Abstract
The activity-regulated cytoskeletal (Arc) gene is implicated in numerous synaptic plasticity paradigms, including long-term potentiation and depression and homeostatic plasticity and is critical for consolidating memory. How Arc facilitates these forms of plasticity is not fully understood. Unlike other neuronal immediate-early genes, Arc encodes a protein that shuttles between the somatodendritic and nuclear compartments to regulate synaptic plasticity. Little attention has been paid to Arc’s role in the nucleus. Here, we highlight the regulatory elements and signaling cascades required to induce Arc transcription and discuss the significance of Arc nuclear localization for synaptic plasticity and scaling. We integrate these findings into the context of cognitive function and disease and propose a model in which Arc mediates an effect on memory as a “chaser” of synaptic activity through homeostatic scaling.
Keywords: Arg3.1, immediate early gene, transcriptional regulation, promoter, homeostatic plasticity, hyperexcitability
1. Introduction
Cognitive functions such as learning and memory, require tight regulation of neuronal gene expression, a prerequisite for long-term synaptic plasticity. The activity-regulated cytoskeletal (Arc, also known as Arg3.1) immediate-early gene (IEG) was discovered as a gene induced by seizures in the hippocampus [1,2] and is implicated in numerous neuronal functions such as synaptic plasticity, including long-term potentiation (LTP, synaptic strengthening), and long-term depression (LTD, synaptic weakening), and homeostatic plasticity [3–9]. Arc is activated during synaptic activity and learning [1,2,10,11] and is essential for memory consolidation [6,12]: Arc knock out (KO) mice fail to form long-lasting memories, whereas short-term memory remains intact [6].
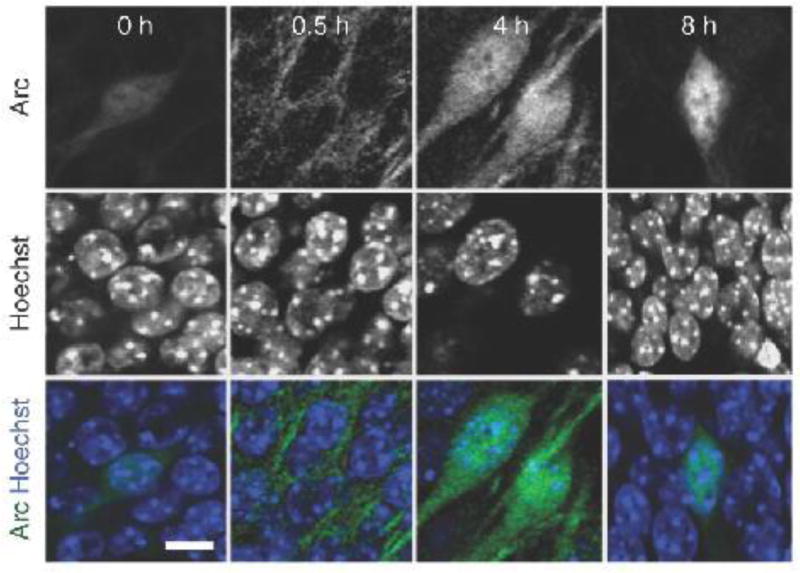
Arc is a single-copy gene, conserved in vertebrates, and predominantly expressed in cortical and hippocampal glutamatergic neurons. Unlike many IEGs, Arc does not encode a classical transcription factor, although it regulates transcription [13]. Arc is involved in numerous neuronal signaling pathways [7,9,14,15] and regulates network stability in vivo [16]. Expression, localization and stability of Arc are tightly regulated [11,17]. Unusual among IEGs, Arc mRNA is quickly transported or stabilized at active synapses upon synaptic activity, suggesting translation of Arc protein near sites of local synaptic activity [11]. At synapses, Arc regulates synaptic strength by promoting AMPA receptor internalization [7] and modulates spine morphology [16]. Half an hour after induction, Arc shuttles to the nucleus where most of it is localized 8 hours after stimulation (Fig. 1) [13], implying that Arc might function as a cytosolic and nuclear protein [13,18].
Fig. 1. Arc becomes enriched in neuronal nuclei after stimulation.
Immunohistochemical staining of Arc and Hoechst nuclear staining in mouse hippocampal sections after exposure to a novel environment for 0–8 hours. Scale bar, 10 µm. Reprinted with permission from Macmillan Publishers Ltd: [Nature Neuroscience] [13], copyright (2013).
Thus, studying Arc offers an exceptional opportunity to explore links among gene expression, synaptic activity and cognitive function. While many studies explored Arc’s role in the somatodendritic compartment (reviewed in this issue), our understanding of Arc induction and its role in the nucleus is incomplete. Here, we will discuss the signaling and regulatory elements that induce Arc transcription, highlight the significance of Arc nuclear localization, and disentangle its roles for cognitive function and disease.
2. Functional response elements required for Arc induction
Activating gene expression in neurons is essential for learning-related long-term changes [19]. Upon neuronal activation, calcium ions rapidly enter the cell via synaptic N-methyl-D-aspartic acid (NMDA) receptors and voltage-gated calcium channels (VGCCs). This activates calcium-dependent signaling cascades that turn on transcription factors to induce transcription of target genes [19,20]. Neuronal activity-regulated gene induction occurs in two waves, based on the latency of their expression after stimulation. First, IEGs, including Arc and transcription factors, are activated rapidly and transiently within minutes of stimulation [21,22]. While induction of IEGs is the result of activation of pre-existing signaling pathways, de novo transcription of IEGs is essential for subsequent induction of the late-response genes (LRGs) [23].
What mechanisms govern rapid expression of the early-response genes? The transcriptional machinery is poised just downstream of the transcription start site (TSS) of IEGs, allowing fast transcriptional activation upon neuronal activity [24]. Further, regulatory genomic sequences, such as promoter and enhancer regions, have been extensively studied to map patterns of neuronal activation in response to distinct stimuli or animal behavior at the cellular level [25–29]. Discovery of these key regulatory elements in the c-fos and other IEGs facilitated the identification of transcription factors that bind these structures, and defined the upstream signaling cascades that lead to activity-dependent modifications of the factors [30–32]. Consequently, monitoring IEG transcription or the activity of a reporter gene constructed from regulatory regions of an IEG can report on the activity of signaling cascades.
To elucidate the transcriptional control of a gene, one must understand how much of the gene locus to evaluate. While many genes have regulatory elements within several kB of the TSS, long-range actions of enhancers are known [33,34]. Presumably, these actions reflect high-order chromatin structures that bring distal DNA elements in physical proximity to the gene in question. A common approach is to search for consensus DNA binding sites for well-known transcription factors in regions adjacent to the studied gene. While this approach can certainly discover regulatory DNA elements, it is inherently biased and bears the caveat that not all cognate sites are active. Thus, it is crucial to directly test function.
Previous Arc reporter gene studies by Kuhl and colleagues identified two serum response elements (SREs) positioned at ~0.9 and ~1.5 kb upstream of the transcription initiation site of the Arc gene. However, their requirement to induce transcription was inconclusive [35]. More recent work by the Bito and Finkbeiner laboratories uncovered regulatory elements in the Arc promoter region that are essential for activity-dependent transcriptional regulation [27,28] (Fig. 2). Using a DNaseI hypersensitivity assay, Pintchovski and colleagues applied an unbiased approach to look for open chromatin regions, structures assumed requisite for active translation [28]. This approach is beneficial as it overcomes the haunting concern associated with reporter gene assays where the DNA may not be fully chromatinized and, thus, may not reflect the physiological conditions of the gene [28]. This study identified two novel enhancer elements located ~6.5 and ~1.4 kb upstream of the Arc TSS and multiple highly conserved regions containing putative binding sites for factors associated with plasticity [28], such as the nuclear factor of activated T cells [36], nuclear factor kB [37] and myocyte-specific enhancement factor 2 (MEF2) [38]. The proximal enhancer region harbors two conserved “Zeste-like” elements that respond to synaptic activity and BDNF and convey transcriptional responses in an NMDAR-, PKA- and ERK-dependent fashion [28]. The distal enhancer bears a functional and highly conserved SRE that binds serum response factor (SRF) to allow regulation of Arc upon stimulation, including synaptic activity, BDNF and forskolin, an activator of adenylyl cyclase that generates cAMP [28] (Fig. 2).
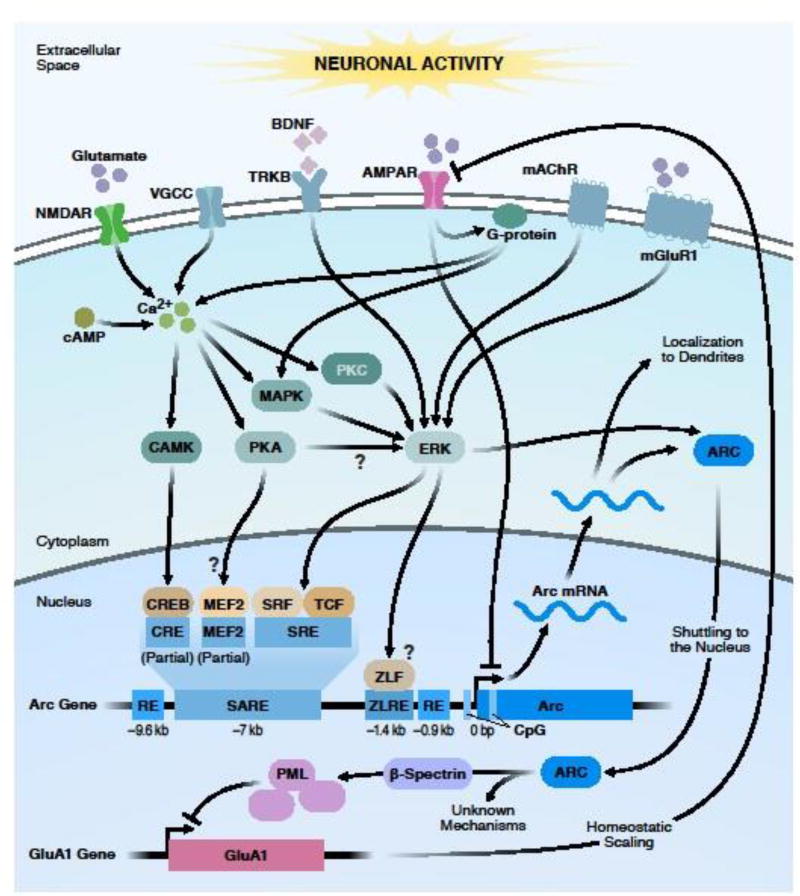
Fig. 2. Signaling cascades regulating Arc expression and its nuclear role.
Neuronal activity promotes intracellular calcium influx via NMDA and voltage gated calcium channels (VGCC). Arc transcription is induced through various signaling cascades. Signaling via NMDA receptors, TrkB and mGluR receptors, induces Arc transcription through one or multiple downstream effector molecules (kinases), such as calcium/calmodulin-dependent kinase (CAMK), protein kinase A (PKA), and protein kinase C (PKC). Several pathways converge upstream of extracellular-signal-regulated kinase (ERK). Signaling is further relayed on nuclear transcription factors and co-activators, including cAMP responsive element binding protein (CREB), myocyte enhancer factor 2 (MEF2), serum-response-factor (SRF), ternary complex factor (TCF), and a putative Zeste-like factor (ZLF) that bind distinct regulatory elements (blue boxes) in the Arc promoter, downstream of the transcription start site (see Sections 2 and 4 for details). Signaling through activation of AMPA receptors impedes Arc transcription by mechanisms involving G-protein signaling. After transcription, Arc mRNA is exported to the cytoplasm. Response elements (REs) in the Arc promoter also negatively regulate Arc transcription. Arc mRNA is localized to dendrites to serve local translation and synaptic function. Arc translocates to the nucleus where it increases the formation of PML nuclear bodies through β-spectrin, leading to reduced transcription of GluA1, indicative of a nuclear role in regulating homeostatic scaling. Nuclear Arc also interacts with the histone acetyltransferase and subunit of a chromatin-remodeling complex, Tip60 (not shown, see Section 5). Abbreviations: AMPAR, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor; bp, base pair; BDNF, brain-derived neurotrophic factor; cAMP, cyclic adenosine monophosphate; GluA1, AMPA-selective glutamate receptor 1; kb, kilobase; mAChR, muscarinic acetylcholine receptor; MAPK, mitogen-activated protein kinase; mGluR1, group 1 metabotropic glutamate receptors; NMDAR, N-methyl-D-aspartic acid receptor; PML, promyelocytic leukemia; TrkB, tropomyosin-receptor kinase B; SARE, synaptic activity-responsive element; SRE, serum response element; ZLRE; zeste-like response element; VGCC, voltage-gated calcium channel.
That SRF/SRE regulates Arc transcription implies a signaling pathway that connects gene expression, synaptic activity and ultimately behavior. Mice deficient in SRF had significant defects in learning and memory [39] and gene expression changes [40]. SRE-dependent transcription can be induced by stimulation with BDNF or synaptic activity via the Ras/ERK pathway (see Section 4). Besides, the Arc promoter contains a partial CRE site near the SRE [28] and an unidentified protein kinase A (PKA)-responsive region [27,28]. Likewise, Arc induction by calcium and cAMP in rat pheochromocytoma (PC12) cells (though not in neuronal cells) depends on signaling through PKA and MAPK-ERK [35], suggesting induction of Arc transcription in neurons is activated through other pathways. Using comparative genome mapping and luciferase reporter assays, Kawashima and colleagues identified a synaptic activity-responsive element (SARE) in the Arc promoter region that enables synapse-to-nucleus signaling in activated neurons [27]. This activity-sensor is a ~100-bp element ~7 kb upstream of the Arc TSS in the mouse genome and holds a cluster of neuronal activity-dependent cis-elements, including binding sites for CREB, SRF and MEF2 [27,41] (Fig. 2). While CREB, SRE, SRF, and MEF2 have been implicated in activity-dependent gene expression [38,40,42–45], the SARE element provides evidence of a clustering and requirement of these cis-elements in such proximity, necessary and sufficient to convey rapid, activity-induced Arc transcription in cultured neurons [27]. These studies identified elements within the Arc promoter region that recapitulate important features of the induction of Arc mRNA [2,27,28,46]. Further studies demonstrated the applicability of the Arc promoter for live imaging of cortical activity [47–50]. Bito and colleagues engineered an activity-dependent promoter, enhanced SARE (E-SARE) that allows activity-dependent, long-distance axonal tracing in vivo, expanding the repertoire of genetic approaches to dissect brain circuits [48].
3. Three-dimensional spatial and epigenetic regulation of Arc transcription
How are regulatory genomic sequences, such as promoter and enhancer regions, spatially regulated in 3D and how might this affect normal and aberrant cognitive function? At low transcriptional activity, the Arc promoter is occupied by the negative elongation factor (NELF) complex, stalling transcriptional machinery. A distal enhancer region of Arc produces regulatory RNA sequences that bind to the NELF complex when the enhancer region moves into proximity of the Arc promoter via distinct loop formations, thus liberating the Arc promoter from the negative regulator and promoting transcription [51,52]. Another intriguing model for enhancer-dependent regulation of transcription was described for other IEGs, including fos and neuronal PAS domain protein 4 (Npas4). Their promoter activities are low at baseline when the genomic sequence is in linear form. Transcription is activated upon topoisomerase IIβ-induced DNA double-strand breaks (DSBs) that mobilize promoter sequences into proximity with enhancer elements via short-range promoter-enhancer DNA loopings [52,53]. Such a model may be applicable to Arc by yet-unknown interactions (see Section 5). What effects do activity-regulated changes in the 3D genome and epigenetic modifications have on transcriptional mechanisms underlying learning, plasticity and cognition? Interestingly, mutations in genes encoding scaffolding proteins for the 3D genome and regulatory non-coding DNA were associated with disease-affecting cognitive functions, such as intellectual disability and autism [52,54,55]. DNA methylation and demethylation are epigenetic processes that underlie long-term changes, including maintenance and persistence of memory by regulating gene transcription [56], and dysregulated DNA methylation may contribute to human cognitive disorders [57]. Arc contains so-called CpG sites in its promoter and intragenic locus that recruit methyl-DNA binding proteins to methylate cytosines [58] (Fig. 2). Penner and colleagues [58] reported changes in the methylation status of Arc in the hippocampus: Aged rats had more methylation than adult rats under resting conditions [58]. Changes in DNA methylation after exploratory behavior suggested a regulatory effect on the transcriptional expression of Arc in response to certain behaviors [58] and an additional epigenetic component regulating Arc.
4. Signaling cascades regulating Arc transcription
Arc expression increases after seizures in the hippocampus and cortex [1,2]. However, additional stimuli induce Arc, including neuronal activity in response to learning [11,59–61]. What downstream signaling cascades relay the changes in neuronal activity to alterations in Arc transcription?
NMDA receptor activation seems to be necessary to induce Arc transcription and important for localizing Arc mRNA [1,2,62]. Although the action of both NMDA and AMPA receptors is required to mediate changes in synaptic efficacy, the potential regulatory role of AMPA receptors underlying Arc function initially remained unexplored. Rao and colleagues demonstrated, low levels of Arc transcription at baseline conditions can be further decreased by selectively activating AMPA receptors [63]. Conversely, inhibiting AMPA receptors strongly potentiated Arc induction, suggesting that AMPA receptors are negative regulators of Arc transcription [63] (Fig. 2). This function is conveyed via a mechanism coupling AMPA receptor function to a pertussis toxin-sensitive G protein, leaving translation and stability of Arc mRNA unaffected [63]. Selectively blocking AMPA or NMDA receptors upon stimulation of Arc transcription via BDNF highlighted a bidirectional regulation of Arc by NMDA and AMPA receptors, whereby the ratio of NMDA-to-AMPA receptor activation may determine Arc expression [63]. This finding is relevant: it uncovered a novel aspect of AMPA receptor function in regulating gene expression and pointed at how Arc may function in LTP and LTD to produce opposing changes in synaptic strength [63,64].
Arc transcription is accelerated by activating muscarinic acetylcholine receptors [65] and BDNF via its receptor TrkB [28,46,63] (Fig 2.). BDNF and synaptic activity are linked: BDNF induces synaptic activity, and synaptic activity stimulates BDNF release [66]. Endogenous BDNF is not required for synaptic activity-induced Arc transcription, but synaptic activity is required for BDNF-induced Arc expression [63]. Arc induction by BDNF depends on mitogen-activated protein kinases (MAPK) [46]. The extracellular signal-regulated protein kinase (ERK) links signaling pathways downstream of the plasma-membrane receptors to Arc expression. This signaling also includes Arc transcriptional activation via group 1 metabotropic glutamate receptors (mGluRs) [8,67,68]. Upon activation, ERK phosphorylates coactivators of the SRF, such as Elk-1, a ternary complex factor (TCF), which binds SREs in the Arc promoter to activate transcription [69]. However, ERK regulates many transcription factors besides TCF and SRF [70]. Induction of BDNF-LTP is coupled to ERK-dependent phosphorylation of calcium-and cAMP-response element binding protein (CREB)[46], a transcription factor that is required for the transcription of CRE-driven genes [71–73]. ERK also signals through a Zeste-like factor that interacts with a Zeste-like response element in the Arc promoter [28] (Fig. 2). Zeste was discovered as an invertebrate transcription factor without a mammalian ortholog [74] nor a role in plasticity-related neuronal gene expression [75]. Pintchovski and colleagues showed the Zeste-like site responded transcriptionally to synaptic activity, depending on NMDA receptor, PKA and ERK [28].
The Arc promoter has a partial MEF site near the SRE [28] that may be regulated by signaling pathways other than those that activate CREB (Fig. 2). MEF-dependent transcription is activated, for instance, by calcium influx that causes calcium-dependent dephosphorylation of MEF2 proteins [38]. Reducing MEF2 expression in hippocampal neurons promotes formation of excitatory synapses [38], suggesting MEF2 target genes affect synaptic scaling. Consistently, Arc induction by MEF2 could prompt AMPA receptor internalization [19,64].
5. The role of Arc in the nucleus
Although Arc was previously detected in the nucleus [76,77], studies addressing its nuclear function have lagged behind. Bloomer and colleagues found substantial expression of Arc in the nuclei of hippocampal neurons and HEK 293T cells where it is associated with promyelocytic leukemia (PML) nuclear bodies [77]. These sites are found in most mammalian cell nuclei and associated with transcriptional regulation [78–80]. Importantly, Arc co-localized with βSpIVΣ5, an isoform of spectrin that associates with PML nuclear bodies and the nuclear matrix [81]. Co-expression of Arc and βSpIVΣ5 increased PML bodies in HEK 293T cells, implying that Arc plays a role in PML nuclear body formation [77]. However, the mechanisms governing Arc nuclear localization were enigmatic. Work by Korb and colleagues [13] suggested Arc nuclear localization affects synaptic plasticity by regulating PML nuclear bodies that are important for neuronal development and associated with neurodegenerative disorders [13,82–84]. After exposure of mice to a novel environment and pharmacological induction of prolonged increased activity, Arc becomes enriched in neuronal nuclei in vivo and in vitro [13] (Fig 1.). Yet, what drives Arc nuclear localization? Korb and coworkers identified multiple cis-acting elements that direct it to nuclei, including a Pat7 nuclear localization signal, a nuclear export signal, and a nuclear retention domain. While short periods of activity decrease Arc nuclear localization, long periods increase it [13]. Moreover, using pharmacology, Korb and colleagues were able to deduce the signaling pathways that regulate Arc nuclear localization upon activity: Inhibition of the MEK-ERK pathway hampered the import of Arc into the nucleus in response to a long BDNF treatment, suggesting this signaling is critical [13]. Nikolaienko and co-workers recently extended these findings by showing that ERK directly phosphorylates Arc and promotes the early cytosolic localization of Arc in stimulated hippocampal neuronal cultures while it does not alter its slow nuclear accumulation [85].
Strikingly, upon activity-dependent translocation to the nucleus, Arc regulates homeostatic plasticity by increasing expression levels of PML, which hampers GluA1 (also called Gria1) transcription and leads to downscaling of synaptic strength [13] (Fig. 2). PML nuclear bodies presumably regulate GluA1 by sequestering and degrading CREB-binding protein (CBP), a well-characterized interaction partner of PML which facilitates transcription of CRE-dependent activity-regulated genes, including GluA1 [13,86]. While Arc modulates synaptic strength by decreasing surface AMPA receptor expression, this effect was primarily associated with its function at the synapse [7,9,16]. However, Arc also reduces total GluA1 levels [7], suggesting an effect additional to regulating endocytosis. Indeed, by expressing Arc that is excluded from the nucleus hampered its ability to decrease surface GluA1 levels [13], implying Arc contributes to the scaling effect in the nucleus.
Others have reported that Arc interacts with other factors that might provide additional mechanisms by which it could regulate gene expression. Nuclear Arc interacts with the histone acetyltransferase and subunit of a chromatin-remodeling complex, Tip60 [87]. Increased synaptic activity promotes recruitment of this complex to the Arc promoter where it favors transcriptional activation by regulating the histone mark H3K9me2 [88]. Additionally, acute cocaine treatment in mice induces accumulation of Arc in striatal medium spiny neurons where it acts as a brake on chromatin remodeling and gene regulation [89].
Remarkably, exploration of a novel environment and increased neuronal activity by sensory or optogenetic stimulation leads to DNA DSBs in neurons of adult wild-type mice, which is augmented in a transgenic mouse model of Alzheimer’s disease [90]. Neuronal activity induces topoisomerase IIβ (Topo IIβ)-mediated DSBs in the promoters of IEGs, such as Fos, Npas4 and Egr1 [53]. Knockdown of Topo IIβ mitigated DSB formation and decreased early-response gene expression upon neuronal stimulation [53], suggesting a novel mechanism for regulating transcriptional induction of IEGs. Although Arc is a relatively short gene and Topo IIβ may preferentially modulate transcription of long genes [91,92], Arc translocation to the nucleus might enable interactions with Topo IIβ, activating transcriptional processes relevant to homeostatic plasticity and cognition. Recurrent DSBs have been allocated to long genes associated with neural circuit formation and accurate cognitive function in neural progenitor cells [93,94], suggesting DSBs are a physiologically relevant and beneficial mechanism of activity-dependent gene regulation in the brain.
6. Dissecting Arc’s role in cognitive function
Proper cognitive function enables animals to respond to environmental stimuli in an appropriate way. Learning and memory are important substrates of cognition and are regulated by experience-driven alterations in neuronal connectivity.
Arc-deficient animals have deficits in memory task performance that require long-term recall [6,10,12,95] which is not evident when animals are tested over shorter times. Conventionally, this has been tightly linked to deficits in memory consolidation. If true, this would point to a critical function of Arc in learning and memory at a late phase in the process.
Furthermore, Arc is induced during sleep [96]. Sleep is perceived to help consolidate memories formed during the day [97,98]. Sleep deprivation leads to a brain-region specific increase of Arc mRNA [99,100], without affecting Arc protein expression [96]. Thus, in addition to Arc’s role during the actual experience, it may act later to convert short-term to long-term memories, raising the question as to how Arc mediates long-term memory formation.
A prevailing view is that activity induces Arc transcription to stabilize specific synapses [5,15,62]. But how would this work? Transcription is rapid [101], however, is it rapid enough to generate Arc mRNA, transport it to specific, particularly distally localized synapses, and translate it in time to strengthen the synapse that initially triggered synaptic potentiation? If this is not true, what is the model? Does activity induce local Arc translation and nuclear Arc transcription in parallel, thus enabling Arc transcription to refill sites where Arc mRNA was depleted after local protein synthesis? Presumably, some neuronal dendrites receive input from different synaptic circuits. Different synapses on the same dendrite could simultaneously be undergoing LTP and LTD, two forms of activity-dependent synaptic plasticity that presumably act in concert to form lasting memories. While synapse-specific changes in efficacy are critical for memory formation, those changes appear to be relative to other synapses within the same neuron, targeted by other circuits and must occur under conditions that keep the overall excitability of the neuron within a certain range [102]. This may be the only way to functionally distinguish a synaptic circuit that represents a memory without spuriously strengthening synapses that are part of other circuits, which would reduce the specificity and resolution of the memory. If the requirement for Arc in memory consolidation was solely due to a critical role in promoting synaptic strength [5,15], Arc KO mice might be hypoexcitable because they lack the critical factor to strengthen synapses in response to excitation. In contrast, Arc may primarily regulate learning and memory through synaptic scaling [13,16]. Rather than being targeted to distinct active synapses to strengthen them, Arc might enable long-term memory formation by scaling synaptic strength after stimulation (Fig. 3). In this case, one would predict the opposite effect. The brains of Arc-deficient mice should be hyperexcitable from lack of negative feedback. This way, Arc might preserve any relative differences in synaptic efficacy that were introduced, while keeping the overall level of synaptic strengths in a range where new learning can occur. Indeed, Peebles and colleagues demonstrated the latter model is true: Behavioral analysis and electroencephalogram (EEG) data showed that Arc KO and mutant mice have increased seizure susceptibility and network hyperexcitability [16]. The capacity of hyperexcitable circuits to store enduring memories is diminished, implying that hyperexcitability or deficits in synaptic homeostasis per se are sufficient for a memory consolidation effect, presumably due to a reduced dynamic range. A model where Arc mediates memory through scaling fits well with the aspect of timing. Instead of producing Arc in time to strengthen the synapse that triggered the memory, Arc could act as a “chaser”, induced by synaptic activity but acting after synaptic strengthening occurred to bring global activity back within an acceptable range, while preserving relative differences in synaptic strength induced by the memory-forming experience (Fig. 3). Thus, if Arc-deficient mice have a fundamental defect in synaptic scaling, the spatial maps they form may lack the resolution or the stability of maps that are formed under normal conditions.
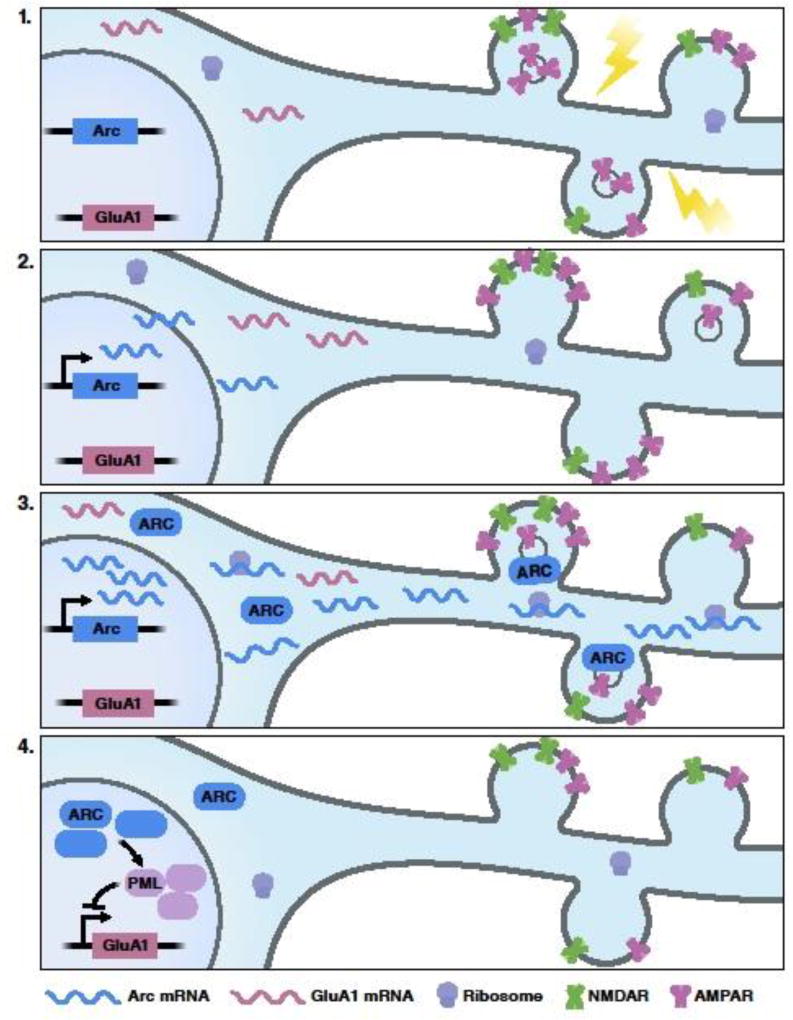
Fig. 3. A dual role for Arc in regulating homeostatic scaling.
The schematic shows a scenario in which a dendritic region receives input from different synaptic circuits (panel 1). Distinct spines on the same dendrite simultaneously undergo synaptic strengthening (LTP, two spines) and synaptic weakening (LTD, one spine), illustrated via insertion or internalization of AMPA receptors to/from the plasma membrane, respectively. In the nucleus, neuronal activity induces Arc transcription approximately within minutes to half an hour after stimulation (panel 2). Subsequently, Arc mRNA is exported to the cytoplasm and transported to dendrites, where it may serve as a template for local protein synthesis (panel 3). Locally available Arc protein decreases synaptic strength by promoting surface AMPA receptor endocytosis (panel 3). Hours after stimulation, Arc is almost exclusively localized to the nucleus where it further regulates homeostatic scaling by downregulating GluA1, the gene encoding the AMPA-selective glutamate receptor 1 (panel 4). After strengthening has occurred, Arc may act to bring the overall excitability of the neuron (depicted by the number of AMPA receptors throughout the panels) back to a baseline range, while preserving relative differences in synaptic efficacy, critical for memory formation (see Section 6). Abbreviations, see Fig. 2.
Activity regulates Arc at almost every conceivable level from the production and trafficking of Arc transcripts, the extent of Arc mRNA translation, the localization, translocation and post-translational modification of Arc protein and the decay of Arc mRNA and protein. The emerging picture underscores the centrality of Arc to the biological mechanisms that convert transient experience into long term adaptive changes in the brain. Unsurprisingly, while the above model explains many of the observations in the field, some studies suggest that Arc contributes to learning and memory via additional mechanisms. For instance, work by Bramham and colleagues [15,103] indicates a direct role for Arc in LTP, suggesting other mechanisms in addition to Arc’s role in scaling.
7. Implications of Arc in homeostatic plasticity and disease
Consistent with the idea of imbalanced excitation and inhibition, work on neurological disorders, including epilepsy, Alzheimer’s disease (AD) and autism spectrum disorder (ASD), linked hyperexcitable circuits with deficits in long-term memory formation [104–106]. Palop and colleagues observed attenuated Arc expression in transgenic mice with neuronal production of human amyloid precursor protein (hAPP) and hAPP-derived amyloid-β (Aβ) peptides, a hallmark of AD [106]. Similar to the Arc KO and mutant mice studied by Peebles and colleagues [16] (see Section 6), the hippocampi of these animals were hyperexcitable, and periodic seizures induced very high expression of Arc in some cells [106]. Consistent with a role for Arc in regulating synaptic homeostasis, insufficient levels of Arc may prevent the network from scaling the excitability of synapses back into a normal range after stimulation. This would lead to hyperexcitability at baseline and a reduced network capacity to store new memories due to a greatly reduced dynamic range. Evidence supporting abnormal Arc levels in AD patient brain autopsies and mouse models of AD is compelling [107], yet controversial [108]. Given that Arc was discovered in a search for genes expressed after seizures [1,2], one explanation may be related to whether an individual had an epileptic event recently. Sparsity of Arc induction in hippocampal neurons is associated with a mouse model of Down syndrome (DS) [109], the leading chromosomal deficit causing intellectual disability. DS yields an extra copy of APP, and the brains of DS patients contain amyloid plaques and other pathology reminiscent of AD [110], suggesting some related mechanisms. Thus, Aβ may alter synaptic plasticity by mitigating Arc levels that lead to dysregulated homeostatic scaling and ultimately, to a hyperexcitable brain that is poorly suited to form memories.
These studies suggest Arc contributes to the pathophysiology of neurological conditions that affect cognitive function. The atomic structure of the Arc N-terminal lobe is inhibited by small chemicals, implying that Arc’s action may be targeted by drugs [18,111] and may open up new avenues to cures.
8. Conclusion
In light of the implications of its regulation of synaptic plasticity and memory consolidation, Arc has been studied from various perspectives (reviewed in this issue). Here, we used a bottom-up approach to describe the regulation of Arc transcription from regulatory elements in the Arc promoter via signal transduction pathways. The multifaceted array of regulatory elements and their interplay with key transcription factors and other effector molecules that govern Arc transcription is complex and not yet fully understood. Although we can now link certain response elements that are necessary for Arc induction to specific stimuli, most of the investigated conditions are fairly non-physiological, often involving tonic application of an agonist. In vivo, however, synapses activate several pathways simultaneously (rather than in isolation), and the temporal pattern of activation is crucial, raising the question as to how distinct signaling pathways work in combination and via different patterns to regulate Arc in vivo. It is intriguing to picture the Arc promoter as a unique combination lock that integrates and processes combinations of different types and patterns of stimuli to activate different factors bound to distinct response elements to induce transcription in a way no factor can by itself.
Once Arc is induced, it further mediates its effects through a variety of mechanisms in a temporally and spatially regulated manner [3,5]. Shortly after stimulation, Arc is localized predominantly to the cytoplasm where it is involved in AMPA receptor endocytosis [7,9]. Within hours of stimulation, however, Arc is almost exclusively localized to the nucleus where it modulates homeostatic scaling through interaction with PML nuclear bodies [13] and other yet unknown mechanisms. A role of Arc in regulating memory at a later time point through homeostatic scaling fits well with data from mouse models of cognitive disorders including AD and epilepsy where hyperexcitability is a prevalent theme [104,106]. Insufficient levels of Arc may hamper the network from scaling the excitability of synapses back to a baseline range following stimulation, leading to hyperexcitability and a diminished network capacity to store new memories.
Ultimately, further research will help to better understand the relationship between the signaling that induces Arc and the complex physiological response that it produces.
Highlights.
Arc induction is regulated by AMPA- and NMDA-receptor signaling
Upstream functional response elements critically regulate Arc transcription
Nuclear Arc regulates homeostatic plasticity through GluA1 transcription
Arc-deficiency may prevent circuits from scaling and lead to hyperexcitability
Acknowledgments
We thank members of the Finkbeiner lab for valuable comments on the manuscript, Giovanni Maki and Teresa Roberts for graphical support, and Gary Howard for editorial input. We apologize to all our colleagues whose excellent work we could not cite due to limitations in space.
This work was supported by the National Institutes of Health R01 NS083390, NSF EAGER BRAIN Initiative, the Taube/Koret Center for Neurodegenerative Disease, and the Gladstone Institutes.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest: none.
References
- 1.Link WW, Konietzko UU, Kauselmann GG, Krug MM, Schwanke BB, Frey UU, Kuhl DD. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc. Natl. Acad. Sci. U. S. A. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 3.Korb E, Finkbeiner S. Arc in synaptic plasticity: From gene to behavior. Trends Neurosci. 2011;34:591–598. doi: 10.1016/j.tins.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shepherd JD, Bear MF. New views of Arc, a master regulator of synaptic plasticity. Nat. Neurosci. 2011;14:279–284. doi: 10.1038/nn.2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bramham CR, Alme MN, Bittins M, Kuipers SD, Nair RR, Pai B, Panja D, Schubert M, Soule J, Tiron A, Wibrand K. The Arc of synaptic memory. Exp. Brain Res. 2010;200:125–140. doi: 10.1007/s00221-009-1959-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SFF, Blanquet V, Wurst W, Salmen B, Bösl MRR, Lipp HP, Grant SGGN, Bliss TVP, Wolfer DP, Kuhl D. Arc/Arg3.1 Is Essential for the Consolidation of Synaptic Plasticity and Memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 Interacts with the Endocytic Machinery to Regulate AMPA Receptor Trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation Factor 2 and Fragile X Mental Retardation Protein Control the Dynamic Translation of Arc/Arg3.1 Essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased Expression of the Immediate-Early Gene Arc/Arg3.1 Reduces AMPA Receptor-Mediated Synaptic Transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guzowski JF, Miyashita T, Chawla MK, Sanderson J, Maes LI, Houston FP, Lipa P, McNaughton BL, Worley PF, Barnes CA. Recent behavioral history modifies coupling between cell activity and Arc gene transcription in hippocampal CA1 neurons. Proc. Natl. Acad. Sci. 2006;103:1077–1082. doi: 10.1073/pnas.0505519103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the leg Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/S0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 12.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, McGaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J. Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. doi:20/11/3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korb E, Wilkinson CL, Delgado RN, Lovero KL, Finkbeiner S. Arc in the nucleus regulates PML-dependent GluA1 transcription and homeostatic plasticity. Nat. Neurosci. 2013;16:874–883. doi: 10.1038/nn.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alberi L, Liu S, Wang Y, Badie R, Smith-Hicks C, Wu J, Pierfelice TJ, Abazyan B, Mattson MP, Kuhl D, Pletnikov M, Worley PF, Gaiano N. Activity-Induced Notch Signaling in Neurons Requires Arc/Arg3.1 and Is Essential for Synaptic Plasticity in Hippocampal Networks. Neuron. 2011;69:437–444. doi: 10.1016/j.neuron.2011.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Messaoudi E, Kanhema T, Soule J, Tiron A, Dagyte G, da Silva B, Bramham CR. Sustained Arc/Arg3.1 Synthesis Controls Long-Term Potentiation Consolidation through Regulation of Local Actin Polymerization in the Dentate Gyrus In Vivo. J. Neurosci. 2007;27:10445–10455. doi: 10.1523/JNEUROSCI.2883-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peebles CL, Yoo J, Thwin MT, Palop JJ, Noebels JL, Finkbeiner S. Arc regulates spine morphology and maintains network stability in vivo. Proc. Natl. Acad. Sci. 2010;107:18173–18178. doi: 10.1073/pnas.1006546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC Factor eIF4AIII Modulates Synaptic Strength and Neuronal Protein Expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 18.Campioni MR, Finkbeiner S. Going retro: Ancient viral origins of cognition. Neuron. 2015;86:346–348. doi: 10.1016/j.neuron.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 19.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bito H, Deisseroth K, Tsien RW. Ca2+-dependent regulation in neuronal gene expression. Curr. Opin. Neurobiol. 1997;7:419–429. doi: 10.1016/S0959-4388(97)80072-4. [DOI] [PubMed] [Google Scholar]
- 21.Flavell SW, Greenberg ME. Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System. Annu. Rev. Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- 23.West AE, Greenberg ME. Neuronal activity-regulated gene transcription in synapse development and cognitive function. Cold Spring Harb. Perspect. Biol. 2011;3:1–21. doi: 10.1101/cshperspect.a005744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang J-Y, Daigle KR, Fenn JD, Adelman K, Dudek SM. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat. Neurosci. 2011;14:848–856. doi: 10.1038/nn.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smeyne RJ, Curran T, Morgan JI. Temporal and spatial expression of a fos-lacZ transgene in the developing nervous system. Mol. Brain Res. 1992;16:158–162. doi: 10.1016/0169-328X(92)90206-Q. [DOI] [PubMed] [Google Scholar]
- 26.Barth RC, Gerkin AL, Dean KL. Alteration of Neuronal Firing Properties after In Vivo Experience in a FosGFP Transgenic Mouse. J. Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc. Natl. Acad. Sci. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pintchovski SA, Peebles CL, Joo Kim H, Verdin E, Finkbeiner S. The Serum Response Factor and a Putative Novel Transcription Factor Regulate Expression of the Immediate-Early Gene Arc/Arg3.1 in Neurons. J. Neurosci. 2009;29:1525–1537. doi: 10.1523/JNEUROSCI.5575-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuno H. Regulation and function of immediate-early genes in the brain: Beyond neuronal activity markers. Neurosci. Res. 2011;69:175–186. doi: 10.1016/j.neures.2010.12.007. [DOI] [PubMed] [Google Scholar]
- 30.Sheng M, Greenberg ME. The regulation and function of c-fos and other immediate early genes in the nervous system. Neuron. 1990;4:477–485. doi: 10.1016/0896-6273(90)90106-P. [DOI] [PubMed] [Google Scholar]
- 31.Sheng M, Dougan ST, McFadden G, Greenberg ME. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol. Cell. Biol. 1988;8:2787–96. doi: 10.1128/MCB.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- 33.Vernimmen D, De Gobbi M, Sloane-Stanley JA, Wood WG, Higgs DR. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 2007;26:2041–2051. doi: 10.1038/sj.emboj.7601654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Noonan JP, McCallion AS. Genomics of Long-Range Regulatory Elements. Annu. Rev. Genomics Hum. Genet. 2010;11:1–23. doi: 10.1146/annurev-genom-082509-141651. [DOI] [PubMed] [Google Scholar]
- 35.Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. J. Neurosci. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. doi:21/15/5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groth RD, Mermelstein PG. Brain-derived neurotrophic factor activation of NFAT (nuclear factor of activated T-cells)-dependent transcription: a role for the transcription factor NFATc4 in neurotrophin-mediated gene expression. J. Neurosci. 2003;23:8125–8134. doi: 10.1523/JNEUROSCI.23-22-08125.2003. doi:23/22/8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-κB functions in synaptic signaling and behavior. Nat. Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 38.Flavell GM, Cowan CW SW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C. Activity-Dependent Regulation of MEF2 Transcription Factors Suppresses Excitatory Synapse Number. Science (80-. ) 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 39.Etkin A, Alarcón JM, Weisberg SP, Touzani K, Huang YY, Nordheim A, Kandel ER. A Role in Learning for SRF: Deletion in the Adult Forebrain Disrupts LTD and the Formation of an Immediate Memory of a Novel Context. Neuron. 2006;50:127–143. doi: 10.1016/j.neuron.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 40.Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schütz G, Linden DJ, Ginty DD. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- 41.Inoue M, Yagishita-Kyo N, Nonaka M, Kawashima T, Okuno H, Bito H. Synaptic activity-responsive element (SARE): A unique genomic structure with an unusual sensitivity to neuronal activity. Commun. Integr. Biol. 2010;3:443–6. doi: 10.4161/cib.3.5.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kandel ER. The molecular biology of memory storage: A dialogue between gene and synapses. Science (80-. ) 2001;294:1030–1038. doi: 10.1126/science.1067020. [DOI] [PubMed] [Google Scholar]
- 43.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 44.Finkbeiner S, Tavazoie SF, Maloratsky a, Jacobs KM, Harris KM, Greenberg ME. CREB: a major mediator of neuronal neurotrophin responses. Neuron. 1997;19:1031–1047. doi: 10.1016/S0896-6273(00)80395-5. [DOI] [PubMed] [Google Scholar]
- 45.Shaywitz AJ, Greenberg ME. CREB: A Stimulus-Induced Transcription Factor Activated by A Diverse Array of Extracellular Signals. Annu. Rev. Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 46.Ying S, Futter M, Rosenblum K, Webber MJ, Hunt SP, Bliss TVP, Bramham CR. Brain-Derived Neurotrophic Factor Induces Long-Term Potentiation in Intact Adult Hippocampus : Requirement for ERK Activation Coupled to CREB and Upregulation of Arc Synthesis. J. Neurosci. 2002;22:1532–1540. doi: 10.1523/JNEUROSCI.22-05-01532.2002. doi:22/5/1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In Vivo Two-Photon Imaging Reveals a Role of Arc in Enhancing Orientation Specificity in Visual Cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- 48.Kawashima T, Kitamura K, Suzuki K, Nonaka M, Kamijo S, Takemoto-Kimura S, Kano M, Okuno H, Ohki K, Bito H. Functional labeling of neurons and their projections using the synthetic activity-dependent promoter E-SARE. Nat. Methods. 2013;10:889–895. doi: 10.1038/nmeth.2559. [DOI] [PubMed] [Google Scholar]
- 49.Eguchi M, Yamaguchi S. In vivo and in vitro visualization of gene expression dynamics over extensive areas of the brain. Neuroimage. 2009;44:1274–1283. doi: 10.1016/j.neuroimage.2008.10.046. [DOI] [PubMed] [Google Scholar]
- 50.Izumi H, Ishimoto T, Yamamoto H, Nishijo H, Mori H. Bioluminescence imaging of Arc expression enables detection of activity-dependent and plastic changes in the visual cortex of adult mice. Brain Struct. Funct. 2011;216:91–104. doi: 10.1007/s00429-010-0297-2. [DOI] [PubMed] [Google Scholar]
- 51.Schaukowitch K, Joo JY, Liu X, Watts JK, Martinez C, Kim TK. Enhancer RNA facilitates NELF release from immediate early genes. Mol. Cell. 2014;56:29–42. doi: 10.1016/j.molcel.2014.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rajarajan P, Gil SE, Brennand KJ, Akbarian S. Spatial genome organization and cognition. Nat. Rev. Neurosci. 2016;17:681–691. doi: 10.1038/nrn.2016.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015;161:1592–1605. doi: 10.1016/j.cell.2015.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gregor A, Oti M, Kouwenhoven EN, Hoyer J, Sticht H, Ekici AB, Kjaergaard S, Rauch A, Stunnenberg HG, Uebe S, Vasileiou G, Reis A, Zhou H, Zweier C. De novo mutations in the genome organizer CTCF cause intellectual disability. Am. J. Hum. Genet. 2013;93:124–131. doi: 10.1016/j.ajhg.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turner TN, Hormozdiari F, Duyzend MH, McClymont SA, Hook PW, Iossifov I, Raja A, Baker C, Hoekzema K, Stessman HA, Zody MC, Nelson BJ, Huddleston J, Sandstrom R, Smith JD, Hanna D, Swanson JM, Faustman EM, Bamshad MJ, Stamatoyannopoulos J, Nickerson DA, McCallion AS, Darnell R, Eichler EE. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am. J. Hum. Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr. Opin. Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gräff J, Mansuy IM. Epigenetic dysregulation in cognitive disorders. Eur. J. Neurosci. 2009;30:1–8. doi: 10.1111/j.1460-9568.2009.06787.x. [DOI] [PubMed] [Google Scholar]
- 58.Penner MR, Roth TL, Chawla MK, Hoang LT, Roth ED, Lubin FD, Sweatt JD, Worley PF, Barnes CA. Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol. Aging. 2011;32:2198–2210. doi: 10.1016/j.neurobiolaging.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ramirez-Amaya BC, Vazdarjanova A V, Mikhael D, Rosi S, Worley PF. Spatial Exploration-Induced Arc mRNA and Protein Expression: Evidence for Selective, Network-Specific Reactivation. J. Neurosci. 2005;25:1761–1768. doi: 10.1523/JNEUROSCI.4342-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Montag-Sallaz MMD. Learning-Induced arg 3.1/arc mRNA Expression in the Mouse Brain. Learn. Mem. 2003;10:99–107. doi: 10.1101/lm.53403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kubik S, Miyashita T, Guzowski JF. Using immediate-early genes to map hippocampal subregional functions. Learn. Mem. 2007;14:758–770. doi: 10.1101/lm.698107. [DOI] [PubMed] [Google Scholar]
- 62.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/S0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 63.Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat. Neurosci. 2006;9:887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- 64.Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 Mediates Homeostatic Synaptic Scaling of AMPA Receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Teber I, Köhling R, Speckmann E-J, Barnekow A, Kremerskothen J. Muscarinic acetylcholine receptor stimulation induces expression of the activity-regulated cytoskeleton-associated gene (ARC) Brain Res. Mol. Brain Res. 2004;121:131–136. doi: 10.1016/j.molbrainres.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 66.Poo M. Neurotrophins as synaptic modulators. Nat. Rev. Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- 67.Wang Y, Zheng F, Zhou X, Sun Z, Wang H. Converging signal on ERK1/2 activity regulates group I mGluR-mediated Arc transcription. Neurosci. Lett. 2009;460:36–40. doi: 10.1016/j.neulet.2009.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid Translation of Arc/Arg3.1 Selectively Mediates mGluR-Dependent LTD through Persistent Increases in AMPAR Endocytosis Rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Posern G, Treisman R. Actin’ together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 2006;16:588–596. doi: 10.1016/j.tcb.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 70.Johnson GL, Lapadat R. Mitogen-activated protein kinase pathways mediated by ERK, JNK, and p38 protein kinases. Science (80-. ) 2002;298:1911–1912. doi: 10.1126/science.1072682. [DOI] [PubMed] [Google Scholar]
- 71.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting ltp in area ca1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/S0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 72.Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, Storm DR. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/S0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- 73.Davis S, Vanhoutte P, Pages C, Caboche J, Laroche S. The MAPK/ERK cascade targets both Elk-1 and cAMP response element-binding protein to control long-term potentiation-dependent gene expression in the dentate gyrus in vivo. J. Neurosci. 2000;20:4563–4572. doi: 10.1523/JNEUROSCI.20-12-04563.2000. doi:20/12/4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Levine SS, Weiss A, Erdjument-Bromage H, Shao Z, Tempst P, Kingston RE. The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol. Cell. Biol. 2002;22:6070–6078. doi: 10.1128/MCB.22.17.6070-6078.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Biggin MD, Bickel S, Benson M, Pirrotta V, Tjian R. Zeste encodes a sequence-specific transcription factor that activates the Ultrabithorax promoter in vitro. Cell. 1988;53:713–722. doi: 10.1016/0092-8674(88)90089-X. [DOI] [PubMed] [Google Scholar]
- 76.Irie Y, Yamagata K, Gan Y, Miyamoto K, Do E, Kuo C-H, Taira E, Miki N. Molecular Cloning and Characterization of Amida, a Novel Protein Which Interacts with a Neuron-specific Immediate Early Gene Product Arc, Contains Novel Nuclear Localization Signals, and Causes Cell Death in Cultured Cells. J. Biol. Chem. 2000;275:2647–2653. doi: 10.1074/jbc.275.4.2647. [DOI] [PubMed] [Google Scholar]
- 77.Bloomer WAC, VanDongen HMA, VanDongen AMJ. Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia (PML) bodies. Brain Res. 2007;1153:20–33. doi: 10.1016/j.brainres.2007.03.079. [DOI] [PubMed] [Google Scholar]
- 78.Wang J, Shiels C, Sasieni P, Wu PJ, Islam SA, Freemont PS, Sheer D. Promyelocytic leukemia nuclear bodies associate with transcriptionally active genomic regions. J. Cell Biol. 2004;164:515–526. doi: 10.1083/jcb.200305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Borden KLB. Pondering the puzzle of PML (promyelocytic leukemia) nuclear bodies: Can we fit the pieces together using an RNA regulon? Biochim. Biophys. Acta - Mol. Cell Res. 2008;1783:2145–2154. doi: 10.1016/j.bbamcr.2008.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 81.Tse WT, Tang J, Jin O, Korsgren C, John KM, Kung AL, Gwynn B, Peters LL, Lux SE. A New Spectrin, βIV, Has a Major Truncated Isoform that Associates with Promyelocytic Leukemia Protein Nuclear Bodies and the Nuclear Matrix. J. Biol. Chem. 2001;276:23974–23985. doi: 10.1074/jbc.M009307200. [DOI] [PubMed] [Google Scholar]
- 82.Regad T, Bellodi C, Nicotera P, Salomoni P. The tumor suppressor Pml regulates cell fate in the developing neocortex. Nat. Neurosci. 2009;12:132–140. doi: 10.1038/nn.2251. [DOI] [PubMed] [Google Scholar]
- 83.Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- 84.Yamada M, Sato T, Shimohata T, Hayashi S, Igarashi S, Tsuji S, Takahashi H. Interaction between Neuronal Intranuclear Inclusions and Promyelocytic Leukemia Protein Nuclear and Coiled Bodies in CAG Repeat Diseases. Am. J. Pathol. 2001;159:1785–1795. doi: 10.1016/S0002-9440(10)63025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nikolaienko O, Eriksen MS, Patil S, Bito H, Bramham CR. Stimulus-evoked ERK-dependent phosphorylation of activity-regulated cytoskeleton-associated protein (Arc) regulates its neuronal subcellular localization. Neuroscience. 2017;360:68–80. doi: 10.1016/j.neuroscience.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 86.St-Germain JR, Chen J, Li Q. Involvement of PML nuclear bodies in CBP degradation through the ubiquitin-proteasome pathway. Epigenetics. 2008;3:342–349. doi: 10.4161/epi.3.6.7203. [DOI] [PubMed] [Google Scholar]
- 87.Wee CL, Teo S, Oey NE, Wright GD, VanDongen HMA, VanDongen AMJ. Nuclear Arc Interacts with the Histone Acetyltransferase Tip60 to Modify H4K12 Acetylation. Neuronal Excit. 2014;0:20. doi: 10.1523/ENEURO.0019-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Oey NE, Leung HW, Ezhilarasan R, Zhou L, Beuerman RW, VanDongen HMA, VanDongen AMJ. A Neuronal Activity-Dependent Dual Function Chromatin-Modifying Complex Regulates Arc Expression(1,2,3) eNeuro. 2015;2 doi: 10.1523/ENEURO.0020-14.2015. ENEURO.0020-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Salery M, Dos Santos M, Saint-Jour E, Moumnè L, Pagès C, Kappès V, Parnaudeau S, Caboche J, Vanhoutte P. Activity-Regulatedcytoskeleton-Associated protein accumulates in the nucleus in response tococaine and acts as a brake on chromatin remodeling and long-Termbehavioral alterations. Biol. Psychiatry. 2017;81:573–584. doi: 10.1016/j.biopsych.2016.05.025. [DOI] [PubMed] [Google Scholar]
- 90.Suberbielle E, Sanchez PE, Kravitz AV, Wang X, Ho K, Eilertson K, Devidze N, Kreitzer AC, Mucke L. Physiologic brain activity causes DNA double-strand breaks in neurons, with exacerbation by amyloid-β. Nat. Neurosci. 2013;16:613–621. doi: 10.1038/nn.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.King IF, Yandava CN, Mabb AM, Hsiao JS, Huang H-S, Pearson BL, Calabrese JM, Starmer J, Parker JS, Magnuson T, Chamberlain SJ, Philpot BD, Zylka MJ. Topoisomerases facilitate transcription of long genes linked to autism. Nature. 2013;501:58–62. doi: 10.1038/nature12504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Joshi RS, Piña B, Roca J. Topoisomerase II is required for the production of long Pol II gene transcripts in yeast. Nucleic Acids Res. 2012;40:7907–7915. doi: 10.1093/nar/gks626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wei PC, Chang AN, Kao J, Du Z, Meyers RM, Alt FW, Schwer B. Long Neural Genes Harbor Recurrent DNA Break Clusters in Neural Stem/Progenitor Cells. Cell. 2016;164:644–655. doi: 10.1016/j.cell.2015.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schwer B, Wei P-C, Chang AN, Kao J, Du Z, Meyers RM, Alt FW. Transcription-associated processes cause DNA double-strand breaks and translocations in neural stem/progenitor cells. Proc. Natl. Acad. Sci. 2016;113:2258–2263. doi: 10.1073/pnas.1525564113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Barry DN, Commins S. Temporal dynamics of Immediate Early Gene expression during cellular consolidation of spatial memory. Behav. Brain Res. 2017;327:44–53. doi: 10.1016/j.bbr.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 96.Soulé J, Alme M, Myrum C, Schubert M, Kanhema T, Bramham CR. Balancing arc synthesis, mRNA decay, and proteasomal degradation: Maximal protein expression triggered by rapid eye movement sleep-like bursts of muscarinic cholinergic receptor stimulation. J. Biol. Chem. 2012;287:22354–22366. doi: 10.1074/jbc.M112.376491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stickgold R. Sleep-dependent memory consolidation. Nature. 2005;437:1272–1278. doi: 10.1038/nature04286. [DOI] [PubMed] [Google Scholar]
- 98.Diekelmann S, Born J. The memory function of sleep. Nat.Rev.Neurosci. 2010;11:114–126. doi: 10.1038/nrn2762. [DOI] [PubMed] [Google Scholar]
- 99.Thompson CL, Wisor JP, Lee CK, Pathak SD, Gerashchenko D, Smith KA, Fischer SR, Kuan CL, Sunkin SM, Ng LL, Lau C, Hawrylycz M, Jones AR, Kilduff TS, Lein ES. Molecular and anatomical signatures of sleep deprivation in the mouse brain. Front. Neurosci. 2010;4 doi: 10.3389/fnins.2010.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cirelli C, Gutierrez CM, Tononi G. Extensive and divergent effects of sleep and wakefulness on brain gene expression. Neuron. 2004;41:35–43. doi: 10.1016/s0896-6273(03)00814-6. [DOI] [PubMed] [Google Scholar]
- 101.Ardehali MB, Lis JT. Tracking rates of transcription and splicing in vivo. Nat. Struct. Mol. Biol. 2009;16:1123–1124. doi: 10.1038/nsmb1109-1123. [DOI] [PubMed] [Google Scholar]
- 102.Turrigiano GG. The Self-Tuning Neuron: Synaptic Scaling of Excitatory Synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Panja D, Bramham CR. BDNF mechanisms in late LTP formation: A synthesis and breakdown. Neuropharmacology. 2014;76:664–676. doi: 10.1016/j.neuropharm.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 104.Goldberg EM, Coulter DA. Mechanisms of epileptogenesis: a convergence on neural circuit dysfunction. Nat. Rev. Neurosci. 2013;14:337–349. doi: 10.1038/nrn3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Nelson SB, Valakh V. Excitatory/Inhibitory Balance and Circuit Homeostasis in Autism Spectrum Disorders. Neuron. 2015;87:684–698. doi: 10.1016/j.neuron.2015.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Palop JJ, Chin J, Bien-Ly N, Massaro C, Yeung BZ, Yu G-Q, Mucke L. Vulnerability of dentate granule cells to disruption of arc expression in human amyloid precursor protein transgenic mice. J. Neurosci. 2005;25:9686–93. doi: 10.1523/JNEUROSCI.2829-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu J, Petralia RS, Kurushima H, Patel H, Jung MY, Volk L, Chowdhury S, Shepherd JD, Dehoff M, Li Y, Kuhl D, Huganir RL, Price DL, Scannevin R, Troncoso JC, Wong PC, Worley PF. Arc/Arg3.1 Regulates an Endosomal Pathway Essential for Activity-Dependent β-Amyloid Generation. Cell. 2011;147:615–628. doi: 10.1016/j.cell.2011.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rudinskiy N, Hawkes JM, Betensky RA, Eguchi M, Yamaguchi S, Spires-Jones TL, Hyman BT. Orchestrated experience-driven Arc responses are disrupted in a mouse model of Alzheimer’s disease. Nat. Neurosci. 2012;15:1422–1429. doi: 10.1038/nn.3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Smith-Hicks CL, Cai P, Savonenko AV, Reeves RH, Worley PF. Increased Sparsity of Hippocampal CA1 Neuronal Ensembles in a Mouse Model of Down Syndrome Assayed by Arc Expression. Front. Neural Circuits. 2017;11 doi: 10.3389/fncir.2017.00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cheon MS, Dierssen M, Kim SH, Lubec G. Protein expression of BACE1, BACE2 and APP in Down syndrome brains. Amino Acids. 2008;35:339–343. doi: 10.1007/s00726-007-0618-9. [DOI] [PubMed] [Google Scholar]
- 111.Zhang W, Wu J, Ward MD, Yang S, Chuang YA, Xiao M, Li R, Leahy DJ, Worley PF. Structural basis of arc binding to synaptic proteins: Implications for cognitive disease. Neuron. 2015;86:490–500. doi: 10.1016/j.neuron.2015.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]