Abstract
The success of new therapies hinges on our ability to understand their molecular and cellular mechanisms of action. We modified BET bromodomain inhibitors, an epigenetic-based therapy, to create functionally conserved compounds that are amenable to click chemistry and can be used as molecular probes in vitro and in vivo. We used click proteomics and click sequencing to explore the gene regulatory function of BRD4 (bromodomain containing protein 4) and the transcriptional changes induced by BET inhibitors. In our studies of mouse models of acute leukemia, we used high-resolution microscopy and flow cytometry to highlight the heterogeneity of drug activity within tumor cells located in different tissue compartments. We also demonstrate the differential distribution and effects of BET inhibitors in normal and malignant cells in vivo. This study provides a potential framework for the preclinical assessment of a wide range of drugs.
Investment and progress in medicinal chemistry has led to the promise of personalized medicine with targeted therapies (1). Although these efforts have seen several novel therapeutic classes emerge and show early promise in the research laboratory, very few of these drugs ultimately make a sustained transition into the clinical arena (1). Underpinning this failure in the clinical domain is a lack of knowledge of the molecular and cellular effects of these therapies. When assessing a newly synthesized small molecule, it is desirable to visualize the cellular localization of the compound (2–4); identify the protein targets that the molecule engages within a cell; and, for drugs that target nuclear proteins, understand where in the genome the drug is located. Similarly, when assessing cancer therapies in animal models, it would be advantageous to assess differential effects of the drug in cancer cells and normal cells within different organs involved in disease.
BET bromodomain inhibitors are drugs that target chromatin-associated proteins. Although they have shown promise in both malignant and nonmalignant conditions (5), the mechanisms that govern sensitivity or resistance to these drugs are poorly understood. We sought to modify chemically distinct BET inhibitors so that they could be used as molecular probes in a manner similar to the way in which antibodies are used in cell and molecular biology research. We and others have previously used small molecules, including BET inhibitors, as an affinity matrix for chemoproteomics (6, 7) and chemical sequencing (4, 8). These approaches, which included coupling of the small molecule to a biotinylated polyethylene glycol, can compromise cellular uptake and intracellular drug-target interactions, thus limiting the ability to accurately delineate mechanisms of action (fig. S1). To preserve the functional integrity of the small molecules, we repurposed the biologically active BET inhibitors to contain distinct chemically reactive moieties amenable to bioorthogonal chemical ligation by click chemistry. This approach allows fluorochromes and/or affinity tags to react with the functionalized drugs in a cellular context (Fig. 1A). Click reactions used in biological applications include the copper-catalyzed and the inverse electron-demand Diels–Alder cycloadditions involving azide-alkyne and tetrazines-trans-cyclooctenes partners, respectively (9–11). Thus, we synthesized derivatives of the BET inhibitors JQ1 and IBET-762—namely, JQ1–propargyl amide (JQ1–PA), JQ1–trans-cyclooctene (JQ1–TCO), and IBET-762–TCO, along with a biotinylated derivative of JQ1 (JQ1–BTN)—for comparison (Fig. 1A and materials and methods).
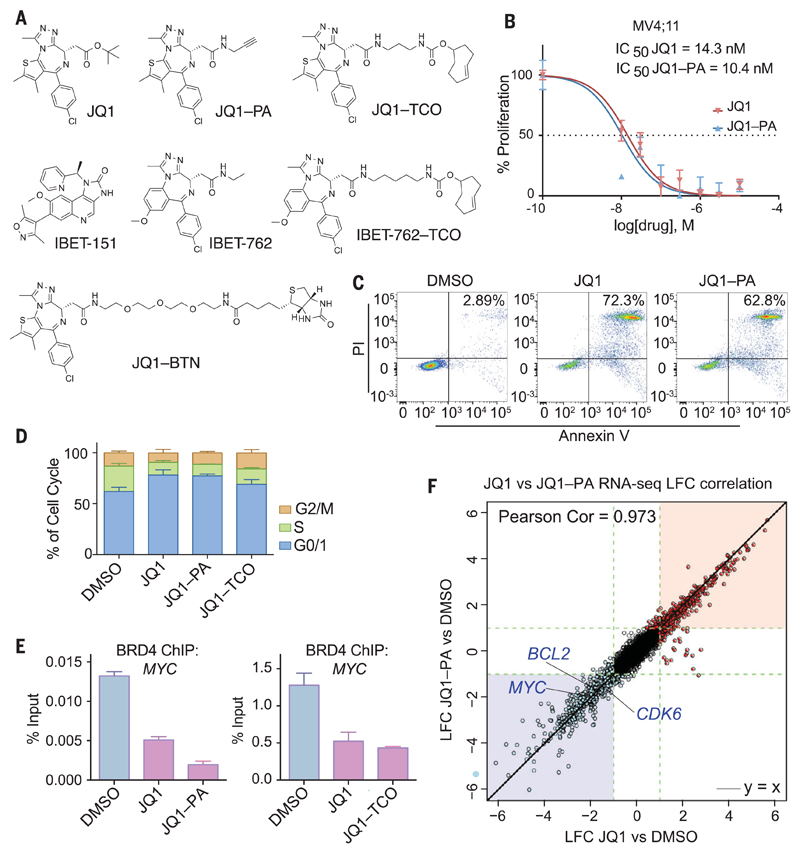
Fig. 1. Clickable compounds phenocopy the parental compounds.
(A) Molecular structures of BET inhibitors and clickable and biotinylated surrogates. (B) Proliferation of MV4;11 cells after 72 hours of treatment, comparing JQ1 versus JQ1–PA. Mean ± SD (error bars) (n = 3 xxxxxxxxxxx). IC50, median inhibitory concentration. (C) Apoptosis assessed by FACS (fluorescence-activated cell sorting) analysis after 72 hours of incubation with dimethyl sulfoxide (DMSO), JQ1 (1 μM), or JQ1–PA (1 μM). PI, propidium iodide. (D) Cell cycle profile of MV4;11 cells after 48 hours of treatment with DMSO, JQ1, JQ1–PA, or JQ1–TCO (all compounds used at 500 nM). Mean ± SD (error bars) (n = 3). (E) qPCR analysis of BRD4 ChIP from MV4;11 cells treated with JQ1 (1 μM) compared with JQ1–PA (1 μM) or JQ1–TCO (1 μM), with primers against MYC. Mean ± SD (error bars), representative graph from three independent experiments. (F) RNA sequencing (RNA-seq) performed in MV4;11 cells treated with JQ1 or JQ1–PA compared with DMSO control. Correlation of log2 fold change of JQ1–PA with log2 fold change of JQ1 is shown. Significantly up- and down-regulated genes are depicted in red and blue, respectively. LFC, xxxxxxxxxxxxxxxxxx.
We compared the effects of the clickable compounds with those of the parental compounds by assessing in vitro cell proliferation, apoptosis, and cell cycle progression (Fig. 1, B to D, and fig. S1), as these are the major cellular phenotypes altered by BET inhibitors (7, 12). In each of these assays, the clickable derivatives phenocopied their unmodified counterparts. The clickable inhibitors also engaged their bromodomain targets and displaced the BET proteins from chromatin, as assessed by chromatin immunoprecipitation (ChIP)–quantitative polymerase chain reaction (qPCR) (Fig. 1E and fig. S1C). In addition, the gene expression programs altered by the clickable inhibitors were virtually identical to those of the parental drugs (Fig. 1F and fig. S1, D and E), thus confirming that the moderate structural alterations we introduced did not affect the functional integrity of the BET inhibitors.
We next explored the molecular activity of the clickable compounds by localizing their chromatin occupancy (Fig. 2A). We used a method we termed “click sequencing” (click-seq), which allowed us to identify specific drug–chromatin target interactions in different cell lines (Fig. 2, B and C, and fig. S2). Although BRD4 occupancy and the influence of BET inhibitors on gene regulation have been studied extensively, it is unclear why only a subset of putative gene targets are down-regulated upon BET bromodomain inhibition. We found that upon BET bromodomain inhibition, chromatin-bound BRD4 was more substantially displaced from enhancer elements compared with the transcriptional start site (TSS) of actively transcribed genes (fig. S3A). Chromatin occupancy of BRD4 around the TSS did not differ between genes that are responsive and unresponsive to BET inhibitors (fig. S3B). As BRD4 can be localized to cis-regulatory elements at chromatin via its tandem bromodomains or in a bromodomain-independent manner via interaction with other chromatin-associated proteins (13), this result prompted us to investigate whether local drug concentration within the transcriptional unit would be a better indicator of BET inhibitor response. Using click-seq, we found that genes that are immediately down-regulated upon BET inhibition have a markedly higher chromatin occupancy of the drug across the transcription unit (Fig. 2D and fig. S3, B and C). Thus, by directly assessing the levels of drug localized across the transcriptional unit, we are able to identify the BET inhibitor–responsive genes by means of a single click-seq experiment.
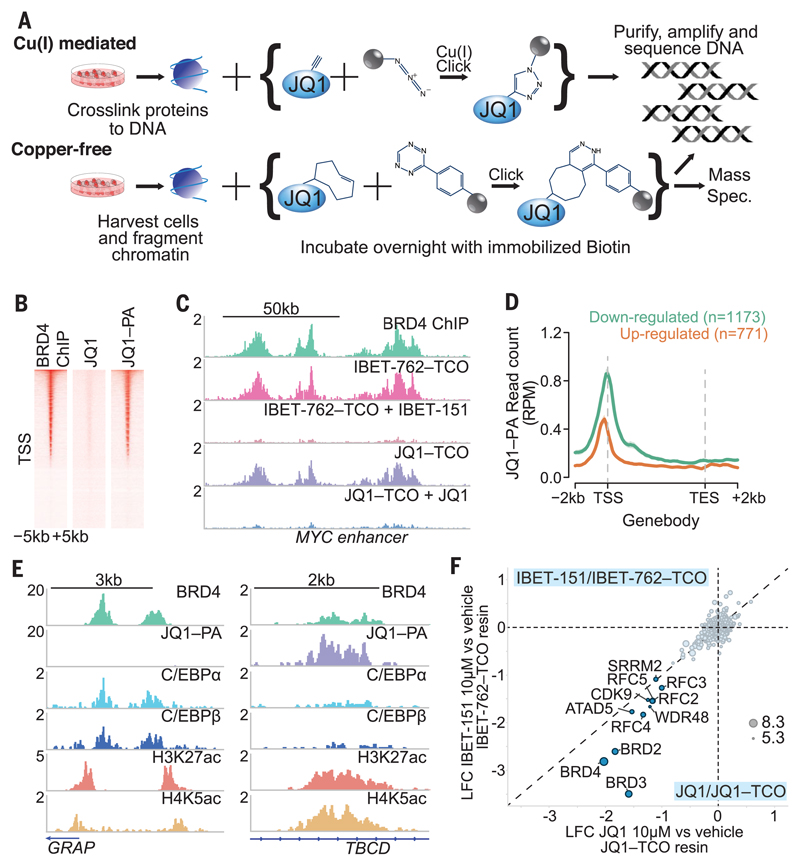
Fig. 2. Click chemistry reveals insights into the binding of BRD4 to chromatin.
(A) Schematic illustration of click-seq and click-proteomics experiments with modified JQ1 and IBET molecules. (B) Heat map of BRD4 ChIP-seq and JQ1–PA click-seq sequencing reads in the 5 kb around the TSS. JQ1 is included as a negative control. (C) Genome browser view of the MYC enhancer, comparing BRD4 ChIP-seq with click-seq using IBET-762–TCO and JQ1–TCO molecules, with competition from unmodified IBET-151 and JQ1. (D) Genes down-regulated or up-regulated after BET inhibitor treatment for 6 hours, assessed for drug occupancy with JQ1-PA click-seq. RPM, reads per million. (E) Genome browser view of two genomic regions with low and high levels of JQ1–PA relative to BRD4 with C/EBPα and C/EBPβ ChIP-seq. (F) Quantitative mass spectrometry of proteins from the lysate of K562 cells captured by click-probes (IBET-762–TCO and JQ1–TCO) in the presence or absence of the respective competitor (IBET-151 and JQ1). Correlation of log2 fold change of abundance of protein captured in the presence of inhibitor relative to vehicle. Circle size represents the quantity of protein from the mass spectrometer.
Together, these findings suggested distinct modes of binding of BRD4 at the BET inhibitor–responsive and –unresponsive genes. It has previously been established that BRD4 associates with chromatin most avidly by binding acetylated (ac) lysines (K), primarily K5ac and K8ac on the tail of histone H4 (14, 15). Consistent with this, we observed increased levels of H4K5ac and H4K8ac spanning the TSS of the down-regulated genes (fig. S3D). To explain the increased drug localization at the down-regulated genes, we hypothesized that chromatin binding at these sites is principally mediated via the first bromodomain (BD1) of BRD4 (16, 17), which would leave the second bromodomain (BD2) free to engage and localize the drug in click-seq studies. In line with this premise, we found that a point mutation that compromises the acetyl-lysine binding ability of BD1 [Tyr97→Ala97 (Y97A)] was sufficient to impair the functional activity of BRD4 in acute myeloid leukemia (AML) cells. In contrast, functional impairment of BD2 (Y390A) was less deleterious (fig. S4). To demonstrate that the clickable compounds primarily localize at chromatin by binding to BD2, we used the BD2-selective drug RVX-208 (18). Although RVX-208 did not displace BRD4 from chromatin or result in functional impairment, the compound’s preferential occupancy of BD2 selectively impaired the ability of the clickable JQ1 to engage BRD4 at chromatin (figs. S5 and S6). These findings demonstrate that at the BET inhibitor–responsive genes, BRD4 is bound to H4K5ac or H4K8ac by its first bromodomain, and these sites can be identified with click-seq as the compound engages its second bromodomain. Binding of BRD4 at the unresponsive genes is not only bromodomain independent but may occur in a manner that precludes access of the drug to either BD1 or BD2. These data also highlight the therapeutic potential of domain-specific inhibitors targeted to BD1 of BRD4.
Next we examined the modes of BRD4 binding at enhancer elements where BRD4 binding is more broadly regulated by bromodomain inhibition (fig. S3A). A direct comparison of JQ1 click-seq with BRD4 ChIP sequencing (ChIP-seq) data suggested that relative to BRD4 binding, some enhancers had greater drug occupancy than others (Fig. 2E and fig. S3E). In contrast to the events at the TSS, BRD4 was equally displaced from chromatin at regions with the highest and lowest drug occupancy, suggesting that at enhancers, BRD4 is localized to chromatin primarily in a bromodomain-dependent manner (fig. S3F). Enhancers with high drug occupancy contained high levels of acetylated histone H4 (fig. S3G) to which BD1 was bound; BD2 was free to engage the clickable compound (fig. S6). In comparison, at regions of low drug occupancy, the inaccessibility of the drug at these sites suggested that either BD2 was also bound to an acetylated lysine residue or was sterically hindered as a result of interactions with a chromatin-associated protein. To further investigate the mode of BRD4 binding at the enhancer elements with low drug occupancy, we performed transcription factor binding site analyses, as BRD4 has recently been shown to associate with transcription factors at chromatin (19, 20). These regions were enriched for binding sites of transcription factors C/EBPα and C/EBPβ, which directly bind BRD4 (19) (fig. S3H). ChIP-seq analyses confirmed that C/EBPα and C/EBPβ colocalized with BRD4 at chromatin and were flanked by acetylated histones (Fig. 2E).
An often unresolved mechanistic issue concerning preclinical assessment of distinct compounds used against the same protein targets is whether their selectivity for the target and associated protein complexes varies, thus conferring differential efficacy. To address this, we performed quantitative click proteomics with IBET-151 and JQ1 and showed that these chemically distinct compounds associated with the same intracellular protein complexes (Fig. 2F and fig. S7), thereby explaining the identical cellular and molecular activity of these compounds and why resistance to one chemical class confers cross resistance (21).
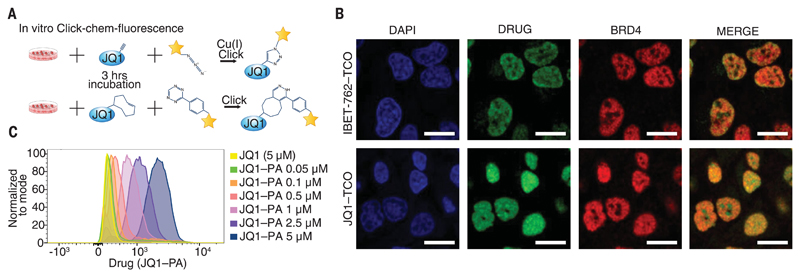
We next assessed the clickable BET inhibitors in cultured cells by immunofluorescence and flow cytometry (fig. S8). Using different clickable BET inhibitors, we demonstrated colocalization of the clickable drugs with their target BRD4 in the nuclei of both hematopoietic and nonhematopoietic cells (Fig. 3, A and B, and fig. S8D). We also used flow cytometry to accurately quantify the intracellular amount of clickable surrogate drugs (Fig. 3C) and showed that we could easily identify the population of cells exposed to the highest amount of drug in mixing experiments (fig. S8E). Furthermore, we also noted that intracellular levels of the drug were not influenced by cell cycle progression or proliferative capacity (fig. S8C).
Fig. 3. Clickable compounds can be visualized and quantified in vitro.
(A) Schematic illustration of the methodology for click fluorescence and flow cytometry. (B) Confocal microscopy of HeLa cells treated with IBET-762–TCO or JQ1–TCO. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI), drug was labeled with Cy5–tetrazine, and BRD4 was stained with an anti-BRD4 antibody. Scale bars, 20 μm. (C) MV4;11 cells treated with JQ1 or increasing concentrations of JQ1–PA, followed by click labeling with 488–azide. Flow cytometry analysis is represented as a histogram.
An assumption sometimes made in preclinical and clinical cancer studies is that drug distribution and efficacy are equal across different tissues involved with the malignancy. Using the same preclinical model in which the efficacy of the BET inhibitors in AML was established (7, 12) (Fig. 4A), we found that these drugs displayed marked differences in tumor cell activity that were dependent on tumor location (fig. S9). These results are consistent with the previous observation that BET inhibitors rapidly clear AML cells circulating in the peripheral blood or localized within the spleen but not those within the bone marrow (7). These findings are potentially clinically important, as they reflect features of tumor cell clearance observed in some patients enrolled on the ongoing clinical trials with BET inhibitors.
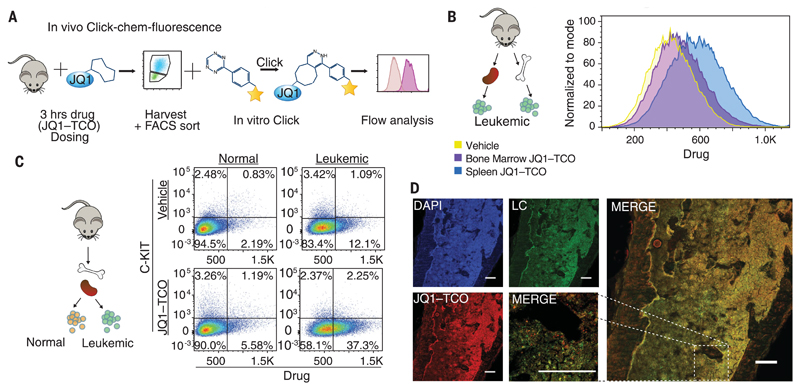
Fig. 4. Preclinical assessment of clickable compounds in vivo.
(A) Schematic illustration of the methodology to detect the clickable small molecules in vivo. (B) Flow cytometry analysis of Venus+ sorted cells from the bone marrow and spleen of a mouse treated with JQ1–TCO for 3.5 hours. (C) Flow cytometry analysis of drug levels within normal hematopoietic cells (Venus−) and leukemia cells (Venus+). (D) Confocal microscopy of mouse femur tissue treated with 100 mg/kg of JQ1–TCO. Leukemia cells (LC) are identified by Venus reporter. The high magnification of the merged image (at right) reveals the ability to discern individual leukemia cells containing drug in situ. Scale bars, 187 μm. All data demonstrated here are representative examples of experiments performed in biological duplicate.
To evaluate whether drug access correlated with the observed tumor cell activity, we assessed the intracellular concentration of BET inhibitors within AML cells localized in the spleen compared with those resident in the bone marrow of animals dosed with a clickable BET inhibitor. Using JQ1-TCO, which had a log P value similar to that of JQ1 (fig. S9A), we found that drug levels and, consequently, repression of BRD4 target genes were significantly higher within the AML cells contained in the spleen of the animals (Fig. 4B and fig. S9, C and E). To show that the clickable compounds accurately reflect the pharmacokinetic properties of the parent drug, we harvested hematopoietic cells from the spleen and bone marrow of mice dosed with JQ1 and analyzed these samples by quantitative intracellular mass spectrometry. These data confirmed that drug concentrations are significantly higher in the spleen compared with the bone marrow (fig. S9C). These findings may help to explain why, when exposed to sustained subtherapeutic levels of BET inhibitors, leukemia stem cells resident within the bone marrow become refractory to treatment (21). They also raise the possibility that compounds with greater bone marrow penetration may improve the efficacy of BET inhibitors.
To further highlight the power of chemically tagging functionally identical small molecules and to address the important preclinical issue of therapeutic window, we next quantitated in vivo the level of drug within normal and malignant hematopoietic cells within the same tissue (Fig. 4C). We found that the relative levels of intracellular drug were greater in AML cells compared with normal hematopoietic cells. Consistent with these data, we found that repression of gene targets of BET inhibitors correlated with intracellular drug levels (fig. S9E). These findings may help to explain why marked cytopenias were not observed in healthy animals treated at drug concentrations that had shown significant therapeutic efficacy (7).
Understanding drug distribution within the architecture of a tissue represents an important asset in evaluating new therapies. We therefore developed the methodology to assess levels of drug at a cellular resolution within any tissue compartment (Fig. 4D). With this technology, one can evaluate drug distribution in various niches, compartments, and cells of interest by costaining cells with the appropriate immunophenotypic markers.
The success of personalized medicine rests on characterizing the mechanisms of action of targeted therapies. We have established an experimental framework that allows clickable drug surrogates to be used as versatile molecular probes. These compounds can be used to examine the benefits and limitations of newly synthesized drugs at the molecular level. Moreover, these probes can be used in model organisms to critically evaluate efficacy, toxicity, and resistance. Together, this knowledge may ultimately improve clinical outcomes.
Supplementary Material
Acknowledgments
Work in the Dawson laboratory is supported by the National Health and Medical Research Council of Australia (grants 1106444, 1085015, and 1106447 to M.A.D.). M.A.D. is currently supported by a Senior Leukaemia Foundation Australia Fellowship and a VESKI Innovation Fellowship. D.S.T. and O.G. are respectively supported by a Ph.D. scholarship and a postdoctoral fellowship from Leukaemia Foundation Australia. E.Y.N.L. and L.M. are supported by fellowships from the Victoria Cancer Agency. Work performed at Cellzome was supported in part by funding from the European Union (FP7 Project BLUEPRINT/282510). We thank V. Benes and the European Molecular Biology Laboratory GeneCore facility for DNA sequencing. Research in the Rodriguez laboratory is supported by the Emergence Ville de Paris Program and funding from the European Research Council under the European Union’s Horizon 2020 research and innovation program (grant agreement 647973). Reagents JQ1-TCO, JQ1-PA, IBET762-TCO, and IBET151 and plasmids for BRD4 are available from the corresponding authors under a material transfer agreement with Institut Curie, GlaxoSmithKline, and Peter MacCallum Cancer Centre. GlaxoSmithKline holds patent WO 2011054846 A1, which covers IBET151. The ChIP-seq, click-seq, and RNA-seq data are available from the National Center for Biotechnology Information Gene Expression Omnibus repository under accession number GSE88751.
References
- 1.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 2.Cañeque T, et al. Nat Chem. 2015;7:744–51. doi: 10.1038/nchem.2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rutkowska A, et al. ACS Chem Biol. 2016;11:2541–2550. doi: 10.1021/acschembio.6b00346. [DOI] [PubMed] [Google Scholar]
- 4.Rodriguez R, Miller KM. Nat Rev Genet. 2014;15:783–796. doi: 10.1038/nrg3796. [DOI] [PubMed] [Google Scholar]
- 5.Dawson MA. Science. 2017;355:1147–1152. doi: 10.1126/science.aam7304. [DOI] [PubMed] [Google Scholar]
- 6.Ong SE, et al. Proc Natl Acad Sci USA. 2009;106:4617–4622. doi: 10.1073/pnas.0900191106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson MA, et al. Nature. 2011;478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Anders L, et al. Nat Biotechnol. 2014;32:92–96. doi: 10.1038/nbt.2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blackman ML, Royzen M, Fox JM. J Am Chem Soc. 2008;130:13518–13519. doi: 10.1021/ja8053805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolb HC, Finn MG, Sharpless KB. Angew Chem Int Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 11.Speers AE, Adam GC, Cravatt BF. J Am Chem Soc. 2003;125:4686–4687. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 12.Zuber J, et al. Nature. 2011;478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, Vakoc CR. Mol Cell. 2014;54:728–736. doi: 10.1016/j.molcel.2014.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gilan O, et al. Nat Struct Mol Biol. 2016;23:673–681. doi: 10.1038/nsmb.3249. [DOI] [PubMed] [Google Scholar]
- 15.LeRoy G, et al. Genome Biol. 2012;13:R68. doi: 10.1186/gb-2012-13-8-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dittmann A, et al. ACS Chem Biol. 2014;9:495–502. doi: 10.1021/cb400789e. [DOI] [PubMed] [Google Scholar]
- 17.Baud MG, et al. Science. 2014;346:638–641. doi: 10.1126/science.1249830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Picaud S, et al. Proc Natl Acad Sci USA. 2013;110:19754–19759. doi: 10.1073/pnas.1310658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu SY, Lee AY, Lai HT, Zhang H, Chiang CM. Mol Cell. 2013;49:843–857. doi: 10.1016/j.molcel.2012.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roe JS, Mercan F, Rivera K, Pappin DJ, Vakoc CR. Mol Cell. 2015;58:1028–1039. doi: 10.1016/j.molcel.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fong CY, et al. Nature. 2015;525:538–542. doi: 10.1038/nature14888. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.