Abstract
Recent studies have identified many genes with rare de novo mutations in autism, but a limited number of these have been conclusively established as disease-susceptibility genes due to lack of recurrence and confounding background mutations. Such extreme genetic heterogeneity severely limits recurrence–based statistical power even in studies with a large sample size. Here we use cell-type specific expression profiles to differentiate mutations in autism patients from those in unaffected siblings. We report a gene expression signature in different neuronal cell types shared by genes with likely gene disrupting (LGD) mutations in autism cases. The signature reflects haploinsufficiency of risk genes enriched in transcriptional and post-transcriptional regulators, with the strongest positive associations with specific types of neurons in different brain regions, including cortical neurons, cerebellar granule cells, and striatal medium spiny neurons. When used to prioritize genes with a single LGD mutation in cases, a D-score derived from the signature achieved a precision of 40% as compared to the 15% baseline with a minimal loss in sensitivity. An ensemble model combining D-score with mutation intolerance metrics from Exome Aggregation Consortium further improved the precision to 60%, resulting in 117 high-priority candidates. These prioritized lists can facilitate identification of additional autism-susceptibility genes.
Keywords: autism spectrum disorders, autism-susceptibility genes, de novo mutations, cell type-specific expression signature, D-score
Introduction
Autism or autism spectrum disorders (ASDs) are common neurodevelopmental diseases characterized by deficits in language, impaired social interaction, and repetitive behaviors with complexes such as seizures and intellectual disability (Devlin and Scherer, 2012; Newschaffer, et al., 2007). Symptom onset is typically early (~3 years old) and the current estimate of incidence is over 1% worldwide (Croning, et al., 2009), underscoring the widespread impact of autism on affected families and for society in general (Abul-Husn, et al., 2009).
Genetic risk factors are believed to play a pivotal role in ASDs, as revealed by a concordance rate up to 90% between monozygotic twins and by over 10-fold increase in the risk for a new born child if a previous sibling is affected (Ronemus, et al., 2014). Some syndromic forms of autisms are known to be monogenic, as represented by mutations of FMR1 (encoding FMRP) in the fragile X syndrome that is comorbid with autism and accounts for up to 5% of ASD cases (Kelleher and Bear, 2008; Verkerk, et al., 1991). However, most of the genomic abnormalities or mutations found in autism patients are extremely rare and frequently de novo. Earlier studies using microarray-based approaches identified hundreds of de novo copy number variants (CNVs) (Itsara, et al., 2010; Levy, et al., 2011; Marshall, et al., 2008; Pinto, et al., 2010; Sanders, et al., 2011; Sebat, et al., 2007). More recently, de novo mutations in individual nucleotides including single nucleotide variations (SNVs) and small insertions and deletions (indels) were identified by exome-sequencing (De Rubeis, et al., 2014; Iossifov, et al., 2014; Iossifov, et al., 2012; Neale, et al., 2012; O’Roak, et al., 2012b; Sanders, et al., 2012) or whole-genome sequencing (Jiang, et al., 2013; Turner, et al., 2016).
The exciting progress in these genetic studies has provided important insights into the etiopathogenesis of autism. First, at least a substantial proportion of autism risk is conferred by individually rare mutations affecting one or more disease-susceptibility genes. The number of risk loci has been estimated to be in the range of several hundred to over 1,000 genes (Iossifov, et al., 2014; Iossifov, et al., 2012; Ronemus, et al., 2014). Second, although the complexity of the genetic landscape underlying autism is still a matter of debate, one theory, supported by several lines of evidence, proposes that there are a large number of autism risk loci, each individually having high penetrance (Gratten, et al., 2013; Ronemus, et al., 2014; Zhao, et al., 2007). Third, analysis of the seemingly isolated candidate autism-susceptibility genes points to disruption in several convergent molecular pathways (Gilman, et al., 2011; Iossifov, et al., 2012; Parikshak, et al., 2013; Ronemus, et al., 2014; Willsey, et al., 2013) that inform the neurobiological underpinnings of autism (reviewed by ref. (Krumm, et al., 2014)).
Strikingly, even the most frequent de novo mutations in single genes can explain no more than 1% of ASD cases (Krumm, et al., 2014). This extreme genetic heterogeneity presents a big challenge for conclusive identification of autism-susceptibility genes, which has impeded further functional studies of autism neurobiology and development of therapeutic strategies. A particular group of de novo mutations identified by whole exome-sequencing (De Rubeis, et al., 2014; Iossifov, et al., 2012; Neale, et al., 2012; O’Roak, et al., 2012b; Sanders, et al., 2012) are “likely gene-disrupting” or LGD mutations, which are a collection of severe mutations introducing frame shift, disrupted splice sites or premature stop codons (these mutations are also named loss-of-function or LOF mutations in the literature). Probands clearly show a higher burden of de novo LGD mutations than their unaffected siblings used as control, indicating enrichment of disease-susceptibility genes disrupted by these genomic lesions. However, although about 4,000 ASD patients and their families have been sequenced so far, only about 40 genes at best have been determined as high-confidence autism-susceptibility genes based on their recurrence. Most of the remaining mutations were observed in only single patient, and ~80% of these are expected to be non-pathogenic (see Material and Methods). The signal-to-noise ratio is even lower for genes with missense mutations (Iossifov, et al., 2014) due to their more moderate effects and high background mutation frequency. Therefore, most autism-susceptibility genes are currently buried among a growing list of potential candidates.
Two general strategies have been used to facilitate the identification of candidate autism-susceptibility genes. One strategy is to sequence larger cohorts of ASD families. For example, the SPARK project aims to recruit and analyze 50,000 individuals of autism and their families (Simons Foundation, 2016). Furthermore, whole-exome or whole-genome sequencing is also complemented by targeted re-sequencing to reduce cost (O’Roak, et al., 2012a). A caveat of this strategy is its prohibitive cost associated with recruitment and sequencing of large cohorts. A second complementary strategy is to stratify already identified mutations based on existing orthogonal information associated with the affected genes. For example, depletion of rare deleterious variants estimated from the general population reflecting severe selection pressure is effective in prioritizing deleterious mutations (Lek, et al., 2016; Petrovski, et al., 2013; Samocha, et al., 2014). Alternatively, gene regulatory and function information can also be used to help distinguish pathogenic vs. neutral mutations. The latter is based on the assumption that the common clinical phenotypes of ASDs originate from certain common features shared by autism risk loci at the molecular level. Along this line, recent studies identified molecular pathways underlying autism etiopathology from analysis of shared genetic phenotypes (Chang, et al., 2015; Gilman, et al., 2011), protein-protein interactions (O’Roak, et al., 2012b), and gene co-expression networks (Parikshak, et al., 2013; Willsey, et al., 2013). Such information has also been used in several recent studies to prioritize autism-susceptibility genes (Krishnan, et al., 2016; Liu, et al., 2015; Tranchevent, et al., 2016).
In this work, we predict autism-susceptibility genes by using gene expression profiles in a wide range of specific neuronal cell types, which has not been systematically investigated in previous studies. Our assumption is that different cell types in the central nervous system (CNS) have different susceptibility or relevance to autism etiopathogy. Due to the heterogeneity among many different cell types in the brain, such an analysis may reveal cell type-specific gene regulation that cannot be detected by analysis of brain tissues used in previous studies. We identified a gene expression signature reflecting haploinsufficiency in the context of autism that was able to effectively predict whether individual LGD mutations confer disease risk. Importantly, the use of cell type-specific expression also allows us to highlight the cellular contexts of the identified risks. Furthermore, this signature is complementary to previous mutation intolerance analysis, and an ensemble model combining multiple scoring metrics results in the optimal prediction accuracy.
Materials and Methods
Data compilation
For the current DAMAGES analysis, we used microarray gene expression profiles in 24 mouse central nervous system (CNS) cell types isolated from six brain regions, as well as unselected RNAs in each of these regions. This dataset was previously generated using a translational profiling approach named TRAP (Doyle, et al., 2008). For each gene, we selected the probeset with the maximum median expression across all 30 samples as a representative, if multiple probesets exist. In total, 20,870 genes with Entrez gene IDs are represented in the dataset. Expression intensities for each gene were first log2-transformed, with a pseudocount of 8 added to the intensities on the original scale for variance stabilization. We determined the one-to-one mouse ortholog of 15,951 human genes (76%) using the HomoloGene database (http://www.ncbi.nlm.nih.gov/homologene), complemented by manual searches.
The initial list of genes with de novo mutations in ASD probands and unaffected siblings were collected from four whole exome-sequencing studies of 952 cases and 594 controls published prior to 2013 (Iossifov, et al., 2012; Neale, et al., 2012; O’Roak, et al., 2012b; Sanders, et al., 2012). A total of 162 genes have de novo LGD mutations either in the probands or siblings. Mouse orthologs were found for 158 genes with LGD mutations, including 123 genes with LGD mutations only in probands and 35 genes with LGD mutations in siblings. Among these, a total of 145 genes, including 112 genes with LGD mutations in probands and 33 genes with LGD mutations in siblings, were represented in the microarray data, and were used for the initial model building and analysis. We also used 1,479 genes with log2 expression intensities ≥ 6 in two or more cell types and a standard deviation ≥ 2 across all cell types for global PCA analysis shown in Figure 3B and C.
For further validation, we compiled an expanded list of genes with LGD mutations in autism patients (cases) or in their unaffected siblings (controls) from more recent exome-sequencing studies of about 3,960 cases and 1911 controls (De Rubeis, et al., 2014; Iossifov, et al., 2014). In total, we obtained a list of 672 genes, including 40 genes with recurrent LGD mutations in patients, 468 genes with singleton LGD mutation in patients, and 173 genes with LGD mutations in controls (note we excluded TTN). Among these, 611 genes (91%) have mouse orthologs and are represented in the microarray data.
De novo copy number variation (CNV) data in ASD probands and annotations of overlapping genes were obtained from (Sanders, et al., 2011). This list is composed of 219 CNVs, and was compiled by the original authors from several previous studies (Itsara, et al., 2010; Marshall, et al., 2008; Pinto, et al., 2010; Sanders, et al., 2011; Sebat, et al., 2007). Technically redundant mutations, due to inclusion of the same patient samples in multiple studies, have already been removed from the list, so that recurrence of CNVs observed in the list is genuine. We similarly identified the mouse orthlogs of these CNV genes, and those (1,571 genes total) represented on the microarrays.
SFARI autism genes were downloaded in July 2013 from https://gene.sfari.org (Basu, et al., 2009). The prioritized gene lists by Krishnan et al. (Krishnan, et al., 2016) and DAWN (Liu, et al., 2015) were obtained directly from the supplementary tables provided by the authors. ExAC metric scores were downloaded from http://exac.broadinstitute.org. De novo mutation calls from the Deciphering Developmental Disorders (DDD) project was obtained from (McRae, et al., 2016).
We listed all data sets used in the analysis in Supp. Figure S1B.
DAMAGES analysis
For the current work, DAMAGES analysis is composed of two major steps. For PCA analysis, log2-transformed expression intensities for each gene were first standardized across the 30 cell/tissue types to obtain zero means and unit standard deviations. PCA was then performed in R using the princomp package. Here we denote Sgi the score of gene g projected onto PC i.
We then used a regularized regression analysis named lasso (Tibshirani, 1996) to evaluate the contribution of each PC to prediction of each gene with respect to the source of mutations (probands versus siblings) with the following representation. Model overfitting was evaluated by a standard leave-one-out cross validation (LOOCV) procedure. For this study, we define the score Dg == Sg2 + 0.135 as the DAMAGES score, denoted D-score, of gene g, in which the constant is determined by LOOCV.
Estimating specificity and sensitivity in predicting autism-susceptibility genes
The specificity and sensitivity of predicting autism-susceptibility genes have to be inferred from the relative enrichment of the mutations in cases versus controls, since the ground truth is unknown for most genes. We first estimated the number of non-disease causing genes hit by random neutral mutations in the initial list of 112 genes with LGD mutations in probands (Supp. Table S1). The five genes with recurrent LGD mutations were considered to be true positives, given the very small chance to observe such recurrent de novo mutations (Iossifov, et al., 2012; Sanders, et al., 2012; Willsey, et al., 2013). For the remaining genes, the number of non-disease causing genes, or false positives, was estimated based on the relative frequency of LGD mutations in siblings. A case-control design was used in three studies, and the number of false positives in each study was estimated separately. For one study (Neale, et al., 2012), no sibling controls were included, so the number of false positives was estimated from the false discovery rate (FDR) of the other three studies pooled together. Overall, 53 genes with non-recurrent LGD mutations (42% of 127 genes) were estimated to be false positives. Therefore, we estimated that among the 112 genes with LGD mutations represented in the microarray dataset, there are ~65 disease-causing genes and ~47 non-disease genes, respectively.
To assess the single-LGD candidate gene prioritization performance of D-score, ExAC metrics, and ensemble score, we used estimated background mutation rate (Samocha, et al., 2014; Ware, et al., 2001) to estimate precision and recall (sensitivity) rate. Specifically, for each gene set (with G genes) defined by various metrics, we estimated the number of true positive (i.e. disease-causing; MT) LGD mutations based on the observed number (M1) of LGD variants in N cases and the expected number of variants (M0) given the background LGD mutation rate (Ri, i indexes genes) and MT = M1 − M0. We denote the total number of true positives in all genes as M, and estimate sensitivity (recall) in each gene set by S = MT/M and precision by P = MT/M1. F-measure combines precision and recall by their harmonic mean F = 2PS/(P+S), which provides a single metric score that balances between precision and recall. We note that genes with recurrent mutations in ASD patients and genes with LGD mutations in controls, which were used to build ensemble regression model, were not used to estimate the precision and recall in this analysis.
Ensemble score by combining D-score and ExAC metrics
We used a logistic regression to combine D-score with direct measurements of mutation intolerance for a better haploinsufficiency prediction. Specifically, we labeled genes with ≥2 LGD de novo mutations in cases as ≥positives”, and the genes with ≥1 LGD de novo mutations in controls and no LGD mutations in cases as “negatives”; we used D-score and ExAC metrics (pLI and mis-Z) as features (xi ), and estimated the effect size (βi ) for each feature by a standard logistic regression. With estimated effect size, including the intercept (β0 ), we calculate a prediction score (“ensemble score”) for each gene by S = 1/[1 + e−(β0+Σiβixi)].
Results
DAMAGES analysis uncovers an expression signature of autism-susceptibility genes
Our study was motivated by a postulation that different cell types in the brain have different susceptibility and impact on autism etiopathology, which is supported by recent studies showing expression bias in candidate autism-susceptibility genes (Chang, et al., 2015; Xu, et al., 2014). However, to the best of our knowledge, the effectiveness of cell type-specific expression in predicting autism-susceptibility genes, alone or in combination with other metrics, has not been systematically explored.
We developed and applied a computational framework for disease-associated mutation analysis using gene expression signatures (DAMAGES) to score the association of human genes with autism, to refine the lists of candidate autism-associated genes currently available, and to uncover features shared by these genes as a means of understanding the molecular underpinnings of the disease (Supp. Figure S1A). We reason that combination of expression and mutation data—two orthogonal types of information—is critical for minimizing “guilt by association” and identifying causal disease risk genes. In contrast to previous network analysis approaches, we adopted a case-control classification framework to optimize and objectively evaluate the accuracy of prediction. To provide a proof of principle in this study, we decided to examine a large microarray dataset that profiles cell type-specific transcripts associated with translating ribosomes in the mouse brain generated by a biochemical assay named TRAP (Doyle, et al., 2008; Heiman, et al., 2008). In total, this dataset is composed of translational profiles of 24 specific mouse CNS cell types, including both neurons and glial cells, isolated from six different regions, together with unselected RNA representing all cell types in each of these regions (Doyle, et al., 2008) (Supp. Figure S1A). This translational profiling approach was previously shown to give robust gene expression measurements, and to effectively identify known and novel cell-type specific markers and provide biological insights into each cell type (Doyle, et al., 2008).
To identify expression signatures of autism-susceptibility genes, we started with a list of 162 genes containing de novo LGD mutations in either ASD probands or unaffected siblings collected from four exome-sequencing studies (Iossifov, et al., 2012; Neale, et al., 2012; O’Roak, et al., 2012b; Sanders, et al., 2012). Our prediction model was built using these mutations representing all information available before 2013, which gave us a chance to use additional genes discovered by large-scale follow-up studies for objective evaluation (Supp. Figure S1B). In total, 145 human genes have mouse orthologs and were represented in the microarray dataset we used (Supp. Table S1). We assume that the 33 genes with LGD mutations in unaffected siblings confer no risk of ASD (non-disease genes). On the other hand, the other 112 genes with LGD mutations in probands were estimated to represent a mixture of about 65 autism -susceptibility genes and 47 non-disease genes (Supp. Table S1 and Material and Methods; a similar estimate provided in (Willsey, et al., 2013)). It is worth noting that we limited our analysis to mutations derived from unbiased genomic screens to build the model, and excluded candidates identified by more targeted or hypothesis-driven approaches or by transcriptomic analysis to avoid potential ascertainment bias. This is particularly critical for an objective assessment of DAMAGES analysis in prediction accuracy.
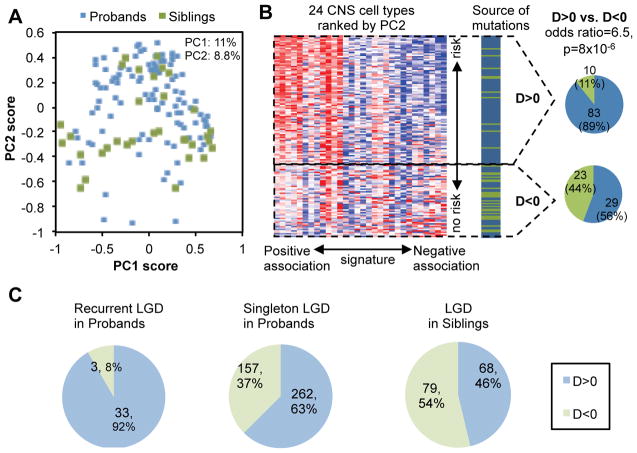
Given the relatively balanced representation of autism-susceptibility genes and non-disease genes (estimated to be ~65 and ~80, respectively) in the dataset, we anticipated that the contrast between these two groups of genes would represent a major axis of expression dynamics in the high-dimensional space. A principal component analysis (PCA) (Duda, et al., 2000) was thus performed to identify the orthogonal axes that explain the most variance. This analysis revealed that the first two principal components explains about 20% of variation and projection of genes to the second principal component (PC) is very predictive of mutations in probands versus controls (Figure 1A–B and Supp. Figure S2A). We note that PCA is an unsupervised method which does not incorporate information on the source of mutations (i.e., patient or sibling) so the prediction performance is not due to data overfitting. To have a more rigorous assessment of the ability of each PC or combination of PCs to differentiate potentially disease-associated mutations from neutral mutations, we performed a regularized linear regression analysis lasso (Tibshirani, 1996) to find the PCs that are most predictive of the source of mutations. We confirmed that the predictive power came almost exclusively from PC2 (Supp. Figure S2 B and C). Therefore, we decided to use the PC2 as a signature of autism-susceptibility genes, and adjusted the threshold according to leave-one-out cross validation (LOOCV), which resulted in the final DAMAGES scores (or D-scores) used for gene ranking (Supp. Figure S2C). As a result, we were able to identify 93 genes with positive D-scores, including 83 genes with LGD mutations in probands and 10 genes with LGD mutations in siblings, respectively (Figure 1B and Supp. Table S1). Proband-specific LGD mutations are strikingly enriched in genes with positive D-scores compared to the remaining genes (Figure 1B; odds ratio=6.5, P=8×10−6, Fisher’s exact test). The ability of the gene expression signature to differentiate mutations in probands from those in siblings suggests that at least some of the CNS cell types included in the microarray dataset are strongly associated with the underlying molecular mechanisms of autism.
Figure 1. A molecular signature differentiates autism-susceptibility genes and non-disease genes.
A. A total of 145 genes, including 112 genes with LGD mutations in probands (blue dots) and 33 genes with LGD mutations in siblings (green dots) are projected onto the two-dimensional space defined by the first two principal components (PCs).
B. The second principal component differentiates autism-susceptibility genes and non-disease genes. In the heatmap on the left, the PC2 score and loading were used to rank genes and arrays respectively. Detail of the cell types is also shown in Figure 3B below. The source of mutation in each gene (i.e., patient or control) is indicated with genes shown in the same order. The number of genes with D-score>0 or <0 is shown on the right.
C. Summary of prediction using an expanded list of genes with LGD mutations in ASD patients and unaffected siblings.
Validation of the expression signature using expanded autism exome sequencing data sets
Based on the signature, we assigned a D-score to all human genes represented in the microarray dataset independent of their mutation status (Supp. Table S2). This allowed us to have an independent, “prospective” evaluation of the performance of the expression signature using an expanded list of genes with LGD mutations from recent large-scale studies after our prediction model was built (Supp. Figure S1B and Supp. Table S1)(De Rubeis, et al., 2014; Iossifov, et al., 2014). In this expanded dataset, almost all genes with recurrent mutations in autism patients (35/38=92%) received a positive D-score (Figure 1C; exceptions are DSCAM, RANBP17, and TCF7L2; two genes not represented on the array were excluded). Among the genes ranked in the top 25% by the D-score, there is a 2.8-fold enrichment (P=3.2×10−12) of LGD mutations in cases comparing to unaffected siblings, whereas there is no significant enrichment in rest of genes (rate enrichment = 1.2, P=0.16) (Table 1). Therefore, DAMAGES analysis prioritized bona fide autism-susceptibility genes with minimal loss.
Table 1.
Enrichment of LGD de novo mutations in cases among genes grouped by D-score.
| D-score rank percentile | Number of LGD mutations in cases | Number of LGD mutations in controls | Rate enrichment | P-value |
|---|---|---|---|---|
| Top 25% (n-genes = 3928) | 257 | 45 | 2.76 | 3.2×10−12 |
| Bottom 75% | 318 | 132 | 1.16 | 0.16 |
P-values were calculated by binomial tests to test a null hypothesis that the rate of LGD mutations is the same in cases as in controls in a set of genes.
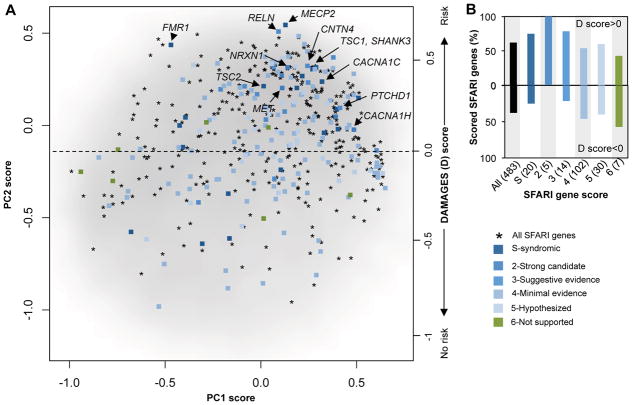
For additional validation, we examined 528 genes compiled in the Simon Foundation Autism Research Initiative (SFARI) autism gene database, a list of potential autism-associated genes manually curated by experts according to various types of evidence available in the literature (https://gene.sfari.org) (Basu, et al., 2009). We used the dataset retrieved in 2013 for comparison, as later releases of database integrated results from sequencing studies. Among the 483 SFARI genes represented in the microarray data, 300 (62%) have a positive D-score (Figure 2A and Supp. Table S3), a very significant enrichment compared to all genes represented in the microarray dataset (odds ratio=2.3, P=6.3×10−20; Fisher’s exact test). In addition, this proportion is higher for genes with more evidence supporting their implication in autism (Figure 2B). For example, genes with positive D-scores include 5/5 (100%) genes that are classified as strong candidates and 15/20 (75%) genes that are classified as syndromic, such as FMR1, MECP2 [Rett syndrome (Amir, et al., 1999)] and TSC1/2 [tuberous sclerosis complex (Crino, et al., 2006)]. Furthermore, SFARI genes received much higher ranks based on the D-score, as compared to ranking by the first PC reflecting the neuron-glial distinction (P=1.2×10−22; Wilcox ranksum test; see below).
Figure 2. The DAMAGES molecular signature refines the list of candidate genes in the SFARI autism gene database.
A. All genes represented in the microarray dataset, except those with average log2 intensity <4.5, are projected onto the first two PCs, and shown as a smoothed scatter plot. The gray-scale intensity reflects the local density of genes. A total of 483 genes from the SFARI autism gene database represented in the microarray dataset (asterisks) are overlaid. A subset of these genes were manually scored by experts by considering strength of existing evidence, and these scored genes are distinguished using different colors. A subset of syndromic ASD genes and the five strong candidate genes are highlighted.
B. The percentage of scored genes in each group with a positive or negative DAMAGES score (D-score) is shown. The color codes are the same as in (A). The number of genes in each group is indicated in the parentheses following the gene categories.
CNS cell types associated with autism
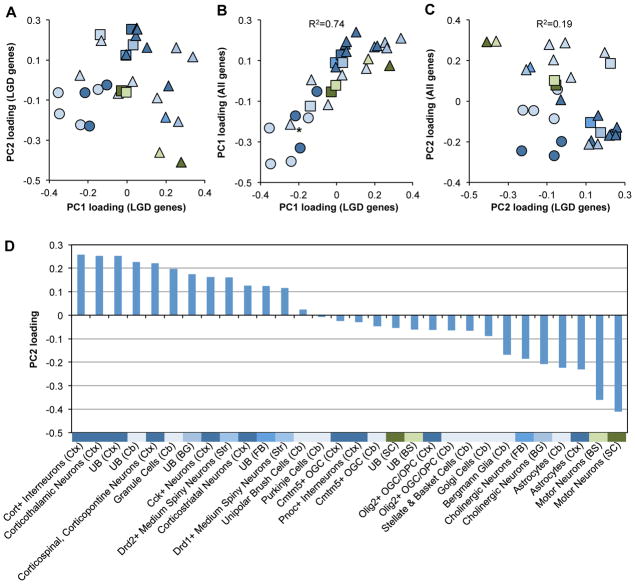
To further confirm this molecular signature and gain more biological insights, we examined the loadings of different cell types on each PC (Supp. Table S4). The first PC essentially differentiates neurons versus glial cells and unselected cell types in different brain regions (Figure 3A). In contrast, the second PC predictive of autism-susceptibility genes appears to give more of a mix of different cell types and regions, although a certain bias is also clear (e.g., cortical neurons have the highest positive loadings; see below). To assess whether this pattern is relevant for the underlying molecular mechanisms of autism and specific for autism-susceptibility genes, we performed another PCA using all genes showing the most variation across different cell types. The first PC of the whole dataset is highly correlated with the one derived from genes with LGD mutations (R2=0.74), and similarly differentiates neurons from glial cells and unselected cell types (Figure 3B). This result is consistent with the notion that even among genes with LGD mutations the distinction of neuronal versus glial genes dominates the expression dynamics. In contrast, the second PC identified in the global PCA has a low correlation with the second PC identified using genes with LGD mutations (R2=0.19; Figure 3C). This observation supports the notion that the molecular signature identified using genes with LGD mutations indeed reflects certain specific features shared by autism-susceptibility genes.
Figure 3. The DAMAGES molecular signature reveals CNS cell types associated with autism.
A. The loadings of different cell types on the first two PCs derived from genes with LGD mutations are shown. Each dot represents a cell type. Different colors represent the brain regions used to isolate the specific types of cells, with the same color codes as shown in Supp. Figure S1A. Neurons, glial cells and unselected RNA samples are represented by triangles, circles, and squares, respectively.
B. The loadings of cell types on PC1 derived from genes with LGD mutations (x-axis) are plotted against that derived from the whole dataset (y-axis). The asterisk indicates cerebellar Grp+ cells that are known to include both unipolar brush cells and Bergmann glial cells (Doyle, et al., 2008). The squared Pearson correlation between the two signatures is indicated.
C. Similar to (B), except that the loadings on PC2 are plotted.
D. Loadings of all cell types on PC2 derived from genes with LGD mutations (DAMAGES signature) are plotted. The color codes and abbreviation of each brain region are the same as shown in Supp. Figure S1A. UB: unbound RNA without selection for specific cell types.
Therefore, the loadings of different cell types on the signature (PC2) likely reflect their association with autism (Figure 3D). In general, none of the glial cell types included for this analysis has a positive association. On the other hand, the association of neurons with the signature varies depending on specific cell types and brain regions. Different types of cortical neurons, including interneurons, corticothalamic neurons, corticospinal and corticopontine neurons, Cck+ neurons, and corticostriatal neurons, have large positive loadings on the signature. However, not all types of cortical neurons have a positive association, and some, such as Pnoc+ interneurons, have a negative loading. Besides cortical neurons, cerebellar granule cells, striatal medium spiny neurons, but not Purkinje cells, cholinergic neurons, or motor neurons, show a strong positive loading. Altogether, these observations are not only consistent with autism being mainly an impairment of high-level cognitive functions, but also suggest that even in a given brain region, specific cell types may play very different roles in the etiopathogenesis of the disease.
Molecular functions associated with the autism-susceptibility gene expression signature
We asked whether the expression signature captures certain molecular functions shared by autism-susceptibility genes. To this end, we performed Gene Ontology (GO) analysis (Dennis, et al., 2003) using the top 500 protein-coding genes ranked by D-scores independent of their mutation status. This analysis revealed very strong enrichment of those involved in “transcription” (Benjamini FDR=3×10−14), “chromatin modification” (Benjamini FDR=7.9×10−6) and “regulation of RNA metabolic process” (Benjamini FDR=2.2×10−4) (Supp. Table S5). It is worth noting that “chromatin organization” is also enriched in the 83 high-priority candidate genes, although the statistical significance is marginal (Benjamini FDR=0.07). Therefore, not only are genes with LGD mutations themselves enriched in those important for transcriptional regulation, as noted previously (Ben-David and Shifman, 2013; Iossifov, et al., 2012; O’Roak, et al., 2012b), but they define a molecular signature represented by a larger set of genes with coherent molecular functions in both transcriptional and post-transcriptional regulation of gene expression.
The expression signature reflects haploinsufficiency of the affected genes
We postulate that the expression signature may reflect haploinsufficiency because it was derived from genes with heterozygous loss of function. To test this hypothesis, we examined genes covered by relatively focal de novo CNV events (≤50 genes) detected in ASD probands (Itsara, et al., 2010; Marshall, et al., 2008; Pinto, et al., 2010; Sanders, et al., 2011; Sebat, et al., 2007). Interestingly, genes with higher D-scores tend to overlap with deletions than amplifications (P<0.04; Spearman correlation test). The significance is relatively marginal, presumably due to the limited spatial resolution of the CNVs. For further confirmation, we examined genes differentially expressed in post-mortem autism brains as compared to controls (Voineagu, et al., 2011), assuming that the dosage-dependent alteration can also be caused by changes at the transcription level. Indeed, genes down-regulated in autism tend to have a positive D-score (P<2.2×10−16), while genes upregulated in autism tend to have a negative D-score (P<2×10−7; Supp. Figure S3A).
Recently, the Exome Aggregation Consortium (ExAC) used large-scale exome sequencing data of general populations without developmental disorders to estimate metrics of haploinsufficiency or mutation intolerance (Lek, et al., 2016), including the probability of being loss-of-function (LoF) intolerant (pLI), LoF-Z score and mis-Z score. Briefly, pLI is an estimate of the probability of a gene being haploinsufficiency based on depletion of rare LGD variants in the population, and LoF-Z and mis-Z are normalized scores measuring the depletion of rare variants comparing to what is expected by chance in a gene. A positive correlation (r = 0.29, P < 10−10) of LoF Z score and D-score was observed among genes with positive D-scores (Supp. Figure S3B). This observation is again consistent with the notion that both metrics are related to haploinsufficiency although they were derived using entirely different approaches with different assumptions.
Prioritizing genes in CNVs including 16p11.2
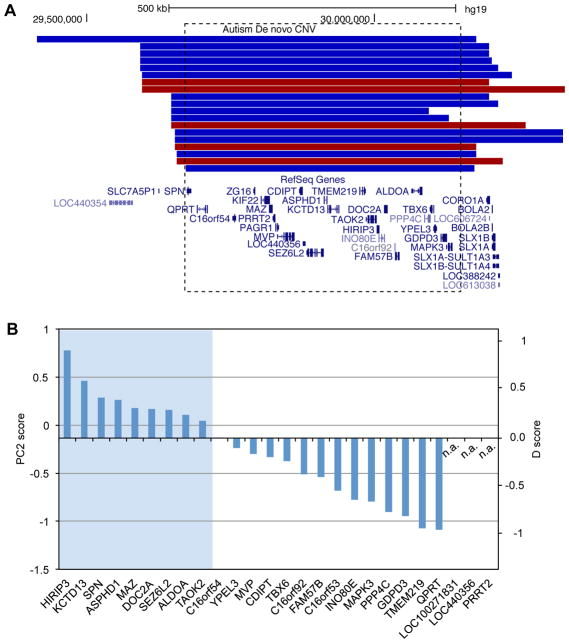
De novo CNVs detected in ASD probands typically span dozens or hundreds of genes (Itsara, et al., 2010; Levy, et al., 2011; Marshall, et al., 2008; Pinto, et al., 2010; Sanders, et al., 2011; Sebat, et al., 2007). Therefore, although over 2,000 genes are covered by at least one CNV identified so far, it is difficult to differentiate bona fide autism-susceptibility genes from other passenger genes. We focused on 58 deletion CNVs for which all overlapping genes have mouse orthologs and are represented in the microarray data. Of these, 30 CNVs each have one and only one gene with a positive D-score. Based on the high sensitivity (~90%, see below) of a positive D-score in predicting autism-susceptibility genes, we argue that if a CNV is pathogenic, the only gene with a positive D-score is most likely the causal gene. We therefore denote the CNV “likely supporting CNV” or LS-CNV of the corresponding gene. This analysis resulted in 19 genes supported by deletion LS-CNV events in one or more patients (Supp. Table S6). Of these genes, five are supported by recurrent LS-CNVs (NRXN1, DPP6, PTPRT, SHANK2 and SLC4A10), and all of these five genes are known to have functional implications in synapse (Clark, et al., 2008; Jacobs, et al., 2008; Lim, et al., 2009; Sudhof, 2008; Won, et al., 2012) and/or autism-related phenotypes (Won, et al., 2012). Remarkably, three genes harbor recurrent LGD mutations in ASD patients (ANKRD11, CHD3 and KMT2C; odd ratio=130, P<3.4×10−6, Fisher’s exact test). Two additional genes (NRXN1 and SHANK2) have singleton LGD mutations from exome sequencing (i.e., recurrent if the LS-CNV is counted).
LS-CNVs tend to span a smaller number of genes than CNVs in general. For a majority of CNVs overlapping with more genes, it is difficult to reliably distinguish susceptibility genes versus passenger genes even with the D-score. Nevertheless, it is still possible to eliminate a substantial fraction of passenger genes. To illustrate this point, we examined the most frequent recurrent de novo CNV located in 16p11.2, which accounts for up to 1% of ASD cases (Marshall, et al., 2008) (14 deletions and 5 duplications in the dataset used for this analysis; Figure 4A). This region spans 26 genes (Sanders, et al., 2011) and all of them have mouse orthologs; deletion of the region in mice phenocopied behavior deficits observed in ASD patients (Horev, et al., 2011). Among the 23 genes represented in the microarray data, nine have a positive D-score (Figure 4B). Interestingly, deletion of a smaller region in this locus also segregates with ASD or ASD traits (Crepel, et al., 2011). This deletion encompasses five genes, including KCTD13, ASPHD1 and SEZ6L2 with a positive D-score. A recent study further demonstrated that KCTD13 is a major driver of the macrocephalic phenotype associated with ASD cases carrying the 16p11.2 CNV (Golzio, et al., 2012). Some of the other genes in this locus, especially the ones with positive D-scores, could contribute to the additional clinical manifestations in ASDs.
Figure 4. Prioritized candidate autism-susceptibility genes with recurrent CNVs in chromosome 16p11.2.
A. A UCSC genome browser view of the region (hg19: chr16:29,350,841–30,433,540) is shown, with de novo CNV events displayed above the RefSeq genes. Duplication and deletion CNVs are shown in red and blue, respectively. The dotted box indicates the region with 26 genes affected in almost all CNVs.
B. The 26 genes are ranked by their D-scores. Three genes not represented in the microarray data are indicated by n.a..
Comparison with other methods and an ensemble model for optimized prediction of autism-susceptibility genes
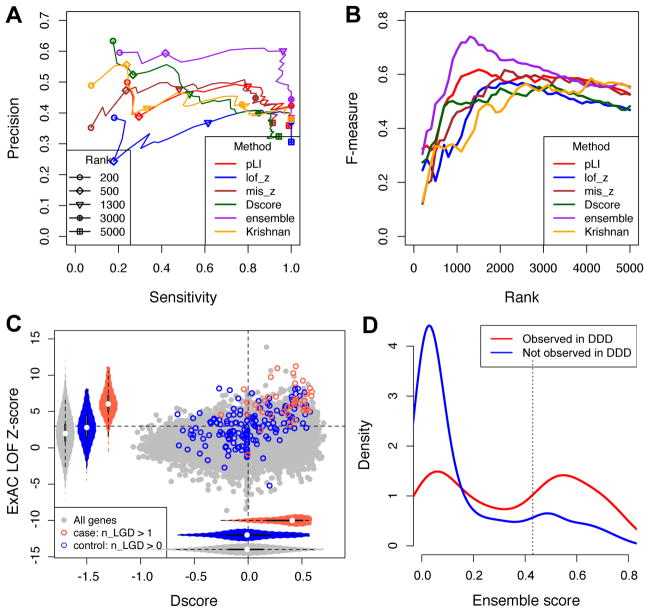
We compared D-score and several other methods in predicting autism-susceptibility genes. A very recent study ranked and predicted autism-associated genes using a human brain-specific functional gene interaction network derived from expression and interaction measurements in thousands of different conditions (Krishnan, et al., 2016). Compared to this method, D-score achieved comparable or favorable results in a wide range of stringency thresholds, as shown in Precision-Recall curves (Figure 5A,B and Supp. Table S2). For example, Krishnan et al. reported enrichment of de novo LGD mutations in the top decile of their ranked gene list. We found the top 2,000 genes ranked by this method (among those with one-to-one mouse orthologs) included 19 of 36 genes with recurrent mutations and 86 of 461 genes with singleton mutations in autism cases (odds ratio=6.8 and 1.4, respectively). For the same number of predictions, D-score predicted 23 of 36 recurrent genes, and 117 of 461 singleton in cases (odds ratio=8.9 and 1.7, respectively).
Figure 5. Comparison D-score with other methods and the ensemble model.
A. Performance of prediction as measured by Precision and Recall (Sensitivity) using different gene prioritizing methods or an ensemble model as a function of varying cutoffs. Note that Precision and Recall were estimated from the relative enrichment of mutations in patients and controls, since the ground truth whether a gene is a true positive is unavailable in most cases.
B. Similar to (A), but the F measure is shown. F measure is the harmonic mean of precision and recall and rewards a balance of the two.
C. Distribution of D-scores and ExAC LOF Z-scores for genes with recurrent LGD mutations in ASD cases and controls.
D. Distribution of ensemble score among singleton genes in which damaging de novo mutations observed (red) in the DDD data set versus the ones not observed (blue). Singleton genes refer to the ones with a single de novo LGD mutation in autism cases.
Similarly, the performance of D-score is also on a par with ExAC scores (Figure 5A–C). In particular, when we focused on genes with single LGD mutations in cases and found ExAC pLI>0.9 or LoF Z-score>3 achieved similar optimal performance as D-score (about 40% precision and 90% sensitivity; Figure 5A, B and Supp. Table S2), as compared to a baseline 15% precision. Importantly, while predictions by the two metrics overlap, small genes with low background mutation rate (such as MECP2, pLI=0.7) tend to be missed by ExAC scores. These results confirmed the effectiveness of cell-type specific expression in prioritizing autism-susceptibility genes compared to the other state-of-the-art approaches.
Since D-score and ExAC scores were derived from very different types of information, we investigated whether the two approaches make independent contributions in gene prioritization. To this end, we performed a logistic regression to classify recurrently mutated genes in cases and genes with LGD mutations in controls, with ExAC pLI, ExAC mis-z, and D-score as features. The coefficients of all three features deviate significantly from zero (Table 2), indicating that D-score is complementary to ExAC scores in determining gene LGD intolerance (interestingly, gene expression in embryonic mouse brain bulk sample at E9.5 (Homsy, et al., 2015) did not any predictive power). Therefore, combining these scores would maximize the performance in candidate gene prioritization. To assess that, we applied the estimated logistic model to all genes to calculate an ensemble score (Supp. Table S2), and estimated precision-recall rates in a range of top rank thresholds, excluding the genes that are recurrently mutated. We found that the ensemble score outperforms all individual methods, with an optimal performance among the top 1,300 genes (Figure 5A, B). With ensemble score, the precision quadruples to 60% with near-maximal sensitivity (estimated to be 97%). Using this threshold, we identified 117 high-priority candidate genes with a singleton LGD mutation (Supp. Table S7).
Table 2.
Logistic regression for classification of recurrently LGD-mutated genes in ASD and genes LGD-mutated in unaffected siblings.
| Features | Estimate | P-value |
|---|---|---|
| pLI | 2.00 | 0.0029 |
| Mis_z | 0.290 | 0.019 |
| D-score | 2.90 | 0.0015 |
We also considered the gene expression rank from brain tissues at mouse embryonic day 9.5 (Brain E9.5; data described in (Homsy, et al., 2015)) as a potential feature. However, in the logistic regression, if we include brain E9.5 together with other features above, the effect size of the feature is not statistically significant from 0 (P-value=0.92). This feature is thus excluded in our final ensemble model.
Since ASD shares substantial number of risk genes with other neurodevelopmental disorders (De Rubeis, et al., 2014; Krumm, et al., 2015), we obtained the de novo mutation calls from the latest released data (McRae, et al., 2016) from the Deciphering Developmental Disorders (DDD) project (The Deciphering Developmental Disorders Study, 2015) to further assess the performance of ensemble score prediction. The DDD data includes 4,293 patients with severe undiagnosed developmental disorders. Overall, among all the genes with a single LGD mutation in ASD data, the ones with at least one de novo LGD or damaging missense (predicted by metaSVM (Dong, et al., 2015) or polyphen-2 (Adzhubei, et al., 2001) and CADD (Kircher, et al., 2014)) mutation in the DDD data have higher ensemble scores than the ones without (P = 1.3×10−11; KS test) (Figure 5D). Importantly, among the 117 high-priority candidate genes predicted by ensemble score, 65 harbor at least one damaging mutation in the DDD data set. This rate is much higher than non-candidate genes with a single LGD mutation in ASD data (odds ratio = 4.6; P = 2×10−11; Fisher’s exact test).
We also compared the ensemble method with the DAWN algorithm, which integrates de novo mutation data with gene co-expression network (Liu, et al., 2015). In total, 113 genes with singleton mutations were among the candidate genes prioritized by DAWN, and 49 genes of them have at least one deleterious de novo mutations in DDD data set. While the proportion is still significantly larger than non-candidate singleton genes (P= 8.3e-4), the odds ratio (2.2) is much lower than that of the ensemble score (4.6). Taken together, the candidate genes with singleton de novo LGD mutations prioritized by the ensemble score are also more likely to be associated with developmental disorders in general.
Gender bias of autism-associated de novo mutations
The incidence of autism has a strong gender bias, with a male:female ratio around 4 overall and even higher for high-functioning cases. This is reflected in the population of participants included in the exome-sequencing studies (M:F=6.4 in the SSC dataset; Supp. Table S8). However, a lower incidence of de novo mutations in males was previously observed (Iossifov, et al., 2014; Iossifov, et al., 2012; Levy, et al., 2011). We examined the gender bias of de novo LGD mutations in different sets of genes, focusing on mutations identified from the Simon SSC dataset, for which the number of male and female patients is known (Supp. Table S8). Consistent with previous observations, the lowest M:F ratio was observed from genes with recurrent LGD mutations and genes with singleton LGD mutations predicted by the ensemble model (M:F~0.5, after correction for the gender bias of the participants; P<0.02, Fisher’s exact test). A more moderate, but significant, gender bias was observed in genes with singleton mutations predicted by D-score alone (M:F=0.62; P=0.02, Fisher’s exact test). No significant gender bias was observed among singleton mutations in genes showing a negative D-score (M:F=0.89; P=0.65, Fisher’s exact test). We also confirmed that there is no correlation between D-score and gender-specific gene expression detected in the mouse brain(Yang, et al., 2006), suggesting that the gender bias of autism-associated mutations predicted by D-score cannot be simply explained by dimorphic gene expression (Supp. Figure S4). These observations provided an independent line of evidence that D-score and the ensemble model can discriminate disease-susceptibility genes from non-disease genes.
Discussion
Here we present the use of cell type-specific gene expression profiles to improve prediction of autism-susceptibility genes. The molecular signature uncovered by DAMAGES analysis has several implications. First, this study echoes recent findings on convergent molecular pathways underlying autism etiopathogenesis including, approximately, three modules: synaptic structure and function, transcriptional regulation and chromatin remodeling, and Wnt signaling (reviewed by ref. (Krumm, et al., 2014) (De Rubeis, et al., 2014)). Conclusions of these studies were drawn from analysis of co-occurrence in genetic phenotypes (Gilman, et al., 2011), protein-protein interactions (O’Roak, et al., 2012b), and gene co-expression networks reflecting developmental dynamics in different brain regions (Parikshak, et al., 2013; Willsey, et al., 2013). This work extended these previous efforts by demonstrating that a robust signature of autism-susceptibility genes can be defined by their expression patterns in a range of specific CNS cell types (Chang, et al., 2015; Xu, et al., 2014). Importantly, this signature reflects genes involved in transcriptional and post-transcriptional regulation and haploinsufficiency caused by LGD mutations in these genes. The importance of transcription factors and chromatin regulators in autism is now well established (Krumm, et al., 2014; O’Roak, et al., 2012b). In addition, the role of post-transcriptional regulation is in line with the observation that several monogenic autism risk loci, including FMRP and MeCP2, are important regulators of RNA metabolism (Smith and Sadee, 2011), and that candidate autism-associated genes show significant overlap with target transcripts of several neuronal RNA-binding proteins including FMRP (Darnell, et al., 2011; Iossifov, et al., 2012) and RBFOX1 (A2BP1) (Voineagu, et al., 2011; Weyn-Vanhentenryck, et al., 2014; Zhang, et al., 2010). Indeed, among the 822 FMRP target genes represented in the microarray dataset (Darnell, et al., 2011), a vast majority (718 or 87%) have a positive D-score.
Second, a key feature of DAMAGES analysis is that it adopts a case-control design using candidate genes derived from genomic DNA screens, which are completely independent of expression data. This design allows rigorous assessment of the biological relevance and predictive power of the uncovered signature by controlling potential confounding factors such as non-uniform mutation rates in different groups of genes. In addition, expression data from model organisms, which represent richer resources compared to the scarcity and non-uniform quality of expression profiles derived from postmortem human brains, can be naturally included in such framework. Our results demonstrated that the cell type-specific expression signature derived from mouse CNS cell types greatly increased the specificity of predicting autism-susceptibility genes with minimal loss of true hits. This is reflected in the observation that DAMAGES analysis predicted 35 out of 40 genes with recurrent LGD mutations identified so far and all 5 non-syndromic candidate genes with the highest confidence in the SFARI autism gene database. Importantly, the information provided by the expression signature is complementary to the scoring metrics based on analysis of mutation intolerance in the general population, and improved performance was achieved by combining the two methods.
Lastly, the cell-type specific signature captures a strong positive association of autism with multiple types of cortical neurons, cerebellar granule cells, and striatal medium spiny neurons. This observation implies haploinsufficiency of genes that are normally highly expressed in these cell types as a converging pathogenic mechanism in autism. The implication of cortical projection neurons (Willsey, et al., 2013) and cells in the granule layer of the cerebellum (Menashe, et al., 2013) has been noted in the last few years. In basal ganglia, striatal medium spiny neurons are known as the primary cell type vulnerable in Huntington’s disease (Ehrlich, 2012). In the context of autism, it has been shown that depletion of SHANK3, a gene highly expressed in striatum and regarded as the cause of the autism-related Phelan-McDermid Syndrome, results in ASD-like features such as impaired social interaction in mouse models (Peca, et al., 2011). SHANK3 has a singleton LGD mutation detected in the current exome-sequencing studies, and ranks among the top 5% genes genome-wide by the D-score (see Figure 4A). In the cerebellum, a reduction of Purkinje cells and granule cells has been found in postmortem autistic brains and in mouse models (Fatemi, et al., 2012; Tsai, et al., 2012). Interestingly, our analysis revealed that cerebellar granule cells show a strong positive loading with a magnitude similar to those observed from cortical neurons, while Purkinje cells in cerebellum show weak loadings. These data suggest an intriguing hypothesis that different molecular mechanisms might underlie the loss of Purkinje and granule cells, although this has to be tested in future work. Finally, glial cells, especially astrocytes, show a strong negative loading. However, this should probably not exclude the contribution of these cells to autism. Instead, an alternative interpretation is that these genes may confer risks through other mechanisms than haploinsufficiency, which is supported by the observation that a subset of immune genes and glia markers are overexpressed in autism brains (Voineagu, et al., 2011).
In summary, this study suggests the potential of utilizing gene expression and regulation information in predicting pathogenic mutations in autism. While we focused on cell type-specific expression in this work to demonstrate the proof of principle, we anticipate that additional spatiotemporal expression profiles and functional annotations of genes, which can be integrated using a machine learning framework, will further improve the performance. Prioritized gene lists from such analysis can facilitate further validation by targeted re-sequencing in large cohorts (O’Roak, et al., 2012a) or more mechanistic studies using model organisms. This application will be particularly useful as the list of mutations are expected to grow steadily as a result of continuing autism exome- and genome-wide sequencing projects (Krumm, et al., 2014; Ronemus, et al., 2014; Simons Foundation, 2016).
Statistical analysis
All statistical tests and logistic regression were performed using the R software.
Supplementary Material
Acknowledgments
We would like to thank members of the Zhang and Shen labs for comments on the manuscript and Joe Dougherty for helpful discussion on TRAP. This work was supported by grants from Simons Foundation (SFARI# 307711 to CZ) and NIH (R00GM95713 to CZ and U01HG008680 to YS). Computation was supported by NIH grants S10OD012351 and S10OD021764.
References
- Abul-Husn NS, Bushlin I, Moron JA, Jenkins SL, Dolios G, Wang R, Iyengar R, Ma’ayan A, Devi LA. Systems approach to explore components and interactions in the presynapse. Proteomics. 2009;9(12):3303–3315. doi: 10.1002/pmic.200800767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Current Protocols in Human Genetics. John Wiley & Sons, Inc; 2001. Predicting functional effect of human missense mutations using PolyPhen-2; pp. 7.20.1–7.20.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37(suppl 1):D832–D836. doi: 10.1093/nar/gkn835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-David E, Shifman S. Combined analysis of exome sequencing points toward a major role for transcription regulation during brain development in autism. Mol Psychiatry. 2013;18(10):1054–1056. doi: 10.1038/mp.2012.148. [DOI] [PubMed] [Google Scholar]
- Chang J, Gilman SR, Chiang AH, Sanders SJ, Vitkup D. Genotype to phenotype relationships in autism spectrum disorders. Nat Neurosci. 2015;18(2):191–198. doi: 10.1038/nn.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BD, Kwon E, Maffie J, Jeong H-Y, Nadal M, Strop P, Rudy B. DPP6 localization in brain supports function as a Kv4 channel associated protein. Front Mol Neurosci. 2008;1 doi: 10.3389/neuro.02.008.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crepel A, Steyaert J, De la Marche W, De Wolf V, Fryns J-P, Noens I, Devriendt K, Peeters H. Narrowing the critical deletion region for autism spectrum disorders on 16p11.2. Am J Med Genet B Neuropsychiatr Genet. 2011;156(2):243–245. doi: 10.1002/ajmg.b.31163. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. [DOI] [PubMed] [Google Scholar]
- Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the Genes to Cognition database. Nucleic Acids Res. 2009;37(Database issue):D846–851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rubeis S, He X, Goldberg AP, Poultney CS, Samocha K, Cicek AE, Kou Y, Liu L, Fromer M, Walker S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515(7526):209–215. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(9):R60. [PubMed] [Google Scholar]
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22(3):229–237. doi: 10.1016/j.gde.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Dong CL, Wei P, Jian XQ, Gibbs R, Boerwinkle E, Wang K, Liu XM. Comparison and integration of deleteriousness prediction methods for nonsynonymous SNVs in whole exome sequencing studies. Hum Mol Genet. 2015;24(8):2125–2137. doi: 10.1093/hmg/ddu733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duda RO, Hart PE, Stork DG. Pattern Classification. Wiley; 2000. [Google Scholar]
- Ehrlich M. Huntington’s disease and the striatal medium spiny neuron: cell-autonomous andnon-cell-autonomous mechanisms of disease. Neurotherapeutics. 2012;9(2):270–284. doi: 10.1007/s13311-012-0112-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatemi SH, Aldinger K, Ashwood P, Bauman M, Blaha C, Blatt G, Chauhan A, Chauhan V, Dager S, Dickson P, et al. Consensus paper: pathological role of the cerebellum in autism. The Cerebellum. 2012;11(3):777–807. doi: 10.1007/s12311-012-0355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman SR, Iossifov I, Levy D, Ronemus M, Wigler M, Vitkup D. Rare de novo variants associated with autism implicate a large functional network of genes involved in formation and function of synapses. Neuron. 2011;70(5):898–907. doi: 10.1016/j.neuron.2011.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golzio C, Willer J, Talkowski ME, Oh EC, Taniguchi Y, Jacquemont S, Reymond A, Sun M, Sawa A, Gusella JF, et al. KCTD13 is a major driver of mirrored neuroanatomical phenotypes of the 16p11.2 copy number variant. Nature. 2012;485(7398):363–367. doi: 10.1038/nature11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratten J, Visscher PM, Mowry BJ, Wray NR. Interpreting the role of de novo protein-coding mutations in neuropsychiatric disease. Nat Genet. 2013;45(3):234–238. doi: 10.1038/ng.2555. [DOI] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135(4):738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homsy J, Zaidi S, Shen Y, Ware JS, Samocha KE, Karczewski KJ, DePalma SR, McKean D, Wakimoto H, Gorham J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350(6265):1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horev G, Ellegood J, Lerch JP, Son Y-EE, Muthuswamy L, Vogel H, Krieger AM, Buja A, Henkelman RM, Wigler M, et al. Dosage-dependent phenotypes in models of 16p11.2 lesions found in autism. Proc Natl Acad Sci U S A. 2011;108(41):17076–17081. doi: 10.1073/pnas.1114042108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, O’Roak BJ, Sanders SJ, Ronemus M, Krumm N, Levy D, Stessman HA, Witherspoon KT, Vives L, Patterson KE, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515(7526):216–221. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iossifov I, Ronemus M, Levy D, Wang Z, Hakker I, Rosenbaum J, Yamrom B, Lee Y-h, Narzisi G, Leotta A, et al. De novo gene disruptions in children on the autistic spectrum. Neuron. 2012;74(2):285–299. doi: 10.1016/j.neuron.2012.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, Eichler EE. De novo rates and selection of large copy number variation. Genome Res. 2010;20(11):1469–1481. doi: 10.1101/gr.107680.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S, Ruusuvuori E, Sipilä ST, Haapanen A, Damkier HH, Kurth I, Hentschke M, Schweizer M, Rudhard Y, Laatikainen LM, et al. Mice with targeted Slc4a10 gene disruption have small brain ventricles and show reduced neuronal excitability. Proc Natl Acad Sci U S A. 2008;105(1):311–316. doi: 10.1073/pnas.0705487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang YH, Yuen RK, Jin X, Wang M, Chen N, Wu X, Ju J, Mei J, Shi Y, He M, et al. Detection of clinically relevant genetic variants in autism spectrum disorder by whole-genome sequencing. Am J Hum Genet. 2013;93(2):249–263. doi: 10.1016/j.ajhg.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46(3):310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan A, Zhang R, Yao V, Theesfeld CL, Wong AK, Tadych A, Volfovsky N, Packer A, Lash A, Troyanskaya OG. Genome-wide prediction and functional characterization of the genetic basis of autism spectrum disorder. Nat Neurosci. 2016 doi: 10.1038/nn.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, O’Roak BJ, Shendure J, Eichler EE. A de novo convergence of autism genetics and molecular neuroscience. Trends Neurosci. 2014;37(2):95–105. doi: 10.1016/j.tins.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumm N, Turner TN, Baker C, Vives L, Mohajeri K, Witherspoon K, Raja A, Coe BP, Stessman HA, He ZX, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47(6):582–588. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536(7616):285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D, Ronemus M, Yamrom B, Lee Y-h, Leotta A, Kendall J, Marks S, Lakshmi B, Pai D, Ye K, et al. Rare de novo and transmitted copy-number variation in autistic spectrum disorders. Neuron. 2011;70(5):886–897. doi: 10.1016/j.neuron.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Lim S-H, Kwon S-K, Lee MK, Moon J, Jeong DG, Park E, Kim SJ, Park BC, Lee SC, Ryu S-E, et al. Synapse formation regulated by protein tyrosine phosphatase receptor T through interaction with cell adhesion molecules and Fyn. EMBO J. 2009;28(22):3564–3578. doi: 10.1038/emboj.2009.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Lei J, Roeder K. Network assisted analysis to reveal the genetic basis of autism. Ann Appl Stat. 2015;9(3):1571–1600. doi: 10.1214/15-AOAS844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, Skaug J, Shago M, Moessner R, Pinto D, Ren Y, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82(2):477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McRae JF, Clayton S, Fitzgerald TW, Kaplanis J, Prigmore E, Rajan D, Sifrim A, Aitken S, Akawi N, Alvi M, et al. Prevalence, phenotype and architecture of developmental disorders caused by de novo mutation. bioRxiv. 2016 doi: 10.1101/049056. [DOI] [Google Scholar]
- Menashe I, Grange P, Larsen EC, Banerjee-Basu S, Mitra PP. Co-expression profiling of autism genes in the mouse brain. PLoS Comput Biol. 2013;9(7):e1003128. doi: 10.1371/journal.pcbi.1003128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale BM, Kou Y, Liu L, Ma/’ayan A, Samocha KE, Sabo A, Lin C-F, Stevens C, Wang L-S, Makarov V, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485(7397):242–245. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newschaffer CJ, Croen LA, Daniels J, Giarelli E, Grether JK, Levy SE, Mandell DS, Miller LA, Pinto-Martin J, Reaven J, et al. The epidemiology of autism spectrum disorders. Annu Rev Public Health. 2007;28(1):235–258. doi: 10.1146/annurev.publhealth.28.021406.144007. [DOI] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Fu W, Egertson JD, Stanaway IB, Phelps IG, Carvill G, Kumar A, Lee C, Ankenman K, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012a;338(6114):1619–1622. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Roak BJ, Vives L, Girirajan S, Karakoc E, Krumm N, Coe BP, Levy R, Ko A, Lee C, Smith JD, et al. Sporadic autism exomes reveal a highly interconnected protein network of de novo mutations. Nature. 2012b;485(7397):246–250. doi: 10.1038/nature10989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parikshak NN, Luo R, Zhang A, Won H, Lowe JK, Chandran V, Horvath S, Geschwind DH. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell. 2013;155(5):1008–1021. doi: 10.1016/j.cell.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovski S, Wang Q, Heinzen EL, Allen AS, Goldstein DB. Genic intolerance to functional variation and the interpretation of personal genomes. PLoS Genet. 2013;9(8):e1003709. doi: 10.1371/journal.pgen.1003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronemus M, Iossifov I, Levy D, Wigler M. The role of de novo mutations in the genetics of autism spectrum disorders. Nat Rev Genet. 2014;15(2):133–141. doi: 10.1038/nrg3585. [DOI] [PubMed] [Google Scholar]
- Samocha KE, Robinson EB, Sanders SJ, Stevens C, Sabo A, McGrath LM, Kosmicki JA, Rehnstrom K, Mallick S, Kirby A, et al. A framework for the interpretation of de novo mutation in human disease. Nat Genet. 2014;46(9):944–950. doi: 10.1038/ng.3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Ercan-Sencicek AG, Hus V, Luo R, Murtha MT, Moreno-De-Luca D, Chu SH, Moreau MP, Gupta AR, Thomson SA, et al. Multiple recurrent de novo CNVs, including duplications of the 7q11.23 Williams syndrome region, are strongly associated with autism. Neuron. 2011;70(5):863–885. doi: 10.1016/j.neuron.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders SJ, Murtha MT, Gupta AR, Murdoch JD, Raubeson MJ, Willsey AJ, Ercan-Sencicek AG, DiLullo NM, Parikshak NN, Stein JL, et al. De novo mutations revealed by whole-exome sequencing are strongly associated with autism. Nature. 2012;485(7397):237–241. doi: 10.1038/nature10945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, et al. Strong association of de novo copy number mutations with autism. Science. 2007;316(5823):445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons Foundation. SPARK project. 2016 https://sparkforautism.org.
- Smith RM, Sadee W. Synaptic signaling and aberrant RNA splicing in autism spectrum disorders. Front Synaptic Neurosci. 2011;3 doi: 10.3389/fnsyn.2011.00001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Deciphering Developmental Disorders Study. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519(7542):223–228. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibshirani R. Regression shrinkage and selection via the lasso. J Royal Statist Soc B. 1996;58(1):267–288. [Google Scholar]
- Tranchevent LC, Ardeshirdavani A, ElShal S, Alcaide D, Aerts J, Auboeuf D, Moreau Y. Candidate gene prioritization with Endeavour. Nucleic Acids Res. 2016;44(W1):W117–121. doi: 10.1093/nar/gkw365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai PT, Hull C, Chu Y, Greene-Colozzi E, Sadowski AR, Leech JM, Steinberg J, Crawley JN, Regehr WG, Sahin M. Autistic-like behaviour and cerebellar dysfunction in Purkinje cell Tsc1 mutant mice. Nature. 2012;488(7413):647–651. doi: 10.1038/nature11310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Hormozdiari F, Duyzend MH, McClymont SA, Hook PW, Iossifov I, Raja A, Baker C, Hoekzema K, Stessman HA, et al. Genome sequencing of autism-affected families reveals disruption of putative noncoding regulatory DNA. Am J Hum Genet. 2016;98(1):58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, et al. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65(5):905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- Voineagu I, Wang X, Johnston P, Lowe JK, Tian Y, Horvath S, Mill J, Cantor RM, Blencowe BJ, Geschwind DH. Transcriptomic analysis of autistic brain reveals convergent molecular pathology. Nature. 2011;474(7351):380–384. doi: 10.1038/nature10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware JS, Samocha KE, Homsy J, Daly MJ. Interpreting de novo variation in human disease using denovolyzeR. Curr Protoc Hum Genet. 2001;87:7.25.1–7.25.15. doi: 10.1002/0471142905.hg0725s87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyn-Vanhentenryck S, Mele A, Sun S, Yan Q, Farny N, Zhang Z, Xue C, Silver PA, Zhang MQ, Krainer AR, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep. 2014;6(6):1139–1152. doi: 10.1016/j.celrep.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willsey AJ, Sanders Stephan J, Li M, Dong S, Tebbenkamp Andrew T, Muhle Rebecca A, Reilly Steven K, Lin L, Fertuzinhos S, Miller Jeremy A, et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell. 2013;155(5):997–1007. doi: 10.1016/j.cell.2013.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Won H, Lee H-R, Gee HY, Mah W, Kim J-I, Lee J, Ha S, Chung C, Jung ES, Cho YS, et al. Autistic-like social behaviour in Shank2-mutant mice improved by restoring NMDA receptor function. Nature. 2012;486(7402):261–265. doi: 10.1038/nature11208. [DOI] [PubMed] [Google Scholar]
- Xu X, Wells AB, O’Brien DR, Nehorai A, Dougherty JD. Cell type-specific expression analysis to identify putative cellular mechanisms for neurogenetic disorders. J Neurosci. 2014;34(4):1420–1431. doi: 10.1523/JNEUROSCI.4488-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Frias MA, Mele A, Ruggiu M, Eom T, Marney CB, Wang H, Licatalosi DD, Fak JJ, Darnell RB. Integrative modeling defines the Nova splicing-regulatory network and its combinatorial controls. Science. 2010;329:439–443. doi: 10.1126/science.1191150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Leotta A, Kustanovich V, Lajonchere C, Geschwind DH, Law K, Law P, Qiu S, Lord C, Sebat J, et al. A unified genetic theory for sporadic and inherited autism. Proc Natl Acad Sci U S A. 2007;104(31):12831–12836. doi: 10.1073/pnas.0705803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.