Abstract
Objectives:
The aim of this research was to study whether plasma microRNAs (miRNA) can be used for early detection of pancreatic cancer (PC) by analyzing prediagnostic plasma samples collected before a PC diagnosis.
Background:
PC has a poor prognosis due to late presenting symptoms and early metastasis. Circulating miRNAs are altered in PC at diagnosis but have not been evaluated in a prediagnostic setting.
Methods:
We first performed an initial screen using a panel of 372 miRNAs in a retrospective case-control cohort that included early-stage PC patients and healthy controls. Significantly altered miRNAs at diagnosis were then measured in an early detection case-control cohort wherein plasma samples in the cases are collected before a PC diagnosis. Carbohydrate antigen 19–9 (Ca 19–9) levels were measured in all samples for comparison.
Results:
Our initial screen, including 23 stage I-II PC cases and 22 controls, revealed 15 candidate miRNAs that were differentially expressed in plasma samples at PC diagnosis. We combined all 15 miRNAs into a multivariate statistical model, which outperformed Ca 19–9 in receiver-operating characteristics analysis. However, none of the candidate miRNAs, individually or in combination, were significantly altered in prediagnostic plasma samples from 67 future PC patients compared with 132 matched controls. In comparison, Ca 19–9 levels were significantly higher in the cases at <5 years before diagnosis.
Conclusion:
Plasma miRNAs are altered in PC patients at diagnosis, but the candidate miRNAs found in this study appear late in the course of the disease and cannot be used for early detection of the disease.
Keywords: blood samples, early detection, micro-RNA, miRNA, pancreatic cancer
Pancreatic cancer (PC) patients have an extremely poor prognosis since the vast majority present with metastatic disease at diagnosis. Moreover, cure is rare even for patients with early-stage disease who undergo surgery with a curative intent, and half of them die within two years.1,2 Improvement in diagnostics and treatment of PC has not been comparable to other cancer forms, and despite its low prevalence, PC is predicted to become the second most common cause of cancer-related death within 4 years.3 Sensitive biomarkers facilitating earlier diagnosis of PC are needed, as the most commonly used PC biomarker, carbohydrate antigen 19–9 (Ca 19–9), lacks sufficient accuracy for early detection of PC.4
Micro-RNAs (miRNAs) are single stranded, noncoding RNA species of ∼22 nucleotides length that participate in post-transcriptional gene regulation. In cancer, miRNAs can act as tumor suppressors or oncogenes by post-transcriptional regulation of genes involved in carcinogenesis.5 miRNAs are surprisingly stable in blood samples, due to high resistance to both temperature changes and endogenous RNase activity.6 This makes them suitable as blood-based biomarkers and several studies have highlighted the potential of circulating miRNAs as biomarkers for various cancer forms, including PC.7 Schultz et al8 screened for >700 miRNAs and validated two whole blood miRNA panels that in combination with Ca 19–9 could accurately differentiate PC patients from controls. Recently, Xu et al9 showed that plasma-miR-486-5p performed equally well as Ca 19–9 in discriminating PC patients from healthy controls. However, all previous studies have examined circulating miRNA levels in patients with an established PC diagnosis, and therefore provide no information on whether miRNA levels are altered before clinical symptoms appear, and thus if they are useful as early detection markers. miRNAs are differentially expressed in PC tissue compared with normal pancreas10–13 and corresponding alterations are also evident in premalignant pancreatic intraepithelial neoplastic lesions (PanINs),14 indicating that miRNA expression changes appear early in PC carcinogenesis. PC is predicted to develop from the initial PanIN lesion into metastatic disease in a time span over 10 years,15 giving a possible window for early detection using miRNAs.
We hypothesized that early miRNA changes in PC might aid in early detection of PC, and thus aimed to find plasma-miRNAs that are altered years before a clinical PC diagnosis.
METHODS
Ethics Statement
All subjects taking part in the study provided written informed consent. The study was approved by the regional research ethics board of northern Sweden and conducted in accord with the ethical standards of the Helsinki Declaration of 1975.
Screening Cohort (Samples Collected Before Surgery)
We retrospectively reviewed the hospital charts for patients who underwent pancreatic surgery for PC between the years 2008 and 2014 at Umeå University Hospital, Sweden. Blood samples were collected from all patients before surgery, and ethylenediaminetetraacetic acid (EDTA) plasma was stored at −80 °C in a prospectively maintained research biobank. We included patients with histopathologically confirmed pancreatic ductal adenocarcinoma at tumor-node-metastasis (TNM) stage I-II with an available preoperative plasma sample. Cases were only included if an age- and sex-matched healthy control was available. Case samples were randomly assigned to matched control samples, collected from patients who either underwent endoscopy without malignant findings or elective surgery for a nonmalignant disease. Controls with a previous history of cancer were excluded.
The following patient characteristics were extracted from hospital charts: age at diagnosis, sex, first clinical sign, preoperative staging (resectability) according to the 7th edition of the American Joint Committee on Cancer staging (AJCC), TNM stage according to AJCC, histopathological grade, tumor size, patient survival, and serum conjugated bilirubin levels (SBR). We only included SBR measurements made on the same day as collection of the plasma samples used for miRNA analysis.
Prediagnostic Cohort (Samples Collected Before Diagnosis)
Prediagnostic plasma samples were derived from a biobank associated with the ongoing population-based Västerbotten Intervention Program (VIP). VIP was launched in 1985 as a primary prevention project for reducing cardiovascular disease in the Swedish county of Västerbotten. Besides offering routine health examinations, participants are asked to donate plasma samples and take part in a large prospective research cohort. Since 1987, the project has covered the entire county, which in 2007 included 258,000 inhabitants. Participation rates have varied between 48% and 67%.16
We included VIP participants who were diagnosed with PC between January 1990 and February 2009, and where EDTA plasma samples collected before the diagnosis date were available. Each case was matched with two healthy controls from the same biobank. Controls were matched by sex, age at sampling, and sampling date (±3 months). Previous history of cancer was an exclusion criterion for both cases and controls. The resulting cohort was randomly split in half into a training set and a validation set. Patient characteristics for all cases were extracted from hospital charts.
miRNA Isolation
miRNA isolation was performed using the Qiagen miRNeasy serum/plasma kit according to the manufacturer's instructions (Qiagen, Hilden, Germany). One hundred microliters of thawed and centrifuged plasma was mixed with 500 μL of QIAzol Lysis Reagent and 0.5 μL of spike-in miRNAs for quality control measurements (Exiqon spike-in kit UniSp2, UniSp4, UniSp5; Exiqon, Vedbaek, Denmark). RNase-free water was run in parallel as a negative control. Phase separation was performed with chloroform; 300 μL of the upper phase was added to 450 μL of 99.5% Ethanol and subsequent RNA cleanup was performed on the provided spin columns. Samples were eluted in 15 μL of RNase-free water, snap-frozen in liquid nitrogen, and stored at −80 °C. Matched cases and controls were isolated together and all match groups were isolated in a randomized order. The case/control designation was blinded.
miRNA Expression by Real-time Quantitative PCR (RT qPCR)
miRNA isolates were shipped on dry ice to Exiqon (Exiqon, Denmark) for subsequent complementary DNA (cDNA) synthesis and RT qPCR. Matched samples were assayed on the same plate, but the match order was randomized. The case/control designation was blinded and all samples were analyzed in duplicates. Samples were reverse transcribed into cDNA using miRCURY LNA Universal RT microRNA PCR. cDNA was then mixed with Exilent SYBR Green mastermix (Exiqon, Vedbaek, Denmark) and RT qPCR was performed on Exiqon miRCURY LNA Universal RT microRNA PCR Human Panel I (V.4, Exiqon, Vedbaek, Denmark) Human Panel I (V.4), with primers covering 372 validated miRNAs. On the basis of the screening results from the patient cohort, we custom-designed a new qPCR panel to be used in the prediagnostic cohort. cDNA amplification, melting points, and raw Ct value acquisition was performed on a Roche LightCycler 480 (Roche Diagnostics, Rotkreuz, Switzerland). Samples had to pass quality control measurements of spike-ins (UniSp2, −4, and −5) before final analysis. Reactions with poor amplification efficacy or multiple melting points were excluded as well as Ct values within 5 Ct values of the negative control (RNase-free water). miRNAs detected in <20 cases or controls were excluded from statistical analysis.
Sample Hemolysis Assessment
Sample hemolysis can affect plasma miRNA levels due to miRNA contamination from red blood cells.17 We therefore measured hemolysis in all samples before isolation, using a previously described spectrophometric method.18 Samples with hemoglobin levels above 25 g/L were excluded. We also controlled for hemolysis by calculating the cycle threshold (Ct) ratio between miR-451 and miR-23a. A Ct ratio >7 was considered indicative of sample hemolysis.19
Plasma Ca 19–9 Measurement
Plasma Ca 19–9 was measured in preoperative samples using the MILLIPLEX MAP Kit and the WideScreen Human Cancer Panel 1 for the prediagnostic samples (Merck KGaA, Darmstadt, Germany) according to the respective manufacturer's instructions. Fluorescence intensities were measured on a Bio-Plex 200 System (Bio-rad, Hercules, California, USA). Results were compared with kit standards using 5-parameter logistic regression. Samples with Ca 19–9 levels below the lowest limit of detection were assigned a value equal to 50% of the lower detection limit.
Statistics
We used ExiqonGenEx 6 Software (Exiqon, Denmark), STATA 12.1 (College Station, Texas, USA), and Prism 5 (GraphPad Software, La Jolla, California, USA) for the statistical analysis. Multivariate statistical analysis was performed using Simca-p 14 (MKS Data Analytics Solutions, Umeå, Sweden). Raw miRNA Ct values were first normalized using the global mean method20 and then compared using Student's t test. False discovery rate for miRNA alterations was controlled at 5% by calculating Benjamini-Hochberg corrected P values.21 The delta mean of normalized Ct values for cases and controls were log2-transformed to calculate the fold change. Other variables were compared using Student's t test for continuous data and Chi-square test for categorical data using P < 0.05 as cutoff level for significance. The correlation between miRNAs and SBR were analyzed using Pearson r.
Multivariate projection analysis to evaluate miRNA combinations was carried out using orthogonal projections to latent structure-discriminative analysis (OPLS-DA).22,23 OPLS-DA regresses the miRNA levels against a binary “dummy vector” carrying the sample class information, in this case patients = 1 and controls = 0, in the search for systematic patterns of miRNAs related to discrimination between the patterns. Hence, in a strong model, controls will cluster around 0 and patients around 1. A 7-fold cross validation of the analysis was performed to estimate the predictive ability of the models.24 Briefly, a model was created using 6/7 of the observations and the remaining 1/7 were predicted by the model. This was repeated 7 times so that all samples were predicted once. The predictive ability was evaluated by cross-validated analysis of variance (CV-ANOVA).25
Receiver-operating characteristic (ROC) curves were generated for the significantly altered miRNAs; the combination of all significant miRNAs and Ca 19–9 and the area under the curve (AUC) was calculated to compare the discriminative performance.
RESULTS
Study Cohorts
Screening Cohort
Twenty-three PC patients and 22 controls were included in our miRNA screening cohort (Fig. 1A). The clinical characteristics of the cases and controls are summarized in Table 1 and Supplementary Table 1. All samples passed hemolysis testing.
FIGURE 1.
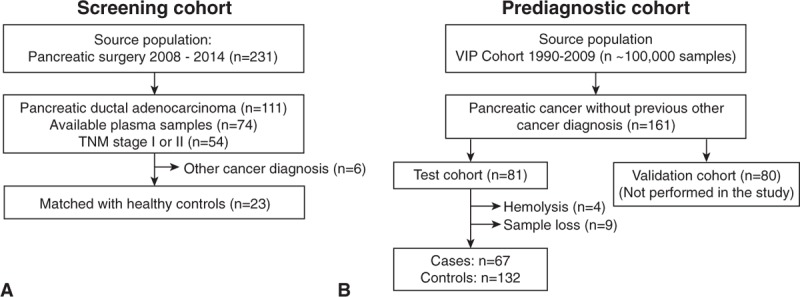
Source population and included cases in the 2 study cohorts. A, The screening cohort consisted of pancreatic cancer patients with TNM stage I-II disease and B, the prediagnostic cohort consisted of individuals who later developed pancreatic cancer. TNM indicates tumor-node-metastasis.
TABLE 1.
Clinical Characteristics of the Screening Cohort
| Variables | Cases (n = 23) | Controls (n = 22) |
| Age, y, Mean (95% CI) | 63.6 (60.3–66.9) | 61.9 (58.8–64.9) |
| Sex | ||
| Men/Women (n) | 12/11 | 12/10 |
| First clinical sign | ||
| Jaundice, n (%) | 17 (73.9%) | |
| Abdominal pain, n (%) | 3 (13.0%) | |
| N.A., n (%) | 3 (13.0%) | |
| TNM stage | ||
| Stage IA–IB, n (%) | 7 (30.4%) | |
| Stage IIA–IIB, n (%) | 16 (69.6%) | |
| Tumor grade | ||
| Grade 1, n (%) | 3 (13.0%) | |
| Grade 2, n (%) | 14 (60.9%) | |
| Grade 3, n (%) | 4 (17.4%) | |
| N.A., n (%) | 2 (8.7%) | |
| Tumor size, cm | ||
| Median (range) | 2.5 (1.5–7) | |
| Survival, mo | ||
| Median (range) | 21 (4–50) |
N.A. indicates not available in hospital charts, TNM, tumor-node-metastasis.
Prediagnostic Cohort
Eighty-one individuals who later developed PC (cases), with a prediagnostic plasma sample available, were randomized to the prediagnostic training cohort (Fig. 1B). During miRNA isolation, 13 case samples and four controls were excluded due to sample hemolysis or insufficient sample volume. The four excluded controls were replaced with controls matched to the excluded cases. Two additional controls were excluded after RT qPCR analysis; one due to low miRNA yield and the other due to uncertain sample identity. The final cohort thus consisted of 67 PC patients and 132 matched controls. The clinical characteristics are summarized in Table 2.
TABLE 2.
Clinical Characteristics of the Prediagnostic Cohort (Training Set)
| Variables | Cases (n = 67) | Controls (n = 132) |
| Age at sampling, y Mean (95% CI) | 54.8 (53.1–56.5) | 54.8 (53.6–56.0) |
| Age at diagnosis, y Mean (95% CI) | 63.6 (61.6–65.5) | |
| Sex | ||
| Men, n (%) | 24 (35.8%) | 49 (37.12%) |
| Women, n (%) | 43 (64.2%) | 83 (62.9%) |
| TNM stage at diagnosis | ||
| Stage IA-IB, n (%) | 3 (4.5%) | |
| Stage IIA-IIB, n (%) | 5 (7.5%) | |
| Stage III, n (%) | 13 (19.4%) | |
| Stage IV, n (%) | 46 (66.7%) | |
| Tumor grade at diagnosis | ||
| Grade 1, n (%) | 1 (1.5%) | |
| Grade 2, n (%) | 18 (26.9%) | |
| Grade 3, n (%) | 10 (14.9%) | |
| N.A., n (%) | 38 (56.7%) | |
| Surgical treatment | ||
| Curative resection | 8 (11.9%) | |
| Palliative surgery | 11 (16.4%) | |
| None | 48 (71.6%) | |
| Time from sampling to diagnosis, y | ||
| Median (range) | 8 (0.4–18.8) |
N.A. indicates not available in hospital charts, TNM, tumor-node-metastasis.
Plasma miRNAs are Altered in Patients at Diagnosis
To identify candidate miRNAs that were altered at PC diagnosis, we analyzed a panel of 372 miRNAs by RT qPCR in the screening cohort. Patients and controls separated somewhat in principle component analysis based on all detectable plasma miRNAs (n = 233), although with a considerable overlap between the groups (Supplementary Figure 1). One hundred ninety miRNAs (51%) were expressed in ≥20 cases or controls and were thus included in the statistical analysis. No miRNA displayed an on/off pattern, meaning that none were solely detected in either cases or controls.
Our screening analysis revealed 15 miRNAs with significant abundance differences between cases and controls after correcting for false discovery rate. Of these 15 candidate miRNAs, 10 were increased and five were decreased in PC patients at diagnosis. miR-885-5p had the highest positive fold change among increased miRNAs, and miR-144-3p the highest negative fold change among decreased miRNAs (Table 3, Supplementary Table 2, Supplementary Figure 2). Among the significantly altered miRNAs, accuracy for patient/control discrimination (AUC) varied from 0.83 [95% confidence interval (95% CI) 0.70–0.95] for miR-574-3p down to 0.75 (95% CI 0.60–0.89) for miR-122-5p (Supplementary Figure 3).
TABLE 3.
Fifteen Significantly Altered miRNAs in Plasma Samples From PC Patients at Diagnosis
| miRNA | ↑↓ | FC | P | FDR (P) |
| miR-574–3p | Up | 1.5 | 0.00008 | 0.0149 |
| miR-885–5p | Up | 3.9 | 0.00013 | 0.0115 |
| miR-144–3p | Down | 0.4 | 0.00014 | 0.0080 |
| miR-130b-3p | Up | 1.5 | 0.00019 | 0.0083 |
| miR-34a-5p | Up | 2.2 | 0.00021 | 0.0073 |
| miR-24–3p | Up | 1.2 | 0.00048 | 0.0121 |
| miR-106b-5p | Down | 0.8 | 0.00060 | 0.0134 |
| miR-22–5p | Up | 1.4 | 0.00067 | 0.0131 |
| miR-451a | Down | 0,5 | 0.00125 | 0.0221 |
| let-7d-3p | Up | 1,3 | 0.00201 | 0.0323 |
| miR-101–3p | Down | 0,7 | 0.00244 | 0.0360 |
| miR-26a-5p | Down | 0,6 | 0.00257 | 0.0350 |
| miR-197–3p | Up | 1,4 | 0.00293 | 0.0370 |
| miR-423–3p | Up | 1,3 | 0.00388 | 0.0458 |
| miR-122–5p | Up | 2,5 | 0.00412 | 0.0455 |
FC indicates fold change; FDR (P), Benjamini-Hochberg corrected P; P, Student t test.
Serum Bilirubin Does Not Correlate With miRNA Levels at Diagnosis
A majority (17/23) of the PC patients suffered from obstructive jaundice at disease presentation (Table 1). Bile duct obstruction can affect liver function,26,27which in turn is associated with plasma miRNA alterations.28 We reasoned that obstructive jaundice might affect levels of circulating miRNA and therefore investigated correlations between candidate miRNAs and SBR. SBR levels were available in-hospital charts from 21 out of 23 cases, with a median SBR level of 23 mmol/L (range 0 to 158 mmol/L). We found no correlation between candidate miRNAs and SBR levels after correcting for multiple testing (Supplementary Table 3).
Plasma miRNAs Are Not Altered Before Diagnosis
Having established that none of our 15 candidate miRNAs were significantly correlated with SBR, we investigated if they could be used to predict a future PC diagnosis. First, we assessed candidate miRNA alterations in the prediagnostic samples independent of time before diagnosis. This clearly showed that none of the 15 miRNAs that were found significantly altered at the time of diagnosis differed before diagnosis (Table 4). We reasoned that one explanation for this negative result could be the time differences in our prediagnostic cohort, where time of sampling varied from 3 months to 18 years before PC diagnosis. We therefore divided the prediagnostic cohort into three groups: 1) >10 years, 2) 5 to 10 years, and 3) <5 years before diagnosis. At <5 years before diagnosis (3 months to 4.9 years), the P value for miR-24-3p was <0.05 and miR-106b was close to being significant, although none passed false discovery rate testing. Of note, most miRNAs had case/control fold changes close to 1 (Table 4).
TABLE 4.
Fold Changes and Corresponding P Values of the 15 Candidate miRNAs in the Prediagnostic Cohort
| All Sample Time Points | >10 y Before Diagnosis | 5–10 y Before Diagnosis | <5 y Before Diagnosis | |||||||||
| miRNA | Cases /Ctrls | FC | P | Cases /Ctrls | FC | P | Cases /Ctrls | FC | P | Cases /Ctrls | FC | P |
| miR-106b-5p | 67/132 | 0.94 | 0.132 | 26/48 | 0.98 | 0.854 | 25/50 | 0.91 | 0.154 | 16/31 | 0.90 | 0.057 |
| miR-574-3p | 67/132 | 0.94 | 0.231 | 26/48 | 0.94 | 0.517 | 25/50 | 1.01 | 0.869 | 16/31 | 0.92 | 0.130 |
| miR-34a-5p | 67/132 | 1.13 | 0.271 | 26/47 | 1.13 | 0.511 | 25/49 | 1.03 | 0.861 | 16/31 | 1.52 | 0.161 |
| miR-451a | 67/132 | 1.12 | 0.377 | 26/48 | 1.04 | 0.856 | 25/50 | 0.95 | 0.818 | 16/31 | 1.20 | 0.086 |
| miR-130b-3p | 67/132 | 0.96 | 0.447 | 26/47 | 0.95 | 0.533 | 25/50 | 1.05 | 0.626 | 16/31 | 0.94 | 0.246 |
| miR-26a-5p | 67/132 | 0.95 | 0.453 | 26/48 | 0.91 | 0.426 | 25/50 | 1.04 | 0.676 | 16/31 | 1.00 | 0.408 |
| miR-144-3p | 67/132 | 1.08 | 0.558 | 26/48 | 1.02 | 0.927 | 25/50 | 0.95 | 0.810 | 16/31 | 1.14 | 0.148 |
| miR-423-3p | 67/132 | 1.03 | 0.660 | 26/48 | 1.12 | 0.276 | 25/50 | 1.05 | 0.621 | 16/31 | 1.02 | 0.250 |
| miR-101-3p | 67/132 | 1.02 | 0.736 | 26/48 | 0.99 | 0.930 | 25/50 | 1.02 | 0.731 | 16/31 | 0.95 | 0.332 |
| miR-122-5p | 67/132 | 0.96 | 0.749 | 26/48 | 0.92 | 0.684 | 25/50 | 0.80 | 0.350 | 16/31 | 1.00 | 0.313 |
| miR-24-3p | 67/132 | 0.99 | 0.877 | 26/48 | 1.05 | 0.478 | 25/50 | 1.06 | 0.402 | 16/31 | 0.97 | 0.046 |
| miR-22-5p | 67/132 | 1.01 | 0.887 | 26/48 | 1.03 | 0.761 | 25/50 | 1.02 | 0.846 | 16/31 | 1.16 | 0.657 |
| let-7d-3p | 67/132 | 1.01 | 0.893 | 26/48 | 1.03 | 0.753 | 25/50 | 1.14 | 0.134 | 16/31 | 0.94 | 0.118 |
| miR-197-3p | 67/132 | 1.00 | 0.949 | 26/48 | 1.04 | 0.613 | 25/50 | 1.09 | 0.292 | 16/31 | 0.99 | 0.159 |
| miR-885-5p | 67/132 | 0.99 | 0.959 | 26/48 | 0.96 | 0.854 | 25/50 | 0.85 | 0.495 | 16/31 | 1.00 | 0.311 |
Significant P values in bold. Ctrls indicates controls; FC, fold change; P, Student t test P (not corrected for false discovery rate).
A Multivariate Statistical Model of Candidate miRNAs Separate Cases and Controls at Diagnosis but not Before
Multiple miRNAs have been shown to act synergistically in gene regulation29 and previous studies have combined several miRNAs to better separate PC patients from controls.8,30,31 We therefore hypothesized that a combination of our 15 candidate miRNAs might perform better to detect early alterations than single miRNAs alone. To test this, we generated a multivariate statistical model on the basis of candidate miRNAs. At diagnosis, the model clearly separated cases from controls, a separation that was consistent and significant after cross validation (Fig. 2B). However, the model failed to separate cases from controls at any time point before diagnosis, although a tendency to separation was noted <5 years before diagnosis (Fig. 2A). The poor group separation before diagnosis was independent of TNM stage and sex (Supplementary Figure 4). By comparison, Ca 19–9 levels were significantly altered <5 years before diagnosis, although only three of these cases presented with levels above 37 U/mL, which is the standard clinical cutoff for Ca 19–9.4 Interestingly, Ca 19–9 levels increased the closer the sampling date was to diagnosis (Figs. 2C, D, Supplementary Table 4).
FIGURE 2.
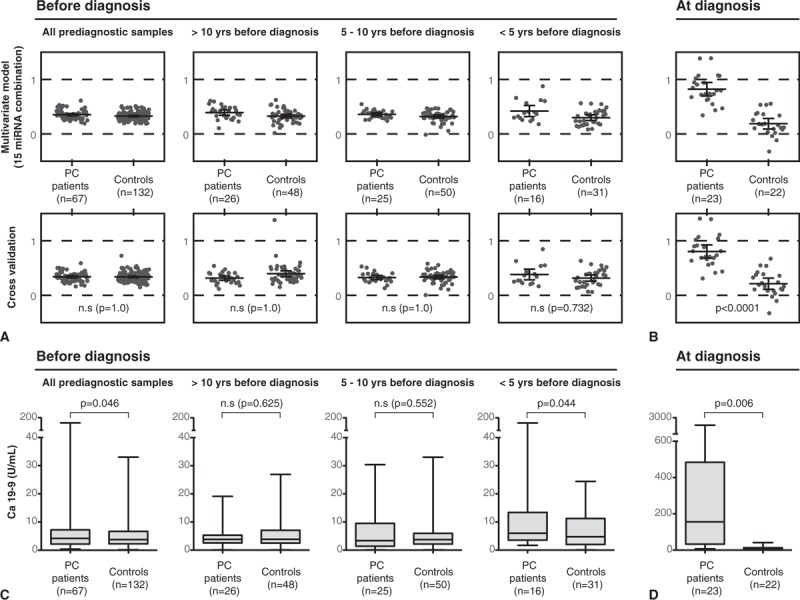
Differences between cases and controls over time. A to B shows the estimated (upper panels) and cross-validated (lower panels) multivariate statistical model (OPLA-DA) of the 15 candidate miRNAs. A, Prediagnostic samples at different time intervals before diagnosis and (B) at diagnosis. Each dot represents an individual. In a strong model, the cases would cluster around 1 and controls around 0. CV-ANOVA P values are derived from the cross validation. C to D shows boxplots of Ca 19–9 levels in (C) prediagnostic samples at different time intervals before diagnosis and (D) at diagnosis.
To compare discriminative performance, we constructed ROC curves for the miRNA model and Ca 19–9. Our miRNA model outperformed Ca 19–9 at diagnosis, but at all time points before PC diagnosis, both the miRNA model and Ca 19–9 performed poorly in discriminating cases from controls (Fig. 3). In the light of the poor performance of miRNAs in the training set of the prediagnostic cohort, we refrained from further analysis in the validation set.
FIGURE 3.
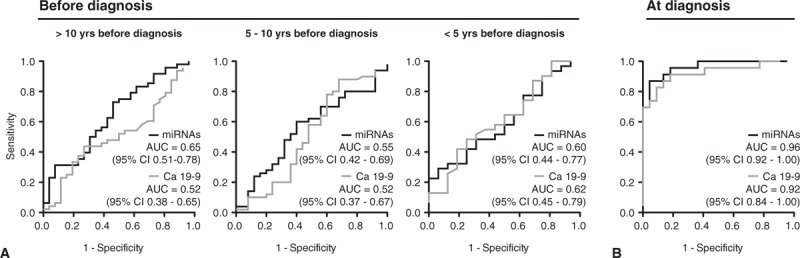
Receiver-operating characteristics (ROC) curves plotting the sensitivity and false-positive rate (1 – specificity) for the cross-validated multivariate model of 15 miRNAs and Ca 19–9 at (A) different time intervals in relation to diagnosis and (B) at diagnosis.
DISCUSSION
Circulating levels of miRNAs are altered in many different cancer forms and various miRNAs and miRNA-combinations have been suggested as potential biomarkers of disease.7 In PC, there is a pressing need for early detection biomarkers, as patients are generally asymptomatic until metastatic disease has developed.1 In the first study of its kind, we evaluated the potential of miRNAs in early PC detection by analyzing candidate miRNAs in plasma samples collected before a PC diagnosis. Although we did identify miRNAs that were altered at diagnosis, they were not suitable for early detection of PC. Early detection performance was unaffected by stratifications for both time to diagnosis and TNM stage. miR-24 was significant <5 years before diagnosis using a permissive significance level at 0.05, but the fold change was minimal and nonsignificant after correcting for false discovery rate, indicative of a false-positive finding.
Nonetheless, we identified 15 miRNAs that were associated with PC at the time of diagnosis and a multivariate model based on these miRNAs outperformed Ca 19–9 in ROC analysis. However, as the model was not validated in an independent PC patient cohort, we refrain from drawing conclusions regarding its clinical potential for PC diagnosis.
Several of the miRNAs we identified have previously been associated with PC, including increased let-7d, miR-22, -24, -34a, -122, -130b, -574, and -885 as well as decreased miR-106b-5p and -1448,9,32–35 (Supplementary Table 5). There are also previous reports of associations for miR-26a, -423, and -451a, although with fold changes in opposite directions9,30,33 (Supplementary Table 5). Of note, miR-451a changes should be interpreted with caution due to its enrichment in red blood cells and its concomitant association with sample hemolysis.17 But as all our screening samples passed hemolysis testing, the decreased levels of miR-451a are more likely to be disease-associated. Importantly, PC association of circulating miR-101 and miR-197 are novel findings. This is of particular interest, as both are implicated in epithelial-mesenchymal transition in PC cells,36,37 suggestive of involvement in tumor invasion and metastasis.
Several of the miRNAs that were altered at diagnosis have been identified in functional in vitro studies on PC cells or found at altered levels in PC tissue (Supplementary Table 5). miRNAs with supporting functional data include miR-24, -197, -26a, -101, -106b, and -144.36–42 Similarly, miRNAs with supporting tissue data include let-7d, miR-24, -26a, and -101.12,37,38,40,43 On the contrary, our results on miR-34a, -122, miR-130b, and -451a contradict previous tissue study findings.11,13,44,45 But the controversy in the latter should not be exaggerated, as it is known that miRNAs in PC serum do not readily correlate with tumor tissue expression.30 We also speculate that circulating miRNA levels could be affected by systemic changes associated with PC.
One contributor to systemic changes during PC is obstructive jaundice. Although often one of the first symptoms of disease, obstructive jaundice is a late complication during PC development and manifest close to diagnosis.1 In our study, some of the candidate miRNAs identified in diagnostic samples trended toward SBR correlation. Although correlations were insignificant after false discovery rate correction, obstructive jaundice cannot be completely ruled out as a potential confounder in samples close to diagnosis. We therefore strongly suggest that future studies should assess SBR correlations for candidate, circulating PC biomarkers.
Ca 19–9 was significantly elevated in PC patients <5 years before diagnosis. However, ROC analysis revealed a poor discriminative performance at this stage. Prediagnostic increases in Ca 19–9 levels have been reported previously in PC patients, with similar AUC at <12 months before diagnosis.46 The poor discriminative performance of Ca 19–9 in prediagnostic samples in that study and in ours may, in part, explain the low positive predictive value of Ca 19–9 evident from prospective cohort studies.4
Almost 90% of the cases in our early detection cohort had stage III or IV cancer at diagnosis. If the temporal development suggested by Yachida et al15 is correct, then a substantial proportion of these patients should have been at stage I or II at sample collection, and thus comparable to patients in the screening cohort. However, it is possible that the transition from stage I to stage IV in fact occurs more rapidly than suggested, which is supported by survival data from nonresected stage I-II patients, showing a median survival of <7 months.2
Although the present screen covered 372 miRNAs, over 2500 miRNA sequences have been annotated in the current miRBase version (www.mirbase.org). In an ideal setting, all known miRNAs should be tested in an unbiased manner using prediagnostic samples. Another powerful approach is to combine results from omics studies on diagnosed patients with a prediagnostic cohort or with animal models. One such metabolomics study demonstrated that branched-chain amino acids are elevated before diagnosis in both patient plasma and in a KRAS-driven mouse PC model.47 In a similar mouse model, early carcinogenic progression was found to correlate with miRNA changes.48 However, to our knowledge, the current study is the first to investigate levels of circulating miRNAs in samples collected before a PC diagnosis.
CONCLUSIONS
A panel of 15 circulating miRNAs can discriminate PC patients from controls at the time of diagnosis. These 15 miRNAs do not hold promise as early detection biomarkers, as the alterations appear late in the disease course.
Supplementary Material
Acknowledgments
We want to thank Anette Berglund for retrieving and keeping track of the frozen patient samples and the personnel at the endoscopy unit at the surgical ward at Umeå University Hospital and Marjo Andersson for helping out with sample collection.
Footnotes
This study was funded by The Swedish Research Council (2011-3089 for M.S., 537-2013-7277 for D.Ö.), the Swedish Cancer society (110679 and 120135 for M.S.) the Swedish Society of Medicine (SLS-591551 for D.Ö.), the County Council of Västerbotten (M.S. and D.Ö.), Cancer research foundation in northern Sweden (AMP15-793 for D.Ö.), JC Kempe Memorial Foundation Scholarship Fund (O.F.), and Grants from the medical faculty at Umeå University (223-1828-13 O.F).
O.F., P.J., O.B., and M.S. planned the study and the study design. O.F., D.Ö., E.L., H.N., and M.S. collected and interpreted patient data from hospital charts. O.F., O.B., C.L., performed the experimental procedures. O.F., P.J., and H.A. performed data analysis. O.F., O.B., P.J., and M.S. wrote the manuscript. All authors read and critically reviewed the intellectual content of the final text, and gave their final approval of the text.
The authors have no conflicts of interest.
REFERENCES
- 1.Vincent A, Herman J, Schulick R, et al. Pancreatic cancer. Lancet 2011; 378:607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bilimoria KY, Bentrem DJ, Ko CY, et al. Validation of the 6th edition AJCC Pancreatic Cancer Staging System: report from the National Cancer Database. Cancer 2007; 110:738–744. [DOI] [PubMed] [Google Scholar]
- 3.Rahib L, Smith BD, Aizenberg R, et al. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res 2014; 74:2913–2921. [DOI] [PubMed] [Google Scholar]
- 4.Poruk KE, Gay DZ, Brown K, et al. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: diagnostic and prognostic updates. Curr Mol Med 2013; 13:340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol 2014; 9:287–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008; 105:10513–10518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jarry J, Schadendorf D, Greenwood C, et al. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol Oncol 2014; 8:819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA 2014; 311:392–404. [DOI] [PubMed] [Google Scholar]
- 9.Xu J, Cao Z, Liu W, et al. Plasma miRNAs effectively distinguish patients with pancreatic cancer from controls: a multicenter study. Ann Surg 2016; 263:1173–1179. [DOI] [PubMed] [Google Scholar]
- 10.Bloomston M, Frankel WL, Petrocca F, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA 2007; 297:1901–1908. [DOI] [PubMed] [Google Scholar]
- 11.Szafranska AE, Davison TS, John J, et al. MicroRNA expression alterations are linked to tumorigenesis and non-neoplastic processes in pancreatic ductal adenocarcinoma. Oncogene 2007; 26:4442–4452. [DOI] [PubMed] [Google Scholar]
- 12.Lee EJ, Gusev Y, Jiang J, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer 2007; 120:1046–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jamieson NB, Morran DC, Morton JP, et al. MicroRNA molecular profiles associated with diagnosis, clinicopathologic criteria, and overall survival in patients with resectable pancreatic ductal adenocarcinoma. Clin Cancer Res 2012; 18:534–545. [DOI] [PubMed] [Google Scholar]
- 14.Yu J, Li A, Hong SM, et al. MicroRNA alterations of pancreatic intraepithelial neoplasias. Clin Cancer Res 2012; 18:981–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010; 467:1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Norberg M, Wall S, Boman K, et al. The Vasterbotten Intervention Programme: background, design and implications. Glob Health Action 2010; 3:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirschner MB, Edelman JJ, Kao SC, et al. The impact of hemolysis on cell-free microRNA biomarkers. Front Genet 2013; 4:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fairbanks VF, Ziesmer SC, O’Brien PC. Methods for measuring plasma hemoglobin in micromolar concentration compared. Clin Chem 1992; 38:132–140. [PubMed] [Google Scholar]
- 19.Shah JS, Soon PS, Marsh DJ. Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One 2016; 11:e0153200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mestdagh P, Van Vlierberghe P, De Weer A, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 2009; 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc B Methodol 1995; 57:289–300. [Google Scholar]
- 22.Bylesjö M, Rantalainen M, Cloarec O, et al. OPLS discriminant analysis: combining the strengths of PLS-DA and SIMCA classification. J Chemometrics 2006; 20:341–351. [Google Scholar]
- 23.Trygg J, Wold S. Orthogonal projections to latent structures (O-PLS). J Chemometrics 2002; 16:119–128. [Google Scholar]
- 24.Wold S. Cross-validatory estimation of number of components in factor and principal components models. Technometrics 1978; 20:397–405. [Google Scholar]
- 25.Eriksson L, Trygg J, Wold S. CV-ANOVA for significance testing of PLS and OPLS (R) models. J Chemometrics 2008; 22:594–600. [Google Scholar]
- 26.Hayat JO, Loew CJ, Asrress KN, et al. Contrasting liver function test patterns in obstructive jaundice due to biliary strictures [corrected] and stones. QJM 2005; 98:35–40. [DOI] [PubMed] [Google Scholar]
- 27.Siriwardena AK, Siriwardena AM. Pancreatic cancer. BMJ 2014; 349:g6385. [DOI] [PubMed] [Google Scholar]
- 28.McGill MR, Jaeschke H. MicroRNAs as signaling mediators and biomarkers of drug- and chemical-induced liver injury. J Clin Med 2015; 4:1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Han YC, Vidigal JA, Mu P, et al. An allelic series of miR-17 approximately 92-mutant mice uncovers functional specialization and cooperation among members of a microRNA polycistron. Nat Genet 2015; 47:766–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kojima M, Sudo H, Kawauchi J, et al. MicroRNA markers for the diagnosis of pancreatic and biliary-tract cancers. PLoS One 2015; 10:e0118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu J, Gao J, Du Y, et al. Combination of plasma microRNAs with serum CA19-9 for early detection of pancreatic cancer. Int J Cancer 2011; 131:683–691. [DOI] [PubMed] [Google Scholar]
- 32.Ali S, Almhanna K, Chen W, et al. Differentially expressed miRNAs in the plasma may provide a molecular signature for aggressive pancreatic cancer. Am J Transl Res 2010; 3:28–47. [PMC free article] [PubMed] [Google Scholar]
- 33.Bauer AS, Keller A, Costello E, et al. Diagnosis of pancreatic ductal adenocarcinoma and chronic pancreatitis by measurement of microRNA abundance in blood and tissue. PLoS One 2012; 7:e34151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ganepola GA, Rutledge JR, Suman P, et al. Novel blood-based microRNA biomarker panel for early diagnosis of pancreatic cancer. World J Gastrointest Oncol 2014; 6:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu R, Chen X, Du Y, et al. Serum microRNA expression profile as a biomarker in the diagnosis and prognosis of pancreatic cancer. Clin Chem 2012; 58:610–618. [DOI] [PubMed] [Google Scholar]
- 36.Hamada S, Satoh K, Miura S, et al. miR-197 induces epithelial-mesenchymal transition in pancreatic cancer cells by targeting p120 catenin. J Cell Physiol 2013; 228:1255–1263. [DOI] [PubMed] [Google Scholar]
- 37.Qazi AM, Gruzdyn O, Semaan A, et al. Restoration of E-cadherin expression in pancreatic ductal adenocarcinoma treated with microRNA-101. Surgery 2012; 152:704–711. discussion 11–13. [DOI] [PubMed] [Google Scholar]
- 38.Liu R, Zhang H, Wang X, et al. The miR-24-Bim pathway promotes tumor growth and angiogenesis in pancreatic carcinoma. Oncotarget 2015; 6:43831–43842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Batchu RB, Gruzdyn OV, Qazi AM, et al. Enhanced phosphorylation of p53 by microRNA-26a leading to growth inhibition of pancreatic cancer. Surgery 2015; 158:981–986. discussion 6–7. [DOI] [PubMed] [Google Scholar]
- 40.Deng J, He M, Chen L, et al. The loss of miR-26a-mediated post-transcriptional regulation of cyclin E2 in pancreatic cancer cell proliferation and decreased patient survival. PLoS One 2013; 8:e76450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo ZL, Luo HJ, Fang C, et al. Negative correlation of ITCH E3 ubiquitin ligase and miRNA-106b dictates metastatic progression in pancreatic cancer. Oncotarget 2016; 7:1477–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sureban SM, May R, Lightfoot SA, et al. DCAMKL-1 regulates epithelial-mesenchymal transition in human pancreatic cells through a miR-200a-dependent mechanism. Cancer Res 2011; 71:2328–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A 2006; 103:2257–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papaconstantinou IG, Manta A, Gazouli M, et al. Expression of microRNAs in patients with pancreatic cancer and its prognostic significance. Pancreas 2013; 42:67–71. [DOI] [PubMed] [Google Scholar]
- 45.Ali S, Saleh H, Sethi S, et al. MicroRNA profiling of diagnostic needle aspirates from patients with pancreatic cancer. Br J Cancer 2012; 107:1354–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nolen BM, Brand RE, Prosser D, et al. Prediagnostic serum biomarkers as early detection tools for pancreatic cancer in a large prospective cohort study. PLoS One 2014; 9:e94928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mayers JR, Wu C, Clish CB, et al. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med 2014; 20:1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.LaConti JJ, Shivapurkar N, Preet A, et al. Tissue and serum microRNAs in the KrasG12D transgenic animal model and in patients with pancreatic cancer. PLoS One 2011; 6:e20687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao G, Zhang JG, Shi Y, et al. MiR-130b is a prognostic marker and inhibits cell proliferation and invasion in pancreatic cancer through targeting STAT3. PLoS One 2013; 8:e73803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang TC, Wentzel EA, Kent OA, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell 2007; 26:745–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cote GA, Gore AJ, McElyea SD, et al. A pilot study to develop a diagnostic test for pancreatic ductal adenocarcinoma based on differential expression of select miRNA in plasma and bile. Am J Gastroenterol 2014; 109:1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


