EVI1 (3q26.2) is a member of the SET/PR domain family of transcription factors and contains two domains of zinc finger (ZF) DNA binding motifs, one at the N-terminus (ZF1) and one at the C-terminus (ZF2), and a C-terminal acidic amino acid cluster domain (AD). The MDS1/EVI1 locus gives rise to alternatively spliced variants, including intergenic MDS1 EVI1 complex (MECOM) splicing. EVI1 is pivotal in leukemogenesis and mammalian development. EVI1 over-expression is associated with the development and progression of high-risk acute myeloid leukemia (AML). EVI1/MECOM is a transcriptional regulator essential for maintaining embryonic and adult hematopoietic stem cells (HSCs) by directly regulating transcription of GATA2. In mouse models, homozygous disruption of Evi1 results in embryonic lethality, with hypocellular bone marrow, reduced body size, small or absent limb buds, and abnormal development of the nervous system. Evi1+/− mice showed an intermediate phenotype compared to Evi1−/− mice both in fetal liver (FL) hematopoiesis as well as in adult hematopoiesis, suggesting a Evi1 gene dosage requirement in the regulation of HSCs.1
To date, two case reports have been published on heterozygous copy number abnormalities (CNA) affecting MECOM. One patient, presenting with congenital thrombocytopenia, had a deletion which included the entire MECOM locus.2 A second newborn presenting with respiratory problems, low birthweight, congenital thrombocytopenia, and slight facial dysmorphisms, had a deletion of the first two exons of MDS1.3 Furthermore, heterozygous missense single nucleotide variations (SNVs) within the eighth zinc finger motif of ZF2 of MECOM were published in three simplex cases with radioulnar synostosis with amegakaryocytic thrombocytopenia (RUSAT [Online Mendelian Inheritance in Man: 065432]), an inherited bone marrow failure (BMF) syndrome with skeletal anomalies.4 Very recently, Ripperger et al. reported on a three generation familial SNV affecting the ninth zinc finger motif of ZF2 in four members with limb dysmorphisms, and thrombocytopenia in one member. Interestingly, two affected family members developed myeloid neoplasia later in life.5 Furthermore, of late Bluteau et al. described six simplex cases; three with mutations affecting ZF2 and three with frameshift mutations, all associated with hypocellular bone marrow, but some also had congenital limb defects.6
We report on the third patient, a neonate born with skeletal and neurological abnormalities and hypocellular bone marrow, with a constitutional copy number alteration in MECOM, particularly the first one with a deletion affecting only the 3′ part of MECOM. The ~854 kb deletion resulted in loss of 3′ MECOM exons, encoding the ZF2 and AD. We will discuss the genetic and clinical heterogeneity of these cases with MECOM abnormalities associated with BMF with or without additional congenital anomalies.
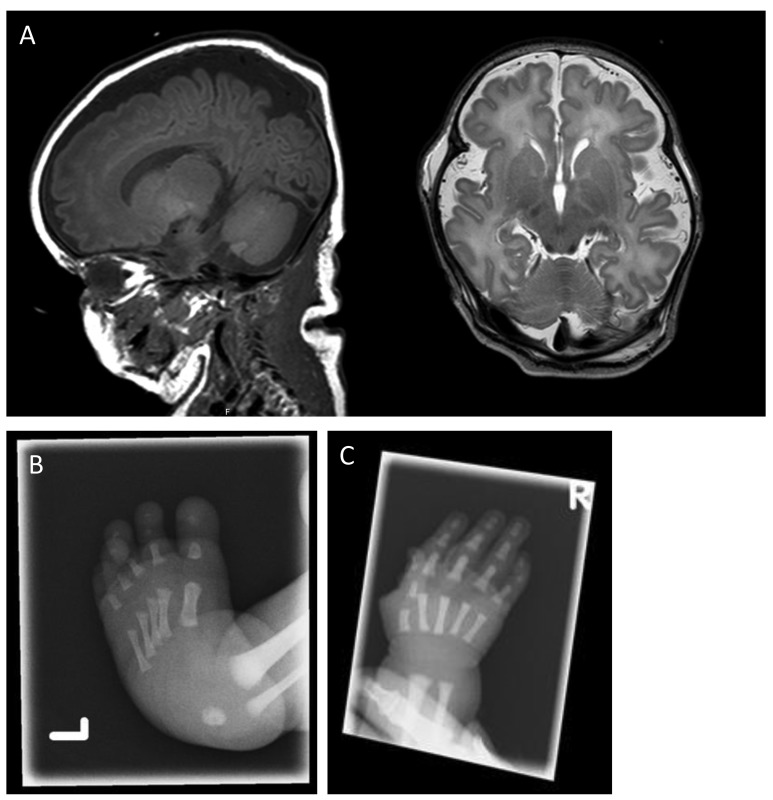
A male neonate was born with severe congenital abnormalities and apparent BMF. He was the fourth child of non-consanguineous parents. The couple had three healthy daughters and no history of miscarriages. Prenatal ultrasounds from 20 weeks gestation onward had shown club feet and polyhydramnios, but normal growth. The patient was born, with respiratory failure, at a gestational age of 38+5 weeks. The placental examination was unremarkable. Clinical examination showed macrocephaly, but otherwise only minor facial dysmorphisms. There was an ulnar deviation of the right hand (the left hand could not be evaluated). The lower extremities showed no abnormalities other than club feet. His hair and nails were normal. Laboratory evaluation showed pancytopenia. Flow cytometry only showed naïve CD4+ and CD8+ T cells with normal T-cell receptor Vβ chain distribution. Therefore, there was no sign of malignant monoclonal hematopoiesis. Bone marrow was hypocellular. A cranial ultrasound performed directly after birth showed subnormal gyrification for the gestational age, wide peripheral cerebrospinal fluid spaces and fissures and a large cavum of the septum pellucidum (Figure 1A). A skeletal evaluation showed club feet and a short fifth midphalanx of the right hand (Figure 1B,C). The cardiac ultrasound examination was normal. The patient died of infections and anemia at 25 days after birth. There was no consent for postmortem evaluation (see Online Supplementary Table S1 for detailed clinical and laboratory characteristics).
Figure 1.
Cranial MRI and X-ray images of hand and foot of a neonate with syndromic BMF associated with interstitial 3′MECOM deletion. (A) T1-weighted sagittal (left) and T2-weighted axial (right) cranial MRI showing enlarged extracerebral cerebrospinal fluid spaces, and reduced frontal gyrification for the developmental stage (gestational age at birth 38+5 weeks, MRI day 6). (B) X-ray images of left foot showing clubfeet, and (C) a short fifth middle phalanx of the right hand.
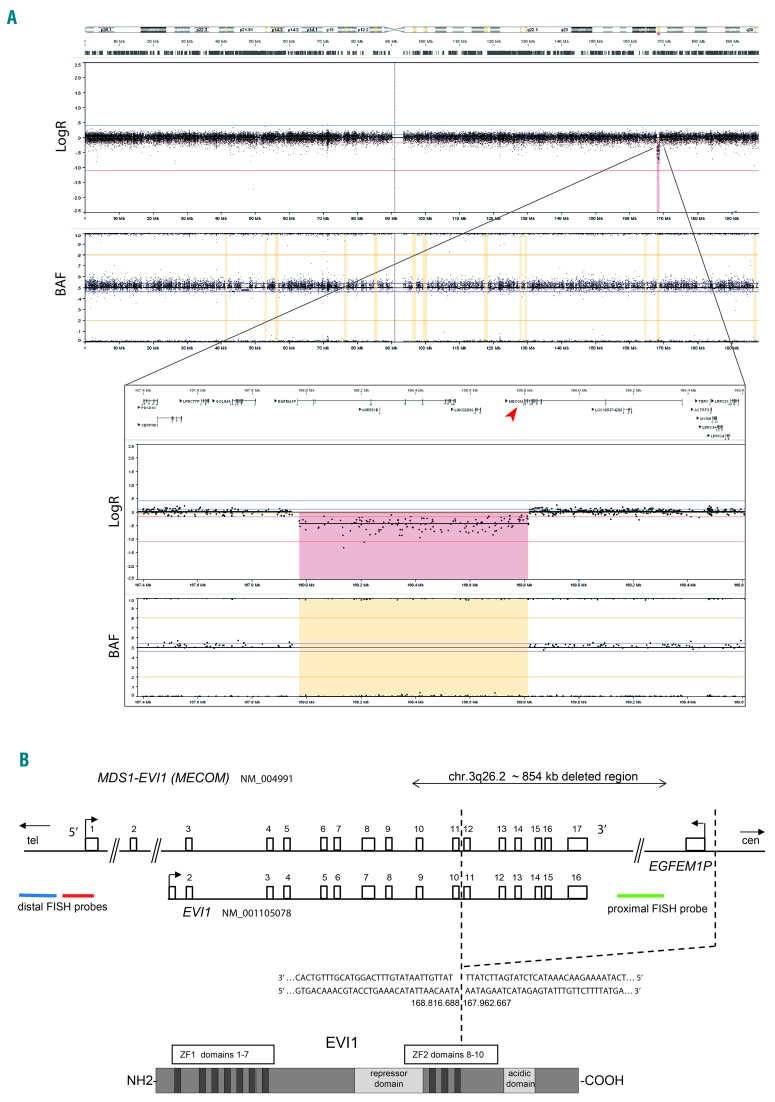
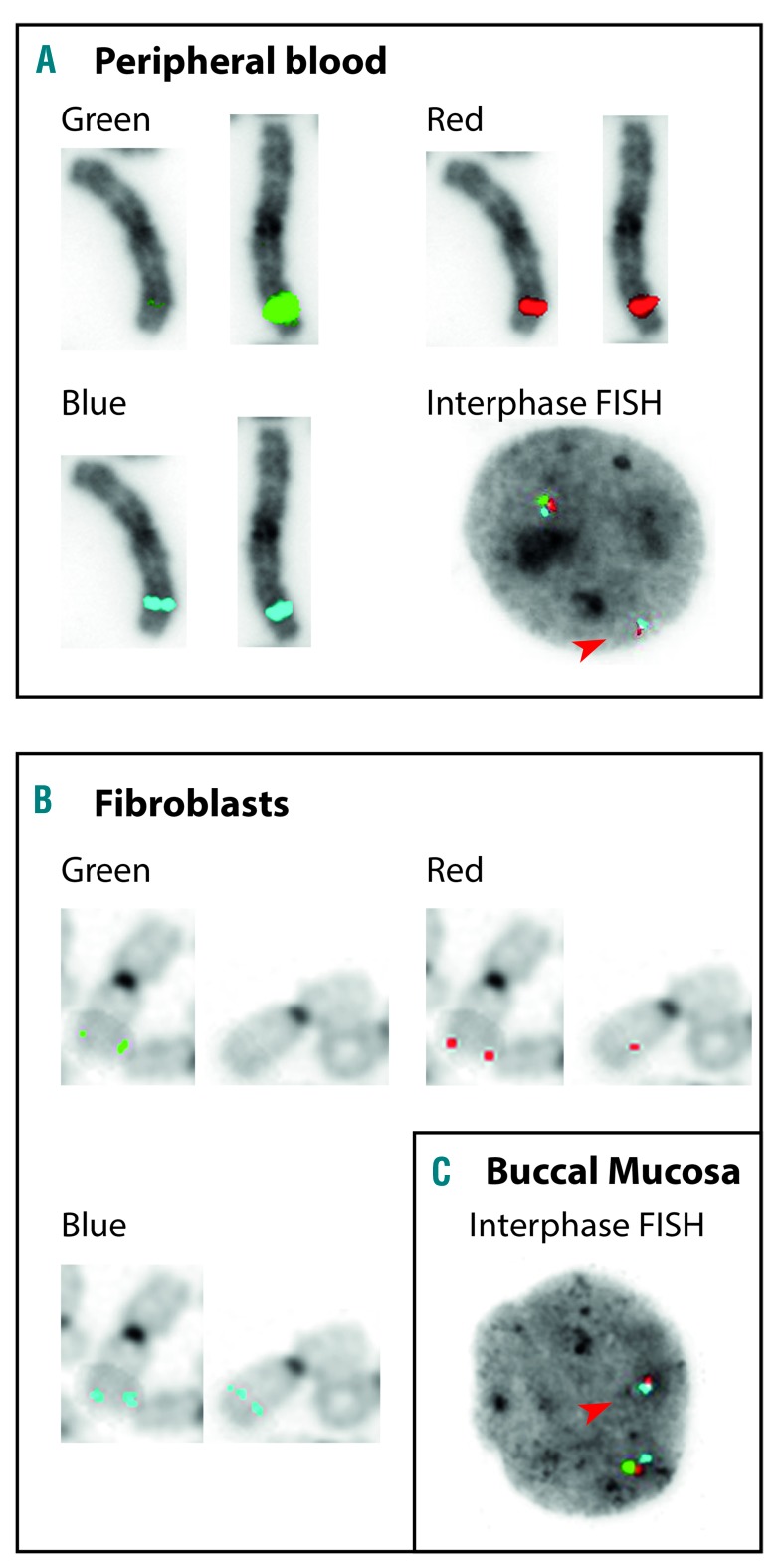
Chromosome analysis on phytohaemaglutin-stimulated T-lymphocytes from peripheral blood resulted in a 46, XY karyotype. Single nucleotide polymorphism (SNP)-array analysis on DNA isolated from peripheral blood revealed a male array profile with heterozygous copy number loss within chromosome 3 band q26.2 of a minimal size of 841 kb, deleting 3′ terminal exons 12 to 17 of MECOM (arr[GRCh37] 3q26.2[167974061_168815099]x1 [International System for Human Cytogenomic Nomenclature [ISCN] 2016]) (Figure 2A). Targeted next-generation sequencing (NGS) of the 3q26.2 region located the exact breakpoints of the 854 kb deletion at chromosomal position 167.962.667 (proximal breakpoint), and at chromosomal position 168.816.688 within intron 11 of MECOM, resulting in the loss of coding exons 12 to 17, affecting ZF2 and AD domains (Figure 2B). Via fluorescence in situ hybridization (FISH) analyses on peripheral blood the deletion was confirmed and its constitutional nature was demonstrated on skin fibroblasts and buccal swabs (46, XY.ish 3 q 26. 2 [3′ MECOM−, 5′ MECOM+]. nuc ish[3′MECOMx1,5′MECOMx2] [ISCN 2016]) (Figure 3). Neither targeted NGS, nor Sanger sequencing of the coding region of MECOM using intronic primers, indicated any additional circulating nucleic acids or sequence variants within MECOM. The coding regions of GATA2 and RUNX1 genes were investigated by Sanger sequencing using intronic primers, revealing no SNV or insertion/deletion. The intronic enhancer of GATA2 was not investigated. Cultured lymphocytes from peripheral blood did not show increased sensitivity for mitomycin C, and as such did not positively identify the patient with Fanconi anemia. Karyotyping and FISH analyses of peripheral blood of both parents did not reveal a structural or numerical abnormality of MECOM, indicating the deletion to be de novo.
Figure 2.
Heterozygous copy number loss of 3′MECOM as a result of an interstitial deletion in 3q26.2 in a neonate with syndromic BMF. (A) Chromosome 3 is shown, and detailed depiction of the affected area, demonstrating heterozygous 3q26.2 3′MECOM copy number loss (in red). SNP-array copy number profiling and analysis of regions of homozygosity were performed on DNA isolated from peripheral blood according to standard procedures using the Infinium Human CytoSNP-850K v1.1 BeadChip (Illumina, San Diego, CA, USA). Subsequently, visualizations of SNP-array results and data analysis were carried out using NxClinical software v3.0 (BioDiscovery, Los Angeles, CA, USA). Human genome build Feb. 2009 GRCh37/hg19 was used. Results were classified with BENCH Lab CNV software (Agilent, Santa Clara, CA, USA). (B) Schematic representation of the chromosome 3q26.2 MECOM region. The bar with whiskers represents the 854 kb deletion encompassing the EGFEM1P and EVI1 (partially) genes. Highlighted is the breakpoint at chromosomal position 167.962.667 upstream of the gene EGFEM1P and at chromosomal position 168.816.688 between exons 11 and 12 of MECOM (hg19/GRCh37) by captured sequencing (as described in10). Breakpoint sequence post-deletion is provided. Localization of the MECOM triple color FISH probe. Representation of EVI1 protein. Map not drawn to scale. Chr: chrosome; FISH: fluorescence in situ hybridization; ZF: zinc finger; BAF: B allele frequency.
Figure 3.
FISH analyses demonstrating constitutional 3′MECOM deletion in syndromic BMF. (A) Triple color FISH analyses demonstrating loss of proximal 3′MECOM signal (green) on one of two paired metaphase chromosome 3 homologues, and one interphase nucleus (phytohaemaglutin-stimulated peripheral blood T-lymphocytes), and (B) on one of two paired metaphase chromosome 3 homologues (cultured skin fibroblasts), and (C) on one interphase nucleus (buccal mucosa), using 3q26.2 MECOM probe on EVI t(3;3); inv(3)(q26) Break, TC (Kreatech, Rijswijk, The Netherlands). For metaphases the individual signals of the triple color probe combination are shown separately. Arrows in interphases point to loss of one green proximal 3′MECOM signal. Images represent all analyzed cells (10 metaphases and 200 interphase nuclei of T-lymphocytes, 10 metaphases of cultured skin fibroblasts, and 200 interphase nuclei of buccal mucosa). FISH: fluorescence in situ hybridization.
Of note are the resemblances of a RUSAT-like phenotypic presentation in the cases as described by Niihori et al., Ripperger et al., Bluteau et al.4–6 and the present case. The constitutional nature of the published SNVs within MECOM, and the deletion in the present case, affecting the ZF2 domain encoding exons of MECOM, are in strong support of their pathogenicity. Of similar distinction is the resemblance in the phenotypic presentation of our case and Evi1-deficit mouse models. Evi1−/− mice die during embryogenesis due to bone marrow hypocellularity. Embryos show defects in the heart, cranial ganglia and the peripheral nervous system with small dorsally positioned forelimb buds.7,8
Notably, the two previously reported cases with a deletion show some differences in the severity of clinical manifestation. The first presented with isolated thrombocytopenia at birth, progressing to pancytopenia with aplastic bone marrow at two months of age, requiring allogeneic stem cell transplantation, but without overt congenital malformations.2 As one entire copy of MECOM was lost, the phenotype can be due to MECOM haploinsufficiency, resembling Mecom+/− mice models in some instantances. The second newborn presented with low birthweight, facial anomalies and a submucosal cleft palate, a small ventricular septal defect and thrombocytopenia.3 There was no information on malformation of the extremities. The patient died 28 days after birth due to respiratory insufficiency. Bouman et al. hypothesized that the hematological phenotype could also be due to haploinsufficiency of MECOM. Nevertheless, as only the first two exons of MDS1 were deleted, the authors stated that this could still allow EVI1 expression because of alternative transcription starting sites. Therefore, one cannot rule out negative interference with normal MECOM function.
Available data do not allow for a conclusion as to whether in the present case the 3′MECOM deletion would result in MECOM haploinsufficiency by messenger ribonucleic acid (mRNA) decay, or in dominant interference by a structurally aberrant protein. It has been reported that EVI1 oligomerization is not impaired by C-terminal truncated EVI1 protein in vitro.9 One could thus hypothesize that in the present case a C-terminally-affected EVI1 oligomerizes with EVI1, thereby tethering EVI1 protein-mediated transcriptional regulation of GATA2 even below the level of 50%, as can be expected in the case of a loss of one MECOM allele as described by Nielsen et al., resulting in a more dramatic perturbation of HSC homeostasis (threshold model). This would be a plausible explanation for the difference in severity of the phenotype of our case, the embryonic lethality in Evi1- or Gata2-deficient mouse models, and the phenotype of Evi1+/- mice or the case with a heterozygous deletion spanning MECOM. One also has to consider the possibility of mosaicism, which could reflect divergence in phenotypic presentation. As EVI1 mRNA expression is not restricted to the hematopoietic system, a direct deleterious effect of the EVI1 anomaly in other tissues seems likely. The data on the cases with MECOM SNV’s as well as the present case indicate that the mutation was present at conception, or occurred during (early) postzygotic isolation. In the two other cases with a deletion affecting MECOM, no data were available on its status in non-hematopoietic cells. Given their phenotypic presentations one could also argue mosaicism in these latter cases.
In conclusion, we report on the third case of BMF syndrome associated with a constitutional CNA in MECOM, particularly the first one with a deletion affecting the 3′ part of MECOM, presenting, however, with multiple congenital anomalies, including limb defects. Together with all reported cases on CNA and SNV of MECOM it clearly establishes MECOM as a recurrent causative gene in (syndromic) BMF. Therefore, one should consider, in case of neonatal thrombocytopenia with or without other dysmorphisms, investigating the MECOM locus for CNAs and SNVs. The syndrome appears to present with variable severity of phenotypic manifestations that might be due to the nature of the MECOM abnormality. Furthermore, investigation of tissues other than the hematopoietic system, together with the consequences both at the transcriptional and post-transcriptional level would greatly contribute to our understanding of the pathogenic mechanism(s).
Supplementary Material
Footnotes
Information on authorship, contributions, and financial & other disclosures was provided by the authors and is available with the online version of this article at www.haematologica.org.
References
- 1.Goyama S, Yamamoto G, Shimabe M, et al. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem Cell. 2008;3(2):207–220. [DOI] [PubMed] [Google Scholar]
- 2.Nielsen M, Vermont CL, Aten E, et al. Deletion of the 3q26 region including the EVI1 and MDS1 genes in a neonate with congenital thrombocytopenia and subsequent aplastic anaemia. J Med Genet. 2012;49(9):598–600. [DOI] [PubMed] [Google Scholar]
- 3.Bouman A, Knegt L, Groschel S, et al. Congenital thrombocytopenia in a neonate with an interstitial microdeletion of 3q26.2q26.31. Am J Med Genet A. 2016;170A(2):504–509. [DOI] [PubMed] [Google Scholar]
- 4.Niihori T, Ouchi-Uchiyama M, Sasahara Y, et al. Mutations in MECOM, encoding oncoprotein EVI1, cause radioulnar synostosis with amegakaryocytic thrombocytopenia. Am J Hum Genet. 2015;97(6):848–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ripperger T, Hofmann W, Koch JC, et al. MDS1 and EVI1 complex locus (MECOM): a novel candidate gene for hereditary hematological malignancies. Haematologica. 2018;103(2):e55–e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bluteau O, Sebert M, Leblanc T, et al. A landscape of germline mutations in a cohort of inherited bone marrow failure patients. Blood. 2018;131(7):717–732. [DOI] [PubMed] [Google Scholar]
- 7.Hoyt PR, Bartholomew C, Davis AJ, et al. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech Dev. 1997;65(1–2):55–70. [DOI] [PubMed] [Google Scholar]
- 8.Parkinson N, Hardisty-Hughes RE, Tateossian H, et al. Mutation at the Evi1 locus in Junbo mice causes susceptibility to otitis media. PLoS Genet. 2006;2(10):e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nitta E, Izutsu K, Yamaguchi Y, et al. Oligomerization of Evi-1 regulated by the PR domain contributes to recruitment of corepressor CtBP. Oncogene. 2005;24(40):6165–6173. [DOI] [PubMed] [Google Scholar]
- 10.Gröschel S, Sanders MA, Hoogenboezem R, et al. A single oncogenic enhancer rearrangement causes concomitant EVI1 and GATA2 deregulation in leukemia. Cell. 2014;157(2):369–381. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.