Abstract
The clinical use of whole-genome sequencing (WGS) is expected to alter pediatric medical management. The study aimed to describe the type and cost of healthcare activities following pediatric WGS compared to chromosome microarray (CMA). Healthcare activities prompted by WGS and CMA were ascertained for 101 children with developmental delay over 1 year. Activities following receipt of non-diagnostic CMA were compared to WGS diagnostic and non-diagnostic results. Activities were costed in 2016 Canadian dollars (CDN). Ongoing care accounted for 88.6% of post-test activities. The mean number of lab tests was greater following CMA than WGS (0.55 vs. 0.09; p = 0.007). The mean number of specialist visits was greater following WGS than CMA (0.41 vs. 0; p = 0.016). WGS results (diagnostic vs. non-diagnostic) modified the effect of test type on mean number of activities (p < 0.001). The cost of activities prompted by diagnostic WGS exceeded $557CDN for 10% of cases. In complex pediatric care, CMA prompted additional diagnostic investigations while WGS prompted tailored care guided by genotypic variants. Costs for prompted activities were low for the majority and constitute a small proportion of total test costs. Optimal use of WGS depends on robust evaluation of downstream care and cost consequences.
Introduction
The potential medical and economic benefits of whole-genome sequencing (WGS) in pediatrics has generated much enthusiasm, in part because anticipated benefits include early detection and tailored management [1–4]. In particular, for children with unexplained developmental delay/intellectual disability, replacement of multiple expensive genetic tests with a single lower cost genome sequence test can shorten a stressful diagnostic odyssey and generate system-level savings [5, 6]. Initial evidence suggests that the diagnostic performance of WGS is superior to conventional genetic tests [7–9]. However, empiric data that reflect upon the downstream care and cost implications remain limited. In a climate of fiscal constraint [10] with evolving systems for evaluating and funding diagnostics [11–13], policymakers require accurate assessments of the downstream consequences of genetic testing to make sound investment, reimbursement, and delivery decisions [14].
Knowledge of genetic status gained through diagnostic investigations is thought to have personal utility for patients and families and clinical utility for practitioners and the health care system. While measurement strategies for capturing utility are being developed and debated [14, 15], preliminary evidence of clinical utility, as indicated by altered medical management, is beginning to emerge [16–25]. However, the known effects of WGS on downstream clinical activities are largely drawn from case reports or case series [16–25]. Comparative studies are needed to systematically determine the health care consequences and costs of WGS in relation to first line conventional tests (e.g., chromosome microarray (CMA)) [26]. The objective of this study was to quantify the incremental change in clinical activity and cost with WGS above and beyond standard care (i.e., CMA) and to contrast the patterns of prompted activity between testing strategies in children with developmental delay/intellectual disability.
Methods Study design
This observational cohort study was embedded within The Hospital for Sick Children (SickKids) Genome Clinic Project [27, 28], a Toronto-based program of translational research in genomics. Since a primary aim of the Genome Clinic was to determine the diagnostic performance of WGS compared to CMA, all children received CMA as part of standard clinical workup and WGS on a research basis [7]. Informed consent was obtained and the study was approved by the SickKids Research Ethics Board.
Sample
Children for whom CMA was indicated because of two or more structural malformations and/or unexplained developmental delay/intellectual disability were eligible; they were recruited prospectively from the Division of Clinical and Metabolic Genetics over a 9-month period (9/2013–5/2014). Only cases for whom post-CMA or WGS clinical follow-up occurred at SickKids were included in this report. Additional details on the study cohort are available [7].
Platforms
CMA was undertaken using the 4 × 180 K CytoSureISCA v2 oligonucleotide microarray platform (Oxford Gene Technology, Oxford, UK), the standard diagnostic service offered by SickKids Clinical Laboratory Improvement Amendments (CLIA)-certified diagnostic laboratory. Variants detected were classified according to the 2011 American College of Medical Genetics’ CMA guidelines [29]. Pathogenic variants and variants of unknown significance-likely pathogenic were categorized as “diagnostic” whereas variants of uncertain clinical significance, uncertain clinical significance-likely benign, and benign significance were classified as “non-diagnostic” [7]. Genomic DNA was sent to Complete Genomics (Mountain View, CA, USA) for WGS as described previously [7, 30]. Raw sequence reads, an average of ~52X coverage, were reassembled against a reference genome (GRCh37) and robust variant calling was completed using Complete Genomics assembly pipeline 2.4 [7, 31]. For six cases, exome sequencing was completed as WGS was not available in a timely manner.
Data collection
Prompted activity included clinical care activity that was either recommended or pursued following CMA result reporting and subsequent WGS result reporting. Activity data were obtained for each participant from hospital medical records and face-to-face physician interviews. An average interval of 15 months (range 5–20 months) elapsed between CMA and WGS result reporting, creating separation in prompted clinical activity and enabling the capture of a full range of activity prompted by CMA. At the outset of the study, physicians were encouraged to maintain the same standard of care that they would have performed outside of a study context. Study data were collected and managed using Research Electronic Data Capture (REDCap) software (Nashville, TN) [32]. From each patient record, we ascertained age at CMA, sex, phenotype, primary CMA and WGS results (i.e., diagnostic or non-diagnostic as per laboratory reports). Phenotype was extracted directly from the patient’s PhenoTips (Toronto, ON) record [33, 34]. Using PhenoTips, we determined the number of organ systems affected in each patient, and used this as a proxy for disease severity and complexity [25].
The type and volume of clinical activity prompted by CMA and WGS results were recorded using a structured, electronic data report form that captured six categories of clinical activity [25]. Activity recommended by medical geneticists at the time CMA (and later WGS) results were disclosed to parents was captured and the actual pursuit of this activity was recorded if indicated in the patient chart (e.g., documentation of laboratory test or specialist consult). All activity occurring up to 1 year following disclosure was recorded. Activity categories included: specialist consults, allied health consults, laboratory tests, imaging tests, medication alterations, and laboratory investigations for family members. In addition, medical geneticists were asked to list any services that were averted due to CMA or WGS results. Each unique activity was counted and was assumed to occur once, unless otherwise specified. Clinical activities related to ongoing clinical care (i.e., phenotype-driven) were distinguished from those prompted by the genetic test result (i.e., genotype-driven). For a given participant, the same ordering physician was involved in the interpretation, reporting and follow-up of CMA and WGS results, thus controlling for variation in practice patterns among practitioners. For each participant, chart extracted data were verified with the consulting physician. While care activities prompted or averted following WGS may have been influenced by care activities prompted or averted following CMA, interviews were conducted with physicians to verify activities that were specifically prompted by CMA or independently by WGS. All findings and categories of activity prompted by the disclosure of primary WGS results were verified by a second post-study chart review.
Costing
Each activity prompted or averted following CMA or WGS was assigned a unit price (Supplementary Table 1). Laboratory test costs were obtained from the Ontario Health Insurance Plan (OHIP) Schedule of Benefits for Laboratory Services [35]. Prices for genetic tests or other tests not found in this Schedule of Benefits were obtained from the laboratories that performed these tests (i.e., SickKids’ Genome Diagnostics Laboratory or external laboratories). Each specialist referral was priced as one assessment visit using the OHIP Schedule of Benefits for Physician Services [36]. Allied health consultations included audiology, dietetics/nutrition, occupational therapy and physiotherapy, speech, and language therapy and psychology. Referrals to allied health professionals were priced using a standard fee per visit obtained from fee schedules provided by professional associations (Ontario Society of Occupational Therapists, Ontario Psychological Association, Ontario Association of Speech-Language Pathologists and Audiologists). For occupational therapy, a fee schedule was not available so a representative average fee was estimated using an Internet search of Canadian providers. Dietician assessment fees were based on an average hourly rate at SickKids. The OHIP Schedule of Benefits—Physician Services was used to obtain prices for medical imaging tests and other tests (e.g., electrocardiogram (ECG) electroencephalogram (EEG), Holter monitor). The actual CMA and WGS tests, follow-up validation testing (i.e., Sanger sequencing), and genetic counseling that occurred following these tests were not counted as separate activities and were not included in the total costs since these costs did not differ by participant.
Analysis
Descriptive statistics were used to describe participant characteristics, as well as the volume, type, and cost of clinical activity that followed CMA and WGS. We examined total volume of activities as well as mean number of activities per participant. First, we characterized the total number and type of activity prompted by ongoing care for the full sample (n = 101). We assessed the association between the mean number of ongoing activities and participant characteristics using a negative binomial model. This model was most appropriate for count data which exhibited over-dispersion, a characteristic of this data set. The model was also compared with ordinary least squares and Poisson models and was found to be the best fit to the data based on Akaike information criterion (AIC) and Bayesian information criterion (BIC). Characteristics included age as a categorical variable: <12 months (reference group), 1–5 years, 6–10 years, >10 years; sex: male (reference group), female; number of organ systems affected: 1–2 (reference group), 3–4, 5–6, 7–8, and 9 or more.
Unless a second genomic diagnosis was suspected [7], WGS would not typically be indicated for children who had received a diagnostic result via CMA. As such, the remainder of our analysis examined activity prompted by WGS compared to activity prompted by CMA non-diagnostic results (n = 93). The data were not independent since each patient received both CMA and WGS. Models for count data were considered, but ultimately linear models were used as they were the most straightforward approach to modelling repeated measures data. We compared the mean number of activities per patient prompted by CMA with the mean number of activities per patient prompted by WGS and compared selected activity types (i.e., specialist and allied health visits, lab tests) based upon descriptive findings using a paired t-test. We used a linear mixed effects regression model to assess whether WGS result type (i.e., diagnostic vs. non-diagnostic) modified the effect that each test (WGS or CMA) had on clinical activity. Three regressions were run; (i) mean number of all activities, (ii) specialist and allied health visits and, and (iii) lab tests. The models used random intercepts to address the non-independence in the data. We also characterized the number and type of services averted as a result of WGS. A total cost per patient was determined by multiplying the volume of resource use for each item by a corresponding unit price in 2016 CDN dollars. The mean cost per patient was calculated for each comparator group. Given the skewed nature of resource use, cost 10th and 90th percentiles were calculated. All statistical analysis was conducted using R (2016, Vienna, Austria) [37].
Results
Sample characteristics
Of 201 potential participants approached, 101 children were included (50.2% participation rate). Most children were under 6 years old and had 3 or more organ systems implicated in their phenotype. The cohort was balanced with respect to sex. Following CMA testing, a diagnostic variant was identified in 8 (7.9%) cases. Among those for whom CMA did not yield a diagnostic result (n = 93), WGS identified a diagnostic variant in 22 (23.7%; Table 1).
Table 1.
Sample characteristics (n = 101)
| Characteristics | n | % | |
|---|---|---|---|
| Sex | |||
| Male | 54 | 53.5 | |
| Female | 47 | 46.5 | |
| Age at CMA | |||
| <12 month | 26 | 25.7 | |
| 1–5 years | 37 | 36.6 | |
| 6–10 years | 15 | 14.9 | |
| >10 years | 23 | 22.8 | |
| Phenotype: number of organ systems affected | |||
| 0–2 | 7 | 6.9 | |
| 3–4 | 25 | 24.8 | |
| 5–6 | 26 | 25.7 | |
| 7–8 | 22 | 21.8 | |
| 9 or more | 21 | 20.8 | |
| CMA result | |||
| Diagnostic | 8 | 7.9 | |
| Non-diagnostic | 93 | 92.1 | |
| WGS result (non-diagnostic CMA only, n = 93) | |||
| Diagnostic | 22 | 23.7 | |
| Non-diagnostic | 71 | 76.3 |
CMA chromosomal microarray analysis, WGS whole-genome sequencing
Activity following genetic testing
Within 1 year following the disclosure of CMA and WGS results, a total of 973 clinical activities were recommended by consulting geneticists for 101 children. The largest proportion of activity was driven by the child’s ongoing care (88.6%; 862 activities) rather than by CMA/WGS. A total of 427 specialist MD visits constituted the largest proportion of ongoing care (49.5%) and 130 allied health consultations (e.g., occupational therapy, physiotherapy) comprised 15.1% of ongoing care. Among specialist MD consults prompted by ongoing care, continued follow-up with genetics was the most common (20.8%), followed by neurology (10.3%). Among allied health providers, the most common referrals were for genetic counseling after result disclosure (29.2%), followed by audiology (16.2%). Medical imaging comprised 24.0% of ongoing care (207 activities), of which ultrasound was the most common. Other investigations comprised 8.2% (e.g., ECG, EEG) and laboratory tests comprised 3.1%, of which other genetic tests (i.e., non-CMA/WGS tests) were the most common (Supplementary Table 2; Supplementary Fig. 1).
With respect to the effect of participant characteristics on mean number of clinical activities, the regression model indicated that sex was not significantly associated with ongoing care whereas certain age groups and number of organ systems involved were (Supplementary Table 3). Relative to infants less than 12 months old, a child aged 6–10 years had a 28% decrease in the estimated mean number of ongoing care activities (p = 0.034) and a child who was 10 years or older had a 31% decrease in the estimated mean number of ongoing care activities (p = 0.008). Compared to 1–2 organ systems affected, having 9 or more affected organ systems increased the estimated mean number of ongoing care activities by 56% (p = 0.042).
Genotype-driven care
A highly skewed distribution of activity was observed; for at least 81.7% of the CMA negative cohort (n = 93), zero activities were prompted by CMA or WGS (Table 2). Of the total volume of care prompted by genetic testing for these patients, the mean number of activities per patient was 0.56 following non-diagnostic CMA and 0.62 following WGS (diagnostic and non-diagnostic combined); these were not significantly different (t = 0.24, df = 92, p = 0.815). However, the mean number of lab tests was significantly greater following CMA (t = −2.78, df = 92, p = 0.007) and the mean number of specialist or allied health visits was significantly greater following WGS (t = 2.46, df = 92, p = 0.016; Table 2 ; Fig. 1)
Table 2.
Distribution of participants by number of activities and mean number of activities per patient, prompted by CMA and WGS for CMA non-diagnostic cases only and stratified by WGS result: all activities, lab tests, specialist and allied health visits
| Prompted by CMA | Prompted by WGS | |||||
|---|---|---|---|---|---|---|
| Category | N (%) | Mean (SD) | Category | N (%) | Mean (SD) | |
| Total (n = 93) | ||||||
| All activities | 0 | 77 (82.8%) | 0.56 | 0 | 76 (81.7%) | 0.62 |
| 1–2 | 10 (10.8%) | (1.66) | 1–2 | 10 (10.8%) | (1.98) | |
| 3–4 | 3 (3.23%) | 3–4 | 2 (2.15%) | |||
| 5 or more | 3 (3.23%) | 5 or more | 5 (5.38%) | |||
| Lab tests | 0 | 77 (82.8%) | 0.55 | 0 | 85 (91.4%) | 0.09 |
| 1–2 | 10 (10.8%) | (1.64) | 1–2 | 8 (8.60%) | (0.28) | |
| 3–4 | 3 (3.23%) | 3–4 | 0 (0%) | |||
| 5 or more | 3 (3.23%) | 5 or more | 0 (0%) | |||
| Specialist and allied health visits | 0 | 93 (100%) | 0 (0) | 0 | 85 (91.4%) | 0.41 |
| 1–2 | 2 (2.15%) | (1.60) | ||||
| 3–4 | 3 (3.23%) | |||||
| 5 or more | 3 (3.23%) | |||||
| WGS non-dx (n = 71) | ||||||
| All activities | 0 | 57 (80.3%) | 0.65 | 0 | 64 (90.1%) | 0.11 |
| 1–2 | 9 (12.7%) | (1.82) | 1–2 | 7 (9.86%) | (0.36) | |
| 3–4 | 2 (2.82%) | 3–4 | 0 (0%) | |||
| 5 or more | 3 (4.23%) | 5 or more | 0 (0%) | |||
| Lab tests | 0 | 57 (80.3%) | 0.65 | 0 | 65 (91.6%) | 0.08 |
| 1–2 | 9 (12.7%) | (1.82) | 1–2 | 6 (8.45%) | (0.28) | |
| 3–4 | 2 (2.82%) | 3–4 | 0 (0%) | |||
| 5 or more | 3 (4.23%) | 5 or more | 0 (0%) | |||
| Specialist and allied health visits | 0 | 71 (100%) | 0 (0) | 0 | 71 (100%) | 0 (0) |
| WGS dx (n = 22) | ||||||
| All activities | 0 | 20 (90.9%) | 0.27 | 0 | 12 (54.5%) | 2.27 |
| 1–2 | 1 (4.55%) | (0.94) | 1–2 | 3 (13.6%) | (3.60) | |
| 3–4 | 1 (4.55%) | 3–4 | 2 (9.09%) | |||
| 5 or more | 0 (0%) | 5 or more | 5 (22.7%) | |||
| Lab tests | 0 | 20 (90.9%) | 0.23 | 0 | 20 (90.9%) | 0.09 |
| 1–2 | 1 (4.55%) | (0.75) | 1–2 | 2 (9.09%) | (0.29) | |
| 3–4 | 1 (4.55%) | 3–4 | 0 (0%) | |||
| 5 or more | 0 (0%) | 5 or more | 0 (0%) | |||
| Specialist and allied health visits | 0 | 22 (100%) | 0 (0) | 0 | 14 (63.6%) | 1.73 |
| 1–2 | 2 (9.09%) | (2.98) | ||||
| 3–4 | 3 (13.6%) | |||||
| 5 or more | 3 (13.6%) | |||||
CMA chromosomal microarray analysis, dx diagnostic, SD standard deviations, WGS whole-genome sequencing
Fig. 1.
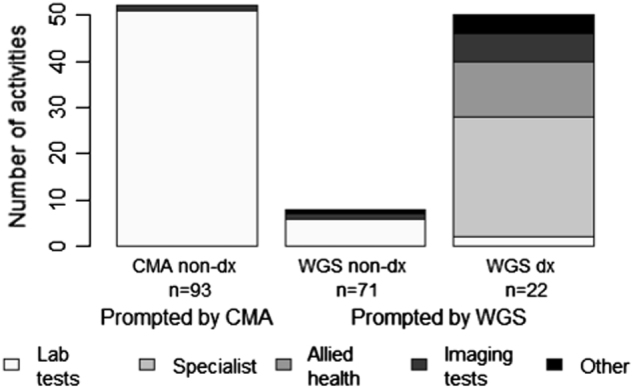
Number of activities by type prompted by sequential testing scenario: non-diagnostic CMA followed by diagnostic and non-diagnostic WGS results (n = 93). CMA chromosomal microarray analysis, WGS whole genome sequencing. 1. Lab tests may include genetic, biochemical and hematogy tests. Allied health may include audiology, dietetics/nutrition, occupational therapy and physiotherapy, spech and language therapy and psychology. Other activities may include electrocardiogram (ECG) electroencephalogram (EEG), Holter monitor, visual evoked potential (VEP), electromyography (EMG), Electroretinography (ERG), evoked potential, monitoring and surveillance and other. 2. The number of times a given test was implemented was included in the activity count. 3. Activities that are standard after CMA or WGS tests, validation testing (Sanger sequencing) and genetic counselling were not included
The mean difference in total number of activities between CMA and WGS was −0.54 when WGS was non-diagnostic, compared to 2.00 when WGS was diagnostic (Table 2). The linear mixed regression results (Table 3) confirm that this difference between WGS and CMA prompted activity depended on the WGS result type (p < 0.001). The regression results also show that the difference in lab activities did not depend on WGS result (p = 0.278) but the difference in specialist and allied health referrals did (p < 0.001). The latter results are highlighted in Table 2, where the mean difference in referrals between CMA and WGS is 0 when WGS is non-diagnostic and 1.73 when WGS is diagnostic. Among WGS diagnostic results (n = 22), the most common specialist referrals were ophthalmology/optometry (19.2%), gastroenterology (15.4%), and cardiology (15.4%). The most common allied health referrals were occupational therapy (25.0%), physiotherapy (25.0%), speech/language therapy (16.7%) and dietetics/nutrition (16.7% Supplementary Table 1).
Table 3.
Linear mixed effects model of mean number of activities based on the test, WGS result and an interaction between the test and WGS result (n = 93)
| Estimate | Standard error | t-value | p-value | |
|---|---|---|---|---|
| All activities | ||||
| Test (WGS vs. CMA) | −0.54 | 0.29 | −1.87 | 0.064 |
| WGS result (dx vs. non-dx.) | −0.38 | 0.42 | −0.90 | 0.370 |
| Interaction (WGS)×WGS result | 2.54 | 0.59 | 4.30 | <0.001 |
| Lab tests | ||||
| Test (WGS vs. CMA) | −0.56 | 0.19 | −2.96 | 0.004 |
| WGS result (dx. vs. non-dx.) | −0.42 | 0.29 | −1.47 | 0.144 |
| Interaction (WGS)×WGS result | 0.43 | 0.39 | 1.09 | 0.278 |
| Visits to specialists and allied health | ||||
| Test (WGS vs. CMA) | 0.00 | 0.17 | 0.00 | 1.000 |
| WGS result (dx. vs. non-dx) | 0.00 | 0.25 | 0.00 | 1.000 |
| Interaction (WGS)×WGS result | 1.73 | 0.35 | 4.95 | <0.001 |
CMA chromosomal microarray analysis, dx diagnostic, WGS whole-genome sequencing
No medication prescriptions/alterations and no cascade family genetic testing outside of parental testing for the purpose of interpreting proband results were observed post CMA or WGS reporting. Physician interviews indicated that following the completion of WGS, six activities were averted among children who received non-diagnostic WGS results (i.e., lab tests and specialist consults) and five activities were averted among children who received diagnostic WGS results that might otherwise have been pursued in the absence of WGS.
Projected cost of genotype-driven care
Given the skewed distribution of activity observed, costs incurred were similarly skewed, but also had heavy tails. The mean per patient cost of activities prompted by a non-diagnostic CMA result alone was $136.20 CDN (range $0–$3595.00) (Table 4). The additional, mean per patient cost of activities prompted by those who went on to receive a diagnostic WGS result was $179.68 CDN (range $0–$1212.00). The additional, mean per patient cost for those who went on to receive a non-diagnostic WGS result was $76.55 CDN (range $0–$4826.00). Some of the very high costs (e.g., >$2000 CDN per activity) were due to comprehensive gene panel tests that were conducted outside of Canada (e.g., ARHGAP31, DOCK6, EOGT, and RBP connective tissue gene panel, performed in USA). The mean savings per patient due to activities averted following WGS was $42.95 CDN (range $0–$583.00) for those who received diagnostic results and $4.17 CDN (range $0–$74.00) for those who received non-diagnostic results. Table 4 also shows that activity costs following non-diagnostic CMA results fell below $168.00 CDN for 90% of patients. For those who went on to receive non-diagnostic WGS results, zero activity costs were incurred for 90% of patients. For those who went on to receive diagnostic WGS results, prompted activity costs fell below $557.30CDN for 90% of patients.
Table 4.
Summary statistics for costs for patients with CMA non-diagnostic result only, for all activities prompted by CMA and WGS
| Mean | Median | SD | Lower quantile (10%) | Upper quantile (90%) | Min | Max | |
|---|---|---|---|---|---|---|---|
| CMA prompted | 136.20 | 0.00 | 530.94 | 0.00 | 168.00 | 0.00 | 3595.00 |
| (n = 93) | |||||||
| WGS prompted | |||||||
| WGS non-dx | |||||||
| (n = 71) | 76.55 | 0.00 | 574.18 | 0.00 | 0.00 | 0.00 | 4826.00 |
| WGS dx | |||||||
| (n = 22) | 179.68 | 0.00 | 327.95 | 0.00 | 557.30 | 0.00 | 1212.00 |
CMA chromosomal microarray analysis, dx diagnostic, WGS whole genome sequencing
Notes: The cost estimates included lab and imaging tests, visits with specialists and allied health professionals, and other activities. The cost estimates excluded the prices of the CMA or WGS test and the cost of activities that are standard after the CMA or WGS test, validation testing (Sanger sequencing) and genetic counseling
Disscussion
In a young, phenotypically complex pediatric cohort that underwent genomic testing, the majority of clinical activities recommended following testing were determined by the child’s clinical presentation, rather than by the child’s genotype. Unsurprisingly, a greater volume of clinical activities was observed in younger and more phenotypically complex children.
While the volumes of activities prompted by CMA and WGS were similar, the nature of the prompted activities was markedly different, highlighting a particular form of clinical utility. Specifically, non-diagnostic CMA results were more likely to lead to additional laboratory investigations, reflecting an ongoing pursuit of a diagnosis, while diagnostic WGS results were more likely to prompt specialist or allied health visits, reflecting clinicians’ shift away from diagnostic activities and toward disease-specific management, likely directed by the organ systems implicated in the genomic variants identified. As such, our data suggest that the incorporation of WGS into the genetic evaluation of children with complex health problems may increase referrals to specific specialty areas. To meet these increasing demands, targeted capacity building and coordination of care may be warranted.
Part of the utility of genomic testing relates to avoiding unnecessary care and associated costs. While diagnostic WGS results prompted specialist visits, these results were also associated with averting other types of care activities, presumably because achieving a diagnosis precluded the need for certain (typically diagnostic) investigations. Importantly, non-diagnostic WGS results were also associated with averted care. This may be due to the clinician’s assumption that WGS is ‘the end of the line’ in terms of our current ability to rule out causal variants; therefore, additional investigations may no longer be justifiable. The reduction in volume of laboratory testing following the receipt of non-diagnostic and (less so) diagnostic WGS results, compared to the volume of laboratory testing following non-diagnostic CMA results supports this premise.
Other studies have investigated the clinical utility of both whole exome and WGS in various pediatric patient populations. Case reports and small case series using WGS report direct links between mutations identified and management changes [16–18]. In the context of whole-exome sequencing (WES), establishing a diagnosis in 37 of 115 (32.2%) patients attending a genetics clinic led to discontinuation of additional diagnostic testing in all whom were diagnosed, screening for additional manifestations in 21.6%, new medication therapy in 5.4%, identification of other familial mutation carriers in 13.5%, and reproductive planning in 16.2% [20]. A more recent study found that of the 23 children who received a diagnosis via WES, clinical management was altered for 6 (26%), additional diagnostic investigations were canceled for 1 (4%), and prenatal and preimplantation genetic diagnosis was initiated for 1 (4%) [21]. In various CMA-based case series, ranging from 48 to 1792 cases, clinical action was prompted for the majority of children who received pathogenic results [22–24]. Finally, in a large pediatric cohort (n = 742), our group found that diagnostic variants and variants of uncertain clinical significance were associated with higher rates of clinical recommendations compared with non-diagnostic results (RR = 1.34; 95% CI 1.22–1.47; RR = 1.23; 95% CI 1.09–1.38) [25]. While findings to date are suggestive of clinical utility, results reported herein represent the first prospective controlled comparative study to address this particular outcome of genomic medicine.
The ultimate value of diagnostic WGS compared to standard care approaches such as CMA cannot be fully considered without an assessment of cost-effectiveness. In the present study, the costs of the genetic tests were omitted to enable examination of the incremental costs of prompted clinical activity. The data are beginning to emerge on the laboratory costs of WGS as well as costs that characterize the entire testing pathway in a clinical rather than research setting [38]. In a separate study, members of our team used a bottom-up micro-costing approach to provide a comprehensive estimate for WGS and CMA. For our institution, the total cost per patient with developmental delay and/or congenital anomalies (including labor, small and large equipment, supplies, bioinformatics, and validation) was $744.00 2015 CDN for CMA and $2851.00 2015 CDN for WGS [39]. Thus while the average per patient costs associated with activity triggered by diagnostic WGS results ($179.68CDN) are greater than those incurred for non-diagnostic CMA or WGS results ($136.20CDN or $76.65CDN, respectively), these costs are a small fraction of the testing pathway of care [39]. These costs are also a small fraction of the total costs associated with often-extensive diagnostic work up [5, 6] and care plan [40] for these complex patients. For example, the diagnostic work up in neurology clinics for patients with a broad range of neurological phenotypes has been reported to range from $5,382CDN to $17,692CDN [5, 6]. In the only other comparative study that reflects on this question, cost of care prompted by WGS among healthy adults did not differ from cost of care prompted by family history alone. In both groups, 6 month post-test care costs ranged between $40 and $200 USD (unpublished data).
It is expected that technical costs associated with high throughput sequencing will continue to decrease and that diagnostic performance will continue to improve. While greater diagnostic yields for primary targeted variants as well as secondary medically actionable variants may translate into improved health outcomes in the long run, in the short term a growing list of reportable variants will prompt additional downstream clinical activities, thereby putting increased pressure on medical geneticists, other clinical sub-specialists, allied health professionals, laboratory scientists, and imaging services, adding costs to the healthcare system. Ultimately, in adjudicating investment and adoption of complex genome diagnostics, decision makers will require rigorous evidence of cost-effectiveness to trade-off these costs with the anticipated value of WGS. The data reported herein begin to characterize the nature and cost of these downstream pathways.
We acknowledge several limitations. While a randomized controlled trial would be the ideal design for assessing the impact of WGS compared to standard care, the Genome Clinic was structured to perform both WGS and CMA on all participants. As an embedded research study, it was not feasible to randomize participants. This limited our ability to directly attribute observed differences to WGS. However, given the heterogeneity of the patient sample, a non-experimental controlled comparison with each subject serving as his/her own control has merit. Moreover, approaching the analysis as a sequence of tests allowed for marginal analysis of additional clinical activity and costs triggered by WGS. Since the same physician advised on clinical activity following CMA and WGS, they were not blind to the CMA results or recommended activity that preceded the receipt of WGS. Thus, it is possible that activity prompted by CMA was affected by the knowledge that WGS results were pending. Similarly, activity prompted post-WGS may have been influenced by activity already prompted by CMA. This bias was mitigated by recording activity recommended after each type of testing strategy as if it was the solo test. This approach also controlled for variation in practice between providers. Although the interval between CMA and WGS reporting was <12 months for a small number of cases, an average of 15 months between CMA and WGS result reporting suggests that practitioners were unlikely to be influenced by the other test in recommending clinical activity. In addition, six cases reported in Stavropoulos et al. [7]. are not reported here because follow-up consultation data were unavailable or because they were siblings of the index case. Six other cases reported here but not in Stavropoulos et al. [7] represent exome sequencing cases. While these exome cases did not meet criteria for Stavropoulos’ technical comparison study, outcome data prompted by WES result disclosure remain relevant to the study reported here. As well, while pharmacogenomics variants were identified in this cohort [41], medical management implications of these variants were not captured because identified variants had not been validated or reported back to patients at the time these data were ascertained. The absence of cascade family testing is surprising, but perhaps attributable to de novo findings or the absence of documentation related to cascade family testing in the proband’s medical record. It is possible that cascade testing was documented in family members’ charts but these records were not reviewed. In addition, while the endpoints (i.e., type, volume and cost of clinical activities triggered by CMA/WGS) are somewhat generic from a clinical perspective, these data assist our efforts to characterize the care pathway and resource demands implicated by genome-wide technologies. Finally, assigned costs represent projected costs as verification that all recommended activities were pursued was not achieved. Although limited in scope, this cost analysis fulfills a necessary first step towards a full economic evaluation that includes both costs and health benefits as a consequence of WGS compared to conventional genetic testing.
In conclusion, this comparative study of the clinical consequences and costs of hospital clinic-based pediatric WGS may inform a decision model to assess cost-effectiveness of WGS compared to standard care. This could be achieved in a variety of ways. The absolute numbers of lab tests, specialist visits and other services prompted by WGS compared to CMA could be used as reference case inputs associated with post-testing health states for the same type of patient population. The relative increase or decrease in use of the various categories of health services observed in the present study could set the boundaries for a “treatment effect” in a model, or be used to validate results obtained from a separate independent observational study or chart review. In addition, we emphasize that more robust study designs that capture the downstream effects of WGS are warranted as are novel measures that capture a more comprehensive notion of clinical utility [15]. However, well-suited to the translational stage of WGS today [42], findings reported herein, as a form of early health technology assessment [43], can begin to inform sound investments in and appropriate use of state of the art genomic technology in pediatrics.
Electronic supplementary material
Acknowledgements
Thank you to Reilly Jones for her assistance with the literature review. This work was funded by an Accelerator Grant in Genomic Medicine, McLaughlin Centre, University of Toronto. S.W.S. was supported by the GlaxoSmithKline-CIHR Chair in Genome Sciences at the University of Toronto and the Hospital for Sick Children.
Funding
This work was funded by Dr. Robin Hayeems’ Accelerator Grant in Genomic Medicine, awarded by the McLaughlin Centre, University of Toronto.
Compliance with ethical standards
Competing interests
The authors declare that they have no competing interests.
Electronic supplementary material
The online version of this article (10.1038/s41431-017-0020-3) contains supplementary material, which is available to authorized users.
References
- 1.Wade C, Tarini BA, Wilfond BS. Growing up in the genomic era: implications of whole-genome sequencing for children, families, and pediatric practice. Annu Rev Genom Hum Genet. 2013;14:535–55. doi: 10.1146/annurev-genom-091212-153425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, Chisholm RL, Ozenberger B, et al. Implementing genomic medicine in the clinic: the future is here. Genet Med. 2013;15:258–67. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johansen Taber KA, Dickinson BD, Wilson M. The promise and challenges of next-generation genome sequencing for clinical care. JAMA Intern Med. 2014;174:275–80. doi: 10.1001/jamainternmed.2013.12048. [DOI] [PubMed] [Google Scholar]
- 4.Next steps in the sequence: the implications of whole genome sequencing for health in the UK. October 2011 © 2011 PHG Foundation. http://www.phgfoundation.org/file/10363. Accessed 10 November 2016.
- 5.van Nimwegen K, Schieving J, Willemsen M, et al. The diagnostic pathway in complex paediatric neurology: a cost analysis. Eur J Paediat Neurol. 2015;19:233–39. doi: 10.1016/j.ejpn.2014.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Richards J, Korgenski EK, Srivastava R, Bonkowsky JL. Costs of the diagnostic odyssey in children with inherited leukodystrophies. Neurology. 2015;85:1167–70. doi: 10.1212/WNL.0000000000001974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stavropoulos DJ, Merico D, Jobling R, et al. Whole genome sequencing expands diagnostic utility and improves clinical management in pediatric medicine. NPJ Genom Med. 1 pii:15012 Epub: 2016. [DOI] [PMC free article] [PubMed]
- 8.Gilissen C, Hehir-Kwa JY, Thung DT, et al. Genome sequencing identifies major causes of severe intellectual disability. Nature. 2014;511:344–7. doi: 10.1038/nature13394. [DOI] [PubMed] [Google Scholar]
- 9.Soden SE, Saunders CJ, Willig LK, et al. Effectiveness of exome and genome sequencing guided by acuity of illness for diagnosis of neurodevelopmental disorders. Sci Transl Med. 2014;6:265ra168. doi: 10.1126/scitranslmed.3010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vozikis A, Cooper DN, Mitropoulou C, et al. Test pricing and reimbursement in genomic medicine: towards a general strategy. Public Health Genom. 2016;19:352–63. doi: 10.1159/000449152. [DOI] [PubMed] [Google Scholar]
- 11.National Institute for Health and Clinical Excellence. Diagnostics Assessment Programme manual December 2011. https://www.nice.org.uk/Media/Default/About/what-we-do/NICE-guidance/NICE-diagnostics-guidance/Diagnostics-assessment-programme-manual.pdf [PubMed]
- 12.Ferrante di Ruffano L, Hyde CJ, McCaffery KJ, Bossuyt PM, Deeks JJ. Assessing the value of diagnostic tests: a framework for designing and evaluating trials. BMJ. 2012;344:e686. doi: 10.1136/bmj.e686. [DOI] [PubMed] [Google Scholar]
- 13.Bossuyt PM, Reitsma JB, Linnet K, Moons KG. Beyond diagnostic accuracy: the clinical utility of diagnostic tests. Clin Chem. 2012;58:1636–43. doi: 10.1373/clinchem.2012.182576. [DOI] [PubMed] [Google Scholar]
- 14.Caulfield T, Evans J, McGuire A, et al. Reflections on the cost of “low-cost” whole genome sequencing: framing the health policy debate. PLoS Biol. 2013;11:e1001699. doi: 10.1371/journal.pbio.1001699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.ACMG Board of Directors. Clinical utility of genetic and genomic services: a position statement of the American College of medical genetics and genomics. Genet Med. 2015;17:505–7. doi: 10.1038/gim.2015.41. [DOI] [PubMed] [Google Scholar]
- 16.Bainbridge MN, Wiszniewski W, Murdock DR, et al. Whole-genome sequencing for optimized patient management. Sci Transl Med. 2011;3:87re3. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Link DC, Schuettpelz LG, Shen D, et al. Identification of a novel TP53 cancer susceptibility mutation through whole-genome sequencing of a patient with therapy-related AML. JAMA. 2011;305:1568–76. doi: 10.1001/jama.2011.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saunders CJ, Miller NA, Soden SE, et al. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci Transl Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Worthey EA, Mayer AN, Syverson GD, et al. Making a definitive diagnosis: successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet Med. 2011;13:255–62. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- 20.Iglesias A, Anyane-Yeboa K, Wynn J, et al. The usefulness of whole-exome sequencing in routine clinical practice. Genet Med. 2014;16:922–31. doi: 10.1038/gim.2014.58. [DOI] [PubMed] [Google Scholar]
- 21.Tan TY, Dillon OJ, Stark Z, et al. Diagnostic impact and cost-effectiveness of whole-exome sequencing for ambulant children with suspected monogenic conditions. JAMA Pediatr. Published online 31 July 2017. 10.1001/jamapediatrics.2017.1755 [DOI] [PMC free article] [PubMed]
- 22.Henderson LB, Applegate CD, Wohler E, Sheridan MB, Hoover-Fong J, Batista DA. The impact of chromosomal microarray on clinical management: a retrospective analysis. Genet Med. 2014;16:657–64. doi: 10.1038/gim.2014.18. [DOI] [PubMed] [Google Scholar]
- 23.Coulter ME, Miller DT, Harris DJ, et al. Chromosomal microarray testing influences medical management. Genet Med. 2011;13:770–776. doi: 10.1097/GIM.0b013e31821dd54a. [DOI] [PubMed] [Google Scholar]
- 24.Saam J, Gudgeon J, Aston E, Brothman AR. How physicians use array comparative genomic hybridization results to guide patient management in children with developmental delay. Genet Med. 2008;10:181–6. doi: 10.1097/GIM.0b013e3181634eca. [DOI] [PubMed] [Google Scholar]
- 25.Hayeems RZ, Hoang N, Chenier S, et al. Capturing the clinical utility of genomic testing: medical management following pediatric microarray. Eur J Hum Gen. 2015;23:1135–41. doi: 10.1038/ejhg.2014.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vassy JL, Lautenbach DM, McLaughlin HM, et al. The MedSeq project: a randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bowdin SC, Hayeems RZ, Monfared N, Cohn RD, Meyn MS. The SickKids genome clinic: developing and evaluating a pediatric model for individualized genomic medicine. Clin Genet. 2016;89:10–9. doi: 10.1111/cge.12579. [DOI] [PubMed] [Google Scholar]
- 28.Bowdin S, Ray PN, Cohn RD, Meyn MS. The genome clinic: a multidisciplinary approach to assessing the opportunities and challenges of integrating genomic analysis into clinical care. Hum Mutat. 2014;35:513–9. doi: 10.1002/humu.22536. [DOI] [PubMed] [Google Scholar]
- 29.Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. Working group of the American College of Medical Genetics Laboratory Quality Assurance C et al. American college of medical genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 2011;13:680–5. [DOI] [PubMed]
- 30.Drmanac R, Sparks AB, Callow MJ, et al. Human genome sequencing using unchained base reads on self-assembling DNA nanoarrays. Science. 2010;327:78–81. doi: 10.1126/science.1181498. [DOI] [PubMed] [Google Scholar]
- 31.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap) - a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–81. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girdea M, Dumitriu S, Fiume M, et al. PhenoTips: patient phenotyping software for clinical and research use. Hum Mutat. 2013;34:1057–65. doi: 10.1002/humu.22347. [DOI] [PubMed] [Google Scholar]
- 34.Robinson PN, Köhler S, Bauer S, et al. The human phenotype ontology: a tool for annotating and analyzing human hereditary disease. Am J Hum Genet. 2008;83:610–15. doi: 10.1016/j.ajhg.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ontario Ministry of Health and Long-Term Care. 1999. OHIP Schedule of Benefits and Fees, Schedule of Benefits for Laboratory Services. http://www.health.gov.on.ca/en/pro/programs/ohip/sob/lab/lab_services_sched_01_19990401.pdf
- 36.Ontario Ministry of Health and Long-Term Care. 2016. OHIP Schedule of Benefits and Fees, Physician Services under the Health Insurance Act. http://www.health.gov.on.ca/en/pro/programs/ohip/sob/physserv/sob_master20160401.pdf.
- 37.R Core Team. R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing; 2016. [Google Scholar]
- 38.Christensen KD, Dukhovny D, Siebert U, Green RC. Assessing the costs and cost-effectiveness of genomic sequencing. J Pers Med. 2015;5:470–86. doi: 10.3390/jpm5040470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsiplova K, Zur RM, Marshall CR, Stavropoulos DJ, Pereira SL, Merico D, Young EJ, Sung WWL, Scherer SW, Ungar WJ. A microcosting and cost-consequence analysis of clinical genomic testing strategies in autism spectrum disorder.Genet Med. 2017 May 4. 10.1038/gim.2017.47. [Epub ahead of print]. [DOI] [PubMed]
- 40.Cohen E, Berry JG, Camacho X, Anderson G, Wodchis W, Guttmann A. Patterns and costs of health care use of children with medical complexity. Pediatrics. 2012;130:e1463. doi: 10.1542/peds.2012-0175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cohn I, Paton TA, Marshall CR, Basran R, Stavropoulos DJ, Ray PN, Monfared N, et al. Genome sequencing as a platform for pharmacogenetic information: a cohort study. npj Genomic Medicine. 2017;9:665–74. doi: 10.1038/s41525-017-0021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khoury MJ, Gwinn M, Yoon PW, Dowling N, Moore CA, Bradley L. The continuum of translation research in genomic medicine: How can we accelerate the appropriate integration of human genome discoveries into health care and disease prevention? Genet Med. 2007;9:665–74. doi: 10.1097/GIM.0b013e31815699d0. [DOI] [PubMed] [Google Scholar]
- 43.Steuten LMG. Early stage health technology assessment for precision biomarkers in oral health and systems medicine. OMICS: J Integ Biol. 2016;20:30–35. doi: 10.1089/omi.2015.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


