Abstract
The developing world is seeing rapid growth in the availability of biological mass spectrometry (MS), particularly through core facilities. As proteomics and metabolomics becomes locally feasible for investigators in these nations, application areas associated with high burden in these nations, such as infectious disease, will see greatly increased research output. This article evaluates the rapid growth of MS in South Africa (currently approaching 20 laboratories) as a model for establishing MS core facilities in other nations of the developing world. Facilities should emphasize new services rather than new instruments. The reduction of the delays associated with reagent and other supply acquisition would benefit both facilities and the users who make use of their services. Instrument maintenance and repair, often mediated by an in-country business for an international vendor, is also likely to operate on a slower schedule than in the wealthiest nations. A key challenge to facilities in the developing world is educating potential facility users in how best to design experiments for proteomics and metabolomics, what reagents are most likely to introduce problematic artifacts, and how to interpret results from the facility. Here, we summarize the experience of 6 different institutions to raise the level of biological MS available to researchers in South Africa.
Keywords: shared instruments, publication standards, capacity development, South Africa
INTRODUCTION
Why biological mass spectrometry?
Mass spectrometry (MS) has become increasingly central to protein and metabolite identification and quantitation during the last decade. In the 1990s, many laboratories used 2-dimensional gels as the “gold standard” for separating and visualizing proteins, using MS in peptide mass fingerprinting to identify protein spots.1 The pairing of liquid chromatography (LC) with fast-scanning, high-resolution tandem MS (MS/MS), ubiquitous in today’s laboratories, makes it possible to identify thousands of proteins in a 90-min experiment.2 The incorporation of fractionation before LC-MS/MS trades more instrument time for deeper identification. The sensitivity and reproducibility available in contemporary proteomics technologies would have been hard to imagine as the year 2000 arrived.
Metabolomics has also gained power through the rise of biological MS. Like proteomics, it has benefited handsomely from the increased diversity of bioinformatics research. At the turn of the century, most MS bioinformatics emphasized the identification of peptides and proteins by database search. Today, the field has diversified to reflect that experiments may be broad-sampling “discovery” designs or more narrowly focused “targeted” designs.3 Whereas identification by database search continues to dominate proteomics, spectral library comparison is the standard in metabolomics.4 Spectral clustering and de novo methods, however, are making it possible to interpret a wider range of MS/MS.5 Algorithms for quantifying analytes have taken on new momentum in the bioinformatics community. The integration of data among genomics, proteomics, and metabolomics increasingly has become its own field.6 Through these advances, biological MS has become a powerful complement to nuclear magnetic resonance for metabolite research.
The role of the core facility
These technological wonders, however, come at a high price, and core facilities exist to lower the cost of access to advanced technologies. Obviously, proteomics and metabolomics have a financial price; acquiring equipment is just the beginning, as reagents, salaries, and service contracts are considerable expenditures over the long run. The human resources price is also significant. A core laboratory needs researchers who excel at navigating experiment design, troubleshooting connections between bits of equipment from different vendors, educating users, keeping a steady hand on the sample queue, and maintaining a viable budget. For a core facility in one of the greatest cities on earth, new employees may be drawn from a large pool of candidates, but the developing world cannot count on a surplus of newly minted Ph.D.s. Retaining personnel, as well, is a substantial challenge. In the developing world, graduate students are often advised that continued training in Europe or the United States will open career paths that would otherwise be closed to them.
Education and training are underappreciated aspects of every core facility. First, the employees of each core laboratory require training with each instrument within their responsibilities. It might not seem remarkable to fly a technician from the University of Washington down to San Jose, California, for training at an instrument vendor facility. Such a trip, however, may well be the first to the United States for a new technician in a developing world core, posing a much higher financial and cultural barrier. To keep current on methods for these fast-moving fields is much more challenging when the conferences at which these ideas are discussed are half a world away. Discussions with a peer core facility may also be more challenging if no comparable laboratory exists in the country. In the end, the same training that produces an excellent staff scientist in a core laboratory also increases his or her ability to move to a country of greater comforts.
The education role must also extend outwards. Potential users of a biological MS facility may never have considered proteomics or metabolomics as an option for their research. Some are unaware of what it can offer, but others display skepticism that newer methods aren’t necessarily better methods. New users will require training so that the samples they deliver to the core are viable for analysis; contaminating reagents are likely for people new to MS, and contaminant proteins may be visible under MS that were invisible before. The encouragement of reasonable expectations of cost, time, and interpretation for core laboratory users is a never-ending challenge.
In South Africa, the educational mission also acquires the name of “capacity development.”7 A large proportion of the population was blocked from opportunities in higher education before the first democratic elections in 1994. The graduate students interacting with core facilities may be the first people in their families to acquire a bachelor of science degree, let alone pursue postgraduate studies. At a concrete level, they may lack reliable transportation to the university (missing meetings during taxi strikes or train-service interruptions). On a more abstract level, many students need additional biostatistical training to avoid leaping to unsubstantiated conclusions from their data. A core facility in the developing world must partner with professors in the training of the new generation of capable scientists.
The unique environment of the developing world
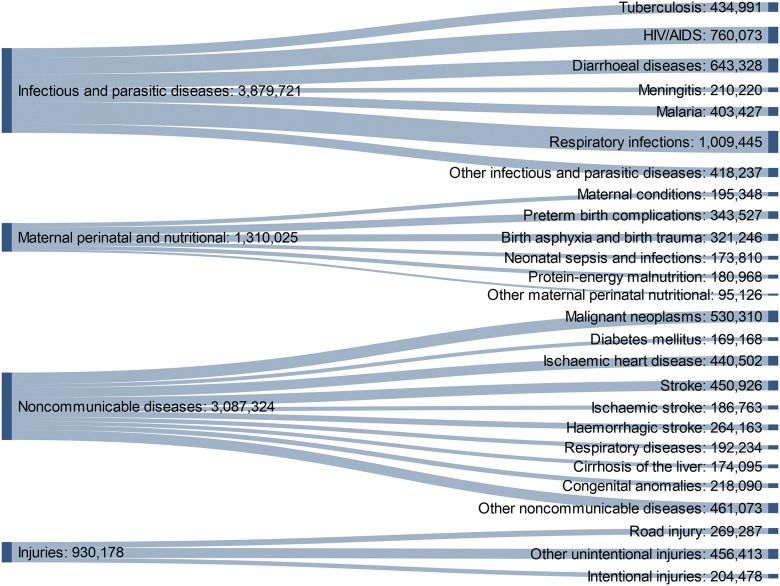
Lord Tennyson once quoted a Quaker farmer as saying, “Doänt thou marry for munny, but goä wheer munny is!” The developing world is where researchers can potentially make the greatest impact, especially for infectious disease. For Africa, the need for tuberculosis (TB), HIV, and malaria research is evident (Fig. 1). The nation is a breeding ground for drug-resistant strains of TB that kill approximately one-half of infected people within 2 yr of diagnosis.8 South Africa hosts one of the largest deployments of anti-retroviral drugs in the world to fight the spread of HIV, with ∼1 in 4 people, aged 25–49, infected with the virus.9 As living conditions improve for the most impoverished sectors of the population, however, the nation is experiencing a rise in noncommunicable diseases, such as diabetes and cardiovascular ailments.10 Health research can translate into public health gains more rapidly in the developing world.
FIGURE 1.
World Health Organization Global Health Estimates of 2015 death statistics for the World Health Organization Africa region. The developing world (here modeled by 47 of the 54 countries on the African continent) is particularly prone to infectious and parasitic diseases. In recent years, however, economic development has caused noncommunicable diseases to increase in these populations.
The developing world faces challenges that are often all but invisible to the developed world, for example, endemic infectious diseases of humans, plants, and animals, which can reach epidemic proportions and impact quality of life quite severely in these regions. These challenges comprise research “hot spots,” as there is intense local pressure to manage these diseases effectively, often with international funding being made available to do so. Access to an MS core facility with high-throughput capacity can open lines of inquiry that would otherwise remain shut. Consequently, researchers in the developing world tend to be enthusiastic about pursuing MS-based research.
In the past, researchers in nations, such as South Africa, were compelled to ship samples to more developed nations to acquire genomic or proteomic data. Core facilities created in the developing world must woo these potential users to use local services, as every sample shipped abroad represents a lost sale to the local core laboratory (and loss of publication credit incentives). Researchers unfamiliar with the developing world may be surprised to hear the term “decolonize” used to encourage use of local laboratories rather than first-world service facilities.11 Researchers in South Africa and other former colonies seek to harvest information from their sample collections themselves rather than function as a conduit for wealthier nations to acquire samples.
The efficiency of regional and national governance has clear impacts on core laboratory operations. During the 2008 “load-shedding” crisis in South Africa,12 rolling blackouts were commonplace as the national electricity provider struggled to meet energy demands. Local core facilities were supported by a backup power generator to maintain the vacuum of instruments, but there was always the looming worry that it would fail. Computational equipment, of course, will routinely require uninterruptible power supplies in environments with questionable power continuity.
Whereas we think of universities as institutions that produce qualified graduates to fuel the economy, it is also true that universities serve as the training ground for civic change. In South Africa, the “fees must fall” movement, starting in 2015, saw students at most campuses engage in mass protest against increases in tuition costs, with some amount of violence taking place on the fringes. As a result, research at some universities was halted for months at a time; core laboratory personnel were unable to access their instruments for long intervals, leading to costly breakdowns. These laboratories occupied buildings that university administrators were most determined to protect through complete lockouts.
Biological MS began in South Africa just before the turn of the century, when Wolf Brandt at the University of Cape Town acquired an AB Sciex Voyager-DE Biospectrometry matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF). A 2008 review of proteomics in South Africa13 details the early growth of this field as the Council for Scientific and Industrial Research in Pretoria established its own proteomics laboratory, and the University of the Western Cape (Cape Town) established a facility in collaboration with the Agricultural Research Council. In this article, we hope to update the growth of biological MS core facilities throughout South Africa as we illustrate this technology in the developing world.
MATERIALS AND METHODS
Acquiring instruments
In the United States, a researcher might contact a sales representative from a vendor for a quote on a mass spectrometer and then write an S10 Shared Instrument Grant to NIH (PAR-17-074) for funding. Part of that application would detail “Institutional Commitment,” confirming that the university was ready to house the instrument and pay for its maintenance over the next 5 yr. This process is similar to that of the “NEP,” or National Equipment Program of the South African National Research Foundation. The National Research Foundation hosts a database of equipment purchased through the National Equipment Program at http://eqdb.nrf.ac.za/.
There are some distinctive features of this process for the developing world, though. First, mass spectrometer manufacturers do not do business directly in every nation of the world. In South Africa, one would work with Anatech Instruments, a distributor for Thermo Scientific, or with Separations (Sciex, Concord, Ontario, Canada), Microsep (Waters, Milford, MA, USA), or Chemetrix (Agilent Technologies, Santa Clara, CA, USA). As a result, much of the subsequent instrument service is performed by a representative of the local company rather than the manufacturer. On the other hand, Bruker and Shimadzu operate directly in South Africa, potentially giving them an advantage for pricing and warranty service.
Ideally, a new core facility might be created by performing a needs assessment for a faculty: the hiring of a director who designs the set of most needed services and then purchases the equipment necessary for those services. In practice, the decision to establish a shared-use facility may take place only after equipment has been purchased by a successful grant application. Likewise, the considerations that lead to the purchase of an instrument may drift from the question of “What is needed?” For example, if current staff is accustomed to using a Bruker instrument, the staff may favor Bruker in the purchase of additional instruments. Instrument distributors sometimes offer deep discounts on additional instruments (particularly older models) that may not have a clear role in performing the services offered by a facility, with the result that they add to maintenance costs without adding capabilities.
Certainly, one can expect that being “first” is a powerful motivation for university administrators. The purchase of the first LTQ Orbitrap Velos and Orbitrap Fusion instruments in South Africa by Stellenbosch University continues to appear prominently on the Central Analytical Facilities: MS website, >2 yr after the LTQ Orbitrap Velos went out of service. Other instruments, such as a Sciex MALDI-TOF/TOF and Agilent Q-TOF at the University of Cape Town, were once highly sought after but now languish in disuse. Core laboratories in all countries must decide whether or not a service that puts an aging instrument to work is worth the cost of maintaining it.
Designing experiments
Each biological MS experiment will offer some combination of separation and measurement [e.g., gas chromatography (GC) × GC-TOF], ideally conducted under a defined Standard Operating Procedure (SOP). Each combination will be capable of a particular degree of sensitivity and reproducibility, and it will have some degree of bias toward particular analytes. Particularly in metabolomics, the characteristics of compounds are so diverse that the separation should be tailored to a particular class of analytes (such as phospholipids, rather than all small molecules). Likewise, proteomics experiments that emphasize membrane proteins would likely run afoul of solubility problems if a separation dependent on gels were used. Many techniques to improve MS sensitivity increase the amount of time required to process each sample and thus the cost to end-users. An initial customer engagement meeting is thus essential to strike the proper balance.
Customer engagement meetings have considerable ground to cover. The request of service users to be explicit about their hypotheses and anticipated effect sizes is a valuable exercise that can forestall the execution of an underpowered experiment with an indeterminate result. “Wet” topics, such as sample collection, handling, storage, and final concentrations, are also essential to these conversations. When all of the required services have been detailed, a reliable cost estimate will enable informed decisionmaking by the user. Cost is generally a larger concern in the developing world than in developed economies. Instruments and reagents are generally no less expensive, but because salaries are generally lower, the fees for biotechnology may be a disproportionately large part of any grant budget (which will also be smaller if only local funding is available). These meetings will be most effective if they are carried out in person, one of the chief advantages of a core laboratory being in the same institution as the end-user. The correct management of end-user expectations is one of the most important facets of running a core facility, not least because satisfied end-users are much more likely to repeat the use of a service and recommend it to others.
It should be clear that biological MS experiments rely very strongly on biostatistical and bioinformatic methods. Every core facility needs personnel who are equipped with these skills. Unfortunately, the developing world is chronically short of these highly mobile researchers, in part, because quantitatively trained personnel have more lucrative opportunities. Helpfully, data are also highly mobile, and collaborations with bioinformatics or biostatistics teams on foreign shores can be mutually beneficial.
Standardizing methods and offering key workflows
The standardization of workflows, for example, via SOPs, is essential for every biological MS core facility. Standardization promotes economies of scale, cost efficiency, and good turnaround times. It also paves the way for future meta-analyses and comparison across different studies. To redesign the entire analytical pipeline for each project is infeasible. When the targets of interest are not known a priori, however, it is difficult to acquire reference standards that may be generalized across studies. This is particularly troublesome in metabolomics, owing to the diversity of target compounds, analytical methods, and data management strategies.14 Nevertheless, the last few years have seen an increasing drive toward standardized methodologies for metabolomics analysis.15, 16 Because of the rapid pace of change, core personnel must remain connected to the international community to stay abreast of international developments.
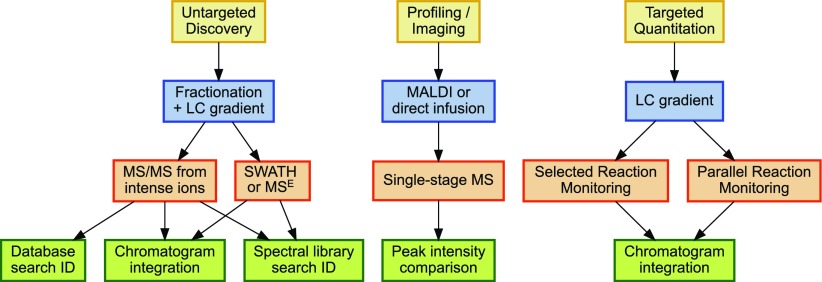
Ideally, each biological MS facility should offer a basket of untargeted discovery methods, profiling techniques, and targeted quantitation capabilities17 (Fig. 2). Of the 3, untargeted experiments generally require the most expensive instruments, generate the largest volumes of data, and require the greatest expertise for interpretation. Their goal is to reveal the identities of as many compounds in a complex mixture as possible. For metabolomics, one might combine data from multiple platforms for this purpose, such as GC × GC-TOF complementing data from a nuclear magnetic resonance instrument. In proteomics, untargeted methods are typified by a “shotgun” experiment using a high-resolution instrument, such as a Thermo OrbiTrap, Thermo Q-Exactive, or SciexTripleTOF, to produce MS/MS from intense ions in MS. These experiments may span many fractions for each sample. Typically, untargeted experiments of high sensitivity have been most challenging for developing countries; core facilities are most needed to enable this type of experiment in these nations.
FIGURE 2.
Biological MS core facilities may specialize in a single workflow or into a greater diversity of services. Untargeted experiments typically consume the most instrument time, but they offer both identification and limited quantitation. Profiling experiments offer extremely high sample throughput, but the differential peaks they discern may subsequently be challenging to identify. Targeted quantitation can yield low coefficients of variation for dozens of prespecified peptides or metabolites.
Where untargeted experiments provide identities for thousands of analytes in a complex mixture, profiling experiments are intended to evaluate which analytes of the thousands are changing among large cohorts. An untargeted experiment spanning 100 samples might easily consume months of instrument time,18 but profiling methods typically require much less instrument time per sample. In proteomic MALDI-TOF profiling, for example, each sample may comprise 1 or more wells on a 96-well MALDI plate, and spectral acquisition is likely to last <1 h. A statistical model would determine which mass-to-charge ratio values are most differential between cohorts, and only then would the user move forward with another experiment to determine the identities of the differential peaks. A similar workflow “profiles” an image of 2-dimensional gels to determine which spots should be subjected to MS/MS for identification. Again, in metabolomics, NMR is likely to complement MS for profiling, although at reduced sensitivity.19 These methods are also applicable to diagnostic applications and within the field of metabolomics, are widely used in the diagnosis of inherited metabolic diseases. In instances where cost or the availability of expertise limits the widespread implementation of profiling methods in diagnostic laboratories, the MS core laboratory in a developing country will have an obligation to make such methods available to diagnostic users.
In a targeted analysis, a particular compound or collection of compounds is analyzed. This approach is usually preferred when very high sensitivity and reproducibility are required, but over the last few years, it has been expanded to include tens to hundreds of compounds in single runs. When compared with an untargeted analysis, this approach is obviously limited to the compounds that are targeted but has the advantage of greatly simplified data analysis and often improved quantification.20 For proteomics, selected reaction monitoring via triple-quadrupole mass spectrometers has represented the standard for targeted quantitation,21 although recent years have seen parallel reaction monitoring and Sequential Windowed Acquisition of All Theoretical Fragment Ion Mass Spectra (SWATH, SCIEX) by high-resolution instruments gaining traction in this space.22, 23 The ability to quantify reproducibly a panel of 100 compounds across hundreds of samples has clear advantages in resource-constrained settings.
Analyzing data and reporting results
Ideally, a facility user drops off a sample that is ready for handling under a clear SOP, the instrument dutifully generates raw data as expected, and those files are analyzed in a turnkey bioinformatics toolkit that outputs a spreadsheet that is easy for end-users to comprehend while producing a standards-compliant package of data that have been annotated fully with metadata for upload to a public repository. The reality, of course, is far from this dream.
For proteomics, one of the most essential resources is the FASTA protein database for the species being analyzed.24 If the user is investigating a metaproteome, such as a digestive tract microbiome, the sequence database must represent an unknown number of species, many of which are likely to be under-represented in the widely used UniProt database. If the user is working in nonmodel organisms, the location and acquisition of a complete proteome database derived from an unpublished genome or from RNA sequencing may require significant detective work. These tasks frequently must be delegated to the facility user, as core laboratory personnel lack the field-specific knowledge to find the most appropriate sequence database.
The amount of information required for publishing a proteome has grown since the 2007 “Paris” Publication Guidelines for Molecular and Cellular Proteomics.25 Core laboratories must support users in publishing their core experiments. For quantitative experiments, cores should be ready to provide calibration curve data to substantiate the values produced in experiments. The publication of post-translational modification data may require the visualization of each annotated MS/MS and/or estimation of localization error. As a result, core facilities need to keep abreast of the requirements for target journals.
The Metabolomics Standards Initiative and the Human Proteomics Organization (HUPO)-Proteomics Standards Initiative have proposed a number of file formats to facilitate interoperability for software tools and establish minimal requirements for chemical analysis, data processing, and metadata reporting.26 Some of these efforts, such as the mzML format for communicating raw MS data,27 are very widely used. Others, such as traML28 or mzTab,29 have been more slowly adopted.30 As the field of metabolome informatics matures, it seems likely that standard formats will become more widely used. At present, the nonstandard reporting implies that the output from 1 tool can be hard to interpret to users of another.
Core laboratories must always evaluate downstream analysis requirements when deciding whether to accept a project. Researchers seeking to determine changes between 2 cohorts of yeast grown under different conditions place a smaller burden of effort on core personnel than those who want to investigate post-translational modifications of a type different than the core has seen before. As mentioned above, users that require customized sequence databases may discover roadblocks before analysis can be completed. Projects that integrate among multiple “omics” disciplines (such as proteogenomics18) can produce high-profile publications, but they frequently require dedicated bioinformatics support, as well. As much as a core facility might favor cutting-edge research, core personnel always need to budget the time they have available.
Finally, the publication of data to public repositories has become a standard publication requirement for biological MS. This process, however, can pose something of a bottleneck for researchers in the developing world. South Africa, for example, has benefited from extensive networking among its many university campuses. Network traffic to the National Center for Biotechnology Information, however, generally passes through undersea optical cables, first to Europe and then to the United States, sharing bandwidth with all other network traffic. The ProteomeXchange Network now has mirrors in the United Kingdom, United States, China, and Japan, but no country in Africa, Oceania, or South America has established a mirror.31 The uploading of even 40 Thermo RAW files (perhaps 1 gigabyte each) can easily be an overnight proposition. With relatively few researchers in the developing world having experience with these repositories, the numbers of data sets of the developing world in these repositories are likely to continue lagging behind the contributions of developed economies.
RESULTS
Biomedical MS has grown in scope throughout South Africa during the last decade (see Table 1 for a current listing of laboratories). As one might expect, the most common type of facility is an individual laboratory with a single instrument that other laboratories at the same institution occasionally use. Other sites, such as the Health Sciences Biological MS Core Facility at the University of Cape Town, feature the powerful trio of quadrupole TOF, triple-quadrupole, and Orbitrap-class instruments. Whereas some research/core laboratories in Europe or North America feature more than 1 dozen instruments, current installations on this scale in South Africa are designed for routine clinical testing and are not available to other biomedical researchers.
TABLE 1.
A current list of biological MS shared instrument laboratories in South Africa
| Institution | Unit | GPS | Proteomics | Metabolomics | URL |
|---|---|---|---|---|---|
| Africa Health Research Institute | Kwa-Zulu Natal Research Institute for TB and HIV | −29.88236,30.988368 | X | X | https://www.ahri.org/research/ |
| Centre for Proteomic and Genomic Research | Proteomics | −33.9415045,18.4645214 | X | X | http://www.cpgr.org.za/services/proteomics/ |
| Council for Scientific and Industrial Research | Biosciences Proteomics Facility | −25.7471931,28.2774823 | X | http://www.acgt.co.za/facilities/proteomics-facility-at-the-csir | |
| North-West University | Technology Innovation Agency Centre for Human Metabolomics | −26.687766,27.0917808 | X | http://natural-sciences.nwu.ac.za/human-metabolomics | |
| Rhodes University | Nanotechnology Innovation Centre, Department of Chemistry | −33.3132727,26.5154525 | X | X | https://www.ru.ac.za/nanotechnology/equipment/ |
| Stellenbosch University | Central Analytical Facilities: Mass Spectrometry | −33.9328078,18.8622583 | X | https://www.sun.ac.za/english/faculty/science/CAF/units/mass-spectrometry | |
| Stellenbosch University-Faculty of Medicine and Health Sciences | Central Analytical Facilities: Proteomics Laboratory | −33.9126528,18.6092176 | X | https://www.sun.ac.za/english/faculty/science/CAF/units/mass-spectrometry | |
| Tshwane University of Technology | Pharmaceutical Sciences | −25.7399957,28.1502218 | X | http://www.alvaroviljoen.com/ | |
| University of Cape Town | Proteomic/Metabolomics Service | −33.957652,18.4590104 | X | X | http://www.mcb.uct.ac.za/mcb/services/msservice |
| University of Cape Town-Faculty of Health Sciences | Health Sciences Biological Mass Spectrometry Core Facility | −33.9425201,18.4655487 | X | X | http://www.chemsysbio.uct.ac.za/blackburn-research-facilities |
| University of Fort Hare | Central Analytical Laboratory | −32.7854241,26.8445361 | X | http://nrfcoms.info/nrfcommunique/triple-tof-mass-spectrometer-for-eastern-cape-researchers/ | |
| University of the Free State | Facility for Genomics and Proteomics | −29.1130299,26.1866028 | X | https://www.ufs.ac.za/research/advanced-biomolecular-research-home/focus-areas/high-throughput-biology | |
| University of Johannesburg | Department of Biochemistry: Plant Metabolomics Facility | −26.1828752,28.0020113 | X | https://www.uj.ac.za/faculties/science/biochemistry/Pages/Molecular-Plant-Microbe-Interactions.aspx | |
| University of KwaZulu-Natal | Catalysis and Peptide Research Unit | −29.817627,30.9391927 | X | X | http://cpru.ukzn.ac.za/ |
| University of Pretoria | Department of Chemistry Separation Sciences Facility | −25.753206,28.2300502 | X | http://www.up.ac.za/en/chemistry/article/2500872/seperation-sciences-facility | |
| University of South Africa | Science Campus, Johannesburg | −26.1584634,27.9038024 | X | X | http://www.unisa.ac.za/sites/corporate/default/Colleges/Science,-Engineering-&-Technology/Research/Nanotechnology-and-Water-Sustainability |
| University of the Western Cape | Agricultural Research Council: Proteomics Unit | −33.9325006,18.6248702 | X | https://www.uwc.ac.za/Faculties/NS/ProteomicsUnit/ | |
| University of Witswatersrand | Mass Spectrometry Service | −26.1921296,28.029106 | X | https://www.wits.ac.za/science/schools/chemistry/research/analytical-services/mass-spectrometry-service/ |
In 20 yr, the use of this technology has spread from a single laboratory to nearly 20 laboratories. GPS, Global Positioning System; URL, Uniform Resource Locator.
Operating under resource constraints
The funding available to create a laboratory is essential context to early decisionmaking. For example, does the university in question qualify for privileged status in grant applications (e.g., South African status of “previously disadvantaged institution”32)? Has the university pledged matching funds for participation in the National Research Foundation National Equipment Program? The funding climate at both national and international levels matters, as well. United Nations Educational, Scientific and Cultural Organization estimates that the United States contributes 28.1% of the global gross expenditure on research and development (https://en.unesco.org/node/252279; United Nations Educational, Scientific and Cultural Organization Science Report: Toward 2030), but it falls behind Switzerland, Singapore, and Sweden for per-capita contribution. In contrast, South Africa has recently released its first Research Infrastructure Roadmap (http://www.hsrc.ac.za/uploads/pageContent/7451/SARIR_2016.pdf) with a total budget of ∼$150 million across 5 y and 13 initiatives (<1% of global gross expenditure on research and development). One interpretation of this disparity is that national grants require complementation with institutional, corporate, or government contributions to establish new facilities. For example, the University of the Western Cape facility was created in partnership between the university and the Agricultural Research Council (similar to the U.S. Department of Agriculture, Agricultural Research Service). The hurdle of initial acquisition costs can be surmounted with good partnerships and a liberal dose of luck.
The acquisition of instruments is no guarantee that a facility will flourish or that future funding will follow the initial investment. Every facility should find mechanisms by which it can approach a full cost-recovery model over time, for example, by accepting samples from external laboratories at a higher price. Resource-limited countries have the advantage that their cost of labor and other local input costs are typically much lower than in developed nations. Thus, when one looks at budgets for a core facility in a resource-limited setting, the typical salary, overhead/space, electricity, and other input-costs portion will be much lower; however, the infrastructure, reagents, and consumables costs will consume a larger proportion of the budget. Sourcing reagents such as sequencing-grade trypsin from domestic suppliers can reduce the effects of currency-exchange fluctuations.
Some of the easiest means to obtain greater efficiencies with a core budget include the following:
Form partnerships with other facilities that have similar needs. This has a host of benefits, including minimizing downtime if one is waiting for a replacement part or reagent (this can often take 3–6 wk); using shared expertise to resolve experimental issues; collective bargaining when it comes to dealing with suppliers; knowledge exchange; and benchmarking exercises to ensure continuous improvement.
Form good relationships with suppliers. A supplier wants its facility consumers to be successful, as that means that the supplier will ultimately get more business. Communication between supplier and facility makes room for a budget that can minimize fluctuations in import costs.
Aspire to high standards. A core facility will live or die by its reputation. Facilities that seek continuous improvement are more likely to increase their portfolios of active projects, improving the economies of scale that drive down overall costs. It may seem costly to pursue International Organization for Standardization 9001 certification, Good Laboratory Practice certification, and other standards, but potential users are swift to avoid further dealings with facilities if outputs are untrustworthy.
Tailor product offerings to client needs. Core facilities cannot waste the attention of personnel by offering “niche” services that are prohibitively costly, require too much time of core personnel, or are uninteresting to most potential users.
Fielding operating costs
Each mass spectrometer can be evaluated as having 2 distinct lifespans; the first relates to the duration during which the instrument is operable, and the second captures the duration before the instrument capabilities become obsolete. Regular maintenance and servicing are essential to extending the operable lifespan of core instruments. As mentioned in instrument acquisition, core laboratories in the developing world will frequently interact with a local distributor rather than the instrument manufacturer. Qualified service engineers are relatively scarce in Sub-Saharan Africa. As a consequence, a single service engineer may cover a region that spans several nations! In our experience, the promptness of service engineer visits claimed in contracts is rather optimistic; after a service request, the delay before an engineer visit may be >1 mo for some vendors. The option to contract with 1 service provider for instruments from several manufacturers does not exist in the developing world. The replacement of broken parts is also problematic, as parts generally must be shipped from the manufacturer (we estimate this delay at 6 wk); the company providing regional service does not generally keep a cache of spares. Without a service contract, each engineer visit is likely to cost ∼$2000, to say nothing of spare-part cost. This heavy burden may be greater than the means of a smaller lab with fewer clients.
The acquisition of the reagents and consumables needed for daily instrument operation requires considerably more forethought in the developing world than in the United States or Europe. Research-intensive universities in the developed world sometimes establish a departmental storeroom from which frequently used items can be acquired on a same-day basis; universities in the developing world do not generally operate at the same scale, and each lab must handle its purchasing separately. When purchasing supplies from a domestic vendor (such as Inqaba, Lasec, or Separations for South Africa), a lab researcher can expect a 3–5 d turnaround time from purchase order to delivery. Many products, however, are not available from domestic sources. If an international supplier does not have a product warehoused inside of the country, one may easily face a delay of 1 mo or more for delivery. In addition, the purchasing of some products from international companies can be hampered by the necessity of import permits, particularly in animal-derived products, such as fetal bovine serum. Some institutions are obligated to purchase reagents or consumables from companies that have been awarded monopolies by the ruling family or party. Finally, the local currency for a developing nation may drift considerably in value against the U.S. dollar, Euro, or British pound. If local political events cause a ratings downgrade for the local currency, then the budget for a grant written before the disruption will bear little relationship to the prices expected at the time the research is performed.
Educating potential facility users
Mass spectrometers in MS core facilities are exposed to samples from a variety of biological sources and from users with a range of experience, from complete novice to expert level. Although all relevant quality control checks may be in place, some dubious samples do inevitably make their way onto the instruments, which can result in LC blockages and downtime, potentially even necessitating service. A core laboratory must therefore prioritize training of new MS users, whether they are graduate students or full professors. If possible, designating an area of the core laboratory for users to complete a sample preparation using the same standard solvents and consumables can eliminate potential sources of contamination. Local experience with clinical samples indicates that most plasticizer or detergent contamination originates during sample acquisition or cell culture. Once a user has been trained, he or she should understand why these contaminants are incompatible with MS analysis.
Each of the proteomics laboratories represented among the authors has standardized the use of a bioinformatics toolkit (such as MaxQuant/Perseus,33 Skyline,3 Protein Metrics ByOnic,34 Nonlinear Dynamics Progenesis QI,35 or Sciex ProteinPilot36) to protect against bioinformatics-induced variability in results. Staff is prepared to help users to learn how to interpret results from these toolkits, and in the case of free software, the staff can help users to reanalyze their data. Frequently, this training takes place in well-established reference data sets. The time investment in training users can reduce the amount of effort staff must contribute at the time of experiment publication, and future users from the same laboratories are likely to have a more nuanced expectation of the results that will come from their experiments.
DISCUSSION
Envisioning a facility scorecard
The evaluation of core laboratory performance extends well beyond whether it was able to “break even” financially. As a start, every laboratory should pursue user feedback to monitor satisfaction with turnaround time, clarity of communication, use of data, and willingness to return for future projects or to recommend the service to others. Funding agencies may specify particular metrics, as well. For example, an instrument for which 90% of run time serves the needs of a single laboratory is unlikely to qualify for shared instrument funding. Whereas each facility should use internal evaluation methods, valuable insights can also be gathered through external audits.
Many laboratories throughout the developed world have benefited from interlaboratory studies driven by the Association for Biomolecular Resource Facilities,37 the Human Proteomics Organization Test Sample Working Group,38 or ProteoRed.39 Initiatives, such as the South African Department of Science and Technology DIPLOMICS program (https://www.cpgr.org.za/cpgr-to-pilot-diplomics-infrastructure-program/), are beginning to organize similar efforts among laboratories in the developing world. Strategies, such as framing standard protocols for trypsin digestion among laboratories, can reduce the differences one would find in repeating an experiment among multiple laboratories. Interlaboratory studies in which a common sample is distributed to different sites for similar processing will help to reveal strengths and weaknesses throughout the network. This article reflects another key goal of DIPLOMICS: raising the visibility of core laboratories in South Africa so that researchers know that they have local options for interrogating their samples.
As mentioned previously (see Operating under resource constraints), some core laboratories will pursue International Organization for Standardization or Good Laboratory Practice certification. These programs require the establishment of quality monitoring systems within core laboratories. These efforts may be aided through the Association of Biomolecular Resource Facilities Workflow Interest Network and by the HUPO-Proteomics Standards Initiative Quality Control Working Group. Quality monitoring can benefit from the use of well-characterized standard samples, such as those from National Institute of Standards and Technology, particularly in support of the National Cancer Institute Clinical Proteomic Tumor Analysis Consortium systems suitability studies,40 or from the European Pharmacopeia. The endorsement of quality and reproducibility, as well as accuracy and precision, is key to long-lived core laboratory management.
Combating isolation
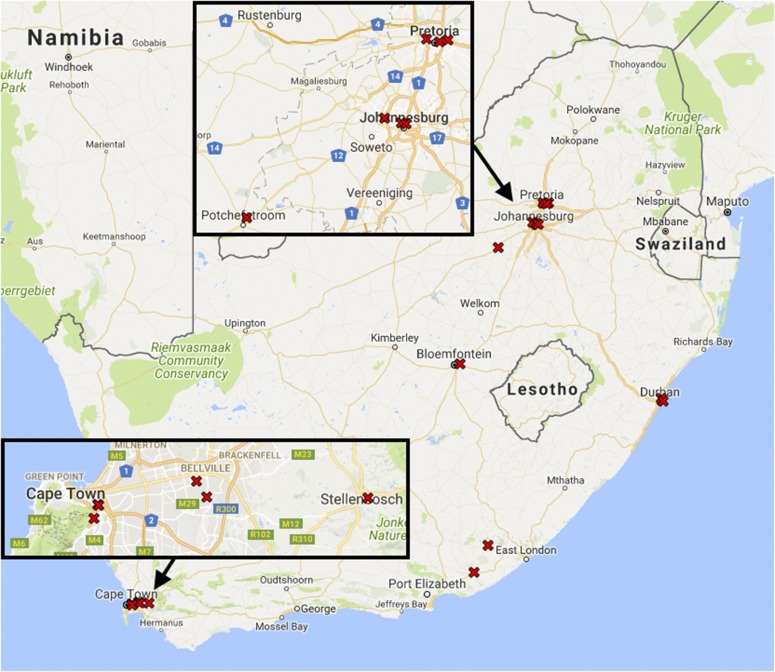
Equitable distribution of resources is a priority for many government-funding agencies in the developing world. One result of this goal is the establishment of core laboratories at universities in less urban areas. A look at Fig. 3 illustrates that existing biological MS facilities are concentrated in Gauteng (Johannesburg and Pretoria) and the Western Cape (Cape Town and Stellenbosch). The South African government has granted funds for the creation of Sol Plaatje University (http://www.spu.ac.za/) in Kimberley, west of Bloemfontein, enrolling its first students in 2014; as research programs mature in the new university, core facilities are likely to follow. New core laboratory personnel are less likely to have peers within the same city, leaving them vulnerable to stagnation without exposure to new ideas in the field of biological MS.
FIGURE 3.
The 3 largest cities in South Africa are Johannesburg, Cape Town, and Durban, with 4.4, 3.7, and 3.4 million inhabitants, respectively, in the corresponding municipalities for the 2011 census. All 3 cities are now home to multiple biological MS facilities.
The development of long-distance links among core laboratories is clearly the answer. This includes collaborative research with influential experts, international presentations, scientific publications, and other marketing and outreach initiatives. The MS core facility in a rural setting will be burdened by additional travel requirements or alternatively, saddled with telephonic or network conferencing. The South African Association for Mass Spectrometry (http://saams.org.za) sponsors an annual Analitika Conference, although its emphasis is currently shaped toward environmental or inorganic analytical chemistry than proteomics and metabolomics. With each Ph.D. granted in biological MS though, South Africa comes closer to the critical mass required to produce, for example, a South African HUPO chapter.
As the network of biological MS grows across South Africa, we may see collaborative relationships forming among core laboratories so that a facility that has instruments to support identification but not quantification may contract for quantification services at another facility. Users need to interact only with their local laboratory but would have the capabilities of both. The creation of a satellite facility is another option; perhaps Nelson Mandela University in Port Elizabeth (https://www.mandela.ac.za/) would staff a local office that counseled users on experimental design and sample preparation, packaged samples for shipment to Rhodes University (http://www.ru.ac.za) in Grahamstown (135 km away), and then explained the resulting data to users at Nelson Mandela University. Both universities in this scenario would benefit from a nationwide organization to support the training of new users.
Conclusions
Ever since its first democratic elections in 1994, South Africa has illustrated many challenges of grafting “first world” technologies into rapidly expanding and evolving universities. The extent of biological MS across the nation is growing at a rate that has surprised even the authors. The developing world is the breeding ground for the most widespread diseases of humanity, and it is here that core laboratories can have the greatest impact.
ACKNOWLEDGMENTS
D.L.T. is funded by the South African Medical Research Council Centre (SAMRC) under the South African Tuberculosis Bioinformatics Initiative. R.H., L.B., S.H.S., and D.L.T. are part of the South African Department of Science and Technology DIPLOMICS program. S.H.S. and D.L.T. are part of Research Collaboration Agreement TP_04_2018 between the Council for Scientific and Industrial Research and Stellenbosch University. The Technology Innovation Agency has been a key proponent of proteomics and metabolomics for South Africa.
DISCLOSURES
The authors declare no conflicts of interest.
REFERENCES
- 1.Cottrell JS. Protein identification by peptide mass fingerprinting. Pept Res 1994;7:115–124. [PubMed] [Google Scholar]
- 2.Peng J, Gygi SP. Proteomics: the move to mixtures. J Mass Spectrom 2001;36:1083–1091. [DOI] [PubMed] [Google Scholar]
- 3.MacLean B, Tomazela DM, Shulman N, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 2010;26:966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith CA, O’Maille G, Want EJ, et al. METLIN: a metabolite mass spectral database. Ther Drug Monit 2005;27:747–751. [DOI] [PubMed] [Google Scholar]
- 5.Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol 2016;34:828–837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ebrahim A, Brunk E, Tan J, et al. Multi-omic data integration enables discovery of hidden biological regularities. Nat Commun 2016;7:13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sewankambo N, Tumwine JK, Tomson G, et al. Enabling dynamic partnerships through joint degrees between low- and high-income countries for capacity development in global health research: experience from the Karolinska Institutet/Makerere University partnership [Erratum in PLoS Med 2015;12:e1001816]. PLoS Med 2015;12:e1001784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pietersen E, Ignatius E, Streicher EM, et al. Long-term outcomes of patients with extensively drug-resistant tuberculosis in South Africa: a cohort study. Lancet 2014;383:1230–1239. [DOI] [PubMed] [Google Scholar]
- 9.Zuma K, Shisana O, Rehle TM, et al. New insights into HIV epidemic in South Africa: key findings from the National HIV Prevalence, Incidence and Behaviour Survey, 2012. Afr J AIDS Res 2016;15:67–75. [DOI] [PubMed] [Google Scholar]
- 10.GBD 2016 Causes of Death Collaborators Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet 2017;390:1151–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fayemi AK, Macaulay-Adeyelure OC. Decolonizing bioethics in Africa. BEOnline 2016;3:68–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Minnaar UJ, Visser W, Crafford J. An economic model for the cost of electricity service interruption in South Africa. Util Policy 2017;48:41–50. [Google Scholar]
- 13.Ndimba BK, Thomas LA. Proteomics in South Africa: current status, challenges and prospects. Biotechnol J 2008;3:1368–1374. [DOI] [PubMed] [Google Scholar]
- 14.Beisken S, Eiden M, Salek RM. Getting the right answers: understanding metabolomics challenges. Expert Rev Mol Diagn 2015;15:97–109. [DOI] [PubMed] [Google Scholar]
- 15.Beckonert O, Keun HC, Ebbels TM, et al. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc 2007;2:2692–2703. [DOI] [PubMed] [Google Scholar]
- 16.Fiehn O. Metabolomics by gas chromatography-mass spectrometry: combined targeted and untargeted profiling. Curr Protoc Mol Biol 2016;114:30.4.1–30.4.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiehn O. Metabolomics--the link between genotypes and phenotypes. Plant Mol Biol 2002;48:155–171. [PubMed] [Google Scholar]
- 18.Zhang B, Wang J, Wang X, et al. Proteogenomic characterization of human colon and rectal cancer. Nature 2014;513:382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shah SH, Kraus WE, Newgard CB. Metabolomic profiling for the identification of novel biomarkers and mechanisms related to common cardiovascular diseases: form and function. Circulation 2012;126:1110–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Z, Yan R. Increase the accessibility and scale of targeted metabolomics: construction of a human urinary metabolome-wide multiple reaction monitoring library using directly-coupled reversed-phase and hydrophilic interaction chromatography. Anal Chim Acta 2015;894:65–75. [DOI] [PubMed] [Google Scholar]
- 21.Addona TA, Abbatiello SE, Schilling B, et al. Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma [Erratum in Nat Biotechnol 2009;27:864]. Nat Biotechnol 2009;27:633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peterson AC, Russell JD, Bailey DJ, Westphall MS, Coon JJ. Parallel reaction monitoring for high resolution and high mass accuracy quantitative, targeted proteomics. Mol Cell Proteomics 2012;11:1475–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gillet LC, Navarro P, Tate S, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics 2012;11:O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jagtap P, Goslinga J, Kooren JA, et al. A two-step database search method improves sensitivity in peptide sequence matches for metaproteomics and proteogenomics studies. Proteomics 2013;13:1352–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deutsch EW, Albar JP, Binz P-A, et al. Development of data representation standards by the human proteome organization proteomics standards initiative. J Am Med Inform Assoc 2015;22:495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sumner LW, Amberg A, Barrett D, et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007;3:211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deutsch E. mzML: a single, unifying data format for mass spectrometer output. Proteomics 2008;8:2776–2777. [DOI] [PubMed] [Google Scholar]
- 28.Deutsch EW, Chambers M, Neumann S, et al. TraML–a standard format for exchange of selected reaction monitoring transition lists. Mol Cell Proteomics 2012;11:R111.015040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griss J, Jones AR, Sachsenberg T, et al. The mzTab data exchange format: communicating mass-spectrometry-based proteomics and metabolomics experimental results to a wider audience. Mol Cell Proteomics 2014;13:2765–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spicer RA, Salek R, Steinbeck C. A decade after the metabolomics standards initiative it’s time for a revision. Sci Data 2017;4:170138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deutsch EW, Csordas A, Sun Z, et al. The ProteomeXchange consortium in 2017: supporting the cultural change in proteomics public data deposition. Nucleic Acids Res 2017;45:D1100–D1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roebken H. The formation and development of co-operations among South African universities. High Educ 2008;56:685–698. [Google Scholar]
- 33.Tyanova S, Temu T, Sinitcyn P, et al. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat Methods 2016;13:731–740. [DOI] [PubMed] [Google Scholar]
- 34.Bern M, Kil YJ, Becker C. Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics 2012;Chapter 13:Unit13.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuharev J, Navarro P, Distler U, Jahn O, Tenzer S. In-depth evaluation of software tools for data-independent acquisition based label-free quantification. Proteomics 2015;15:3140–3151. [DOI] [PubMed] [Google Scholar]
- 36.Shilov IV, Seymour SL, Patel AA, et al. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics 2007;6:1638–1655. [DOI] [PubMed] [Google Scholar]
- 37.Ivanov AR, Colangelo CM, Dufresne CP, et al. Interlaboratory studies and initiatives developing standards for proteomics. Proteomics 2013;13:904–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bell AW, Deutsch EW, Au CE, et al. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics [Comment in Nat Methods 2009;6:411–412]. Nat Methods 2009;6:423–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paradela A, Escuredo P-R, Albar J-P. Geographical focus. Proteomics initiatives in Spain: ProteoRed. Proteomics 2006;6(Suppl 2):73–76. [DOI] [PubMed] [Google Scholar]
- 40.Abbatiello SE, Mani DR, Schilling B, et al. Design, implementation and multisite evaluation of a system suitability protocol for the quantitative assessment of instrument performance in liquid chromatography-multiple reaction monitoring-MS (LC-MRM-MS). Mol Cell Proteomics 2013;12:2623–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]