Abstract
Staphylococcus aureus is the major cause of foodborne diseases worldwide. In this retrospective study, 84 S. aureus strains were characterized. The collection comprises 78 strains isolated during 1998 and 2014 from dairy products and tissue samples from livestock bred for dairy production in Sicily. One isolate was obtained from a pet (dog), one from an exotic animal (a circus elephant), and four human isolates were obtained during a severe food poisoning outbreak that occurred in Sicily in 2015. All the strains were characterized by pulsed-field gel electrophoresis (PFGE), for antibiotic resistance and presence of toxin genes. PFGE results showed 10 different pulsotypes, with three relatively frequent and three unique. The antibiotic resistance profiling showed that penicillin G (35.7%) and tetracycline (20.2%) resistance is largely spread. Most isolates contained at least one toxin gene making them a potential threat for public health. Enterotoxin sec gene was observed in 28.6% and seg in 23.8% of the strains, respectively; the human isolates were the only ones to concurrently harbor both seg and sei genes. In addition, 24 isolates were randomly selected and analyzed by multilocus sequence typing. Interestingly, the analysis showed the presence of 12 sequence types (STs), of which 6 were novel. One of them, ST700, was detected in 29% of the isolates and was found to be spread throughout Sicily. ST700 has been present in the island for almost 16 years (1998–2014) and it shows no host preference since it was isolated from different ruminant species. Four human isolates shared both the pulsotype (PT10) and the sequence type (ST9), as well as the virulence genes (seg-sei); this observation suggests that the isolates originated from a single clone, although they were obtained from two different individuals.
Keywords: : antibiotic resistance, MLST, PFGE, Staphylococcus aureus, MRSA, toxin genes
Introduction
Staphylococcus aureus is a major resident or transient colonizer of the skin and the mucosa of humans and primates. S. aureus can cause a variety of infections, from superficial skin infections to severe, and potentially fatal, invasive diseases (Wang et al., 2014; Aires-de-Sousa, 2017; Sergelidis and Angelidis, 2017). S. aureus is also a common pathogen of ruminants such as cattle, goats, and sheep that may lead to clinical and subclinical mastitis. The pathogen can spread from the udder of the infected animal into raw milk and dairy products, affecting the quality and quantity of the products; therefore, the pathogen can become a significant economic burden for farmers and a serious problem for the dairy industry (Seegers et al., 2003).
In the last years, different multidrug-resistant strains have emerged making S. aureus a major concern for public health. The multidrug-resistant phenotype is a particular characteristic of the methicillin-resistant S. aureus (MRSA) strains (Gould et al., 2012; Rodvold and McConeghy, 2014). The mecA gene, present in the Staphylococcal cassette chromosome mec (SCCmec), is a biomarker gene responsible for resistance to methicillin and other β-lactam antibiotics (Peacock and Paterson, 2015; Liu et al., 2016). Methicillin-resistant S. aureus isolates have been widely observed not only in hospitals but also in human communities with no risk factors for MRSA acquisition (Aires-de-Sousa, 2017). In recent years, several cases of MRSA transmission from pets or animals of the food chain to humans have been reported. The adaptation of MRSA clones of human origin to animal hosts has also been observed (Pomba et al., 2016).
Some S. aureus strains produce toxins, such as superantigen staphylococcal toxic shock syndrome toxin (TSST-1), staphylococcal enterotoxins (SEs), or enterotoxin-like proteins (SEl).
Up to now, more than 20 SEs or enterotoxin-like proteins have been identified (Mehrotra et al., 2000; De Buyser et al., 2009; Argudín et al., 2010).
The presence of SEs in food can lead to staphylococcal food poisoning, one of the most common in the world. In Sicily, food poisoning cases occur sporadically (Kadariya et al., 2014). A previous screening for enterotoxigenic S. aureus strains isolated from food samples showed that some pathogenic S. aureus strains were circulating in farms with apparently healthy animals. A high percentage of the isolates (46%) carried a toxin gene, creating significant concern that pathogenic S. aureus strains can be transmitted through food (Vitale et al., 2015).
The molecular characterization of bacterial strains is important for the detection of transmission routes and infection sources and for the monitoring of bacterial strain circulation among animal populations (Lange et al., 1999; Rodriguez et al., 2015; Macori et al., 2017). Pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) techniques are excellent methods for epidemiological studies and for the identification of sources and transmission routes for control improvement (Golding et al., 2015).
The aim of this work was the molecular characterization and the analysis of antibiotic resistance and the detection of toxin genes in S. aureus isolates derived from food and livestock. Four human isolates, one isolate from a dog, and one from a circus elephant were also characterized.
Materials and Methods
Origin and biochemical analysis of the bacterial isolates
Eighty-four S. aureus strains collected between 1998 and 2015 and isolated from food, animals, and humans were analyzed. Seventy-eight strains were isolated from dairy products and from animal tissue samples such as cow milk and cheese, sheep milk and cheese, goat milk, sheep skin flakes, and sheep udder. Two isolates obtained from skin flakes of other animals (elephant and dog) were used for comparison. Four human isolates were obtained from clinical samples of two individuals deceased after a food poisoning episode, which also affected four other patients (who fully recovered after severe gastroenteric symptoms). Single hemolytic colonies were inoculated in the brain/heart infusion broth (BHI) agar at 37°C. The strains were subjected to Gram staining and biochemical analysis, including coagulase, catalase, and Voges-Proskauer (VP) tests (BioMérieux), oxidase test (Oxoid), glucose and mannitol acidification in red phenol broth (Difco). The colonies had been identified as S. aureus by the API STAPH test (BioMérieux). Bacteria were maintained as frozen cell glycerol stocks as described elsewhere (Giardina et al., 2010; Lo Grasso et al., 2015).
PFGE analysis
Plug preparation, genomic restriction, and PFGE analysis of isolates were carried out as described in Alduina and Pisciotta (2015). In short, a single colony was inoculated into 5 mL of BHI broth and incubated at 37°C for 24 h. Cells were harvested and suspended in TE buffer (10 mM Tris HCl, 1 mM EDTA, pH 8). Three microliter lysostaphin solution (Sigma-Aldrich) (1 mg/mL in 20 mM sodium acetate) and 2% (wt/vol) SeaKem Gold agarose (Cambrex, Rockland, Maine) in TE buffer were added. The mixtures were dispensed into the wells of a small mold. Once solidified, the plugs were incubated in EC lysis buffer (6 mM Tris HCl, 1 M NaCl, 100 mM EDTA, 0.2% sodium deoxycholate, 0.5% sodium lauroyl sarcosine) at 37°C for 4 h. Plugs were washed with TE buffer three times and stored at 4°C.
The DNA was digested with 20 U SmaI (New England Biolabs) at room temperature for 4 h. Macrorestriction fragments were separated using a BioRad CHEF System (30" 12 h, 15" 6 h, 1% gel in Tris-borate-EDTA [TBE] 0.5 × , 200 V) and a PFGE size standard (CHEF DNA Size Standard, Lambda [λ] ladder; BioRad) was added.
After run, the gels were stained with ethidium bromide (0.5 μg/mL) and viewed under UV light. Gel images were captured by Molecular Imager Gel Doc XR (BioRad) and the banding patterns were used to establish isolate relatedness. Identical PFGE profiles (100% similarity) were defined as a pulsotype. The pulsotypes identified were given customized names PT1-10. PFGE pulsotypes were classified on the basis of the number of isolates sharing the same PT as major (more than six isolates/PFGE types), intermediate (between two and six isolates/PFGE types), or unique pulsotypes.
Multilocus sequence typing
MLST was carried out using the protocol described in Enright et al. (2000) on 24 isolates of the collection. The selection was performed in such a way that at least one isolate for each pulsotype and different year, if available, could be analyzed. Polymerase chain reaction (PCR) was performed in a 30 μL volume reaction containing 1.5 U of recombinant Taq DNA polymerase (Invitrogen, Life Technologies) as described in Randazzo et al. (2015).
PCR products derived from the seven housekeeping genes (arcc, aroe, glpf, gmk, pta, tpi, yqil) were treated with HT ExoSAP-IT (Affymetrix) following the manufacturer's instruction. The purified samples were used for sequencing using the BigDye Terminator v1.1 Cycle Sequencing Kit (Applied Biosystems) followed by capillary electrophoresis on the ABI Prism 310 Genetic Analyzer (Applied Biosystems) as described in D'Andrea et al. (2012). The sequences were then analyzed using the ABI3130 Genetic Analyzer (Applied Biosystems). The allelic profile for these genes was obtained from the MLST website (www.mlst.net). The combination of the seven allele numbers defines the allelic profile of the strain and each different allelic profile was assigned as a sequence type (ST), which is used to describe the strain (Aanensen and Spratt, 2005). All STs described in the study were compared with the major international S. aureus STs published in the MLST website www.mlst.net/databases/default.asp. The sequence of the new alleles was deposited in the MLST website.
Antimicrobial susceptibility tests
The antimicrobial susceptibility profiles to the main classes of antibiotics were determined by using the Kirby–Bauer method using Mueller–Hinton agar (MHA) medium, as described by the NCCLS (CLSI, 2015). Bacterial suspensions in BHI broth with a turbidity equivalent to a 0.5 McFarland standard were prepared and spread on the surface of MHA plates. Antibiotic disks containing the aminoglycosides gentamycin (CN, 10 μg) and kanamycin (K, 30 μg), the lincosamide lincomycin (MY, 2 μ g), the macrolide erythromycin (E, 15 μg), tetracycline (TE, 30 μg), and the β-lactam antibiotics cefoperazone (CFP, 75 μg) and penicillin G (P, 10 U.I) were used. Antimicrobial disks were obtained from Oxoid (United Kingdom). The results were interpreted in accordance with the standards for inhibition zone diameters for Staphylococcus spp. (CLSI, 2015). S. aureus ATCC 25923 was used as a reference strain for antimicrobial susceptibility testing.
Detection of SE (sea-see, seg-sei, sej, sep), tsst-1, eta, etb, and mecA genes
Total DNA was extracted from each isolate by boiling the samples for 20 min in 1 mL of TE buffer. Two multiplex PCR assays described in Mehrotra et al. (2000) were used to amplify sea- see and tsst-1, eta, etb, mecA genes, respectively. For detection of seg, seh, sei, sej, and sep, a multiplex PCR assay described by De Buyser et al. (2009) was followed. Detection of femA was used as an internal positive control and S. aureus ATCC 25923 as quality control. For the multiplex reactions, the 25 μL reaction mixture contained 1 U of AmpliTaq Gold 360 (Thermo Fisher Scientific), 2.5 mM MgCl2, 0.2 mM dNTPs, 1 × PCR buffer, 0.2 μM of each primer, and 1 μL of total DNA. PCR was performed on a 9700 Thermo cycler (Applied Biosystems). The thermal cycle for the amplification of sea- see, tsst-1, eta, etb, and mecA genes included the following: an initial denaturation at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 57°C for 1 min, extension at 72°C for 1 min 30s, and a final extension at 72°C for 7 min. The conditions for the multiplex PCR of seg, seh, sei, sej, and sep genes were as described above, except that the annealing step was performed at 52°C for 30 s. Positive strains carrying enterotoxin genes, kindly provided by the Italian reference laboratory for Staphylococcus spp., were used as controls. The following reference strains with relative genes in parenthesis were used: FRIS6 (sea, seb); FRI137 (sec, seg, seh, sei); HMPL280 (sed, seg, sei, sej, sep, ser); and FRI326 (see). The amplicons were detected using a 2.5% agarose gel containing ethidium bromide and visualized under ultraviolet light. The presence of a band of the expected size was considered as positivity to the presence of the corresponding gene.
Results
Molecular typing of S. aureus isolates
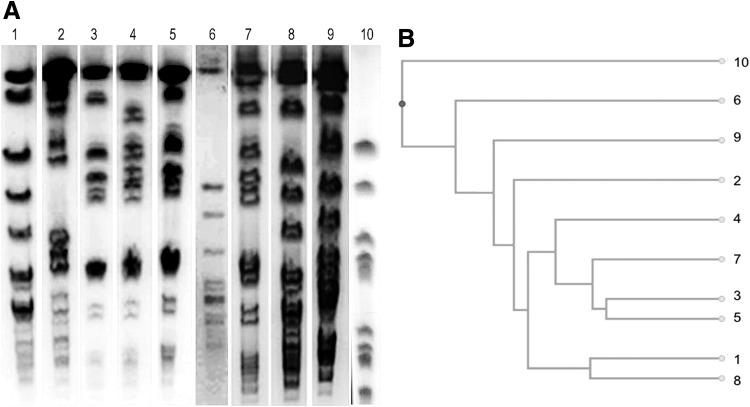
Eighty S. aureus strains isolated in Sicily from 1998 to 2014 from a collection of milk, cheese, and animal tissue were analyzed; in addition, four human samples recovered from a food poisoning episode were added (Table 1). PFGE analysis resulted in the detection of 10 pulsotypes (PT1-PT10, Fig. 1A). Sixty-five of the isolates (PT3, −4, and −5) grouped into 3 major pulsotypes (77.4%), 15 isolates (PT7, −8, and −10) grouped into 3 intermediate pulsotypes (17.9%), and 4 isolates (PT1, −2, −6, and −9) showed unique restriction profiles.
Table 1.
Staphylococcus aureus Strains Isolated in Sicily Analyzed by Multilocus Sequence Typing and Pulsed-Field Gel Electrophoresis
| Typing by PFGE and MLST | ||||
|---|---|---|---|---|
| Isolate | Year | Sample | PT | ST |
| 1 | 1998 | Elephant skin | 9 | ST1614 |
| 2 | 1998 | Sheep milk | 4 | ST700 |
| 3 | 1999 | Dog skin | 5 | ST522 |
| 4 | 2001 | Sheep skin | 7 | ST700 |
| 5 | 2001 | Sheep skin | 6 | ST425 |
| 6 | 2002 | Sheep skin | 5 | ST1616 |
| 7 | 2003 | Sheep milk | 2 | ST1626 |
| 8 | 2005 | Sheep cheese | 3 | ST700 |
| 9 | 2006 | Sheep skin | 3 | ST700 |
| 10 | 2007 | Goat milk | 1 | ST1 |
| 11 | 2008 | Goat milk | 5 | ST130 |
| 12 | 2008 | Sheep milk | 4 | ST700 |
| 13 | 2008 | Goat cheese | 4 | ST130 |
| 14 | 2008 | Cow cheese | 4 | ST97 |
| 15 | 2009 | Sheep milk | 4 | ST130 |
| 16 | 2010 | Sheep udder | 4 | ST700 |
| 17 | 2010 | Cow milk | 7 | ST1615 |
| 18 | 2012 | Cow cheese | 3 | ST1627 |
| 19 | 2012 | Sheep udder | 8 | ST522 |
| 20 | 2014 | Goat milk | 4 | ST700 |
| A | 2015 | Human | 10 | ST9 |
| B | 2015 | Human | 10 | ST9 |
| C | 2015 | Human | 10 | ST9 |
| D | 2015 | Human | 10 | ST9 |
| 21 | 2008 | Cow milk | 3 | NA |
| 22 | 2008 | Cow milk | 3 | NA |
| 23 | 2008 | Cow milk | 3 | NA |
| 24 | 2008 | Cow milk | 3 | NA |
| 25 | 2008 | Cow milk | 3 | NA |
| 26 | 2008 | Cow milk | 3 | NA |
| 27 | 2008 | Cow milk | 7 | NA |
| 28 | 2008 | Goat cheese | 3 | NA |
| 29 | 2008 | Cow milk | 3 | NA |
| 30 | 2009 | Cow milk | 7 | NA |
| 31 | 2009 | Sheep milk | 4 | NA |
| 32 | 2010 | Goat milk | 3 | NA |
| 33 | 2010 | Sheep skin | 7 | NA |
| 34 | 2014 | Sheep milk | 7 | NA |
| 35 | 2010 | Sheep milk | 3 | NA |
| 36 | 2010 | Sheep skin | 5 | NA |
| 37 | 2010 | Sheep milk | 5 | NA |
| 38 | 2010 | Sheep milk | 3 | NA |
| 39 | 2010 | Sheep skin | 5 | NA |
| 40 | 2010 | Sheep milk | 5 | NA |
| 41 | 2010 | Sheep milk | 5 | NA |
| 42 | 2010 | Sheep milk | 5 | NA |
| 43 | 2010 | Sheep milk | 5 | NA |
| 44 | 2010 | Sheep milk | 5 | NA |
| 45 | 2010 | Sheep milk | 5 | NA |
| 46 | 2010 | Sheep milk | 4 | NA |
| 47 | 2010 | Sheep milk | 3 | NA |
| 48 | 2010 | Sheep milk | 8 | NA |
| 49 | 2010 | Goat udder | 4 | NA |
| 50 | 2008 | Sheep cheese | 3 | NA |
| 51 | 1998 | Sheep udder | 4 | NA |
| 52 | 2004 | Goat cheese | 4 | NA |
| 53 | 2007 | Goat skin | 8 | NA |
| 54 | 2008 | Goat cheese | 4 | NA |
| 55 | 2005 | Sheep milk | 8 | NA |
| 56 | 2005 | Sheep skin | 8 | NA |
| 57 | 2007 | Sheep milk | 4 | NA |
| 58 | 2005 | Sheep milk | 4 | NA |
| 59 | 2005 | Sheep milk | 3 | NA |
| 60 | 2005 | Sheep milk | 5 | NA |
| 61 | 2005 | Sheep milk | 4 | NA |
| 62 | 2005 | Sheep milk | 5 | NA |
| 63 | 2005 | Sheep milk | 4 | NA |
| 64 | 2005 | Sheep udder | 4 | NA |
| 65 | 2006 | Sheep milk | 5 | NA |
| 66 | 2006 | Sheep milk | 5 | NA |
| 67 | 2010 | Sheep milk | 3 | NA |
| 68 | 2002 | Goat cheese | 3 | NA |
| 69 | 2002 | Goat cheese | 3 | NA |
| 70 | 2012 | Sheep milk | 4 | NA |
| 71 | 2012 | Sheep milk | 4 | NA |
| 72 | 2012 | Sheep udder | 4 | NA |
| 73 | 2012 | Sheep milk | 4 | NA |
| 74 | 2012 | Goat skin | 4 | NA |
| 75 | 2012 | Sheep skin | 4 | NA |
| 76 | 2012 | Sheep milk | 4 | NA |
| 77 | 2012 | Sheep milk | 3 | NA |
| 78 | 2012 | Sheep milk | 4 | NA |
| 79 | 2012 | Sheep milk | 4 | NA |
| 80 | 2012 | Goat milk | 4 | NA |
Numbers 1–80 indicate isolates from animals or dairy products, letters A–D indicate the human isolates. All the strains were typed by PFGE, the first 20 randomly chosen isolates and the 4 human isolates were additionally typed using MLST. The choice of the 20 isolates was done using at least an isolate per year.
MLST, multilocus sequence typing; NA, not analyzed; PFGE, pulsed-field gel electrophoresis; PT, pulsotype; ST, sequence type.
FIG. 1.
Pulsotypes of Staphylococcus aureus isolates. (A) Example of the PFGE profiles after SmaI digestion of genomic DNA, detected in this study. (B) Dendrogram of the ten PFGE profiles.
The dendrogram (Fig. 1B) clearly shows that the human isolates (PT10) are more distant from the other isolates that share provenience and that appear to be evolved from the same clone.
MLST analysis was carried out on 18 of the 78 isolates obtained from dairy products and livestock randomly selected (including at least one for each pulsotype and each year, if available), together with the four human isolates, the isolate from the pet, and that from the exotic animal (Table 1). The analysis revealed that 12 S. aureus isolates belonged to 5 known allelic profiles: ST9 (four human isolates), ST130 (three isolates), ST522 (two), ST1 (one), ST97 (one), and ST425 (one). The remaining 12 isolates showed 6 new MLST combinations: ST700, ST1614, ST1615, ST1616, ST1626, and ST1627. In particular, 29% of the isolates belonged to the ST700 allelic profile. The isolate collected in 1998 from the skin sample of a circus elephant showed the new pulsotype ST1614. The isolate from dog skin belonged to the ST522 allelic profile (Table 1).
In addition, PFGE and MLST, carried out on human isolates after a severe food poisoning episode occurred in 2015, revealed that they belonged to the same pulsotype PT10 and to the same sequence type ST9 (Table 1, marked as A–D).
Antimicrobial susceptibility
Thirty isolates (35.7%) were found to be resistant to penicillin G, 17 (20.2%) to tetracycline, 4 (5%) to gentamycin, 3 (3.75%) to lincomycin, 3 (3.75%) to cefoperazone, 2 (2.5%) to erythromycin, and 1 (1.25%) to kanamycin (Table 2). All isolates belonging to the ST700 profile resulted sensitive to gentamycin and intermediate to kanamycin, one was resistant to CFP, one to tetracycline, and one to penicillin G. Six isolates (7.1%) were classified as potentially dangerous, in that they display resistance to three different classes of antibiotics (Table 2). The human isolates showed an intermediate resistance to erythromycin, tetracycline, and penicillin G.
Table 2.
Antibiotic Profile and Detection of Virulence Genes of 84 Staphylococcus aureus Isolates
| Antibiotic profile | Presence of virulence genes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Isolate |
CN S ≥15 I 13–14 R/≤12 |
K S ≥18 I 14–17 R/≤13 |
MY S ≥21 I 15–20 R/≤14 |
E S ≥23 I 14–22 R/≤13 |
TE S ≥19 I 15–18 R/≤14 |
CFP S ≥21 I 16–20 R/≤15 |
P S ≥29 I — R/≤28 |
sea-see | seg-i, sej, sep | tsst, eta, etb, mecA |
| 1 | S | I | S | I | S | I | R | ND | seg | ND |
| 2 | S | I | S | S | S | S | S | sec | seg | tsst1 |
| 3 | S | I | R | I | R | I | R | ND | seg | ND |
| 4 | S | I | S | I | S | I | S | sec | ND | tsst1 |
| 5 | S | S | S | I | S | S | S | ND | seg | ND |
| 6 | S | S | S | S | R | I | R | ND | seg | ND |
| 7 | S | I | S | I | R | I | R | ND | ND | ND |
| 8 | S | I | I | I | I | S | S | ND | ND | ND |
| 9 | S | I | S | S | S | S | S | sec | ND | tsst1 |
| 10 | S | I | S | I | I | S | S | ND | seg | ND |
| 11 | S | S | S | I | S | S | S | ND | seg, sep | ND |
| 12 | S | I | I | S | S | S | S | ND | sep | ND |
| 13 | S | S | S | I | S | S | S | sec | seg | tsst1 |
| 14 | S | I | S | S | S | S | R | ND | ND | ND |
| 15 | S | S | S | I | S | S | S | ND | seg | ND |
| 16 | S | I | S | I | S | S | R | ND | seg | ND |
| 17 | I | I | S | I | S | S | S | ND | seg | ND |
| 18 | S | I | S | I | R | I | R | sec | ND | tsst1 |
| 19 | S | S | S | I | S | I | R | ND | seg | ND |
| 20 | S | I | S | I | S | S | S | sec | seg | ND |
| A | S | S | S | I | I | S | I | ND | seg, sei | ND |
| B | S | S | S | I | I | S | I | ND | seg, sei | ND |
| C | S | S | S | I | I | S | I | ND | seg, sei | ND |
| D | S | S | S | I | I | S | I | ND | seg, sei | ND |
| 21 | S | S | S | S | S | S | S | sea | ND | ND |
| 22 | S | I | S | S | S | S | S | ND | ND | ND |
| 23 | S | S | S | S | S | S | S | ND | ND | ND |
| 24 | S | S | S | I | S | I | R | ND | ND | ND |
| 25 | S | S | S | I | S | S | S | ND | ND | ND |
| 26 | S | S | S | S | S | S | S | see | ND | ND |
| 27 | S | S | S | I | S | S | S | ND | sej | ND |
| 28 | S | S | S | S | S | S | R | ND | sej | ND |
| 29 | S | I | S | I | S | S | S | ND | seg, sej | tsst1 |
| 30 | S | I | I | I | R | I | R | sec, sed | ND | eta, tsst1 |
| 31 | S | S | S | S | S | S | S | sea | ND | ND |
| 32 | S | S | S | S | S | S | S | ND | seh | ND |
| 33 | I | I | R | I | R | I | R | ND | seh | eta |
| 34 | S | S | S | S | S | S | S | ND | ND | ND |
| 35 | S | S | S | S | S | S | S | ND | ND | ND |
| 36 | S | S | S | S | S | S | S | ND | ND | ND |
| 37 | S | S | S | I | S | S | S | ND | sej | ND |
| 38 | S | S | S | S | S | S | S | ND | seh | ND |
| 39 | S | S | S | I | S | I | S | ND | seh | ND |
| 40 | S | S | S | S | S | S | S | ND | ND | ND |
| 41 | S | S | S | S | S | S | S | ND | ND | ND |
| 42 | S | I | S | S | S | S | R | ND | sej | ND |
| 43 | S | S | S | S | S | S | S | ND | sej | ND |
| 44 | S | I | S | S | S | S | S | ND | ND | ND |
| 45 | S | I | S | R | R | S | S | sea | ND | ND |
| 46 | S | S | S | S | S | S | S | ND | ND | ND |
| 47 | S | S | S | S | R | S | R | ND | seg, sep | ND |
| 48 | S | S | S | S | S | S | S | ND | seg, sep | ND |
| 49 | S | S | S | S | S | S | S | ND | ND | ND |
| 50 | S | I | S | I | I | R | R | ND | ND | ND |
| 51 | S | S | S | S | S | S | S | sea, see | ND | ND |
| 52 | S | S | S | S | S | S | S | ND | ND | ND |
| 53 | S | S | S | S | S | S | S | ND | ND | ND |
| 54 | R | I | I | I | R | I | R | ND | ND | tsst1 |
| 55 | S | I | S | S | S | S | S | sec | ND | tsst1 |
| 56 | S | S | S | S | S | S | S | ND | ND | eta, tsst1 |
| 57 | S | S | S | I | R | S | R | ND | ND | mecA |
| 58 | S | I | S | S | S | S | S | sec | ND | tsst1 |
| 59 | S | I | I | I | I | I | R | ND | seh | mecA |
| 60 | S | I | S | R | R | S | R | sec | ND | tsst1 |
| 61 | S | I | I | I | R | I | R | sec | ND | tsst1 |
| 62 | S | S | S | I | S | S | S | sec | ND | ND |
| 63 | I | S | S | S | S | I | R | sec | ND | tsst1 |
| 64 | S | S | S | S | S | S | S | sec | ND | tsst1 |
| 65 | S | S | S | S | S | S | R | ND | sej | mecA |
| 66 | S | S | S | S | R | S | R | ND | seh | mecA |
| 67 | R | I | R | I | S | I | R | sec | ND | ND |
| 68 | S | S | S | S | R | S | R | sec | ND | tsst1 |
| 69 | S | S | S | S | S | S | S | sec | sej | ND |
| 70 | I | I | I | I | R | I | R | sec | ND | tsst1 |
| 71 | S | S | S | S | S | S | S | sec | ND | tsst1 |
| 72 | S | I | S | S | S | S | S | sec | ND | tsst1 |
| 73 | S | S | S | S | S | S | S | sec | ND | tsst1 |
| 74 | S | I | S | I | R | S | R | see | ND | ND |
| 75 | S | S | S | S | S | S | S | sec, see | ND | tsst1 |
| 76 | S | I | S | I | S | S | S | see | ND | ND |
| 77 | I | I | I | I | I | R | R | sec, see | ND | tsst1 |
| 78 | R | S | S | S | S | S | R | see | ND | ND |
| 79 | R | R | S | I | S | S | R | sec, see | ND | tsst1 |
| 80 | S | I | S | S | R | R | S | ND | ND | etb |
| A | S | S | S | I | I | S | I | ND | seg, sei | ND |
| B | S | S | S | I | I | S | I | ND | seg, sei | ND |
| C | S | S | S | I | I | S | I | ND | seg, sei | ND |
| D | S | S | S | I | I | S | I | ND | seg, sei | ND |
Numbers 1–80 indicate isolates from animals or dairy products, letters A–D indicate the human isolates. For antibiotic profile, gentamycin (CN), kanamycin (K), lincomycin (MY), erythromycin (E), tetracycline (TE), cefoperazone (CFP), penicillin G (P) were tested. Antibiotic disk diffusion ranges (mm) for susceptible (S), intermediate (I), and resistant (R) phenotypes are given under the name of the antibiotic. For the presence of virulence genes, three different multiplex polymerase chain reactions were used to detect the genes indicated. ND indicates the analyzed genes were not detected. se: staphylococcal enterotoxins, tsst: toxic shock syndrome toxin, eta and etb: exfoliative toxins, mecA: encodes the low-affinity penicillin-binding protein 2A (PBP 2A) and it determines resistance to methicillin.
Detection of virulence genes
The presence of enterotoxin, tsst-1, exfoliative toxins (eta and etb), and mecA genes was investigated in the 84 isolates by using multiplex PCRs (Table 2). The sec gene was the most frequently detected (n = 24, 28.6%), followed by tsst-1 (n = 23, 27.4%), seg (n = 20, 23.8%), sej (n = 8, 9.5%), see (n = 7, 8.3%), seh (n = 6, 7.1%), sea, sei, sep, and mecA (n = 4, 4.8%), eta (n = 3, 3.5%), sed and etb (n = 1, 1.2%). The simultaneous presence of several toxin genes was detected in 27 isolates (Table 2). Interestingly, the four human isolates carried both seg and sei toxin genes.
Discussion
This is the first report of molecular genotyping, evaluation of resistance profiles, and analysis of toxin genes of S. aureus in bacterial isolates from dairy animals and dairy food in Sicily. Our study was carried out on a collection of S. aureus isolates obtained during the years 1998–2014, and it demonstrates the existence of 9 pulsotypes (Fig. 1) and 11 sequence types with high heterogeneity. MLST analysis demonstrated the presence of six new sequence types. Seven ST700 isolates were found in sheep, cow, and goat milk and udder, from different areas in Sicily. This allelic profile has been observed in Sicily for the last 16 years; in 1998 in sheep udder and in 2014 in cow milk (Table 1). The detection of a new major clone among all isolates evidenced no host preference for animal species (sheep, cattle, and goat) and its distribution was spread all over Sicily. Among the novel profiles, ST1614 contains a new aroe allele that had never been previously detected in Sicily; however, it was isolated from an Indian elephant present in 1998 in an Italian circus, and so, the actual origin is unknown and we could not have any further information on the animal at this time. The ST425 and ST522 types had never been isolated in Sicily before; ST522 was found in both dog skin and in sheep udder, but with different pulsotypes, antimicrobial susceptibilities, and enterotoxin genes. Another study carried out in Spain suggested that ST522 is the most common S. aureus clone associated with small ruminants (Porrero et al., 2012). Allelic profiles ST1 and ST97 had already been isolated as hospital-associated methicillin-resistant strains in Catania (Campanile et al., 2009) and in Italian pig finishing holdings (Battisti et al., 2009).
Antibiotic profiling showed a high level of penicillin (35.7%) and tetracycline (20.2%) resistance (Table 2). Resistance to penicillin remains the most common, as observed in other studies (Spanu et al., 2014; Jamali et al., 2015; Ferreira et al., 2016). The prevalence of resistance to β-lactam antibiotics is frequent in S. aureus strains obtained from milk and related products worldwide, as reported by Daka et al. (2012), Hu et al. (2013), and Xu et al. (2014).
The tetracycline resistance observed in this study is more significant than that found in Italy by Spanu et al. (2014) in strains isolated from cheese (10.6%) or that found by Ferreira et al. (2016) in strains isolated from artisanal cheese (10.3%). In Sicilian farms, tetracyclines or a mix of clavulanic acid and amoxicillin is frequently used to fight infections, often without veterinarian prescriptions, thus, antibiotic resistance is likely to have increased over the years.
Molecular analysis (Table 2) showed that only four isolates contained the mecA gene, hinting at the circulation of methicillin resistance in dairy products. The mecA gene is related to methicillin resistance (Liu et al., 2016).
The ST522 isolates were resistant to penicillin G, but the isolate from dog skin flakes also resulted resistant to lincomycin and tetracycline. The fact that the strain isolated from a pet showed a multiresistant phenotype (i.e., resistance to three different classes of antibiotics) confirms that multidrug resistance is easily spread among pets (Davis et al., 2014). This could be due to close physical contact between pets and humans, which may allow strain transmission, or to the fact that many pets are often treated with antibiotics used in human medicine (Boost et al., 2008; van Duijkeren et al., 2008; Knox et al., 2015).
In this study, we found that 78.5% of the isolates contained at least a toxin gene. The highest frequency was observed for the sec gene among classical SEs, and for the seg gene among the new SEs. Toxigenic strains of S. aureus were isolated in Sicilian healthy farms (Vitale et al., 2015) and from sheep and goat cheese in Southern Italy (Basanisi et al., 2016).
PFGE and MLST analyses showed the same pulsotype (PT10) and sequence type (ST9) for the four human isolates hinting that they probably belong to a single clone although they were obtained from two different individuals. ST9 was shown to be frequently spread among animals, whereas it appears to be rare among S. aureus isolates from human infections (Kehrenberg et al., 2009). In our study, the leftover food that probably caused the case of food poisoning resulted negative to the isolation procedures and it was not possible to identify any food source. The human isolates did not show any novel profile in the genetic analysis, however, they all showed the concurrent presence of sei and seg, not found in the other isolates. The human isolates likely carry the operon ecg containing seg and sei (Smyth et al., 2005). The SEI toxin was shown to have a high pathogenicity, in that only 10 ng of SEI is sufficient for a lethal effect in rabbits (Roetzer et al., 2016).
Conclusions
This study showed for the first time a high heterogeneity and novelty of sequence types of S. aureus isolates collected in Sicily from tissues and/or dairy products from different animals between the years 1998 and 2014. Moreover, our analysis showed which S. aureus strains circulate in Sicily as well as a high diffusion of penicillin G and tetracycline resistance and toxin genes among the isolates. In addition, we showed that isolates obtained from patients involved in a food poisoning episode in 2015 belonged to the same allelic type ST9, and contained sei and seg toxin genes.
Acknowledgments
This study was supported by research grants provided by the Italian Ministry of Health to M.V. (IZS SI 13/15 RC) and by the University of Palermo, Fondo Finalizzato alla Ricerca to R.A. We thank the Italian Reference Laboratory for Staphylococci for the enterotoxin-positive strains. We thank Luca Dolce for critical reading of the article.
Disclosure Statement
No competing financial interests exist.
References
- Aanensen DM, Spratt BG. The multilocus sequence typing network: mlst.net. Nucleic Acids Res 2005;33(Web Server issue):W728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aires-de-Sousa M. MRSA among animals: Current overview. Clin Microbiol Infect 2017;23:373–380 [DOI] [PubMed] [Google Scholar]
- Alduina R, Pisciotta A. Pulsed field gel electrophoresis and genome size estimates. Methods Mol Biol 2015;1231:1. [DOI] [PubMed] [Google Scholar]
- Argudín MÁ, Mendoza MC, Rodicio MR. Food poisoning and Staphylococcus aureus enterotoxins. Toxins (Basel) 2010;2:1751–1773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basanisi MG, Nobilia G, La Bella G, Russo R, Spano G, Normanno G, La Salandra G. Molecular characterization of Staphylococcus aureus isolated from sheep and goat cheeses in southern Italy. Small Rumin Res 2016;135:17 [Google Scholar]
- Battisti A, Franco A, Merialdi G, Hasman H, Iurescia M, Lorenzetti R, Feltrin F, Zini M, Aarestrup FM. Heterogeneity among methicillin-resistant Staphylococcus aureus from Italian pig finishing holdings. Vet Microbiol 2009;142:361. [DOI] [PubMed] [Google Scholar]
- Boost MV, O'Donoghue MM, James A. Prevalence of Staphylococcus aureus carriage among dogs and their owners. Epidemiol Infect 2008;136:953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campanile F, Bongiorno D, Borbone S, Stefani S. Hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) in Italy. Ann Clin Microbiol Antimicrob 2009;8:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Fifth Informational Supplement. CLSI document M100-S25. Wayne, PA: CLSI, 2015 [Google Scholar]
- Daka D, Solomon GS, Yihdego D. Antibiotic-resistance Staphylococcus aureus isolated from cow's milk in the Hawassa area, South Ethiopia. Ann Clin Microbiol Antimicrob 2012;11:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea A, Martinez YZ, Alduina R, Monteverde V, Molina CF, Vitale M. Comparison of two PCR methods for detection of Leptospira interrogans in formalin-fixed and paraffin-embedded tissues. Mem Inst Oswaldo Cruz 2012;107:85–88 [DOI] [PubMed] [Google Scholar]
- Davis JA, Jackson CR, Fedorka-Cray PJ, Barrett JB, Brousse JH, Gustafson J, Kucher M. Carriage of methicillin-resistant staphylococci by healthy companion animals in the US. Lett Appl Microbiol 2014;59:1. [DOI] [PubMed] [Google Scholar]
- De Buyser ML, Grout J, Brisabois A, Assere A, Lombard B. Detection of Genes Encoding Staphylococcal Enterotoxins. Multiplex PCR for sea to see and ser. Method of the crl for Coagulase Positive Staphylococci Including Staphylococcus aureus, 1st ed. Maisons-Alfort, France: CRL CPS, AFSSA, 2009; pp. 1–5 [Google Scholar]
- Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000;38:1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, Bernardo LG, Neves LS, Campos MR, Lamaro-Cardoso J, André MC. Virulence profile and genetic variability of Staphylococcus aureus isolated from artisanal cheese. J Dairy Sci 2016;99:8589. [DOI] [PubMed] [Google Scholar]
- Giardina A, Alduina R, Gottardi E, Di Caro V, Süssmuth RD, Puglia AM. Two heterologously expressed Planobispora rosea proteins cooperatively induce Streptomyces lividans thiostrepton uptake and storage from the extracellular medium. Microb Cell Fact 2010;9:9–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golding GR, Campbell J, Spreitzer D, Chui L. Pulsed-field gel electrophoresis of Staphylococcus aureus. Methods Mol Biol 2015;1301:85. [DOI] [PubMed] [Google Scholar]
- Gould IM, David MZ, Esposito S, Garau J, Lina G, Mazzei T, Peters G. New insights into meticillin-resistant Staphylococcus aureus (MRSA) pathogenesis, treatment and resistance. Int J Antimicrob Agents 2012;39:96–104 [DOI] [PubMed] [Google Scholar]
- Hu SK, Liu SY, Hu WF, Zheng TL, Xu JG. Molecular biological characteristics of Staphylococcus aureus isolated from food. Eur Food Res Technol 2013;236:285 [Google Scholar]
- Jamali H, Paydar M, Radmehr B, Ismail S, Dadrasnia A. Prevalence and antimicrobial resistance of Staphylococcus aureus isolated from raw milk and dairy products. Food Control 2015;54:383 [Google Scholar]
- Kadariya J, Smith TC, Thapaliya D. Staphylococcus aureus and staphylococcal food-borne disease: An ongoing challenge in public health. Biomed Res Int 2014;2014:827965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrenberg C, Cuny C, Strommenger B, Schwarz S, Witte W. Methicillin-resistant and -susceptible Staphylococcus aureus strains of clonal lineages ST398 and ST9 from swine carry the multidrug resistance gene cfr. Antimicrob Agents Chemother 2009;2:779–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox J, Uhlemann AC, Lowy FD. Staphylococcus aureus infections: Transmission within households and the community. Trends Microbiol 2015;23:437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Cardoso M, Senczek D, Schwarz S. Molecular subtyping of Staphylococcus aureus isolates from cases of bovine mastitis in Brazil. Vet Microbiol 1999;67:127. [DOI] [PubMed] [Google Scholar]
- Liu J, Chen D, Peters BM, Li L, Li B, Xu Z, Shirliff ME. Staphylococcal chromosomal cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog 2016;101:56–67 [DOI] [PubMed] [Google Scholar]
- Lo Grasso L, Maffioli S, Sosio M, Bibb M, Puglia AM, Alduina R. Two master switch regulators trigger A40926 biosynthesis in Nonomuraea sp. Strain ATCC 39727. J Bacteriol 2015;197:2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macori G, Giacinti G, Bellio A, Gallina S, Bianchi DM, Sagrafoli D, Marri N, Giangolini G, Amatiste S, Decastelli L. Molecular epidemiology of methicillin-resistant and methicillin-susceptible Staphylococcus aureus in the ovine dairy chain and in farm-related humans. Toxins (Basel) 2017;9:161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrotra M, Wang G, Johnson WM. Multiplex PCR for detection of genes for Staphylococcus aureus enterotoxins, exfoliative toxins, toxic shock syndrome toxin 1, and methicillin resistance. J Clin Microbiol 2000;38:1032–1035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock SJ, Paterson GK. Mechanisms of methicillin resistance in Staphylococcus aureus. Annu Rev Biochem 2015;84:577–601 [DOI] [PubMed] [Google Scholar]
- Pomba C, Rantala M, Greko C, Baptiste KE, Catry B, van Duijkeren E, Mateus A, Moreno MA, Pyörälä S, Ružauskas M, Sanders P, Teale C, Threlfall EJ, Kunsagi Z, Torren-Edo J, Jukes H, Törneke K. Public health risk of antimicrobial resistance transfer from companion animals. J Antimicrob Chemother 2016;72:957–968 [DOI] [PubMed] [Google Scholar]
- Porrero MC, Hasman H, Vela AI, Fernández-Garayzábal JF, Domínguez L, Aarestrup FM. Clonal diversity of Staphylococcus aureus originating from the small ruminants goats and sheep. Vet Microbiol 2012;156:157–161 [DOI] [PubMed] [Google Scholar]
- Randazzo L, Montana G, Alduina R, Quatrini P, Tsantini E, Salemi B. Flos Tectorii degradation of mortars: An example of synergistic action between soluble salts and biodeteriogens. J Cult Herit 2015;16:838–847 [Google Scholar]
- Rodriguez M, Hogan PG, Satola SW, Crispell E, Wylie T, Gao H, Sodergren E, Weinstock GM, Burnham CA, Fritz SA. Discriminatory indices of typing methods for epidemiologic analysis of contemporary Staphylococcus aureus strains. Medicine (Baltimore) 2015;94:e1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodvold KA, McConeghy KW. Methicillin-resistant Staphylococcus aureus therapy: Past, present, and future. Clin Infect Dis 2014;58 Suppl 1:S20–S27 [DOI] [PubMed] [Google Scholar]
- Roetzer A, Gruener CS, Haller G, Beyerly J, Model N, Eibl MM. Enterotoxin gene cluster-encoded SEI and SElN from Staphylococcus aureus isolates are crucial for the induction of human blood cell proliferation and pathogenicity in rabbits. Toxins (Basel) 2016;pii:E314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seegers H, Fourichon C, Beaudeau F. Production effects related to mastitis and mastitis economics in dairy cattle herds. Vet Res 2003;34:475. [DOI] [PubMed] [Google Scholar]
- Sergelidis D, Angelidis AS. Methicillin-resistant Staphylococcus aureus: A controversial food-borne pathogen. Lett Appl Microbiol 2017;64:409–418 [DOI] [PubMed] [Google Scholar]
- Smyth DS, Hartigan PJ, Meaney WJ, Fitzgerald JR, Deobald CF, Bohach GA, Smyth CJ. Superantigen genes encoded by the egc cluster and SaPIbov are predominant among Staphylococcus aureus isolates from cows, goats, sheep, rabbits and poultry. J Med Microbiol 2005;54:401–411 [DOI] [PubMed] [Google Scholar]
- Spanu V, Scarano C, Cossu F, Pala C, Spanu C, De Santis EP. Antibiotic resistance traits and molecular subtyping of Staphylococcus aureus isolated from raw sheep milk cheese. J Food Sci 2014;79:M2066–M2071 [DOI] [PubMed] [Google Scholar]
- van Duijkeren E, Houwers DJ, Schoormans A, Broekhuizen-Stins MJ, Ikawaty R, Fluit AC, Wagenaar JA. Transmission of methicillin-resistant Staphylococcus intermedius between humans and animals. Vet Microbiol 2008;128:213. [DOI] [PubMed] [Google Scholar]
- Vitale M, Scatassa ML, Cardamone C, Oliveri G, Piraino C, Alduina R, Napoli C. Staphylococcal food poisoning case and molecular analysis of toxin genes in Staphylococcus aureus strains isolated from food in Sicily, Italy. Foodborne Pathog Dis 2015;12:21. [DOI] [PubMed] [Google Scholar]
- Wang X, Li G, Xia X, Yang B, Xi M, Meng J. Antimicrobial susceptibility and molecular typing of methicillin-resistant Staphylococcus aureus in retail foods in Shaanxi, China. Foodborne Pathog Dis 2014;11:281–286 [DOI] [PubMed] [Google Scholar]
- Xu J, Shi C, Song M, Xu X, Yang P, Paoli G, Shi X. Phenotypic and genotypic antimicrobial resistance traits of foodborne Staphylococcus aureus isolates from Shanghai. J Food Sci 2014;79:M635. [DOI] [PubMed] [Google Scholar]