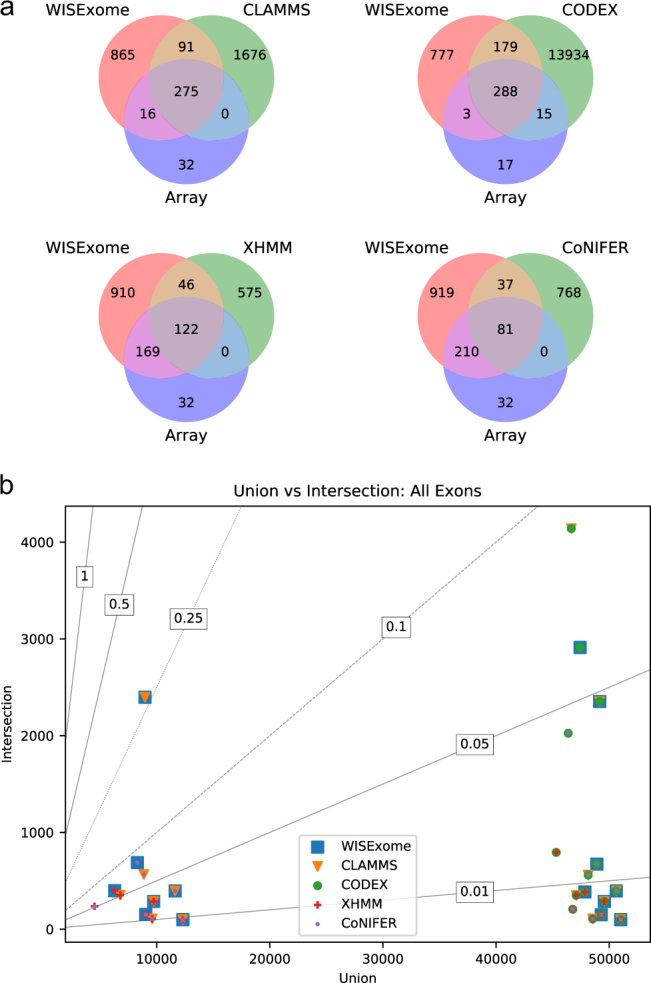
a Venn diagrams showing overlap based on the detected aberrated genes between WISExome and any of the four other CNV detection tools, as well as overlap with the known CNVs according to the array analysis. For known CNVs, CLAMMS shows a strong overlap with WISExome but adds no unique calls that overlap with the array, whereas WISExome shows 15 unique calls wrt CLAMMS that do overlap with the array results. CODEX does find 24 more affected genes (than WISExome) overlapping with the array data, but has a significantly larger set of affected genes in total. XHMM misses half of the genes called by the array analysis, and CoNIFER even misses three quarters of the known aberrated genes. An exon-level-based equivalent of this figure is shown in Supplementary Fig. S6. b Plot showing the overlap in detected exons between any combination of tools. The horizontal axis shows the size of the union of the exons detected by a set of tools, and the vertical axis shows the size of the intersection of the exons detected by the same set of tools. Lines marked with numbers show y = ax, where a is the number shown. When the set of tools agree the size of the intersection should equal the size of the union, which would put the marker on the line marked with 1. A unique color and shape is used for every tool, and plotted on top of each other for every tool involved in a combination. For example, the top right is marked by both CLAMMS and CODEX, meaning that the unions and intersection of these two tools are considered. The bottom right shows the result when comparing all five tools. Due to the number of affected exons called by CODEX, two main groups can be observed: the combinations on the right of the plot that include CODEX, and the combinations on the left of the plot that exclude CODEX. Next, vertically another two groups can be observed: all combinations including CoNIFER or XHMM below an intersection of size 1000, and combinations between WISExome, CLAMMS, and CODEX above this line. WISExome and CLAMMS are most similar since the size of their intersection is ¼ of the size of their union, which is considerably larger than any other combination. A similar plot where overlap among genes is considered is shown in Supplementary Fig. S11.
Differences in detected genes and exons
Figure 4a shows the comparisons between WISExome and any of the other tools with respect to the number of genes detected as aberrated, whereas Supplementary Fig. S6 shows these comparisons for aberrated exons. A gene/exon is called aberrated when a call of a tool shows at least one base pair overlap with that gene/exon. It immediately becomes clear that WISExome and CLAMMS behave relatively similar, although CLAMMS misses a few more genes found detected by array analysis (Fig. 4a). CODEX found several of the array-based detected genes missed by WISExome, but at the cost of a huge set of unique calls. Both CoNIFER and XHMM miss out on a lot of genes detected by the array analysis. Fragmentation by CLAMMS and XHMM can also be seen in the relatively small number of detected genes and exons (shown in Supplementary Fig. S7) compared to the total number of calls as shown in Fig. 3b.
To further zoom in on differences, Supplementary Fig. S8 shows the overlap between WISExome, CLAMMS, and CODEX. This figure again shows that CODEX calls significantly more genes/exons aberrated than other tools. WISExome and CLAMMS show a larger overlap, but WISExome’s unique calls are more overlapping with array results than CLAMMS’ unique calls (also visible in Fig. 4a).
Prioritizing WISExome calls
Within WISExome, we made it possible to prioritize calls based on its scoring scheme that measures the difference between the amount of reliable and unreliable target regions (“Methods” section, and Supplementary Fig. S9 shows a distribution of scores). Prioritizing calls based on this score puts all 20 known pathogenic CNVs at the top of the list with minor exceptions; one CNV was ranked second (CNV 9 in Supplementary Fig. S4), two CNVs were ranked 5th (CNV 15 and 18), and the smallest CNV was ranked 25th (CNV 20). We further annotated calls with their potential pathogenicity based on the OMIM phenotype key of the underlying genes (“Methods” section). By lowering CNVs that have no genes with a high OMIM phenotype key, and thus are not known to be involved in any syndrome, a large fraction of the calls can be ignored (Supplementary Fig. S10), leaving only a few calls (5 calls on average) per sample to be inspected in more detail. Note that this prioritization will also rank common CNVs lower as those are not expected to harbor pathogenic genes.
Conclusion
We developed a new CNV detection methodology for WES data that uses a within-sample comparison approach to capture the expected read count distributions across the genome. The benefit of this approach is that we only need to describe experimental variation and not in-between sample variation, making the method more accurate than previous approaches. We have shown that this new methodology, called WISExome, reliably reproduces array results, opening the possibility to perform genome diagnoses using WES data exclusively, in contrast to current practice where WES analysis still has to be combined with array analysis.
We compared WISExome to four existing CNV detection tools: CoNIFER, XHMM, CODEX, and CLAMMS. From these tools, CoNIFER deviates the most, calling few CNVs in general, and it detects only a few CNVs that result from clinical interpretation of the array analysis. XHMM breaks calls made by other tools into smaller calls (fragmentation) and misses a few CNVs detected by array analysis. Note that fragmentation of calls might lead to overlooking pathogenic genes.
CODEX found all CNVs detected by array analysis, but made significantly more calls than any of the other tools, resulting in the detection of many more genes and exons than other tools. CLAMMS showed results similar to WISExome, both in the start and stop positions of the CNVs resulting from array analysis, as well as the numbers of detected genes and exons. Yet, CLAMMS shows more fragmentation, which can be clearly seen in the size distribution of the calls. Additionally, CLAMMS missed one of the CNVs detected by array analysis that was found by all other tools (sample 18, Fig. 2a).
From this comparison, we conclude that WISExome performs consistently well over the different analysis that we investigated, i.e., it shows a proper balance between accuracy and specificity, without fragmentation of regions. These are favorable aspects when applying a CNV detection tool within the clinic. Nevertheless, we have observed considerable variability between tools, and we recommend to run multiple tools in parallel for clinical practice.
Note that the within-comparison approach might suggest that WISExome is independent for different sequencing or enrichment protocols, and that it would be possible to run WISExome without (re)training in different centers. Our experiences with WISExome and WISECONDOR (the first method in which we introduced the within-comparison approach, although designed for detecting chromosomal aberrations in cell-free DNA) indicates that even the within-comparison scheme is influenced by differences between centers. That is, although the within-comparison scheme works across different centers and sequencing technologies, we have seen performance improvements when reference bins are determined using training samples that were processed similar to the test cases. This should, however, be tested more thoroughly by setting up an inter-center comparison. For now, we advocate to create a new reference set table when there is a change in center or protocol. However, we suspect WISExome can be trained with a relatively small training set (until now we typically used 200–300 samples). Furthermore, we would like to stress that by using a within-comparison scheme, our tool is likely still capable of detecting CNVs in cases where the read distribution of a sample deviates strongly from the reference data. More so than when the sample is compared to the reference set directly, as the reference targets from the within-comparison scheme vary accordingly within the sample.
Finally, we introduced two ways to prioritize calls made by WISExome. One that expresses the quality of the call, which is directly influenced by the size of the call, the number of genes and the amount of probe targets, and another that annotates the call with OMIM phenotype keys. The prioritization of calls based on their score and annotation, allows geneticists to quickly zoom in on the most likely candidate genes, making WISExome an extremely useful diagnostic tool. To even further reduce the overall time spent per sample for a geneticist additional filtering of calls might be done by, for example, filtering common CNVs, or annotating calls using other databases, such as the CNV in disease database [25] or the database of genomic variants [26].
Taken together, WISExome provides an alternative to array analysis with a quick and easy workflow for geneticists that includes a prioritization scheme for calls that improves the diagnostic relevance.
Electronic Supplementary Material
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41431-017-0005-2) contains supplementary material, which is available to authorized users.
References
- 1.Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nat Rev Genet. 2013;14:415–426. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hester SD, Reid L, Nowak N, et al. Comparison of comparative genomic hybridization technologies across microarray platforms. J Biomol Tech. 2009;20:135–151. [PMC free article] [PubMed] [Google Scholar]
- 3.Cheung SW, Shaw CA, Yu W, et al. Development and validation of a {CGH} microarray for clinical cytogenetic diagnosis. Genet Med. 2005;7:422–432. doi: 10.1097/01.GIM.0000170992.63691.32. [DOI] [PubMed] [Google Scholar]
- 4.Boone PM, Bacino CA, Shaw CA, et al. Detection of clinically relevant exonic copy-number changes by array {CGH} Hum Mutat. 2010;31:1326–1342. doi: 10.1002/humu.21360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ng SB, Turner EH, Robertson PD, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chiang DY, Getz G, Jaffe DB, et al. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoon S, Xuan Z, Makarov V, Ye K, Sebat J. Sensitive and accurate detection of copy number variants using read depth of coverage. Genome Res. 2009;19:1586–1592. doi: 10.1101/gr.092981.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rapley R. Polymerase chain reaction. Totowa, NJ: Springer; 1998. [Google Scholar]
- 9.Aird D, Ross MG, Chen WS, et al. Analyzing and minimizing {PCR} amplification bias in Illumina sequencing libraries. Genome Biol. 2011;12:R18. doi: 10.1186/gb-2011-12-2-r18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao M, Wang Q, Wang Q, Jia P, Zhao Z. Computational tools for copy number variation ({CNV}) detection using next-generation sequencing data: features and perspectives. BMC Bioinformatics. 2013;14(Suppl 1):S1. doi: 10.1186/1471-2105-14-S11-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benjamini Y, Speed TP. Summarizing and correcting the {GC} content bias in high-throughput sequencing. Nucleic Acids Res. 2012;40:e72. doi: 10.1093/nar/gks001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rigaill GJ, Cadot S, Kluin RJC, et al. A regression model for estimating {DNA} copy number applied to capture sequencing data. Bioinformatics. 2012;28:2357–2365. doi: 10.1093/bioinformatics/bts448. [DOI] [PubMed] [Google Scholar]
- 13.Sathirapongsasuti JF, Lee H, Horst BAJ, et al. Exome sequencing-based copy-number variation and loss of heterozygosity detection: {ExomeCNV} Bioinformatics. 2011;27:2648–2654. doi: 10.1093/bioinformatics/btr462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansson LF, van Dijk F, de Boer EN, et al. {CoNVaDING}: single exon variation detection in targeted {NGS} data. Hum Mutat. 2016;37:457–464. doi: 10.1002/humu.22969. [DOI] [PubMed] [Google Scholar]
- 15.Krumm N, Sudmant PH, Ko A, et al. Copy number variation detection and genotyping from exome sequence data. Genome Res. 2012;22:1525–1532. doi: 10.1101/gr.138115.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91:597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Ligt J, Boone PM, Pfundt R, et al. Platform comparison of detecting copy number variants with microarrays and whole-exome sequencing. Genom Data. 2014;2:144–146. doi: 10.1016/j.gdata.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legault MA, Girard S, Lemieux Perreault LP, Rouleau GA, Dubé MP. Comparison of sequencing based {CNV} discovery methods using monozygotic twin quartets. PLoS One. 2015;10:e0122287. doi: 10.1371/journal.pone.0122287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Packer JS, Maxwell EK, O’Dushlaine C, et al. CLAMMS: a scalable algorithm for calling common and rare copy number variants from exome sequencing data. Bioinformatics. 2015;32:133–135. doi: 10.1093/bioinformatics/btv547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jiang Y, Oldridge DA, Diskin SJ, Zhang NR. CODEX: a normalization and copy number variation detection method for whole exome sequencing. Nucleic Acids Res. 2015;43:e39. doi: 10.1093/nar/gku1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Straver R, Sistermans EA, Holstege H, Visser A, Oudejans CBM, Reinders MJT. WISECONDOR: Detection of fetal aberrations from shallow sequencing maternal plasma based on a within-sample comparison scheme. Nucleic Acids Res. 2014;42:e31. doi: 10.1093/nar/gkt992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wolf NI, Salomons GS, Rodenburg RJ, et al. Mutations in {RARS} cause hypomyelination. Ann Neurol. 2014;76:134–139. doi: 10.1002/ana.24167. [DOI] [PubMed] [Google Scholar]
- 23.Li H, Durbin R. Fast and accurate short read alignment with {Burrows-Wheeler} transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man ({OMIM)}, a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qiu F, Xu Y, Li K, et al. CNVD: Text mining-based copy number variation in disease database. Hum Mutat. 2012;33:E2375–E2381. doi: 10.1002/humu.22163. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald JR, Ziman R, Yuen RKC, Feuk L, Scherer SW. The database of genomic variants: a curated collection of structural variation in the human genome. Nucleic Acids Res. 2014;42:D986–D992. doi: 10.1093/nar/gkt958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.