Abstract
Kabuki syndrome is a monogenic disorder caused by loss of function variants in either of two genes encoding histone-modifying enzymes. We performed targeted sequencing in a cohort of 27 probands with a clinical diagnosis of Kabuki syndrome. Of these, 12 had causative variants in the two known Kabuki syndrome genes. In 2, we identified presumptive loss of function de novo variants in KMT2A (missense and splice site variants), a gene that encodes another histone modifying enzyme previously exclusively associated with Wiedermann-Steiner syndrome. Although Kabuki syndrome is a disorder of histone modification, we also find alterations in DNA methylation among individuals with a Kabuki syndrome diagnosis relative to matched normal controls, regardless of whether they carry a variant in KMT2A or KMT2D or not. Furthermore, we observed characteristic global abnormalities of DNA methylation that distinguished patients with a loss of function variant in KMT2D or missense or splice site variants in either KMT2D or KMT2A from normal controls. Our results provide new insights into the relationship of genotype to epigenotype and phenotype and indicate cross-talk between histone and DNA methylation machineries exposed by inborn errors of the epigenetic apparatus.
Introduction
Kabuki syndrome (MIM 147920, 300867) is a pleiotropic disorder characterized by intellectual disability, postnatal growth retardation and dysmorphic facial features [1, 2]. Immune defects including defects of B cell differentiation are also frequent [3]. In about 60% of Kabuki syndrome individuals, the disorder is inherited as an autosomal dominant trait caused by loss of function (LOF) variants in lysine (K)-specific methyltransferase 2D (KMT2D previously known as MLL2) [1, 4]; in another 4% of the cases, the disorder shows X-linked dominant inheritance caused by LOF variants in lysine (K)-specific demethylase 6A (KDM6A previously known as UTX) [2, 4]. The explanation for the remaining ~36% of Kabuki syndrome cases is unknown [4].
KMT2D encodes a lysine-specific histone methyltransferase (KMT2D) that catalyzes methylation of H3K4. KDM6A encodes a lysine-specific demethylase (KDM6A) that catalyzes removal of methyl groups from H3K27me3. Thus, both Kabuki syndrome associated genes regulate histone tail methylation and mice with a deletion of KMT2D reveal global abnormalities of H3K4 methylation [5, 6]. Little is known about the target genes for KMT2D and KDM6A in normal human cells and their relevance to the Kabuki syndrome phenotype [7].
Wiedemann-Steiner syndrome (MIM 605130) is a rare autosomal dominant disorder caused by heterozygous loss of function variants in the KMT2A gene [8]. Jones et al. identified heterozygous, de novo nonsense or indel variants in KMT2A in 5 out of 6 individuals with a phenotype characterized by hypertrichosis cubiti, excessive hair on the back with a whorl-like distribution, long eyelashes, thick or arched eyebrows with a lateral flare, and down-slanting and vertically narrow palpebral fissures, sacral dimple, height below the 10th centile, mild to moderate intellectual disability and behavioral difficulties [8]. Since the original study by Jones, there have been several reports that suggest that the Wiedemann-Steiner syndrome phenotype may be broader [9].
Given the interdependence of epigenetic modifications which has been established by study of cultured cells [10] we asked if DNA methylation was altered in a large cohort of Kabuki syndrome patients with genetic defects in the histone modification machinery to determine epigenotype interactions in patients with Kabuki syndrome. We describe novel variants in both KMT2D and KMT2A in these patients and demonstrate that patients with a Kabuki syndrome phenotype exhibit abnormalities of DNA methylation.
Materials and methods
Patient samples
The study cohort consisted of 27 probands with a Kabuki syndrome phenotype seen at Escola Paulista de Medicina, Sao Paulo and at Universidade de Sao Paulo, Brazil submitted to the Baylor-Hopkins Center for Mendelian Genomics (BHCMG) through the online submission portal PhenoDB [11] as well as 9 samples from non-affected control individuals that were matched on age, sex and ethnicity. Control samples originated from the same centers and DNA was isolated from peripheral blood cells in an identical manner in cases and controls. The clinical diagnosis was originally based on the presence of the most common features seen in Kabuki syndrome [12, 13], but we verified that these diagnoses were compatible with more recently described molecularly confirmed cohorts [4]. Our study was approved by the Johns Hopkins Medicine Institutional Review Board and by the IRBs of the other participating institutions. We obtained informed consent from responsible individuals in all families.
Targeted next generation sequencing
We designed probes for targeted sequencing (TruSeq Custom Amplicon kit, Illumina) with the online Illumina DesignStudio software for all candidate genes including exon-intron boundaries. We prepared the sequencing library according to the manufacturer´s protocol and sequenced one library pool of 28 samples (27 individuals and 1 control) in a single run on a MiSeq sequencer (Illumina, 2 × 251 bp paired end reads). Alignment of NGS data to the human reference genome and variant calling were performed using software provided by Illumina. All variant calls were based on RefSeq transcript and NCBI human genome assembly build 37. We then used the Analysis Tool of PhenoDB [14] to prioritize rare heterozygous and homozygous functional variants (missense, nonsense, splice site variants and indels) and excluded variants with a MAF > 0.01 in the Exome Variant Server (release ESP6500SI-V2) or 1000 Genomes Project [15] and variants present in dbSNP 126, 129, or 131. Next we generated a heterozygous, homozygous and a compound heterozygous variant list for each subject and every candidate single nucleotide variant and indel was verified by inspection in Integrative Genomics Viewer [16].
Infinium human methylation 450 BeadChip arrays
Genomic DNA (1125 ng) was prepared, for each sample, in a total volume of 45 μl and bisulfite treated using the EZ DNA Methylation kit (Zymo Research Corp, Orange, CA, USA), as specified by the manufacturer for use with the Infinium Human Methylation 450 BeadChip arrays. To obtain genome-scale methylation measurements, bisulfite treated DNA was processed on the Infinium HumanMethylation450 BeadChip (Illumina, San Diego, CA, USA) at the Johns Hopkins SNP Center in accordance with the manufacturer’s recommendation. The Infinium HumanMethylation450 BeadChip measures DNA methylation levels at 485,512 loci across the genome. All samples were processed in parallel. A total of 36 samples, including 27 Kabuki syndrome samples and 9 non-Kabuki syndrome controls were randomized across four 12-sample array BeadChips that were run in parallel to minimize potential confounding batch effects related to hybridization date or laboratory conditions. For quality control purposes we also included technical control samples with known amounts of DNA methylation (0, 50, 75, and 100% methylated). These were created using mixtures of 100 and 0% methylated HCT116 DKO (D5014-1/2, Zymo Research, Irvine, CA, USA).
Analysis of DNA methylation data
All data preprocessing steps and statistical analyses were performed using R-3.1.2 and Bioconductor 3.0 [17]. Using the ‘minfi’ package [18], we applied several quality control measures including removal of samples with low overall array intensities (n = 0), as well as removal of poorly performing probes (n = 2633), defined as having a detection p value greater than 0.01 in any sample. Two of the samples had a predicted sex that was discordant with the sample annotation, which was reconciled with clinical notes before proceeding. All samples that passed quality control (n = 38), were retained for calculation of principle components for ancestry adjustment although not all of these were included in our analytical set. All samples then underwent quantile normalization. Beta values, which represent the proportion of methylation at a given CpG site and range from 0 to 1, were logit transformed to create M-values. This logit transformation is mathematically equivalent to taking the log of the ratio of the methylated to unmethylated signal; the generated M-values are normally distributed and therefore more appropriate for statistical testing [19]. Because the DNA methylation measurements were obtained from whole blood samples with a heterogeneous mix of nucleated white cells, we estimated the proportion of B cells, CD4 and CD8 positive T cells, natural killer cells, monocytes, and granulocytes [20]. Although we did not find significant differences (p < 0.05) in cell composition between Kabuki syndrome and control individuals, we adjusted for cell composition estimates in our analyses to identify Kabuki syndrome-associated methylation changes.
Pyrosequencing validation
PCR Primers and sequencing primers were designed by MethPrimer [21] and are available by request. Bisulfite treated [22] genomic DNA was PCR amplified (50-cycles) in a total of 25 μl volume and 5 μl were loaded onto agarose gel to verify single, strong product and an absence of any unused primers. The biotinylated PCR product was captured on streptavidin-coated sepharose beads (GE Healthcare, Milwaukee, WI) and pyrosequencing reactions were set up using the PyroMark Gold Q24 kit (Qiagen), according to the manufacturer’s instructions. Each individual pyrosequencing assay was designed with PyroMark Q24 software and the percentage of methylation at each CpG site was analyzed with PyroMark Q24 software.
Hierarchical clustering
For sample clustering based on global DNA methylation, we first restricted the 482,879 Infinium HumanMethylation450 BeadChip probes that passed QC measures to autosomal probes to prevent clustering solely on the basis of patient sex. 11,587 probes on the X and Y chromosomes were removed to leave 471,292 autosomal probes. Hierarchical clustering was performed with dist() and hclust() functions using the complete linkage method and the 10% most variably methylated probes (n = 47,130).
Results
We performed targeted sequencing and genome-wide DNA methylation analyses to (1) identify novel genetic variants associated with Kabuki syndrome, and (2) characterize Kabuki syndrome-associated epigenomic patterns, generally, as well as the relationship between specific genetic variants identified in Kabuki syndrome patients and epigenomic profiles. We performed targeted next-generation sequencing on a cohort of 27 patients with a clinical diagnosis of Kabuki syndrome (Supplementary Table 1). Targeted sequencing was performed on 9 genes (Supplementary Table 2), including the 2 genes known to cause Kabuki syndrome (KMT2D and KDM6A); 2 genes known to cause ICF syndrome (DNMT3B and ZBTB24), a recognized genocopy of Kabuki syndrome with facial flattening and immune dysfunction [23] (Supplementary Table 3); and 5 candidate genes (KDM6B, MEN1, KMT2A, KMT2B, HCFC1) known to interact with or have overlapping function with known Kabuki syndrome genes [24].
In 12 Kabuki syndrome probands, we identified KMT2D variants: 3 with unique de novo heterozygous variants (Table 1). Ten of the 12 KMT2D variants we identified are not present in the Genome Aggregation Database (gnomAD) which currently contains 123,136 exomes and 15,496 genomes (one variant is present as heterozygous variants in two alleles, and one is present as a heterozygous variant in three alleles). In one individual (KS11) we found a missense variant in KMT2D p.(N4572S) and a missense variant in KMT2B p.(E2354K). The KMT2D variant is rare (8.142e-6 in gnomAD), does not involve a known protein domain and has not been previously associated with Kabuki syndrome, while the KMT2B variant is novel and alters a conserved residue p.(E2354). KMT2B is a histone methyltransferase that also targets H3K4, raising the possibility that in KS11 either or both of these variants contribute to the Kabuki syndrome phenotype. The phenotype of individual KS11 is not atypical and fits the clinical diagnosis of Kabuki syndrome (Supplementary Table 1).
Table 1.
Summary of variants identified with targeted next-generation sequencing among 27 individuals with clinically defined Kabuki syndrome
| Subject | Gene | Variant | Parents sequenced | gnomAD MAF | DNA methylationa | ||
|---|---|---|---|---|---|---|---|
| Nucleotide change | Amino acid change | Exon | |||||
| Known KS genes | |||||||
| KS4 | KMT2D | c.14515+1G>T | 47 | N | 0 | + | |
| KS7 | KMT2D | c.12268C>T | p.(Q4090Ter) | 39 | N | 0 | + |
| KS9 | KMT2D | c.5124_5125del | p.(T1708fs) | 21 | N | 0 | + |
| KS11 | KMT2D | c.13715A>G | p.(N4572S) | 41 | N | 8.142e-6 | − |
| KS12 | KMT2D | c.1582_11584dup | p.(Q3861delinsQQ) | 39 | N | 0 | + |
| KS13 | KMT2D | c.185C>T | p.(P62L) | 3 | Mother (not present) | 1.228e-5 | + |
| KS14 | KMT2D | c.1329_1332del | p.(P443fs) | 10 | N | 0 | + |
| KS15 | KMT2D | c.6595delT | p.(Y2199fs) | 31 | Y (de novo) | 0 | + |
| KS19 | KMT2D | c.15920_15921insT | p.(S5307fs) | 49 | Mother (not present) | 0 | + |
| KS21 | KMT2D | c.13207_13208del | p.(N4403fs) | 39 | Y (de novo) | 0 | + |
| KS22 | KMT2D | c.12304C>T | p.(Q4102Ter) | 39 | Y (de novo) | 0 | + |
| KS24 | KMT2D | c.11800C>T | p.(Q3934Ter) | 39 | N | 0 | + |
| Other candidate genes sequenced | |||||||
| KS8 | KMT2A | c.3019G>T | p.(G1007C) | 3 | Y (de novo) | 0 | + |
| KS29 | KMT2A | c.5803-1G>A | 22 | Y (de novo) | 0 | + | |
| KS5 | HCFC1 | c.3794C>T | p.(S1265L) | 17 | N | 0.08% | − |
| KS10 | ZBTB24 | c.146G>A | p.(R49Q) | 2 | Mother (not present) | 0.6% | − |
| KS11 | KMT2B | c.7060G>A | p.(E2354K) | 30 | N | 0 | − |
| KS18 | DNMT3B | c.1211A>G | p.(Y404C) | 11 | N | 0.7% | − |
KS Kabuki syndrome, KMT2A lysine (K)-specific methyltransferase 2A, KMT2D lysine (K)-specific methyltransferase 2D, HCFC1 host cell factor C1, DNMT3B DNA methyltransferase 3 beta, ZBTB24 zinc finger and BTB domain containing. All variants have been submitted to ClinVar Database (ClinVar accessions SCV000583531 – SCV000583548)
aKS11 was removed from the methylation analysis because it had variants in both KMT2B and KMT2D, and was unique in that the KMT2D variant was a missense variant of unknown functional consequence
bFor KS1-3, 6, 16–17, 20, 26–28 no mutation was found
cPlus and minus signs indicate a sample was included or excluded from DNA methylation analyses, respectively. KMT2D transcript identifier is NM_003482.3, KMT2D genomic identifier (for introns) is NG_027827.1, KMT2A transcript identifier is NM_001197104.1 and genomic identifier (for introns) is NG_027813.1, HCFC1 transcript identifier is NM_005334.2, ZBTB24 transcript identifier is NM_001164313.1, KMT2B transcript identifier is NM_014727.2, DNMT3B transcript identifier is NM_006892.3
dIn one female individual (KS5) we identified a heterozygous variant in HCFC1 (MIM 309541), a gene known to be responsible for an X-linked disorder phenotypically distinct from Kabuki syndrome (Table S3)
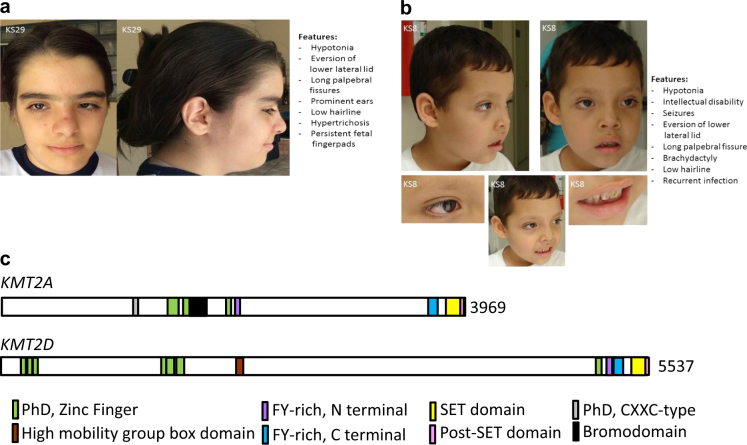
We also found two individuals who had carried a clinical diagnosis of Kabuki syndrome (KS8 and 29) who had normal KMT2D and KDM6A sequence but had novel de novo heterozygous missense or donor splice site variants in KMT2A, a histone methyltransferase gene previously associated with Wiedemann-Steiner syndrome [8]. Wiedemann-Steiner syndrome is a disorder with overlapping features with Kabuki syndrome [8] such as vertically narrow palpebral fissures, strabismus, broad nasal bridge/tip, external ear deformity, short stature, hypotonia, small hands, hip abnormalities, developmental delay, intellectual disabilities, seizures, feeding difficulties, hypertrichosis, heart anomalies, urological anomalies, and recurrent infections (Supplementary Table 3). Review of the phenotype of the two individuals with the KMT2A variants and the clinical diagnosis of Kabuki syndrome revealed some features consistent with what can be seen in Kabuki syndrome such as the presence of fetal fingerpads, brachydactyly and elongated palpebral fissures with eversion of the lower lateral lid (Figs. 1a and b). The comparison of their facial features to the patients diagnosed with Wiedemann-Steiner syndrome revealed significant facial similarities with the patients described by Jones et al. (WSS-5) [8], Mendelssohn et al. (2014) [25] and Stellacci et al. (2016) [26]. KMT2A and KMT2D share many domains (Fig. 1c). We also identified an individual (KS18) with a heterozygous variant in DNMT3B and one individual (KS10) with a heterozygous variant in ZBTB24. Both of these genes have previously been associated with ICF syndrome, an autosomal recessive genocopy of Kabuki syndrome (Supplementary Table 3). These patients have a typical Kabuki syndrome phenotype without any atypical features. Individual KS7 had a KMT2D p.(Q4090Ter) variant and an atypical feature, hypoplasia/aplasia of the medial and/or distal phalanges of the toes bilaterally. Similarly, individual KS15, with the KMT2D p.(Y2199fs) variant, had shortening of the medial phalange of the 2nd and 5th fingers bilaterally and syndactyly of the 4th and 5th toes bilaterally. In thirteen Kabuki syndrome probands (48%) we failed to identify variants likely to affect function in any of the 9 genes. This negative result is similar to that (36%) of a recent large sequencing study of patients with Kabuki syndrome [4]. One explanation for the unexplained cases is that we did not search for copy number variants that could account for some fraction of our negative results. Direct Sanger sequencing of PCR amplified products validated all variants and confirmed appropriate Mendelian segregation in available family members (Table 1).
Fig. 1.
Facial features in patients with variants in KMT2A, a Trithorax ortholog (like KMT2D). a, b Facial features (left) in two patients identified as KS29 a and KS8 b led to a clinical diagnosis of Kabuki syndrome. The observed phenotype is summarized (right) and in more detail in Supplementary Table 1. These two patients suggest that there is sometimes phenotypic overlap between Kabuki and Wiedemann-Steiner syndromes, two disorders that are caused by variants in two independent Trithorax orthologs (KMT2A and KMT2D). c The domain structure of the two orthologous proteins. Domain structure is based on InterPro (https://www.ebi.ac.uk/interpro/)
To further elucidate potential mechanisms of how the primary histone alterations contribute to disease in Kabuki syndrome, we measured DNA methylation levels across the genome in a total of 36 samples including 27 Kabuki syndrome and 9 control samples. Our supervised analyses were restricted to the 9 patients with KMT2D variants thought to affect function and 9 controls (Supplementary Figure 1) to identify differential methylation in genetically determined Kabuki syndrome. Differentially methylated positions (DMPs) were identified using limma [27] and linear regression models were adjusted for sex, blood cell composition, and ancestral population [28]. Differentially methylated regions (DMRs) were identified using bumphunter [29]; the analytic model was adjusted for sex, blood cell composition, and ancestral population [28]. DMR significance was assessed using bootstrapping as available in minfi [18] and a family-wise error rate threshold of 0.05 was applied.
While technical sources of variation in our data were minimized by running the samples in parallel, on the same date and by the same technician, we wanted to confirm that our results were not artifacts. Specifically, since position on array is a potential source of bias we explored the location of our samples. We noted that many of the control samples were processed in rows 5 and 6 and many of the Kabuki syndrome samples were processed in rows 1–4 of the Illumina BeadChip. To confirm that the DNA methylation changes related to Kabuki syndrome were not solely attributable to array position, we plotted DNA methylation values and array row position for the DMRs reported here (Supplementary Fig. 2). We observed overlapping DNA methylation levels among samples processed in rows 1–6; thus, DNA methylation values at these Kabuki syndrome-related sites are not purely driven by row alone. Furthermore, our findings for these regions were validated using bisulfite pyrosequencing, an independent and highly reliable method, where array row and location is irrelevant.
When we compared DNA methylation in 9 Kabuki syndrome individuals with a KMT2D variant expected to alter function with 9 age and sex-matched controls from the same population (Supplementary Figure 1), we found 590 differentially methylated loci, at a false discovery rate (FDR) < 0.05 (Supplementary Table 4). The 590 differentially methylated loci included 414 sites that occurred within genes and 176 outside of genes. Of the ones that occurred within genes, there were 6 genes that had five or more DMPs (ESR1, TSPAN4, GARS, MYO1F, SOX18 and AGAP2). Of these ESR1 and TSPAN4 had 7 and 6 DMPs, other had 5 each. Similarly, of the 176 loci that we identified outside of gene bodies, several (n = 6) were located 5’ of and in close proximity to (within 1 KB) TJP1. To illustrate these genotype specific differences, here we show two CpG sites in two independent genes (CIRBP, FEM1B) with relative hypomethylation among individuals with Kabuki syndrome and histone machinery variants compared to controls (Figs. 2a and b). Similarly, Figs. 2c and d show an example of two genes (c10orf11, SOX18) that are hypermethylated in Kabuki syndrome relative to controls. We performed a GO analysis with the DMP targets using the Gene Ontology Database (release 052717). We found enrichment of several categories including nervous system development, cell development and cell adhesion, however, none of these passed Bonferroni correction.
Fig. 2.
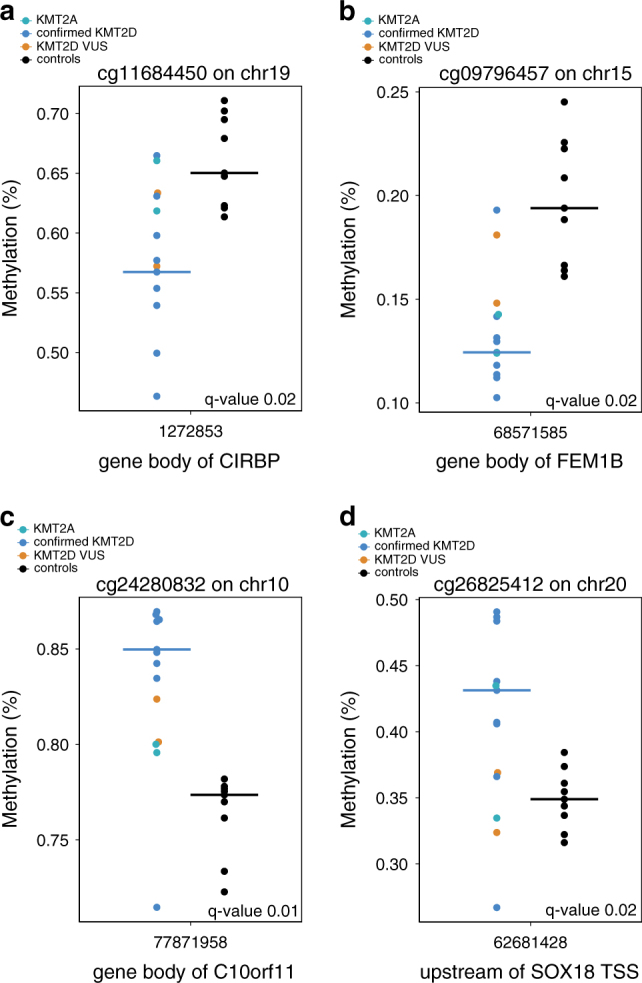
Individuals with Kabuki syndrome (KS) phenotype and a variant that alters function in KMT2D show site-specific hyper- and hypo-methylation relative to non-Kabuki syndrome controls. (a, b) Genomic loci showing significant relative loss of methylation (CIRBP, FEM1B) in samples from individuals with KMT2D variants that change function compared to controls. (c, d) Genomic loci showing significant gain of methylation (c10orf11, SOX18) in samples from individuals with KMT2D variants that alter function compared to controls. Black points denote non-KS controls. Blue points denote KS patients with KMT2D variants that alter function. Although not included in our main analysis, we also plot methylation values for individuals with Kabuki syndrome and variants of unknown significance (orange) and KMT2A variants (aqua)
We also used bumphunter [29], as an alternative analysis to identify differentially methylated regions (DMRs) and found several genomic regions showing Kabuki syndrome-related differences in methylation (Supplementary Table 5); one of these DMRs achieved genome-wide significance and this was located in the 5’ region of HOXA4. Supplementary Figure 3 illustrates several of the best DMR’s identified by this approach including HOXA4. Since our cutoff was stringent we decided to validate a number of the genes found on this list that did not reach genome wide significance. To validate these results with an independent assay and to determine whether similar changes in DNA methylation exist in Kabuki syndrome samples without a known histone machinery variant, we performed bisulfite pyrosequencing for 36 individuals including 9 patients with KMT2D variants thought to affect function, 17 individuals with clinical features of Kabuki syndrome but no variant that expected to alter function (including VUS in KMT2D and variants in KMT2A) and 10 controls. We bisulfite treated all samples used for the genome-scale DNA methylation measurement and available parents; however, after bisulfite treatment, only a subset of these samples had the minimal amount needed for PCR-based validation studies (Controls = 7, No KMT2D variant = 9, and others = 16) and these were used for the validation studies. We set the p-value threshold for significance to <0.05. Consistent with our array-based findings, individuals with a KMT2D variant show relative hypomethylation at the MYO1F locus (Fig. 3a). Similarly, we also identified hypermethylated DMRs in Kabuki syndrome. For example, we observed a 20% increase in methylation in individuals with a variant in KMT2D expected to alter function on average, at a DMR located in the 5’ UTR region of the LAMB2 gene (Fig. 3b and Supplementary Table 5) which was similarly validated by pyrosequencing in patients with variants in KMT2D (Fig. 3b). Although we did not include KS patients lacking a clear functional KMT2D variant in our main analyses, we sought to further characterize the KMT2D DNA methylation changes among KS patients without a KMT2D variant. For some of the loci, we observed a similar methylation pattern in patients with a Kabuki syndrome phenotype with no discovered variants in KMT2D (Figs. 2b–c). These results imply there may be shared downstream targets among individuals with a Kabuki syndrome phenotype despite locus heterogeneity for the causative variant.
Fig. 3.
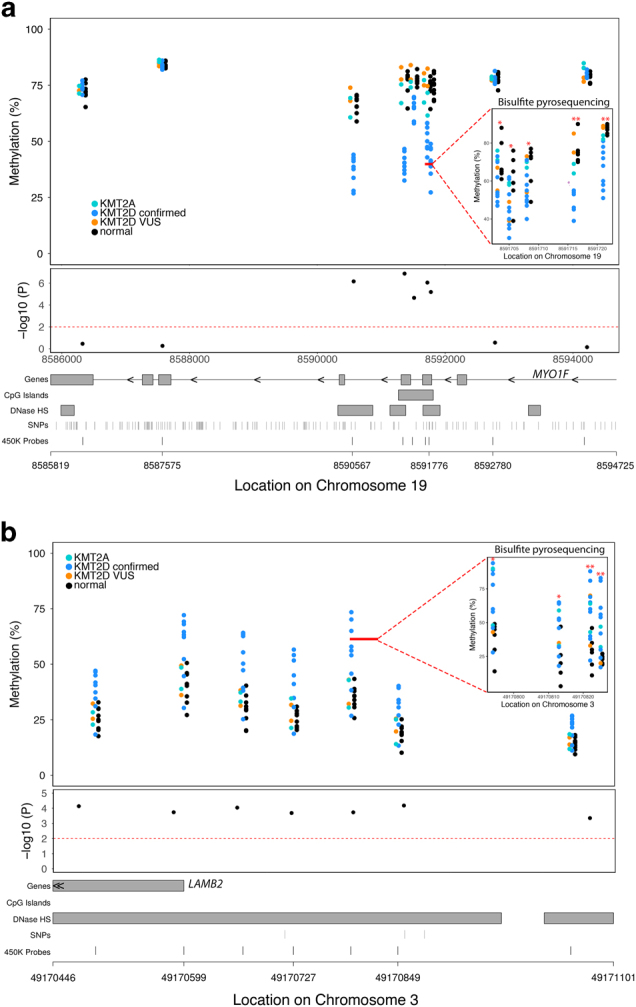
Examples of differentially methylated regions (DMRs) associated with Kabuki syndrome with KMT2D variants (KS-KMT2D) that alter function. (a) An example of a DMR within the MYO1F gene showing KS-KMT2D individuals have relative hypomethylation compared to non-Kabuki controls that we validated via bisulfite pyrosequencing. (b) Relative hypermethylation at the LAMB2 locus is associated with KMT2D variant that alters function that we validated with bisulfite pyrosequencing. Each of the upper panels displays genomic location on the x-axis and percent methylation on the y-axis. Inset top panel shows methylation values assessed using bisulfite pyrosequencing for the CpG sites in the region denoted by the solid red line. Black points denote non-KS controls. Blue points denote KS patients with KMT2D variants that alter function. Although not included in our main analysis, we also plot methylation values for individuals with Kabuki syndrome and variants of unknown significance (orange) and KMT2A variants (aqua) for comparison. Red asterisks denote significant differential methylation (*p < 0.05; **p < 0.01), via bisulfite pyrosequencing, between KS-KMT2D and control samples. The middle panel shows individual probe-based nominal p-values for differences in methylation between the two groups, with the dashed red line at a level of p = 0.01. The bottom panel provides gene annotation information for the differentially methylated region
We also performed unsupervised analyses, using hierarchical clustering, to evaluate global epigenetic patterns across all samples, regardless of variant type, using the top 10% most variably methylated probes on the Infinium Human Methylation 450 BeadChip arrays. As shown in Fig. 4, we observed two main dendrogram branches. One branch showed strong clustering of the Kabuki syndrome samples with KMT2D loss of function variants. The second branch contained samples from KS patients with a missense KMT2D variant. Thus, patients with KMT2D loss of function and missense variants show distinct DNA methylation patterns. Interestingly, KMT2A missense and splice site variants clustered with the KMT2D missense variants, indicating that epigenetic similarities may account for phenotypic overlap. Clustering patterns did not correspond to other covariates such as patient sex or sample cell type composition (Fig. 4; Supplementary Figure. 4a–f). DNA methylation patterns appear to be related to the type of histone methylation machinery variants present in KMT2D and KMT2A.
Fig. 4.
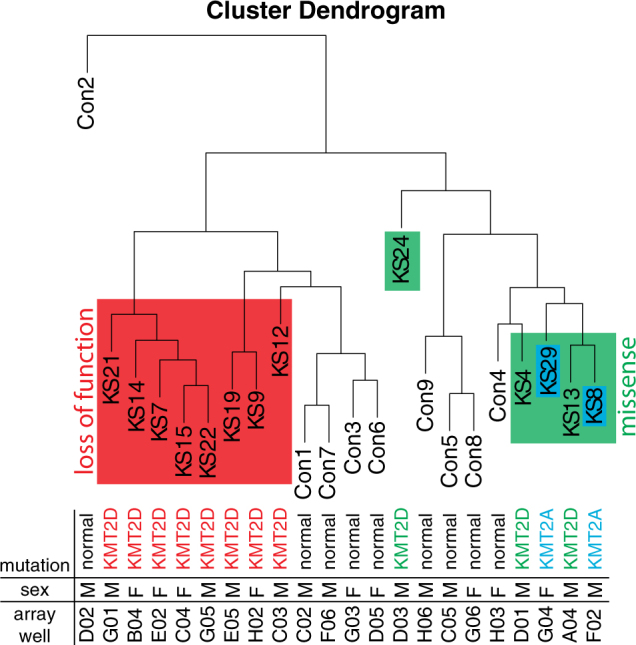
Unsupervised analysis reveals clustering of Kabuki syndrome cases based on variant type. Hierarchical clustering dendrogram, based on the 10% most variably methylated probes (n = 47,130), shows differences in DNA methylation patterns based on type of genetic variation within the histone machinery genes KMT2A and KMT2D. We depict loss of function KMT2D variants in red, KMT2D missense changes in green, and KMT2A missense and splice site variants in blue
Discussion
We find disease causing variants in KMT2D, the gene most commonly found to harbor disease causing variants in patients with a classical Kabuki syndrome phenotype. Skeletal anomalies are characteristic of Kabuki syndrome but they have traditionally been thought to affect the hands, not the feet and toes but here we also describe several individuals with lower limb anomalies. In addition, we find variants in another component of the epigenetic machinery (KMT2A); this was surprising, because previously variants in KMT2A have been exclusively associated with Wiedemann-Steiner syndrome. In summary these results indicate phenotypic overlap between Kabuki and Wiedemann-Steiner syndrome suggesting that pathological variants in KMT2A should be considered in Kabuki syndrome individuals who lack variants in KMT2D or KDM6A. Interestingly, the proteins encoded by KMT2A and KMT2D (Wiedemann-Steiner and Kabuki syndromes, respectively) have many of the same domains (Fig. 1c). These results also suggest that focusing on components of the histone methylation machinery may be more productive than sequencing components that interact with the known Kabuki syndrome genes.
Two recent publications also found disease-causing KMT2A variants in a cohort of patients with Cornelia De Lange syndrome [30, 31]. These findings highlight an overlap in clinical features of Kabuki, Wiedemann-Steiner and Cornelia De Lange syndromes and suggest that an expanded set of genes should be considered during genetic testing for these syndromes. Also, in two patients (KS10, KS18), we found single variants likely to alter function in ZBTB24 and DNMT3B. Without further study it is unclear whether these individuals have autosomal recessive ICF syndrome (in which case we did not identify the second allele) or are merely heterozygotes.
Although the primary defect in Kabuki syndrome is an abnormality in the histone machinery, we observe robust site-specific DNA methylation changes in patients with a clinical diagnosis of Kabuki syndrome relative to controls. These changes were first identified by a global assay, but 9 sites in two differentially methylated regions associated with Kabuki syndrome features with KMT2D/A variants were then validated with an independent assay (bisulfite pyrosequencing). These two regions demonstrated large magnitude differences (20–28%) and it is likely that we would have found additional regions with a larger sample size. We also found that Kabuki syndrome patients without KMT2D/A variants had similar DNA methylation profiles at these 2 regions, indicating that there may be additional unknown genetic causes (other genes) that also lead to overlapping methylation changes or that some of our patients have changes of these genes that we did not detect. One of the genes that were recurrently found to demonstrate DNA methylation changes (TSPAN4) is a known target of KMT2D [32] and ESR1 is a known binding partner of KMT2D [33]. However, further studies will be required to see which fraction of genes harboring DNA methylation changes are direct targets of KMT2D compared to those that are secondary to dysregulation of other regulators.
We also performed unsupervised analyses of the global methylation patterns and found that Kabuki and Wiedemann-Steiner syndrome patients fell into two distinct clusters, one containing those with loss of function KMT2D variants and the other with missense or splice site variants in KMT2D or KMT2A. This raises the possibility that specific classes of variants may have unique epigenetic signatures which could have potential as a biomarker of the disease state or therapeutic efficiency; however, further studies will be needed to clarify this. Furthermore, future studies should be designed to include sufficient numbers of patients containing variants of a particular type as opposed to pooling all patients with Kabuki syndrome in an effort to maximize the potential for epigenotype–phenotype exploration.
Although we adjusted for cell type heterogeneity in all of our DNA methylation analyses, it is possible that these measures did not account for all differences in cell type composition between patients and controls. Future studies examining specific homogenous cell types would help address this concern. However, even if the changes in DNA methylation reflect a shift in the blood cell composition of Kabuki syndrome vs. control patients, this would be of interest and could lead to novel insights into the pathological basis of this rare disease.
DNA methylation abnormalities have also recently been found in Kabuki syndrome [34] and Sotos syndrome (117,550, 614,753), another histone methylation machinery disorder [35]. The DNA methylation changes found in Kabuki syndrome [34] showed similar magnitude and some overlapping target genes such as the hypermethylation in HOXA5 (Supplementary Table 5) and hypomethylation in MYO1F (Fig. 3). In contrast, the Sotos syndrome changes [35] appear more extensive, although the published results do not correct for cell composition or other relevant covariates (sex and ancestry). NSD1, the protein product of the gene most frequently defective in Sotos syndrome, adds both open and closed chromatin modifications (H3K36 and H4K20) whereas KMT2A/D only adds open chromatin modifications (H3K4). Therefore, it is possible that the increased number of DNA methylation changes identified in Sotos syndrome compared to Kabuki syndrome reflect biologically meaningful differences related to the function of the different histone modifying proteins involved in each disorder. Similarly, specific DNA methylation abnormalities have been found in Floating-Harbor syndrome, a disorder caused by variants in SRCAP, a chromatin remodeler [36]. Interestingly, at least two of the genes found to be differentially methylated in patients with Floating-Harbor syndrome are the same as we find to be differentially methylated in Kabuki syndrome (MYO1F and SOX18) although here the abnormality is in the opposite direction. The DNA methylation abnormalities found in Floating-Harbor, Sotos, Wiedemann-Steiner and Kabuki syndrome suggest that disruption of the chromatin machinery often leads to disruption of DNA methylation levels. Additionally, they provide a potential explanation for phenotypic overlap among the Mendelian disorders of the epigenetic machinery (Table S3) [7] as shared DNA methylation abnormalities could be one possible disease mechanism.
There has been long standing debate regarding the directionality of epigenetic information flow, i.e., whether DNA methylation dictates histone modification [37] or whether histone modification leads to downstream DNA methylation changes [38, 39]. Cells from patients with ICF syndrome (MIM 242860, 614069, 616910, 616911), a condition caused by variants in a de novo DNA methyltransferase and interacting proteins, demonstrate global DNA hypomethylation [39] but also secondary abnormalities of histone modification [40]. Our data show that individuals with KMT2A/D variants also have site-specific DNA methylation changes, thus, providing support for histone modification leading to downstream DNA methylation changes. These results together with those published for Sotos and ICF syndromes suggest that the information flow is bi-directional forming a feedback loop among individual epigenetic machineries.
Electronic supplementary material
Acknowledgements
We would like to thank all the families that participated in this study. We also thank Maggie Baker for assistance in selecting the candidate genes for the amplicon study. This work was supported by a grant to H.T.B. by the NIH Director’s Early Independence Award (DP5OD017877) and a grant to D.V. from the National Human Genome Research Institute (1U54HG006493).
Compliance with ethical standards
Conflict of interest
The authors declare that they have no competing interests.
Footnotes
Nara Sobreira, Martha Brucato, Li Zhang, and Christine Ladd-Acosta contributed equally to this work.
Vera A Meloni and Hans T Bjornsson contributed equally to this work.
Electronic supplementary material
The online version of this article (10.1038/s41431-017-0023-0) contains supplementary material, which is available to authorized users.
References
- 1.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42:790–3. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Miyake N, Mizuno S, Okamoto N, et al. KDM6A point mutations cause Kabuki syndrome. Hum Mutat. 2013;34:108–10. doi: 10.1002/humu.22229. [DOI] [PubMed] [Google Scholar]
- 3.Lindsley AW, Saal HM, Burrow TA, et al. Defects of B-cell terminal differentiation in patients with type-1 Kabuki syndrome. J Allerg Clin Immunol. 2016;137:179–87. doi: 10.1016/j.jaci.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bögershausen N, Gatinois V, Riehmer V, et al. Mutation update for Kabuki syndrome genes KMT2D and KDM6A and further delineation of X-Linked Kabuki syndrome subtype 2. Hum Mutat. 2016;37:847–64. doi: 10.1002/humu.23026. [DOI] [PubMed] [Google Scholar]
- 5.Bjornsson HT, Benjamin JS, Zhang L, et al. Histone deacetylase inhibition rescues structural and functional brain deficits in a mouse model of Kabuki syndrome. Sci Transl Med. 2014;6:256ra135. doi: 10.1126/scitranslmed.3009278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Dominguez-Sola D, Hussein S, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagnesis. Nat Med. 2015;21:1190–8. doi: 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bjornsson HT. The Mendelian disorders of the epigenetic machinery. Genome Res. 2015;25:1473–81. doi: 10.1101/gr.190629.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones WD, Dafou D, McEntagart M, et al. De novo mutations in MLL cause Wiedemann-Steiner syndrome. Am J Hum Genet. 2012;91:358–64. doi: 10.1016/j.ajhg.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Miyake N, Tsurusaki Y, Koshimizu E, et al. Delineation of clinical features in Wiedemann-Steiner syndrome caused by KMT2A mutations. Clin Genet. 2016;89:115–9. doi: 10.1111/cge.12586. [DOI] [PubMed] [Google Scholar]
- 10.Murr R. Interplay between different epigenetic modifications and mechanisms. Adv Genet. 2010;70:101–41. doi: 10.1016/B978-0-12-380866-0.60005-8. [DOI] [PubMed] [Google Scholar]
- 11.Hamosh A, Sobreira N, Hoover-Fong J, et al. PhenoDB: a new web-based tool for the collection, storage, and analysis of phenotypic features. Hum Mutat. 2013;34:566–71. doi: 10.1002/humu.22283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Adam MP, Hudgins L. Kabuki syndrome: a review. Clin Genet. 2005;67:209–19. doi: 10.1111/j.1399-0004.2004.00348.x. [DOI] [PubMed] [Google Scholar]
- 13.Dentici ML, Di Pede A, Lepri FR, et al. Kabuki syndrome: clinical and molecular diagnosis in the first year of life. Arch Dis Child. 2015;100:158–64. doi: 10.1136/archdischild-2013-305858. [DOI] [PubMed] [Google Scholar]
- 14.Sobreira N, Schiettecatte F, Boehm C, Valle D, Hamosh A. New tools for Mendelian disease gene identification: PhenoDB variant analysis module; and GeneMatcher, a web-based tool for linking investigators with an interest in the same gene. Hum Mutat. 2015;36:425–31. doi: 10.1002/humu.22769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robinson JT, Thorvaldsdóttir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aryee MJ, Jaffe AE, Corrada-Bravo H, et al. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014;30:1363–9. doi: 10.1093/bioinformatics/btu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Du P, Zhang X, Huang CC, et al. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinformatics. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houseman EA, Accomando WP, Koestler DC, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. 2002;18:1427–31. doi: 10.1093/bioinformatics/18.11.1427. [DOI] [PubMed] [Google Scholar]
- 22.Tost J, Gut IG. DNA methylation analysis by pyrosequencing. Nat Protoc. 2007;2:2265–75. doi: 10.1038/nprot.2007.314. [DOI] [PubMed] [Google Scholar]
- 23.Ehrlich M, Jackson K, Weemaes C. Immunodeficiency, centromeric region instability, facial anomalies syndrome (ICF) Orphanet J Rare Dis. 2006;1:2. doi: 10.1186/1750-1172-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Nuland R, Smits AH, Pallaki P, Jansen PW, Vermeulen M, Timmers HT. Quantitative dissection and stoichiometry determination of the human SET1/MLL histone methyltransferase complexes. Mol Cell Biol. 2013;33:2067–77. doi: 10.1128/MCB.01742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mendelsohn BA, Pronold M, Long R, Smaoui N, Slavotinek AM. Advanced bone age in a girl with Wiedemann-Steiner syndrome and an exonic deletion in KMT2A (MLL) Am J Med Genet A. 2014;8:2079–83. doi: 10.1002/ajmg.a.36590. [DOI] [PubMed] [Google Scholar]
- 26.Stellacci E, Onesimo R, Bruselles A, et al. Congenital immunodeficiency in an individual with Wiedemann-Steiner syndrome due to a novel missense mutation in KMT2A. Am J Med Genet A. 2016;170:2389–93. doi: 10.1002/ajmg.a.37681. [DOI] [PubMed] [Google Scholar]
- 27.Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucl Acid Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barfield RT, Almli LM, Kilaru V, et al. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–41. doi: 10.1002/gepi.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaffe AE, Murakami P, Lee H, et al. Bump hunting to identify differentially methylated regions in epigenetic epidemiology studies. Int J Epidemiol. 2012;41:200–9. doi: 10.1093/ije/dyr238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan B, Pehlivan D, Karaca E, et al. Global transcriptional disturbances underlie Cornelia de Lange syndrome and related phenotypes. J Clin Invest. 2015;125:636–51. doi: 10.1172/JCI77435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parenti I, Teresa-Rodrigo ME, Pozojevic J, et al. Mutations in chromatin regulators functionally link Cornelia de Lange syndrome and clinically overlapping phenotypes. Hum Genet. 2017;136:307–20. doi: 10.1007/s00439-017-1758-y. [DOI] [PubMed] [Google Scholar]
- 32.Guo C, Chang CC, Wortham M, et al. Global identification of MLL2-targeted loci reveals MLL2’s role in diverse signaling pathways. Proc Natl Acad Sci USA. 2012;109:17603–8. doi: 10.1073/pnas.1208807109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mo R, Rao SM, Zhu YJ. Identification of the MLL2 complex as a coactivator for estrogen receptor alpha. J Biol Chem. 2006;281:15714–20. doi: 10.1074/jbc.M513245200. [DOI] [PubMed] [Google Scholar]
- 34.Butcher DT, Cytrynbaum C, Turinsky AL, et al. CHARGE and Kabuki Syndromes: Gene-specific DNA Methylation signatures identify epigenetic mechanisms linking these clinically overlapping conditions. Am J Hum Genet. 2017;100:773–88. doi: 10.1016/j.ajhg.2017.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choufani S, Cytrynbaum C, Chung BH, et al. NSD1 mutations generate a genome-wide DNA methylation signature. Nat Commun. 2015;6:10207. doi: 10.1038/ncomms10207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hood RL, Schenkel LC, Nikkel SM, et al. The defining DNA methylation signature of Floating-Harbor syndrome. Sci Rep. 2016;38803. 10.1038/srep38803. [DOI] [PMC free article] [PubMed]
- 37.Okitsu CY, Hsieh CL. DNA methylation dictates histone H3K4 methylation. Mol Cell Biol. 2007;27:2746–57. doi: 10.1128/MCB.02291-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hu JL, Zhou BO, Zhang RR, et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci USA. 2009;106:22187–92. doi: 10.1073/pnas.0905767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Hevi S, Kurash JK, et al. The lysine demethylase LSD1 (KDM1) is required for maintenance of global DNA methylation. Nat Genet. 2009;41:125–9. doi: 10.1038/ng.268. [DOI] [PubMed] [Google Scholar]
- 40.Jin B, Tao Q, Peng J, et al. DNA methyltransferase 3B (DNMT3B) mutations in ICF syndrome lead to altered epigenetic modifications and aberrant expression of genes regulating development, neurogenesis and immune function. Hum Mol Genet. 2008;17:690–709. doi: 10.1093/hmg/ddm341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.