Abstract
Epigenetic modulators play critical roles in reprogramming of cellular functions, emerging as a new class of promising therapeutic targets. Nuclear receptor binding SET domain protein 3 (NSD3) is a member of the lysine methyltransferase family. Interestingly, the short isoform of NSD3 without the methyltransferase fragment, NSD3S, exhibits oncogenic activity in a wide range of cancers. We recently showed that NSD3S interacts with MYC, a central regulator of tumorigenesis, suggesting a mechanism by which NSD3S regulates cell proliferation through engaging MYC. Thus, small molecule inhibitors of the NSD3S/MYC interaction will be valuable tools for understanding the function of NSD3 in tumorigenesis for potential cancer therapeutic discovery. Here we report the development of a cell lysate-based time-resolved fluorescence resonance energy transfer (TR-FRET) assay in an ultrahigh-throughput screening (uHTS) format to monitor the interaction of NSD3S with MYC. In our TR-FRET assay, anti-Flag-terbium and anti-glutathione S-transferase (GST)-d2, a paired fluorophores, were used to indirectly label Flag-tagged NSD3 and GST-MYC in HEK293T cell lysates. This TR-FRET assay is robust in a 1,536-well uHTS format, with signal-to-background >8 and a Z′ factor >0.7. A pilot screening with the Spectrum library of 2,000 compounds identified several positive hits. One positive compound was confirmed to disrupt the NSD3/MYC interaction in an orthogonal protein–protein interaction assay. Thus, our optimized uHTS assay could be applied to future scaling up of a screening campaign to identify small molecule inhibitors targeting the NSD3/MYC interaction.
Keywords: : NSD3, MYC, TR-FRET, screening, protein–protein interaction, epigenetics
Introduction
C-MYC (MYC) is a well-validated oncoprotein.1 It has been extensively studied as a potential target for cancer therapy due to its demonstrated functions in processes that drive cell transformation, including cell proliferation, cell metabolism, migration, and apoptosis.2–5 In normal cells, MYC is strictly regulated at multiple levels, such as mRNA expression and protein stability.6 However, MYC is deregulated in a wide range of cancer types, including lymphoma, leukemia, breast cancer, hepatocellular carcinoma, and others.7–9 Thus, understanding how MYC is regulated may provide critical insights into effective strategies to target MYC for therapeutic intervention.
MYC has been functionally associated with epigenetic regulation. Our recent study has revealed nuclear receptor binding SET domain protein 3 (NSD3), an epigenetic modulator, as an interaction partner for MYC, offering a potential mechanism to control MYC-driven tumorigenesis.10 NSD3, nuclear receptor binding Suppressor of variegation 3–9, Enhancer of zeste and Trithorax (SET) domain Protein 3, is encoded by Wolf–Hirschhorn Syndrome Candidate 1-Like 1 (WHSC1L1) gene. It is a member of the NSD protein histone lysine methyltransferase (HKMT) family, which also includes NSD1 (KMT3B) and NSD2 (WHSC1/MMSET).11 The NSD HKMT family proteins are responsible for methylation of lysine 36 on histone H3 (H3K36me2) and works as an important epigenetic factor with implicated roles in breast and lung cancer.12,13 NSD3, located on chromosome 8p12, was first discovered to be amplified in breast cancer, suggesting its potential oncogenic ability.14 In a genomic and transcriptomic analysis of 958 breast cancer samples, NSD3 stood out as one of the top four most frequently amplified methyltransferases. NSD3 is present in cells in three isoforms: long, short, and whistle isoforms.14,15 The long isoform (NSD3L) contains two Pro-Trp-Trp-Pro (PWWP) domains and five Plant homeodomains that serve as chromatin reader domains and the H3K36 methyltransferase SET domain at the C-terminal region of the protein. The short isoform (NSD3S) is produced by alternative splicing and only has one PWWP domain, and lacks six chromatin reader domains and the SET domain with the catalytic activity.11 The whistle isoform shares the C-terminal region with the NSD3L, containing the SET domain.15,16 Although NSD3S does not have the methylation catalytic activity, it is required for the acute myeloid leukemia maintenance through bridging BRD4 to CHD8, working as an oncoprotein,17 suggesting its crucial role in leukemia. NSD3 is also found to translocate in cancer. The NSD3–NUT oncofusion was discovered in a patient with NUT midline carcinoma (NMC) playing a key role in sustaining the proliferation in NMC cells.18 Interestingly, only amino acid 1–569 of NSD3 without SET domain, which is similar to NSD3S (645 amino acids), was involved in this novel NSD3–NUT fusion,18 implying NSD3S could independently contribute to cancer without methyltransferase activity. NSD3 has emerged as a new potential oncoprotein for further mechanistic and therapeutic investigations.
Our data suggest that MYC interacts with NSD3 and the MYC/NSD3 interaction was validated in cancer cells under physiological conditions.10 This interaction is mapped to the NSD3S fragment. NSD3S is sufficient to increase MYC transcriptional activity and stabilizes MYC protein levels, indicating a potential oncogenic role for NSD3/MYC protein–protein interaction (PPI). This interaction further links epigenetics to transcription regulation. To understand the functional roles of the NSD3/MYC interaction in normal physiological processes and in tumorigenesis, we began to search for small molecule inhibitors of NSD3/MYC as molecular probes. However, there is no known inhibitor for the NSD3/MYC PPI.
To discover inhibitors for the NSD3/MYC PPI, here, we report the development of a time-resolved fluorescence resonance energy transfer (TR-FRET) assay for high-throughput screening (HTS) using cell lysate with overexpressed target proteins. After extensive optimization for DNA constructs and TR-FRET antibodies, a TR-FRET configuration that generates robust signals was determined (Fig. 1A). The assay system contains cell lysates with overexpressed Venus-Flag-tagged NSD3 (VF-NSD3) and glutathione S-transferase (GST)-tagged MYC (GST-MYC) proteins, anti-Flag-terbium (Tb), and anti-GST-d2 antibodies for indirectly labeling of NSD3 and MYC proteins with paired TR-FRET donor (Tb) and acceptor (d2). The interaction of NSD3 and MYC in cell lysates brings Tb and d2 into proximity and generates TR-FRET signal (Fig. 1B). We have optimized and validated this cell lysate-based TR-FRET assay to achieve robust assay performance in a 1,536-well ultra-HTS (uHTS) format. The assay can be applied for large-scale screening campaign in the future to discover NSD3 and MYC PPI inhibitors.
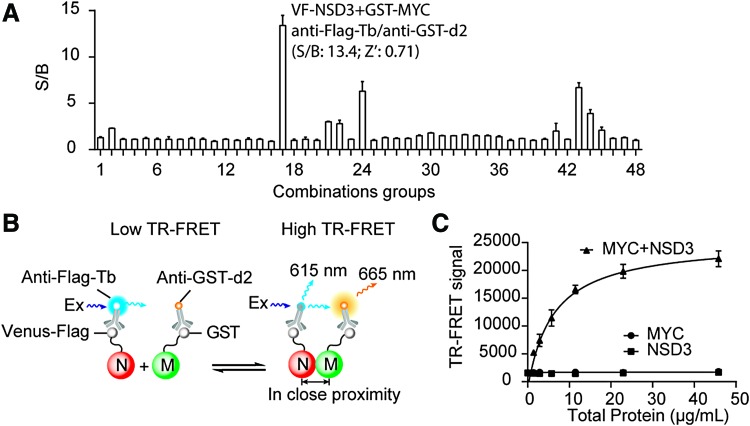
Fig. 1.
TR-FRET assay development for NSD3/MYC interaction. (A) Selection of constructs and TR-FRET antibodies for NSD3/MYC TR-FRET assay development. HEK293T cells were seeded into a 384-well plate and transfected with various tagged DNA constructs. After 48 h, TR-FRET assay was carried out directly in the plate as described in “Materials and Methods” section. A total of 48 combinations were tested as indicated in Table 1. S/B ratio was calculated for each condition. The data are presented as mean ± SD from triplicate samples. (B) The principle of our selected system for NSD3/MYC TR-FRET assay development. In cell lysate with overexpression proteins, VF-NSD3 protein was labeled with Tb through anti-Flag-Tb (N) to serve as a TR-FRET donor. GST-MYC (M) was coupled to TR-FRET acceptor d2 using anti-GST-d2. Interaction between NSD3 and MYC brings Tb and d2 into proximity, leading to the generation of TR-FRET signals. (C) TR-FRET signal from titration of stock cell lysate in a 384-well format. Transfection was performed in a six-well plate and stock cell lysate was collected after 48 h. Increasing amount of stock cell lysate with overexpressing VF-NSD3 and GST-MYC (15 μL) was mixed with diluted anti-Flag-Tb and anti-GST-d2 antibodies (15 μL). The TR-FRET signal was measured. The data shown are average from triplicate samples with SD. GST, glutathione S-transferase; NSD3, nuclear receptor binding SET domain protein 3; S/B, signal-to-background; SD, standard deviation; Tb, terbium; TR-FRET, time-resolved fluorescence resonance energy transfer; VF, Venus-Flag. Color images available online at www.liebertpub.com/adt
Materials and Methods
Reagents
Tb cryptate-conjugated mouse monoclonal anti-Flag antibody (anti-Flag-Tb, #61FG2TLB) and d2-conjugated anti-GST antibody (anti-GST-d2, #61GSTDLB) were purchased from Cisbio Bioassays (Bedford, MA). Europium anti-His (anti-His-Eu) and allophycocyanin (APC)-anti-Flag (anti-Flag-APC) were purchased from PerkinElmer (Waltham, MA). Polyethylenimine (PEI; Polysciences, Inc.), dissolved in sterile water at a concentration of 1 mg/mL, was used as transfection reagent. Anti-Flag mouse monoclonal antibody (#F3165), anti-β-actin mouse monoclonal antibody (#A2228), and protease inhibitor (#P8340) were purchased from Sigma-Aldrich. Anti-GST rabbit polyclonal antibody (#sc-459), goat antimouse immunoglobulin G (IgG)-horserdish peroxidase (HRP, #sc-2005), and goat antirabbit IgG-HRP (#sc-2004) were purchased from Santa Cruz. Glutathione Sepharose™ 4B (#17-0756-05) was obtained from GE Healthcare.
Cell Line and Culture Conditions
Human embryonic kidney 293T cells (HEK 293T; American Type Culture Collection) were cultured in Dulbecco's modified Eagle's medium (Corning; #10-013-CV) medium supplemented with 10% fetal bovine serum (Sigma-Aldrich; #F2442), 100 IU/mL penicillin, and 100 μg/mL streptomycin (Corning; #30-001-CI) in a humid environment with 5% CO2 at 37°C.
DNA Constructs
All GST-, VF-, Flag- or hexa-His (His6)-tagged human NSD3S and MYC plasmids for mammalian expression were generated using Gateway® cloning kit (Invitrogen). pDEST26 vector for 6XHis-tag and pDEST27 vector for GST-tag were purchased from Invitrogen. pFUW-VF vector for N-VF-tag was generated and described as before.10 The HindIII/NheI and XhoI/KpnI sites of the modified pcDNA3.2/V5-DEST vector (g894t, t908g, a3182c, and c3243g)19 were used, respectively, to generate N- and C-terminal epitope-tagged Gateway-compatible destination vectors, including N- or C-Flag-, C-VF, and C-His6-tagged plasmids. A 10-amino acids flexible linker sequence [(Gly4Ser)2] was inserted between the Flag (GACTACAAGGACGACGATGACAAG), His6 (CATCACCATCACCATCAC), or VF and the gene cDNA. The pDONR vector containing the NSD3S or MYC was purchased from DNASU or cloned by PCR. The DNA was purified using ZymoPURE™ Plasmid Maxiprep Kit (Zymo Research; #D4203).
TR-FRET Measurements
The FRET buffer used throughout the assay contains 20 mM Tris-HCl, pH 7.0, 50 mM NaCl, and 0.01% nonidet P-40 (NP-40). FRET signals were measured using Envision Multilablel plate reader (PerkinElmer). Tb or Eu was used as TR-FRET donor and excited at 337 nm laser. Venus, d2, or APC served as TR-FRET acceptors, respectively, for Tb/Venus, Tb/d2, or APC, Eu/d2, or APC. Venus is a variant of yellow fluorescent protein with fast and efficient maturation for cell biological applications.20 In our study, VF-tagged DNA was used to monitor transfection efficiency. Venus has excitation at 515 nm and emission at 528 nm and also serves as an acceptor for Tb in a TR-FRET setting for PPI. When Venus served as an acceptor for Tb, the emissions for Tb and Venus were measured at 486 and 520 nm, respectively. When d2 or APC served as acceptor for Tb or Eu, the emissions for Tb or Eu and d2 or APC were measured at 615 and 665 nm, respectively. The delay time was set as 50 μs and total time of windows was 150 μs. All FRET signals were expressed as a TR-FRET ratio:
F665nm/F615nm × 104 (when d2 or APC was used as acceptor)
F520nm/F486nm × 104 (when Venus was used as acceptor)
Selection of the DNA Constructs and TR-FRET Antibodies for NSD3/MYC Interaction
To obtain the robust TR-FRET signal for NSD3/MYC interaction, we first tested different combinations of various tagged DNA constructs for NSD3 and MYC with different corresponding paired TR-FRET antibodies, as summarized in Table 1. For this purpose, the transfection and TR-FRET assay were carried out in a 384-well format for large number of samples. HEK 293T cells (6,000 cells/well in a 40 μL/well) were seeded in clear bottom 384-well cell culture plate (Corning Costar, #3712). After incubating overnight, transfection was carried out using PEI at a 1:3 ratio of DNA (μg): PEI (μg). In brief, a 5 μL/well of DNA:PEI mixture containing 50 ng of MYC DNA, 50 ng of NSD3 DNA, and 300 nL (1 mg/mL in stock) PEI was incubated at room temperature (RT) for 15 min and directly added to the cells without washing cells or replacing medium. Forty-eight hours after transfection, the medium were removed by centrifuging the plate upside down on paper towels at 130 g for 1 min. Cells were then lysed by adding 15 μL per well of 0.5% NP-40 lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 5% glycerol, and 5 mM NaF). The plate was frozen at −80°C freezer for 1 h and then thawed to RT, which was defined as one freeze-and-thaw cycle. After three freeze-and-thaw cycles, 15 μL per well of FRET buffer with indicated TR-FRET antibodies (Eu-conjugated antibodies at 1 nM final concentration and the rest antibodies at 1:500 final dilution) was added to the corresponding wells. The plate was incubated at RT for 1 h and TR-FRET signal was recorded by EnVision Multilabel plate reader.
Table 1.
Combinations of Constructs and Time-Resolved Fluorescence Resonance Energy Transfer Antibody Pairs
| No. | NSD3 Constructs | MYC Constructs | S/B | TR-FRET Antibody Pair (Donor/Acceptor) |
|---|---|---|---|---|
| 1 | N-VF-NSD3 | N-His-MYC | 1.3 | Anti-Flag-Tb/anti-His-d2 |
| 2 | N-VF-NSD3 | C-His-MYC | 2.3 | |
| 3 | C-VF-NSD3 | N-His-MYC | 1.1 | |
| 4 | C-VF-NSD3 | C-His-MYC | 1.1 | |
| 5 | N-His-NSD3 | N-VF-MYC | 1.2 | |
| 6 | N-His-NSD3 | C-VF-MYC | 1.1 | |
| 7 | N-His-NSD3 | N-Flag-MYC | 1.1 | |
| 8 | N-His-NSD3 | C-Flag-MYC | 1.1 | |
| 9 | C-His-NSD3 | N-VF-MYC | 1.2 | |
| 10 | C-His-NSD3 | C-VF-MYC | 1.1 | |
| 11 | C-His-NSD3 | N-Flag-MYC | 0.9 | |
| 12 | C-His-NSD3 | C-Flag-MYC | 1.1 | |
| 13 | N-Flag-NSD3 | N-His-MYC | 1.0 | |
| 14 | N-Flag-NSD3 | C-His-MYC | 1.1 | |
| 15 | C-Flag-NSD3 | N-His-MYC | 1.1 | |
| 16 | C-Flag-NSD3 | C-His-MYC | 0.9 | |
| 17 | N-VF-NSD3 | GST-MYC | 13.4 | Anti-Flag-Tb/anti-GST-d2 |
| 18 | C-VF-NSD3 | GST-MYC | 1.0 | |
| 19 | N-Flag-NSD3 | GST-MYC | 1.1 | |
| 20 | C-Flag-NSD3 | GST-MYC | 1.0 | |
| 21 | GST-NSD3 | N-VF-MYC | 3.0 | |
| 22 | GST-NSD3 | C-VF-MYC | 2.8 | |
| 23 | GST-NSD3 | N-Flag-MYC | 1.1 | |
| 24 | GST-NSD3 | C-Flag-MYC | 6.3 | |
| 25 | GST-NSD3 | N-His-MYC | 1.0 | Anti-His-Eu/anti-GST-d2 |
| 26 | GST-NSD3 | C-His-MYC | 1.3 | |
| 27 | N-His-NSD3 | GST-MYC | 1.2 | |
| 28 | C-His-NSD3 | GST-MYC | 1.2 | |
| 29 | N-VF-NSD3 | N-His-MYC | 1.5 | Anti-His-Tb/Venus |
| 30 | N-VF-NSD3 | C-His-MYC | 1.8 | |
| 31 | C-VF-NSD3 | N-His-MYC | 1.5 | |
| 32 | C-VF-NSD3 | C-His-MYC | 1.5 | |
| 33 | N-His-NSD3 | N-VF-MYC | 1.6 | |
| 34 | N-His-NSD3 | C-VF-MYC | 1.5 | |
| 35 | C-His-NSD3 | N-VF-MYC | 1.5 | |
| 36 | C-His-NSD3 | C-VF-MYC | 1.4 | |
| 37 | N-His-NSD3 | GST-MYC | 1.0 | Anti-GST-Tb/anti-His-d2 |
| 38 | C-His-NSD3 | GST-MYC | 1.2 | |
| 39 | GST-NSD3 | N-His-MYC | 1.0 | |
| 40 | GST-NSD3 | C-His-MYC | 1.1 | |
| 41 | N-VF-NSD3 | GST-MYC | 2.0 | Anti-GST-Tb/Venus |
| 42 | C-VF-NSD3 | GST-MYC | 1.1 | |
| 43 | GST-NSD3 | N-VF-MYC | 6.7 | |
| 44 | GST-NSD3 | C-VF-MYC | 3.9 | |
| 45 | N-VF-NSD3 | GST-MYC | 2.1 | Anti-GST-Tb/anti-Flag-APC |
| 46 | C-VF-NSD3 | GST-MYC | 1.2 | |
| 47 | N-Flag-NSD3 | GST-MYC | 1.3 | |
| 48 | C-Flag-NSD3 | GST-MYC | 1.0 |
APC, allophycocyanin; Eu, europium; GST, glutathione S-transferase; NSD3, nuclear receptor binding SET domain protein 3; S/B, signal-to-background; Tb, terbium; TR-FRET, time-resolved fluorescence resonance energy transfer; VF, Venus-Flag.
Development of NSD3/MYC TR-FRET Assay in a 384-Well HTS Format
Among various combinations of DNAs and TR-FRET antibodies tested as summarized in Table 1, the combination of VF-NSD3 (N-VF-tagged NSD3S), GST-MYC, anti-Flag-Tb, and anti-GST-d2 gave rise to the highest TR-FRET signal. Therefore, we selected this combination for developing NSD3/MYC TR-FRET assay for HTS.
To obtain sufficient cell lysate for assay development and screening, all the following transfections were carried out in six-well plates. HEK 293T cells (1.0 × 106 cells/well) were seeded in six-well plates and incubated overnight to allow the cells reach 80%–90% confluency. Cells were cotransfected with GST-MYC (1 μg) and VF-NSD3 (1 μg) using PEI at a 1:3 ratio of DNA (μg):PEI (μg) per well. A 100 μL/well of transfection mixture containing 1.5 μg of GST-MYC DNA, 1.5 μg of VF-NSD3 DNA, 9 μL of PEI (1 mg/mL in stock), and cell culture medium was incubated at RT for 15 min and added directly to the cells in a six-well plate. After incubating for 48 h, the cells were harvested and lysed using 200 μL per well of 0.5% NP-40 lysis buffer. Cell lysates were then sonicated at 36 W for 5 s at 4°C using Fisher Scientific™ Model FB120 (Pittsburgh). After sonication at 4°C, cell lysates were centrifuged at 12,000 g for 10 min at 4°C and supernatant was collected as stock cell lysate for the following TR-FRET assay optimization.
To determine the optimal cell lysate amount required to generate robust TR-FRET signal for HTS, the TR-FRET assay was carried out using various amounts of cell lysate in black 384-well plates (Corning Costar, #3573). A 15 μL of stock cell lysate was serially diluted in FRET buffer and mixed with 15 μL of TR-FRET antibodies mixture (anti-Flag Tb and anti-GST-d2) diluted at 1:500 in FRET buffer. The total volume for each well was 30 μL. The plate was centrifuged at 1,000 rpm for 5 min and incubated at RT for 1 h or as indicated. TR-FRET signals were then measured with EnVision Multilabel plate reader (PerkinElmer) as described with laser excitation at 337 nm and the emissions at 615 and 665 nm for Tb and d2 emission signals, respectively. TR-FRET signals were expressed as ratio: F665/F615 × 104.
Evaluation of Assay Performance for HTS
The cell lysate with coexpression of VF-NSD3 and GST-MYC proteins defines the interaction TR-FRET signal, whereas cell lysate with single protein expression is served as background control. To evaluate the performance of the assay for HTS, Z′ factor and signal-to-background (S/B) ratio of the assay were calculated using the following equations:
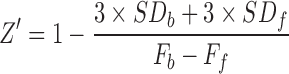 |
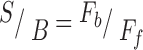 |
where Fb is mean TR-FRET signal from VF-NSD3/GST-MYC coexpression lysate, which defines the highest signal from PPI. Ff is mean TR-FRET signal from VF-NSD3 only expression lysate, which defines the minimal or background signal. SDb and SDf are standard deviations. S/B reflects the signal window of the assay and Z′ factor determines the robustness of the assay for HTS. A Z′ factor between 0.5 and 1 indicates an assay is robust and suitable for HTS.
The stability of the assay was evaluated by recording TR-FRET signals after incubation for 1–96 h. The effect of dimethyl sulfoxide (DMSO) on the TR-FRET signal was tested out in 384-well plate. Five microliters of serial diluted DMSO in FRET buffer was mixed with 25 μL reaction buffer containing optimized assay components for NSD3/MYC interaction signal. TR-FRET signal was measured after 1 h of incubation at RT. DMSO final concentration tested was up to 17%. All experiments were carried out in triplicates per sample and standard deviations were calculated.
Miniaturization of the Assay into a 1,536-Well uHTS Format
The TR-FRET assay for uHTS was performed in a black 1,536-well plate (Corning Costar, #3724) with a total volume of 5 μL in each well. As summarized in protocol table Table 2, 5 μL of the reaction mixture containing optimal amount of NSD3/MYC coexpression lysate and anti-Flag-Tb and anti-GST-d2 antibodies was dispensed to black 1,536-well plates using multiple-drop Combi dispenser (Thermo; #5840320). The VF-NSD3 lysate only without GST-MYC coexpression was used as background control. The TR-FRET signals were measured using Envision Multilabel plate reader. The S/B and Z′ were calculated as already described.
Table 2.
Ultrahigh-Throughput Screening Protocol
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Dispense controls | 5 μL/well | Cell lysate (22.9 μg/mL) expressing VF-NSD3S mixed with TR-FRET antibodies (anti-Flag-Tb and anti-GST-d2) |
| 2 | Dispense reaction buffer | 5 μL/well | A mixture of cell lysate (22.9 μg/mL) coexpressing VF-NSD3S and GST-MYC with TR-FRET antibodies (anti-Flag-Tb and anti-GST-d2) |
| 3 | Library compounds | 100 nL/well | 19.6 μM final from 1 mM stock in DMSO |
| 4 | Incubation time | 16 h | Plates were incubated at 4°C for 16 h |
| 5 | Assay readout | Excitation 337 nm (laser), Emission 1 615 nm, Emission 2 665 nm; dual mirror D400/D630 | Time-resolved fluorescence; Envision Multilabel plate reader |
Step Notes
1. 1,536-well black solid bottom plates (Corning Costar, #3724) were used. FRET buffer (20 mM Tris-HCl, pH 7.0, 50 mM NaCl, and 0.01% nonidet P-40) was used to mix with assay components. One column (32 wells) was used for controls. MultiDrop Combi dispenser (Thermo Scientific) was used for dispensing.
2. Reaction buffer was prepared in parallel with controls.
3. 0.1 μL compounds (1 mM stock in DMSO in 384-well plates) were added using a Pintool (VP Scientific) integrated with a Beckman NX (Beckman Coulter, Brea, CA).
4. Plates were incubated at 4°C for 16 h with top plates lidded.
5. Plates were read using EnVision Multilabel plate reader with 50 μs delay in the TR-FRET signal measurement.
DMSO, dimethyl sulfoxide.
Validation of the Assay for uHTS in a 1,536-Well Format with Pilot Screening
To validate the assay for screening in a 1,536-well uHTS format, a pilot screening was carried out using the Spectrum library containing 2,000 pharmacologically active compounds. In brief, the reaction mixture was dispensed at 5 μL per well into 1,536-well black plates. Then 0.1 μL of library compound (1 mM) dissolved in DMSO was added using pintool integrated with Beckman NX (Beckman Coulter, Brea, CA). The final compound concentration was 20 μM and the final DMSO concentration was 2%. After incubating at 4°C for 16 h, TR-FRET signals were measured using Envision Multilabel plate reader as already described.
Screening data were analyzed using Bioassay software from CambridgeSoft (Cambridge, MA). The S/B and Z′ in uHTS format were calculated. The effect of compound on the interaction TR-FRET signal was expressed as percentage of inhibition and calculated as the following equation:
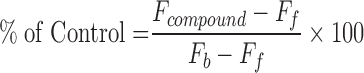 |
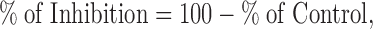 |
where Fb and Ff are the average TR-FRET signal from PPI (NSD3/MYC coexpressing lysate) and background (VF-NSD3 only expressing lysate), respectively. Fcompound is the TR-FRET signal from PPI in the presence of library compound. Compounds that caused percentage inhibition >50 were defined as positives.
GST Pull-Down Assay
To validate the hits from the pilot screening, we developed and optimized a GST pull-down assay using cell lysate with coexpression VF-NSD3 and GST-MYC. In a 1.5 mL Eppendorf tube, 0.5 μL of increasing concentrations of compound diluted in DMSO was added to 100 μL of the diluted cell lysates in 0.25% triton lysis buffer. The mixtures were rotated for 1 h at 4°C. Then 15 μL of 50% prewashed GST beads was added to the 100 μL incubation system. After incubating on a rotator at a low speed for 1.5 h at 4°C, the GST beads were washed three times with 1 mL 0.25% triton lysis buffer on the rotator for 3 min at 4°C. Twenty-five microliters of 1.5 × sodium dodecyl sulfate (SDS) sample buffer was added to beads, followed by boiling for 5 min.
Western Blot
Proteins in sample buffer were separated by 10% SDS-polyacrylamide gel electrophoresis (10% acrylamide gels) and were transferred to nitrocellulose filter membranes at 100 V for 2 h at 4°C. After blocking the membranes in 5% nonfat dry milk in 1 × TBST (20 mM Tris-base, 150 mM NaCl, and 0.05% Tween 20) for 1 h at RT, membranes were blotted with the indicated antibodies overnight at 4°C. Membranes were washed by 1 × TBST for three times, 10 min for each time. SuperSignal west Pico Chemiluminescent substrate (Thermo; #34080) and Dura Extended Duration substrate (Thermo; #34076) were used for developing membranes.
The bands of Western blot were quantified using Image J software (NIH). The amount of VF-NSD3 in the pull-down sample was normalized to the amount of GST-MYC in the same lane. The effect of the compound on the interaction was then normalized to the vehicle control and expressed as % of Control. IC50 was analyzed using GraphPad Prism software.
Results
Optimize the NSD3/MYC Interaction TR-FRET Assay in a 384-Well HTS Format
TR-FRET is a widely used technology for monitoring PPI.21 It comprises two important components: a paired donor (D) and acceptor (A) fluorophores with overlapping emission (D) and excitation (A) spectrum. Fusion proteins overexpressed in cell lysate can be indirectly labeled with donor and acceptor through fluorophore-conjugated antifusion antibodies. Binding of the two interacting proteins brings D and A fluorophores together, leading to the energy transfer from D to A to generate FRET signal. The use of a long-lifetime (e.g., >100 s) fluorescence donor, such as Tb or Eu, allows for a temporal delay between donor excitation and detection of acceptor emission, termed TR-FRET
In order for TR-FRET signal to occur, donor and acceptor have to be in proximity. Indirectly labeling of two interacting proteins with donor- and acceptor-conjugated antifusion-tag antibodies may present challenges to generate TR-FRET signal due to the orientations of the fluorophores. Therefore, to select the optimal paired DNA constructs and TR-FRET antibodies for generating sufficient NSD3/MYC interaction TR-FRET signal, we first tested the TR-FRET signal using different combinations of various epitope-tagged NSD3 and MYC DNA constructs with corresponding paired donor- and acceptor-conjugated TR-FRET antibodies as summarized in Table 1.
Indeed, for the same pair of PPI, different combinations of fusion-tagged proteins and corresponding TR-FRET antibody pairs did give rise to different TR-FRET signals (Fig. 1A). Among all the conditions tested, VF-NSD3 and GST-MYC with the use of anti-Flag-Tb and anti-GST-d2 as TR-FRET donor and acceptor generated the highest TR-FRET signal with S/B of 13.4. Therefore, we chose this combination for further assay development. In this system, as illustrated in Figure 1B, the cell lysate with coexpressing VF-NSD3/GST-MYC is mixed with TR-FRET antibodies (anti-Flag-Tb and anti-GST-d2) for indirectly labeling proteins with TR-FRET donor and acceptor. The interaction of NSD3 and MYC in cell lysate brings Tb and d2 into proximity and generates TR-FRET signal.
To optimize the assay for HTS, we did a twofold serial dilution of stock cell lysate with TR-FRET antibodies in a 384-well plate. As shown in Figure 1C, the TR-FRET signal from cell lysate with coexpression of GST-MYC and VF-NSD3 increased in a dose-dependent manner with increasing amount of cell lysate (0.7–45 μg/mL). There was no significant TR-FRET signal from the same amount of cell lysate with only GST-MYC or VF-NSD3 expression. These data indicate that the TR-FRET signal is specific for NSD3 and MYC interaction.
Evaluate Performance of the TR-FRET Assay for HTS
To monitor the day-to-day transfection efficiency as protein expression control before collecting cell lysate for TR-FRET assay, the expression of the VF-NSD3, which has the Venus fusion tag, was routinely monitored using green fluorescence image of the cells. As a representative image shown in Figure 2A, almost all cells have exhibited the green fluorescence, indicating the excellent expression of the transfected proteins.
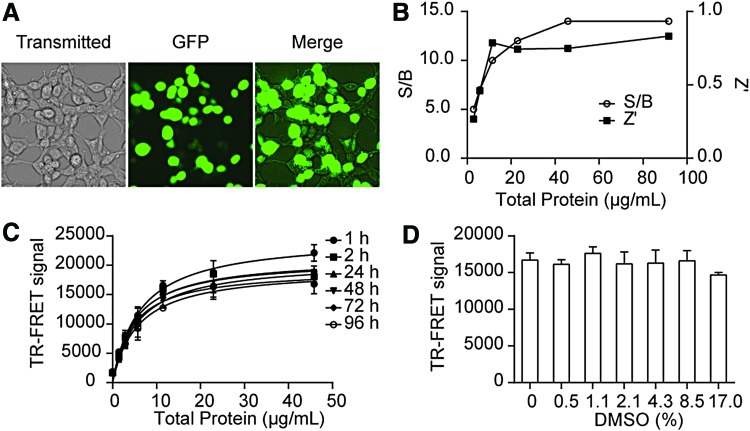
Fig. 2.
Assay performance evaluation in 384-well HTS format. (A) Transfection efficiency monitored by images of HEK 293T cells cotransfected (green) with VF-NSD3 and GST-MYC. Images were taken by ImageXpressmicro automated imaging system (Molecule Devices). (B) Z′ factor and S/B of the assay with increasing amount of cell lysate. (C) Temporal stability of TR-FRET signal. The TR-FRET signal with increasing amount of cell lysate was measured in over a time course of 96 h. (D) The TR-FRET signal in the presence of increasing amount of DMSO. DMSO, dimethyl sulfoxide; HTS, high-throughput screening. Color images available online at www.liebertpub.com/adt
To evaluate the assay performance for HTS, S/B ratio and Z′ were calculated for the TR-FRET titration experiment. This assay exhibited an excellent S/B (>10) and Z′ values (>0.7) when total protein concentration was >40 μg/mL, indicating a robust assay for HTS (Fig. 2B).
To determine the temporal stability of the assay, we monitored the TR-FRET signals for a period of 96 h. As shown in Figure 2C, the TR-FRET assay signals at different time points were not changed significantly for 96 h. We also compared the TR-FRET signal between fresh and frozen cell lysate. Although frozen lysate showed a slightly lower S/B than fresh lysate, it is still robust enough for HTS with S/B >10 and Z′ >0.8 (data not shown). The good stability of the assay offers the advantage for scheduling large-scale screening. Given that all the compounds in the libraries are dissolved in DMSO, whether DMSO itself can affect the TR-FRET signal needs to be evaluated. We performed a DMSO tolerance test in a wide range of DMSO concentrations. As shown in Figure 2D, the TR-FRET signals did not change significantly in the presence of up to 17% of DMSO.
Miniaturization of the Assay into a 1,536-Well uHTS Format
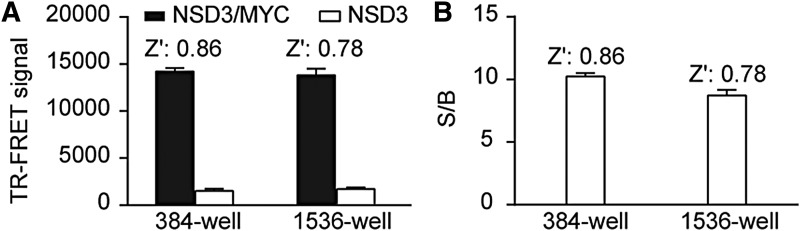
To reduce the regent cost for large-scale screening, especially the high cost of commercial TR-FRET antibodies, we further miniaturized the assay into a 1,536-well uHTS format. The same TR-FRET reaction buffer was added to 384-well (30 μL/well) and 1,536-well (5 μL/well) plates. The TR-FRET signal was then measured and compared. As shown in Figure 3A and B, the TR-FRET signal is similar between a 384- and 1,536-well format. The S/B was 8 and Z′ was 0.78 in a 1,536-well format, indicating a robust TR-FRET assay for MYC/NSD3 interaction in a 1,536-well uHTS format.
Fig. 3.
Miniaturization of the NSD3/MYC TR-FRET assay into a 1,536-well uHTS format. (A) TR-FRET signals of the same reaction buffer in 384-well and 1,536-well format. (B) S/B ratios and Z′ factors in 384-well and 1,536-well format. Data are presented as mean ± SD from triplicate samples. uHTS, ultrahigh-throughput screening.
Pilot Screening
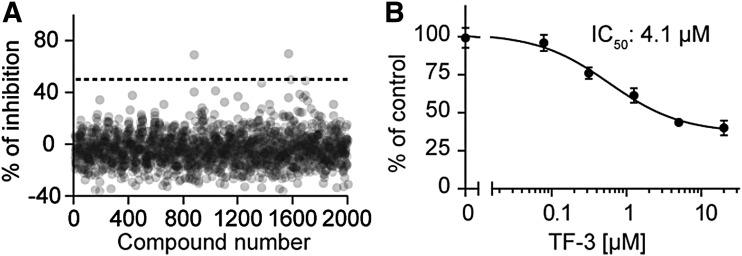
To validate the assay performance for uHTS, we performed a pilot screening using the Spectrum library of 2,000 compounds with a final compound concentration at 20 μM in a 1,536-well uHTS format. Z′ and S/B were calculated for each plate. The Z′ factors were >0.7 and S/B was >8 across two 1,536-well plates, demonstrating the excellent assay performance for uHTS. Screening results are shown in Figure 4A. With a hit cutoff at 50% inhibition, two positive compounds, Brazlin (69.4% inhibition) and theaflavin (50% of inhibition), were identified and the resulting hit rate was 0.1%. The results indicate that the uHTS assay is robust and sensitive to identify primary positives.
Fig. 4.
Pilot screening in a 1,536-well uHTS format. (A) uHTS with the Spectrum library of 2,000 compounds. Percentage of inhibition was calculated for each compound. (B) Dose–response curve of one positive compound, Theaflavin 3,3′-digallate (TF-3), on NDS3/MYC TR-FRET signal.
To confirm the primary positives, dose–response (DR) test was carried out for those two primary hits, Brazlin and theaflavin. Increased concentrations of compounds at five doses (0.08, 0.74, 2.22. 6.67, and 20 μM) were tested in triplicates for each sample in the TR-FRET assay. The % of Control was calculated and dose curves were established for each positive compound. Brazlin failed to decrease the TR-FRET signal in this DR test and was a false primary hit. Theaflavin decreased the TR-FRET signal in a dose-dependent manner with an IC50 of 4.1 μM (Fig. 4B). From this pilot screening, we have validated that our cell lysate-based TR-FRET uHTS assay is sensitive and robust for future large-scale screening.
Hit Validation with an Orthogonal GST Pull-Down Assay
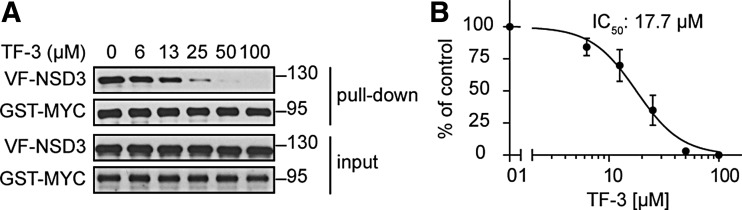
To validate the positive compounds identified in the pilot screening, we used an optimized GST pull-down assay. Cell lysate with coexpression of GST-MYC and VF-NSD3 was mixed with increasing concentrations of compounds (final at 6, 13, 25, 50, and 100 μM) for 1 h before GST pull-down assay. One compound, theaflavin 3,3′-digallate (TF-3), a natural product extracted from black tea, exhibited the disruption on the interaction of NSD3 with MYC in a dose-dependent manner with an IC50 at 17.7 μM (Fig. 5A, B).
Fig. 5.
Validation of a positive compound, TF-3, in GST pull-down assay. (A) Dose-dependent inhibition of TF-3 on NSD3/MYC interaction in GST pull-down assay. Cell lysates were incubated with increasing concentrations of TF-3 for 2 h at 4°C before performing GST pull-down, and then incubated with GST beads for another 2 h. The amount of VF-NSD3 in the GST-MYC complex was detected by Western blotting with antibodies against Flag and GST. (B) Quantification of the Western blot data in (A). The amount of Flag-NSD3 in the GST-MYC complex was normalized with signals of GST-MYC. Data are presented as mean ± SD (n = 3).
These results have demonstrated that our optimized TR-FRET assay using cell lysate is robust and sensitive for screening NSD3/MYC interaction inhibitors. Secondary GST pull-down assay has been optimized and in place for further validation of positive compounds from the primary screening. These results provide the foundation in support of future large-scale screening to discover MYC and NSD3 interaction inhibitors as tools to study the function of NSD3 and for therapeutics development.
Discussion
NSD3 has emerged as an oncoprotein that may serve as a potential target for therapeutic intervention. In a PPI network mapping study, our group has recently identified MYC as one of the interaction proteins of NSD3.10 Our discovery links NSD3 to an important transcription factor, MYC, implying a potential function of NSD3. The small molecule inhibitors of NSD3/MYC interaction could be a useful tool to understand the function of NSD3 protein under physiological and pathophysiological conditions for therapeutic development. Currently there is no small molecule inhibitor for the NSD3/MYC interaction. Here, we present a well-designed, optimized, and validated uHTS TR-FRET assay using cell lysates to monitor the interaction of the NSD3 and MYC proteins. We have optimized the TR-FRET assay for HTS in a 384-well format. The assay has been further miniaturized into a 1,536-well uHTS format with S/B >8 and Z′ >0.7, which is well suited for large-scale screens.
With the use of long-live donor fluorophore, such as Tb, it allows a time-delayed measurement in a TR-FRET format, presenting less fluorescence interference from the compounds themselves.21,22 Compared with some other commonly used methods in HTS for PPI, TR-FRET has demonstrated advantage with reduced interwell variations due to the ratio metric measurement and analysis. In addition, here we used cell lysates from cells with overexpressed target proteins. This not only eliminates the need for labor-intensive protein purification, but also offers a more physiologically relevant screening platform by maintaining the native conformation of PPI in a cellular environment. From the pilot screening with the Spectrum library, we have validated the robustness and sensitivity of this assay for uHTS. There are several factors that may cause the false positives in uHTS, such as compound fluorescence interference and impact of the nonspecific binding between target proteins and TR-FRET antibodies.23 To address this issue, we have developed a nonfluorescence-based GST pull-down assay as a secondary confirmatory assay to validate primary positives and identify true hits. The primary TR-FRET uHTS assay and the secondary GST pull-down validation assay are now in place to support future large-scale library compounds screening for NSD3/MYC interaction inhibitors
From the pilot screening, we have confirmed one compound, TF-3, that can inhibit the interaction of NSD3/MYC in a dose-dependent manner in the GST pull-down assay. TF-3, a member of TF family that also includes TF-1, TF-2a, and TF-2b, is a product from catechins after a fermentative oxidation process during the production of black tea. Recently, it is reported to exhibit anticancer effects in various cancer types by inhibiting cell growth and inducing apoptosis as well as some anti-inflammatory effects through downregulating proinflammatory cytokines.24–27 Mechanically, TF-3 prevents tumorigenesis by decreasing MYC expression level.28 Interestingly, TF-3 is identified as an inhibitor of NSD3/MYC interaction in our assay. We previously demonstrated that NSD3 stabilizes MYC through direct interaction.10 It is possible that TF-3 may destabilize MYC through disrupting the NSD3/MYC interaction. Therefore, our results may reveal a potential mechanism underlying the regulation of MYC by TF-3.
In summary, we have developed and validated a cell lysate-based TR-FRET assay for screening small molecule inhibitors for the NSD3/MYC interaction. To our knowledge, this is the first developed TR-FRET uHTS assay to target the NSD3/MYC interaction. Further large-scale screening is needed to discover promising inhibitors of NSD3/MYC interaction for studying the function of NSD3 as well as potential therapeutic development.
Abbreviations Used
- APC
allophycocyanin
- DMSO
dimethyl sulfoxide
- DR
dose–response
- Eu
europium
- GST
glutathione S-transferase
- HKMT
histone lysine methyltransferase
- HRP
horseradish peroxidase
- HTS
high-throughput screening
- IgG
immunoglobulin G
- NMC
NUT midline carcinoma
- NP-40
nonidet P-40
- NSD3
nuclear receptor binding SET domain protein 3
- PEI
polyethylenimine
- PPI
protein–protein interaction
- RT
room temperature
- S/B
signal-to-background
- SD
standard deviation
- SDS
sodium dodecyl sulfate
- Tb
terbium
- TR-FRET
time-resolved fluorescence resonance energy transfer
- uHTS
ultrahigh-throughput screening
- VF
Venus-Flag
- WHSC1L1
Wolf–Hirschhorn Syndrome Candidate 1-Like 1
Acknowledgments
This research was supported, in part, by NIH NCI U01CA168449 (H.F) and Winship Cancer Institute of Emory University (NIH 5P30CA138292). J.X. is supported by the Emory-Xiangya Medical School collaborative research program.
Disclosure Statement
No competing financial interests exist.
References
- 1.Nie Z, Hu G, Wei G, et al. : c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell 2012;151:68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA: The hallmarks of cancer. Cell 2000;100:57–70 [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg RA: Hallmarks of cancer: The next generation. Cell 2011;144:646–674 [DOI] [PubMed] [Google Scholar]
- 4.Dang CV: MYC on the path to cancer. Cell 2012;149:22–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kress TR, Sabo A, Amati B: MYC: Connecting selective transcriptional control to global RNA production. Nat Rev Cancer 2015;15:593–607 [DOI] [PubMed] [Google Scholar]
- 6.Conacci-Sorrell M, McFerrin L, Eisenman RN: An overview of MYC and its interactome. Cold Spring Harb Perspect Med 2014;4:a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meyer N, Penn LZ: Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976–990 [DOI] [PubMed] [Google Scholar]
- 8.Fletcher S, Prochownik EV: Small-molecule inhibitors of the Myc oncoprotein. Biochim Biophys Acta 2015;1849:525–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalkat M, De Melo J, Hickman KA, et al. : MYC deregulation in primary human cancers. Genes (Basel) 2017;8:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Z, Ivanov AA, Su R, et al. : The OncoPPi network of cancer-focused protein-protein interactions to inform biological insights and therapeutic strategies. Nat Commun 2017;8:14356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vougiouklakis T, Hamamoto R, Nakamura Y, Saloura V: The NSD family of protein methyltransferases in human cancer. Epigenomics 2015;7:863–874 [DOI] [PubMed] [Google Scholar]
- 12.Morishita M, di Luccio E: Cancers and the NSD family of histone lysine methyltransferases. Biochim Biophys Acta 2011;1816:158–163 [DOI] [PubMed] [Google Scholar]
- 13.Bernard-Pierrot I, Gruel N, Stransky N, et al. : Characterization of the recurrent 8p11-12 amplicon identifies PPAPDC1B, a phosphatase protein, as a new therapeutic target in breast cancer. Cancer Res 2008;68:7165–7175 [DOI] [PubMed] [Google Scholar]
- 14.Angrand PO, Apiou F, Stewart AF, Dutrillaux B, Losson R, Chambon P: NSD3, a new SET domain-containing gene, maps to 8p12 and is amplified in human breast cancer cell lines. Genomics 2001;74:79–88 [DOI] [PubMed] [Google Scholar]
- 15.Kim SM, Kee HJ, Eom GH, et al. : Characterization of a novel WHSC1-associated SET domain protein with H3K4 and H3K27 methyltransferase activity. Biochem Biophys Res Commun 2006;345:318–323 [DOI] [PubMed] [Google Scholar]
- 16.Kim SM, Kee HJ, Choe N, et al. : The histone methyltransferase activity of WHISTLE is important for the induction of apoptosis and HDAC1-mediated transcriptional repression. Exp Cell Res 2007;313:975–983 [DOI] [PubMed] [Google Scholar]
- 17.Shen C, Ipsaro JJ, Shi J, et al. : NSD3-short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol Cell 2015;60:847–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.French CA, Rahman S, Walsh EM, et al. : NSD3-NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov 2014;4:928–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mo X, Qi Q, Ivanov AA, et al. : AKT1, LKB1, and YAP1 revealed as MYC interactors with NanoLuc-based protein-fragment complementation assay. Mol Pharmacol 2017;91:339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A: A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol 2002;20:87–90 [DOI] [PubMed] [Google Scholar]
- 21.Inglese J, Johnson RL, Simeonov A, et al. : High-throughput screening assays for the identification of chemical probes. Nat Chem Biol 2007;3:466–479 [DOI] [PubMed] [Google Scholar]
- 22.Du YH, Havel JJ: Time-resolved fluorescence resonance energy transfer technologies in HTS. Chem Genomics 2012:198–214 [Google Scholar]
- 23.Li L, Du Y, Chen W, Fu H, Harrison DG: A novel high-throughput screening assay for discovery of molecules that increase cellular tetrahydrobiopterin. J Biomol Screen 2011;16:836–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu Y, Jin F, Wang Y, et al. : In vitro and in vivo anti-inflammatory effects of theaflavin-3,3′-digallate on lipopolysaccharide-induced inflammation. Eur J Pharmacol 2017;794:52–60 [DOI] [PubMed] [Google Scholar]
- 25.Tu YY, Tang AB, Watanabe N: The theaflavin monomers inhibit the cancer cells growth in vitro. Acta Biochim Biophys Sin (Shanghai) 2004;36:508–512 [DOI] [PubMed] [Google Scholar]
- 26.Liang YC, Chen YC, Lin YL, Lin-Shiau SY, Ho CT, Lin JK: Suppression of extracellular signals and cell proliferation by the black tea polyphenol, theaflavin-3,3′-digallate. Carcinogenesis 1999;20:733–736 [DOI] [PubMed] [Google Scholar]
- 27.Schuck AG, Ausubel MB, Zuckerbraun HL, Babich H: Theaflavin-3,3′-digallate, a component of black tea: An inducer of oxidative stress and apoptosis. Toxicol In Vitro 2008;22:598–609 [DOI] [PubMed] [Google Scholar]
- 28.Gao Y, Rankin GO, Tu Y, Chen YC: Theaflavin-3,3′-digallate decreases human ovarian carcinoma OVCAR-3 cell-induced angiogenesis via Akt and Notch-1 pathways, not via MAPK pathways. Int J Oncol 2016;48:281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]