Abstract
Background
High expression of collagen type X alpha 1 chain (COL10A1), a member of the collagen family, had been observed in various human cancers, but the detailed function and molecular mechanism of COL10A1 were largely unclear.
Aim
The aim of this study was to investigate the expression of COL10A1 in colorectal cancer (CRC) tissues and cells and to reveal its biological function and mechanism in CRC.
Materials and methods
Immunohistochemistry (IHC), real-time quantitative polymerase chain reaction (QPCR) and Western blot experiments were used to determine the clinical relevance between expression levels of COL10A1 and CRC.
Results
Compared with normal tissues, COL10A1 expression was significantly higher in CRC tissues. Biological functional experiments showed that overexpression of COL10A1 enhanced proliferation, migration, and invasion of CRC cells, and knockdown of COL10A1 inhibited tumorigenesis in vivo. Western blot assays showed that COL10A1 promoted the process of epithelial–mesenchymal transition (EMT). The overexpression of COL10A1 was associated with adverse prognosis in CRC by tissue microarray (TMA) analysis.
Conclusion
Our findings had provided evidences to support the fact that COL10A1 was abnormally up-expressed in CRC and involved in the progression of CRC and the process of EMT. Furthermore, we demonstrated that the high-level expression of COL10A1 was an independent risk factor of prognosis and overall survival in CRC patients. These suggested that COL10A1 might be a new potential target for cancer therapy in the future.
Keywords: collagen type X alpha 1 chain, colorectal cancer, proliferation and invasion, epithelial-mesenchymal transition
Introduction
Colorectal cancer (CRC) is one of the most common malignant cancers in the world, with about 1.4 million new cases and more than 690,000 deaths per year.1 Many patients with CRC were diagnosed at an advanced stage, because of a lack of mass screening. Distant metastasis, especially peritoneal and liver metastasis, is the main cause of death in CRC. CRC metastasis directly leads to poor prognosis in patients, and the median survival time was only 5–9 months.2,3 Because of early screening and treatment, decreasing CRC mortality rates have been observed in some countries worldwide.4,5 Therefore, if we can fully apply tumor molecular biological research, and select some novel molecular markers for early screening, the positive predictive value for early diagnosis and the clinical prognosis of CRC will obviously be improved.6,7
In CRC, there are a large number of differentially expressed genes between primary tumors and metastases. Further clarifying the molecular mechanism of CRC distant metastasis is of great significance.8 Therefore, we used gene chip technology to compare the difference of genome-wide expression levels in four pairs of CRC primary tumor tissues and corresponding peritoneal metastasis tissue, and determined the differentially expressed genes through multiple methods, and performed functional annotation cluster analysis for these genes. Ultimately, 217 differentially expressed genes were selected (Figure S1).9 Through bioinformatics analysis, quantitative polymerase chain reaction (QPCR) and immunohistochemistry (IHC) technology verification, we found two genes, CTHRC1 and COL10A1, as the metastasis relevant candidate genes of CRC.9 In the previous studies, we had confirmed that CTHRC1 was abnormally high in CRC and it was an independent risk factor for CRC prognosis.9,10
COL10A1, a protein in humans, is a member of the collagen family. The COL10A1 gene “encodes the alpha chain of type X collagen, a short chain collagen expressed by hypertrophic chondrocytes during endochondral ossification”.11 COL10A1 gene mutation is associated with Japanese type spondylometaphyseal dysplasia and Schmid type metaphyseal chondrodysplasia.12–15 We found that COL10A1 mRNA expression level in CRC was increased through a meta-analysis of about 12 studies of CRC transcriptome sequencing in the Oncomine database (Figure S2). Meanwhile, all available online data on the Prognoscan website showed that the expression of COL10A1 was connected with overall survival (OS) and disease free survival (DFS) (Figure S3). High COL10A1 expression was observed in various solid human tumor tissues, and it was likely to be associated with tumor angiogenesis.16 Additionally, in the early diagnosis of CRC, the concentration of COL10A1 in the peripheral blood could be considered as a potential biomarker.17 However, the relationship between COL10A1 and CRC remains unknown, and the specific role of COL10A1 in cancer is unclear.
Our aim was to further research the expression characteristics of COL10A1, to explore its mechanism to promote proliferation and invasion in CRC, and to preliminarily clarify the clinical interrelation between COL10A1 and CRC.
Materials and methods
Clinical tissue samples and cell lines
Human CRC samples were obtained from the Department of General Surgery at Nanfang Hospital (Guangzhou, China). Tissue samples collected from CRC patients who underwent surgical resection were immediately transported in liquid nitrogen and stored at −80°C for further use. Adjacent normal tissues, as normal control in this study, were at least 5 cm away from the tumor border and ensured that they were free of tumor deposits.18 All tissues were obtained for scientific research. Written informed consent was obtained from all patients included in the study. The study protocol was reviewed and approved by Medical Research Ethics Committees of Nanfang Hospital, Southern Medical University. Five CRC cell lines, HCT116, Caco2, SW480, SW620, and LoVo, and normal colorectal mucosa cells, FHC, were purchased from the American Type Culture Collection (ATCC) Cell Biology Collection. All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) (HyClone, Logan, UT, USA) with 10% fetal bovine serum (FBS; Gibco, Brazil) at 37°C in a 5% CO2 incubator.
Immunohistochemistry, Western blot, and real-time QPCR
The standard two-step IHC technique was performed to qualitatively and quantitatively detect the expression of COL10A1 protein in CRC samples.
Total proteins, extracted from tissue samples and cells by using a RIPA lysis buffer, were separated on SDS-PAGE gels and transferred to polyvinylidene fluoride membranes. The membranes were blocked in 5% skim milk for 1 h and then probed with primary antibodies against COL10A1, E-cadherin, N-cadherin, Beta-catenin, Slug, Snail, and GAPDH (Cell Signaling Technology, CST, Danvers, MA, USA) at 4°C overnight, followed by the secondary rabbit or mouse antibody (CST) for 1 h. Enhanced chemiluminescence (ECL) kit (Millipore, Billerica, MA, USA) was used to visualize the protein bands.
Total RNA, including miRNAs, was extracted using RNAiso Plus reagent (Takara, Japan). To compare the different expression of COL10A1 between tumor and normal tissues, we performed reverse transcription–polymerase chain reaction (RT-PCR) and QPCR, respectively, by using a PrimeScript RT Master Mix Perfect Real Time Kit (Takara) and SYBR Premix Ex Taq II Kit (Takara) according to the manufacturer’s instructions.
Cell transfection experiment
HCT116 and LoVo cells were cultured in 6-well plates. When cells convergence degrees reached about 70% and were in good condition, COL10A1 overexpressed (OE) plasmids (GeneChem, Shanghai, China) mixed with serum free medium were transfected by using Lipofectamine3000 as a transfection reagent, according to the corresponding instruction. The same vectors with an empty sequence were transfected into HCT116 and LoVo cells as a negative control (NC). After 48~72 h transfection, the cells could be used for further study. Small RNA interference (siRNA) technology was used to interfere the expression of COL10A1 in cells. Through the verification of Western blot and QPCR assays, the siRNA (GenePharma) with highest effect was used to design lentivirus expressing COL10A1-siRNA. We transfected the lentivirus into SW480, SW620 cells to establish CRC cell lines with stable knockdown (KD) of COL10A1 (SW480-COL10A1-KD, SW620-COL10A1-KD). SW480-NC and SW620-NC cells were constructed by transfection of negative control siRNA with a random sequence.
Wound healing assay
Cells were seeded into 6-well plates to 80%–85% confluence and scratched with a 200 μL pipette tip to create a straight scratch. After the scratch, images were taken at 0, 24, 36, and 48 h to assess the migration abilities of different treatment cells.
Cell migration and invasion assays
After transfection with the COL10A1-OE plasmids or lentivirus for 48 h, 5×104 cells were seeded in 200 μL serum-free medium, and added to the top chambers in 24-well plates (Corning, Corning, NY, USA); 500 μL 10%–20% FBS were added to the lower chamber to induce the cells migration. Each chamber was coated freshly with 50 μg Matrigel (Becton, Dickinson and Company, Franklin Lakes, NJ, USA) before performing the invasion assay. After 24 h, the cells were fixed by using 4% paraformaldehyde and stained with 0.5% crystal violet. The numbers of migrated and invasive cells were calculated in five random fields per well with an inverted light microscope.
Cell counting kit 8 (CCK-8) assay and clone formation assay
SW480-COL10A1-KD/NC cells were seeded into 96-well plates at a density of 3,000 cells per well and cultured for 24 h in DMEM plus 10% FBS at 37°C and 5% CO2. After 6, 18, 30, 42, 54, and 66 h, 10 μL CCK-8 solution (Donjindo, Kumamoto, Japan) was added to each well, then the plates were incubated for 2 h in the incubator. The absorbance data (OD value) at a wavelength of 450 nm was collected for cell viability analysis. This experiment was repeated three times. SW480-COL10A1-KD/NC cells were seeded and incubated into six-well plates for 2~3 weeks at 37°C. When the clones were visible to the naked eye, the culture was terminated and the cells were carefully washed with phosphate buffered saline twice and stained with 0.5% crystal violet solution. The number of colonies was counted directly by the naked eye or by a microscope (low magnification). Plate clone formation efficiency=(number of colonies/number of cells inoculated)×100%.
Tumor growth assay in vivo
Nude mice aged 4–6 weeks were purchased from the Experimental Animal Center of Southern Medical University. To conduct an animal tumorigenesis assay, SW480-COL10A1-KD cells and the SW480-NC cells in 0.2 mL serum-free medium were subcutaneously injected into the right flank of nude mice (six mice in each group). Six days after injection, the tumor volumes were continuously measured every 4 days until 30 days after injection by the formula: 0.5×length×width2. All nude mice were specific pathogen free and housed in a clean environment. All procedures involving animals and their care in this study were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Southern Medical University Institutional Animal Care and Use Committee.
Tissue microarray (TMA)
The TMA, involving a total of 197 CRC patients who underwent colorectal resections from November 2013 to June 2014 in Nanfang Hospital, Southern Medical University (Guangzhou, China), was applied to analyze the clinical prognostic correlation between COL10A1 expression and CRC patients. The analyses were base on the data of tumor, node, and metastasis stage, tumor size, OS, and so on of patients. IHC staining of COL10A1 was assessed by three observers using the same microscope (OLYMPUS BX51, Olympus Corporation, Tokyo, Japan). We classified the COL10A1 staining into four degrees: negative, weak, moderate, and intense staining. We defined that tissues with negative and weak staining were low COL10A1 expression, while moderate and intense staining were high COL10A1 expression.
Statistical analysis
Data were presented as mean ± SD and were analyzed by SPSS 22.0 and GraphPad Prism 5.0 for windows. Data statistical evaluation was performed by using the t-test. Survival curves were calculated using the Kaplan–Meier method and log-rank test. Cox proportional hazards regression was applied to confirm independent factors with a significant influence on CRC patient survival. Significant differences were considered when P-values<0.05.
Results
COL10A1 is significantly upregulated in CRC tissues compared with normal tissues
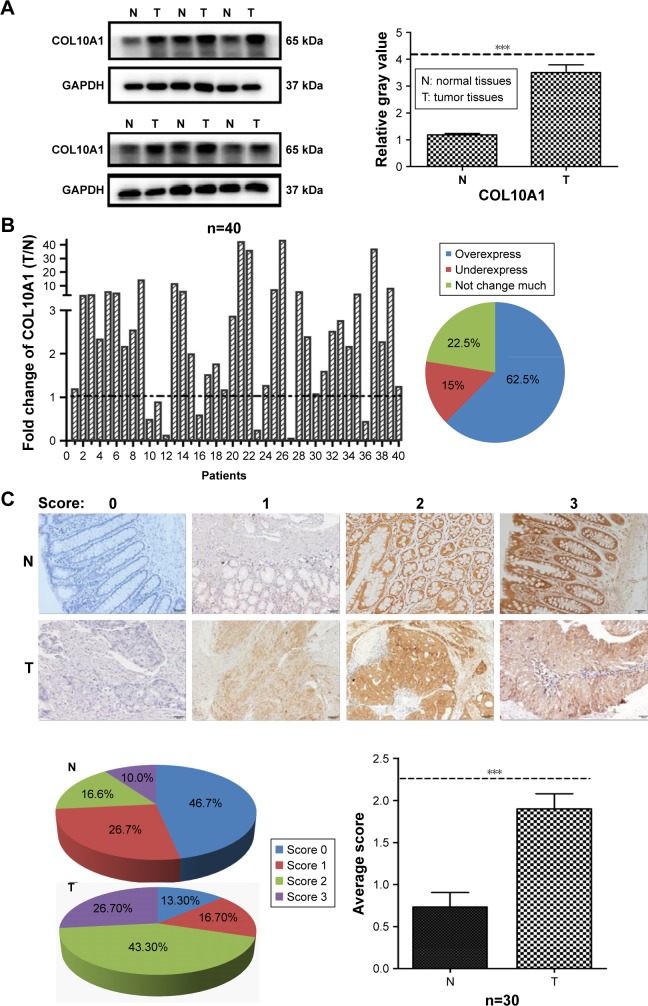
The expression level of COL10A1 was detected in 40 paired CRC tissues and adjacent normal tissues by QPCR and Western blot assays. The results of Western blot analysis showed that COL10A1 was frequently up-regulated (33 of 40, 82.5%) in CRC tissues (T) compared to adjacent normal tissues (N) (P<0.001; Figure 1A). The QPCR results showed that COL10A1 expression was up-regulated in 25 of 40 (62.5%) CRC tissues, compared with corresponding nontumorous tissues (Figure 1B). Moreover, we performed the further verification through IHC assay with 30 CRC patient specimens. We scored them according to the intensity of the staining (negative: 0; weak: 1; moderate: 2; intense: 3). The same result was observed that CRC tissues staining were obviously deeper (higher scores) than the corresponding normal tissues, indicating that COL10A1 was stably high expressed in CRC tissues (P<0.001; Figure 1C).
Figure 1.
COL10A1 is overexpressed in CRC tissues. (A) Expression of COL10A1 in CRC patients was detected by Western blot assays, followed by gray value analysis. (B) COL10A1 expression levels of CRC and corresponding normal tissues detected by QPCR were shown using a histogram and pie chart. The fold change of COL10A1 expression (T/N).2 was defined as overexpressed. (C) IHC staining of COL10A1 protein in 30 pairs of CRC and nontumorous tissues and followed scores pie chart and average score analysis. ***P<0.001.
Abbreviations: COL10A1, collagen type X alpha 1 chain; CRC, colorectal cancer; QPCR, quantitative polymerase chain reaction; T, tumor tissues; N, normal tissues; IHC, immunohistochemistry.
Overexpression of COL10A1 motivates migration and invasion of CRC cell in vitro
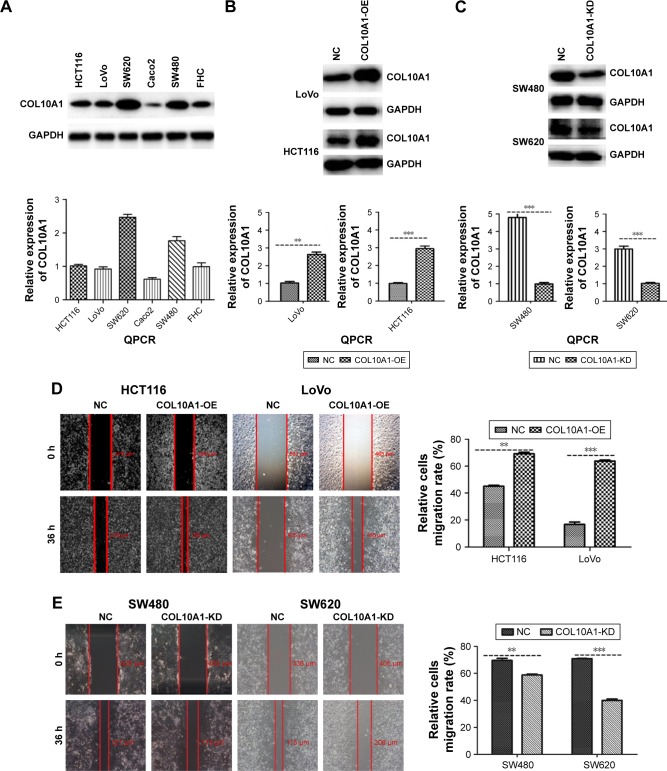
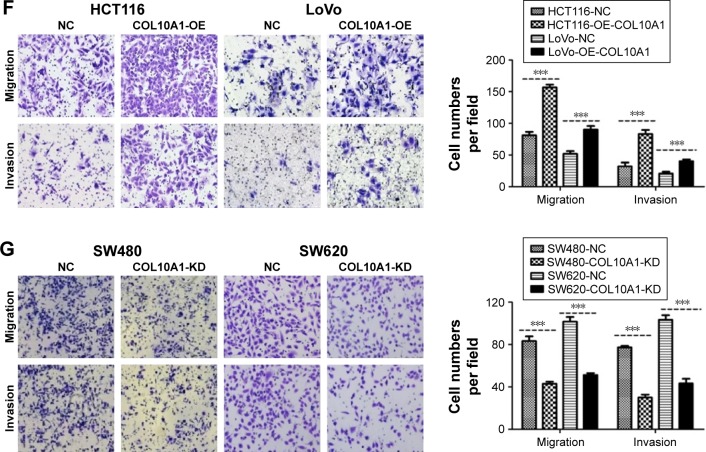
The expression of COL10A1 in six cell lines – HCT116, Caco2, SW480, SW620, LoVo, and FHC – was detected by QPCR and Western blot assays. The results were accordant that COL10A1 was high expressed in SW480, SW620 cells and relatively low expressed in the others cell lines (Figure 2A). Therefore, we chose HCT116 and LoVo cells to structure HCT116-COL10A1-OE and LoVo-COL10A1-OE cell lines, while SW480 and SW620 cells were used to generate SW480-COL10A1-KD and SW620-COL10A1-KD cell lines by transfecting with COL10A1 siRNA lentivirus. The transfection efficiency was detected by QPCR and Western blot assays (all P<0.01; Figure 2B and C). The cell migration rate was measured by wound healing assay. The migration rate and ability was markedly increased in HCT116-COL10A1-OE (P<0.01) and LoVo-COL10A1-OE (P<0.001) cells, and, inversely, reduced in SW480 (P<0.01), SW620 (P<0.001) cells when COL10A1 was knockdown compared with respective NC groups (Figure 2D and E). The same results were observed by Transwell chamber migration assay and invasion assay (all P<0.001, Figure 2F and G). Consequently, it was shown that COL10A1 dramatically increased the ability of migration and invasion ability in CRC cells.
Figure 2.
COL10A1 promotes the migration and invasion of CRC cells in vitro. (A) The expression of COL10A1 in six cell lines was detected by QPCR and Western blot assays. (B, C) Transfection efficiency of LoVo, HCT116, SW480, and SW620 was detected by QPCR and Western blot assays. (D, E) Cell migration was tested using wound healing assay. (F, G) The cells’ migration and invasion changes after transfection were tested by Transwell chamber migration assay and invasion assay. **P<0.01, ***P<0.001.
Abbreviations: COL10A1, collagen type X alpha 1 chain; CRC, colorectal cancer; QPCR, quantitative polymerase chain reaction; NC, normal control.
COL10A1 promotes tumor cells proliferation in vitro
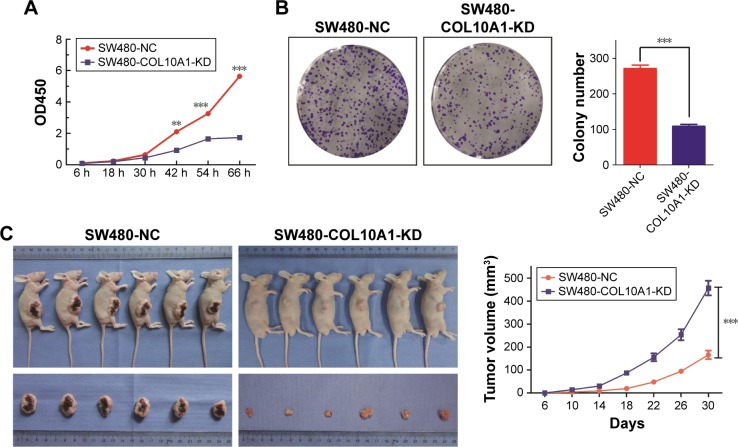
CCK-8 assay and clone formation assay were performed to investigate the effect of COL10A1 on CRC cells proliferation. In CCK-8 assay, the proliferation ability was markedly reduced in SW480-COL10A1-KD cells compared with SW480-NC cells (P<0.01, Figure 3A). Moreover, the number of colonies was significantly reduced in SW480 cells when COL10A1 was knocked down compared with the NC groups (P<0.001, Figure 3B). Therefore, knockdown of COL10A1 inhibited CRC cells proliferation.
Figure 3.
Knockdown of COL10A1 inhibits cells proliferation in vitro and tumorigenesis in vivo. (A, B) Knockdown of COL10A1 repressed SW480 cells proliferation by CCK-8 assay and colony formation assay. (C) Cells were subcutaneously injected into the right flank of nude mice. Tumor volumes were repeatedly measured on the indicated days to evaluate the effects of COL10A1 on tumor growth. Data points are showed as the mean ± SD tumor volume. **P<0.01, ***P<0.001.
Abbreviations: COL10A1, collagen type X alpha 1 chain; CCK-8, Cell counting kit 8; SD, standard deviation; NC, normal control.
COL10A1 promotes tumor cells growth in vivo
For exploring the effect of COL10A1 on CRC cells growth in vivo, the stable transfection cells SW480-COL10A1-KD and SW480-NC cells were injected subcutaneously into the right flank of nude mice. After 30 days’ repeated observation and measure, we found that the tumor growth rate of the SW480-COL10A1-KD group was evidently slower than that of the SW480-NC group (P<0.001, Figure 3C). The results showed that knockdown of COL10A1 repressed the CRC cells growth in vivo.
COL10A1 accelerates EMT in CRC
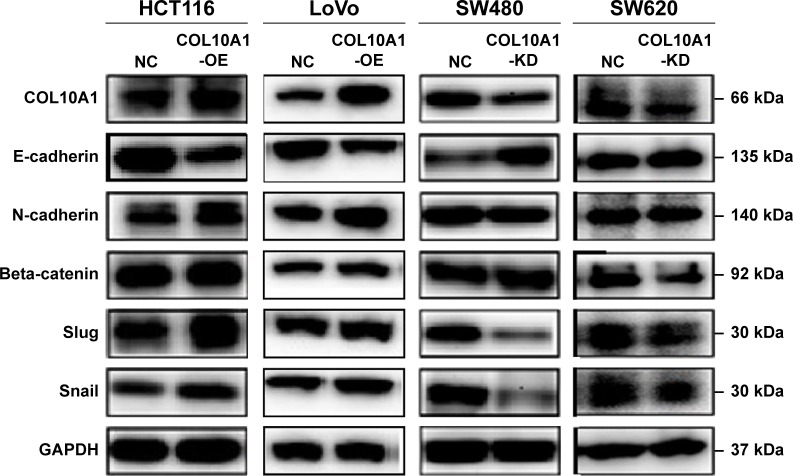
Western blotting was performed to explore the potential relation between COL10A1 and EMT phenomenon. EMT indicators were detected including E-cadherin, N-cadherin, Beta-catenin, Slug, and Snail. Western blot analysis showed that COL10A1 overexpression promoted EMT in LoVo and HCT116 cell lines, and, on the contrary, EMT was suppressed in SW480, SW620 cell lines when COL10A1 was knocked down (Figure 4). Therefore, COL10A1 was a positive regulator for the EMT process.
Figure 4.
COL10A1 promotes the EMT process. The effect of COL10A1 in the EMT was detected by Western blot assay. E-cadherin, N-cadherin, Beta-catenin, Slug, and Snail were detected after successful transfection.
Abbreviations: COL10A1, collagen type X alpha 1 chain; EMT, epithelial–mesenchymal transition; NC, normal control.
COL10A1 overexpression relates to adverse prognosis in CRC
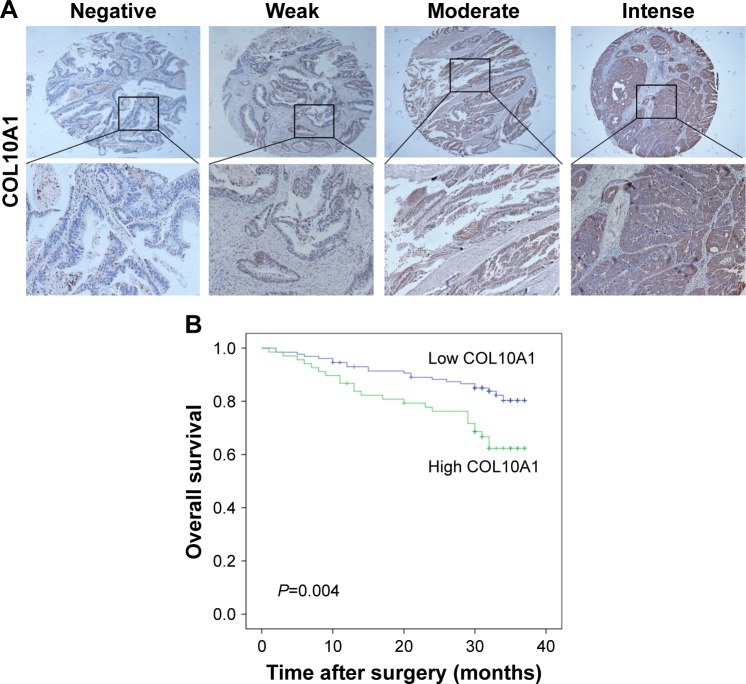
The TMA involving a total of 197 CRC patients with clinical baseline data (Table S1) was used to research the clinical prognostic relevance of COL10A1 expression in CRC by IHC analysis (Figure 5A). The results showed that the expression of COL10A1 in CRC had a high relevance with prognosis and OS. According to the Kaplan–Meier Log-rank test, abnormal upregulated expression of COL10A1 (P=0.004) related to poor OS (Figure 5B). Univariate Cox regression model analysis showed that poor survival was significantly correlated with tumor size (hazard ratio, HR=2.10, 95% confidence interval, 95% CI=1.18–3.76; P=0.012), differentiation (HR=2.22, 95% CI=1.13–4.38; P=0.021), invasion degree (HR=3.20, 95% CI=1.26–8.09; P=0.014), nodal metastasis (HR=3.25, 95% CI=1.77–5.96; P<0.001), M (metastasis) stage (HR=7.20, 95% CI=3.92–13.21; P<0.001), American Joint Committee on Cancer (AJCC) stage (HR=5.13, 95% CI=2.60–10.11; P<0.001), and COL10A1 expression (HR=2.28, 95% CI=1.28–4.06; P=0.005). Multivariate survival analysis was performed based on the results of the univariate survival analysis. Through adjustment, tumor size, differentiation, AJCC stage, and COL10A1 expression were confirmed as covariates in multivariate analyses. Because of the interactions with AJCC stage, colorectal wall invasion degree, nodal metastasis, and M stage were excluded from the multivariate survival analysis. According to the results of multivariate analysis, AJCC stage (HR=4.71, 95% CI=2.36–9.39; P<0.001) and COL10A1 expression (HR=2.04, 95% CI=1.11–3.74; P=0.022) were considered to be independent risk predictors for poor OS (Table 1). The analysis results of clinical samples and TMA showed that COL10A1 was associated with prognosis and survival of patients with CRC.
Figure 5.
COL10A1 overexpression is correlated with adverse prognosis in CRC. (A) Tissue microarray analysis of COL10A1 in human CRC tissues. Original magnification, 100× and 200×. (B) Kaplan–Meier survival analysis of COL10A1 expression in patients with CRC (log-rank test).
Abbreviations: COL10A1, collagen type X alpha 1 chain; CRC, colorectal cancer.
Table 1.
Univariate and multivariate analyses of different prognostic factors in 197 patients with CRC in tissue microarray
| Variables | Cases | Univariate analysis*
|
Multivariate analysis*
|
||
|---|---|---|---|---|---|
| HR (95% CI) | P-value | HR (95% CI) | P-value | ||
| Gender | 1.02 (0.56–1.85) | 0.951 | |||
| Male | 121 | ||||
| Female | 76 | ||||
| Age (years) | 1.19 (0.67–2.13) | 0.554 | |||
| <60 | 104 | ||||
| ≥60 | 93 | ||||
| BMI | 1.09 (0.61–1.95) | 0.764 | |||
| <23 | 108 | ||||
| ≥23 | 89 | ||||
| Tumor size (diameter) | 2.10 (1.18–3.76) | 0.012 | 1.50 (0.81–2.77) | 0.195 | |
| ≤4 cm | 122 | ||||
| >4 cm | 75 | ||||
| Differentiation | 2.22 (1.13–4.38) | 0.021 | 1.38 (0.69–2.74) | 0.365 | |
| Well/moderate | 169 | ||||
| Poor | 28 | ||||
| Invasion degree | 3.20 (1.26–8.09) | 0.014 | |||
| Within the serosa | 49 | ||||
| Breakthrough serosa | 148 | ||||
| Nodal metastasis | 3.25 (1.77–5.96) | <0.001 | |||
| N0 | 119 | ||||
| N1/N2 | 78 | ||||
| M stage | 7.20 (3.92–13.21) | <0.001 | |||
| M0 | 175 | ||||
| M1 | 22 | ||||
| AJCC stage | 5.13 (2.60–10.11) | <0.001 | 4.71 (2.36–9.39) | <0.001 | |
| I/II | 113 | ||||
| III/IV | 84 | ||||
| COL10A1 expression | 2.28 (1.28–4.06) | 0.005 | 2.04 (1.11–3.74) | 0.022 | |
| Decreased expression | 129 | ||||
| High expression | 68 | ||||
Note:
Cox regression model (method = Enter).
Abbreviations: HR, hazard ratio; CI, confidence interval; AJCC, American Joint Committee on Cancer; COL10A1, collagen type X alpha 1 chain.
Discussion
Even though COL10A1 was reported to be highly expressed in tumors by high throughput sequencing, such as gene chip,8,17 the specific role of COL10A1 in CRC was unknown. A preliminary understanding was that COL10A1 expression was restricted in most normal tissues and elevated in many diverse tumor types, such as colon cancer,17 esophagus cancer,19 and breast cancer,20 and was associated with tumor vasculature by immunofluorescence studies.16 COL10A1 protein levels in serum might be a potential diagnostic predictor for early detection of colon cancer. This means it has potential value in clinical application.17
The study in this article was conducive to learn the molecular mechanism of COL10A1, which promoted CRC progression. Through IHC, Western blot, and real-time Q-PCR assays, we have confirmed that COL10A1 was significantly elevated in CRC tissues compared with adjacent normal tissues. To further explore the biological function of COL10A1 in CRC, we constructed stable CRC cell lines with overexpression or knockdown of COL10A1 by cell transfection experiments. Through a series of functional experiments in vivo and in vitro, we gradually understood the role of COL10A1 in CRC. The results showed that the invasion and migration ability of LoVo and HCT116 cells with overexpressing COL10A1 were significantly upregulated compared with LoVo-NC and HCT116-NC cells. On the contrary, the invasion and migration ability were significantly downregulated in SW480 and SW620 cells with COL10A1 knockdown compared with SW480-NC and SW620-NC cells. In tumor growth assay of nude mice, the tumor growth speed of the SW480-COL10A1-KD group was evidently slower than that in the NC group.
Collagen is one of the major components of the tumor microenvironment. EMT plays an important role in the process of tumor invasion and metastasis.21 Collagen, tumor microenvironment, and EMT interacted with each other and were closely related to the tumor progression.22,23 It had been reported that Collagen Type I suppressed E-cadherin and promoted proliferation of pancreatic carcinoma cells,24 and COL11A1 was associated with pancreatic and ovarian cancer by inducing EMT.25,26 According to our results, we hypothesized that high expression of COL10A1 promoting proliferation and invasion of CRC might relate to the EMT phenomenon. However, the mechanism of COL10A1 influenced on the EMT process is not clear and needs further study.
Of course, the limitations of this research are obvious. There is no research on the upstream transcription factor or downstream interaction protein of COL10A1, so the detailed molecular mechanism of COL10A1 in CRC has not been studied thoroughly. It had been reported that the Run227 and MEF2C,28 even non-steroidal anti-inflammatory drugs,29 might be involved in the regulation of COL10A1 expression, but the specific mechanism in CRC is unclear. This is also the focus and direction of our next research. The predictive value and impact on long-term outcomes of COL10A1 require further studies to confirm. Meanwhile, whether COL10A1 exerts a direct promoting effect on the CRC cells or plays a role by regulating other factors remains to be determined.
Conclusion
Our study demonstrates that COL10A1 is overexpressed in CRC tissues and involves the proliferation, migration, invasion, and tumorigenesis process of CRC cells. Meanwhile, COL10A1 promotes the EMT process. High-level expression of COL10A1 can be considered as an independent risk factor of prognosis and OS in CRC. COL10A1 is likely to be a potential target gene for CRC therapy.
Acknowledgments
This work was supported by the grants from the State’s Key Project of Research and Development Plan (2017YFC0108300, 2017YFC0108301), National Natural Science Foundation of China (81672446, 81270565), Natural Science Foundation of Guangdong Province (2016A030313843), Guangdong Provincial Science and Technology Key Project (2014A020215014), Scientific Research Foundation for the Returned Overseas Chinese Scholars, Ministry of Education (series 48), Research Fund of Public Welfare in the Health Industry, the National Health and Family Planning Commission of China (201402015), the Southern Medical University Clinical Research Start-Up Project (LC2016ZD003), Guangzhou Science and Technology Project (201400000004-5), and the Key Clinical Specialty Discipline Construction Program ([2011]170).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Sato H, Kotake K, Sugihara K, Takahashi H, Maeda K, Uyama I. Clinicopathological factors associated with recurrence and prognosis after R0 resection for stage IV colorectal cancer with peritoneal metastasis. Dig Surg. 2016;33(5):382–391. doi: 10.1159/000444097. [DOI] [PubMed] [Google Scholar]
- 3.Zhou ZX, Zhao LY, Lin T, et al. Long-term oncologic outcomes of laparoscopic vs open surgery for stages II and III rectal cancer: a retrospective cohort study. World J Gastroenterol. 2015;21(18):5505–5512. doi: 10.3748/wjg.v21.i18.5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Center MM, Jemal A, Ward E. International trends in colorectal cancer incidence rates. Cancer Epidemiol Biomarkers Prev. 2009;18(6):1688–1694. doi: 10.1158/1055-9965.EPI-09-0090. [DOI] [PubMed] [Google Scholar]
- 5.Center MM, Jemal A, Smith RA, Ward E. Worldwide variations in colorectal cancer. CA Cancer J Clin. 2009;59(6):366–378. doi: 10.3322/caac.20038. [DOI] [PubMed] [Google Scholar]
- 6.Martin DB, Nelson PS. From genomics to proteomics: techniques and applications in cancer research. Trends Cell Biol. 2001;11(11):S60–S65. doi: 10.1016/s0962-8924(01)02123-7. [DOI] [PubMed] [Google Scholar]
- 7.Michener CM, Ardekani AM, Petricoin ER, Liotta LA, Kohn EC. Genomics and proteomics: application of novel technology to early detection and prevention of cancer. Cancer Detect Prev. 2002;26(4):249–255. doi: 10.1016/s0361-090x(02)00092-2. [DOI] [PubMed] [Google Scholar]
- 8.Ki DH, Jeung HC, Park CH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121(9):2005–2012. doi: 10.1002/ijc.22975. [DOI] [PubMed] [Google Scholar]
- 9.Tan F, Liu F, Liu H, Hu Y, Liu D, Li G. CTHRC1 is associated with peritoneal carcinomatosis in colorectal cancer: a new predictor for prognosis. Med Oncol. 2013;30(1):473. doi: 10.1007/s12032-013-0473-3. [DOI] [PubMed] [Google Scholar]
- 10.Yan L, Yu J, Tan F, et al. SP1-mediated microRNA-520d-5p suppresses tumor growth and metastasis in colorectal cancer by targeting CTHRC1. Am J Cancer Res. 2015;5(4):1447–1459. [PMC free article] [PubMed] [Google Scholar]
- 11.Kielty CM, Kwan AP, Holmes DF, Schor SL, Grant ME. Type X collagen, a product of hypertrophic chondrocytes. Biochem J. 1985;227(2):545–554. doi: 10.1042/bj2270545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Makitie O, Susic M, Ward L, Barclay C, Glorieux FH, Cole WG. Schmid type of metaphyseal chondrodysplasia and COL10A1 mutations – findings in 10 patients. Am J Med Genet A. 2005;137A(3):241–248. doi: 10.1002/ajmg.a.30855. [DOI] [PubMed] [Google Scholar]
- 13.Gu J, Lu Y, Li F, et al. Identification and characterization of the novel Col10a1 regulatory mechanism during chondrocyte hypertrophic differentiation. Cell Death Dis. 2014;5:e1469. doi: 10.1038/cddis.2014.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hasegawa K, Higuchi Y, Yamashita M, Tanaka H. Japanese familial case with metaphyseal dysplasia, Schmid Type caused by the p.T555P mutation in the COL10A1 gene. Clin Pediatr Endocrinol. 2015;24(1):33–36. doi: 10.1297/cpe.24.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebhard S, Hattori T, Bauer E, et al. BAC constructs in transgenic reporter mouse lines control efficient and specific LacZ expression in hypertrophic chondrocytes under the complete Col10a1 promoter. Histochem Cell Biol. 2007;127(2):183–194. doi: 10.1007/s00418-006-0236-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman KB, Prendes MJ, Sternberg H, et al. COL10A1 expression is elevated in diverse solid tumor types and is associated with tumor vasculature. Future Oncol. 2012;8(8):1031–1040. doi: 10.2217/fon.12.79. [DOI] [PubMed] [Google Scholar]
- 17.Sole X, Crous-Bou M, Cordero D, et al. Discovery and validation of new potential biomarkers for early detection of colon cancer. PLoS One. 2014;9(9):e106748. doi: 10.1371/journal.pone.0106748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sternberg A. Carcinoma of the colon: margins of resection. J Surg Oncol. 2008;98(8):603–606. doi: 10.1002/jso.21032. [DOI] [PubMed] [Google Scholar]
- 19.Karagoz K, Lehman HL, Stairs DB, Sinha R, Arga KY. Proteomic and metabolic signatures of esophageal squamous cell carcinoma. Curr Cancer Drug Targets. 2016;16(8):721–736. [PubMed] [Google Scholar]
- 20.Brodsky AS, Xiong J, Yang D, et al. Identification of stromal ColXalpha1 and tumor-infiltrating lymphocytes as putative predictive markers of neoadjuvant therapy in estrogen receptor-positive/HER2-positive breast cancer. BMC Cancer. 2016;16:274. doi: 10.1186/s12885-016-2302-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28(1–2):15–33. doi: 10.1007/s10555-008-9169-0. [DOI] [PubMed] [Google Scholar]
- 22.Aguilera KY, Rivera LB, Hur H, et al. Collagen signaling enhances tumor progression after anti-VEGF therapy in a murine model of pancreatic ductal adenocarcinoma. Cancer Res. 2014;74(4):1032–1044. doi: 10.1158/0008-5472.CAN-13-2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Mendoza MG, Inman DR, Ponik SM, et al. Neutrophils drive accelerated tumor progression in the collagen-dense mammary tumor microenvironment. Breast Cancer Res. 2016;18(1):49. doi: 10.1186/s13058-016-0703-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koenig A, Mueller C, Hasel C, Adler G, Menke A. Collagen type I induces disruption of E-cadherin-mediated cell-cell contacts and promotes proliferation of pancreatic carcinoma cells. Cancer Res. 2006;66(9):4662–4671. doi: 10.1158/0008-5472.CAN-05-2804. [DOI] [PubMed] [Google Scholar]
- 25.Wu YH, Chang TH, Huang YF, Huang HD, Chou CY. COL11A1 promotes tumor progression and predicts poor clinical outcome in ovarian cancer. Oncogene. 2014;33(26):3432–3440. doi: 10.1038/onc.2013.307. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Pravia C, Galvan JA, Gutierrez-Corral N, et al. Overexpression of COL11A1 by cancer-associated fibroblasts: clinical relevance of a stromal marker in pancreatic cancer. PLoS One. 2013;8(10):e78327. doi: 10.1371/journal.pone.0078327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Lu Y, Ding M, et al. Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J Bone Miner Res. 2011;26(12):2899–2910. doi: 10.1002/jbmr.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnold MA, Kim Y, Czubryt MP, et al. MEF2C transcription factor controls chondrocyte hypertrophy and bone development. Dev Cell. 2007;12(3):377–389. doi: 10.1016/j.devcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 29.Almaawi A, Wang HT, Ciobanu O, et al. Effect of acetaminophen and nonsteroidal anti-inflammatory drugs on gene expression of mesenchymal stem cells. Tissue Eng Part A. 2013;19(7–8):1039–1046. doi: 10.1089/ten.TEA.2012.0129. [DOI] [PubMed] [Google Scholar]