Abstract
An exome-wide association study (EWAS) was performed to identify genetic variants, particularly low-frequency or rare coding variants with a moderate to large effect size, that confer susceptibility to atrial fibrillation in Japanese. The EWAS for atrial fibrillation was performed with 13,166 subjects (884 patients with atrial fibrillation and 12,282 controls) using an Illumina HumanExome-12 DNA Analysis BeadChip or Infinium Exome-24 BeadChip arrays. The association of atrial fibrillation with allele frequencies of 41,243 single nucleotide polymorphisms (SNPs) that passed quality control was examined with Fisher's exact test. Based on Bonferroni's correction, a P<1.21×10−6 was considered statistically significant. The EWAS for atrial fibrillation revealed that 122 SNPs were significantly associated with this condition. The association of the identified SNPs to atrial fibrillation was further examined by multivariable logistic regression analysis with adjustment for age, sex and the prevalence of hypertension. Eight SNPs were related (P<0.01) to atrial fibrillation, among which three polymorphisms, rs11552708 [G/A (G67R)]of TNF superfamily member 13 (TNFSF13; dominant model; P=9.36×10−9; odds ratio, 0.58), rs113710653 [C/T (E231 K)] of spermatogenesis and centriole associated 1 like (SPATC1L; dominant model; P=1.09×10−5; odds ratio, 3.27), and rs11231397 [G/C (R300T)] of solute carrier family 22 member 25 (SLC22A25; additive model; P=3.71×10−5; odds ratio, 1.77), were significantly (P<1.02×10−4) associated with this condition. The minor T allele of rs113710653 and the minor C allele of rs11231397 were risk factors for atrial fibrillation, whereas the minor A allele of rs11552708 was protective against this condition. In addition, rs77538589 [C/T (G117R)] of SALL4 exhibited a tendency to be associated with atrial fibrillation (dominant model; P=0.0002; odds ratio, 1.88), with the minor T allele representing a risk factor for this condition. TNFSF13, SPATC1L, SLC22A25 and SALL4 may thus be novel susceptibility loci for atrial fibrillation in the Japanese population.
Keywords: atrial fibrillation, arrhythmia, genetics, exome-wide association study, polymorphism
Introduction
Atrial fibrillation (AF) is the most common type of cardiac arrhythmia and a major public health problem. The estimated number of individuals with AF worldwide was 33.5 million in 2010 (1). The prevalence of AF is increasing and is estimated to double in the United States by 2050 (2). AF is associated with an increased risk of heart failure, thromboembolic diseases, such as cardioembolic stroke, and mortality (3). Although the molecular mechanism underlying AF is complex and has not been determined definitively, several risk factors, including age, smoking, obesity, hypertension, diabetes mellitus, valvular heart disease, coronary artery disease and heart failure, have been identified clinically (4). In addition to these conventional risk factors, recent studies have reported the importance of genetic factors in the development of AF (5). Genome-wide association studies (GWASs) and subsequent meta-analyses of such studies have identified several genes and loci that confer susceptibility to AF in populations of European ancestry (6–10). However, susceptibility genes for AF have not been determined definitively in Japanese populations.
The majority of genetic variants previously reported to be associated with AF have a minor allele frequency (MAF) of ≥5% and a small individual effect size. Given that these common variants explain only a fraction of the heritability of AF, it is likely that low-frequency (0.5%≤ MAF <5%) or rare (MAF <0.5%) variants with larger effect sizes contribute to the genetic architecture of AF (11).
An exome-wide association study (EWAS) with exome array-based genotyping was performed to identify single nucleotide polymorphisms (SNPs), especially low-frequency or rare coding variants with moderate to large effect sizes, that confer susceptibility to AF in Japanese individuals. Given that GWAS arrays of previous studies did not include most low-frequency or rare variants, Illumina human arrays that cover functional SNPs in entire exons including such variants were used in the current study.
Materials and methods
Study subjects
A total of 13,166 Japanese individuals (884 patients with AF and 12,282 controls) were examined. The subjects were recruited either from individuals who visited outpatient clinics of or were admitted to participating hospitals (Gifu Prefectural Tajimi Hospital, Tajimi; Gifu Prefectural General Medical Center, Gifu; Japanese Red Cross Nagoya First Hospital, Nagoya; Inabe General Hospital, Inabe; Hirosaki University Hospital and Hirosaki Stroke and Rehabilitation Center, Hirosaki, Japan) because of various symptoms or for an annual health checkup between 2002 and 2014; from community-dwelling individuals recruited to a population-based cohort study in Inabe between 2010 and 2014 (Health Care Center of Inabe General Hospital) or in Tokyo or Kusatsu between 2011 and 2015 (Tokyo Metropolitan Institute of Gerontology); or from cases of autopsy performed at the Tokyo Metropolitan Geriatric Hospital from 1995 to 2012.
Individuals with persistent or paroxysmal AF or with a history of AF were included in the AF patient group regardless of medical treatment. Patients with AF who had severe valvular heart disease, hypertrophic or dilated cardiomyopathy, or congenital heart disease were excluded from the study. The control individuals had no history of AF or other significant supraventricular or ventricular arrhythmias or of taking antiarrhythmic medication. Autopsy cases were not used as controls.
The study protocol complied with the Declaration of Helsinki and was approved by the Committees on the Ethics of Human Research of Mie University Graduate School of Medicine (Tsu, Japan), Hirosaki University Graduate School of Medicine (Hirosaki, Japan), Tokyo Metropolitan Institute of Gerontology (Tokyo, Japan) and participating hospitals. Written informed consent was obtained from each participant or families of the deceased subjects.
EWAS
Venous blood (5–7 ml) was collected into tubes containing 50 mmol/l ethylenediaminetetraacetic acid (disodium salt), peripheral blood leukocytes were isolated, and genomic DNA was extracted from the cells with the use of a DNA extraction kit (Genomix supplied by Talent Srl, Trieste, Italy; or SMITEST EX-R&D supplied by Medical & Biological Laboratories, Co., Ltd., Nagoya, Japan) or by standard protocols based on phenol-chloroform extraction and spin columns. Lysed blood cells were mixed with an equal volume of a phenol-chloroform mixture. Following centrifugation at 16,000 × g for 5 min at room temperature, two distinct phases were formed. The upper aqueous phase was collected, and genomic DNA was further purified by a spin column-based DNA extraction method that is based on the fact that DNA binds to the solid phase of silica under certain conditions. A buffer solution (Buffer B3; Takara Bio, Inc., Otsu, Japan) was added to the DNA sample along with 100% ethanol. This solution was transferred to a spin column, and the column was centrifuged at 11,000 × g for 1 min at room temperature. A wash buffer [Buffer BW (first wash) or Buffer B5 (second wash); Takara Bio, Inc.] was added to the column, and the column was centrifuged at 11,000 × g for 1 min at room temperature again. An elution buffer (Buffer BE; Takara Bio, Inc.) was added to the column, and the column was centrifuged at 11,000 × g for 1 min at room temperature again. Genomic DNA was collected from the bottom of the column. In autopsy cases, genomic DNA was extracted from kidneys. The EWAS was performed for 884 subjects with AF and 12,282 control individuals with the use of a HumanExome-12 v1.1 or v1.2 DNA Analysis BeadChip or Infinium Exome-24 v1.0 BeadChip (Illumina, Inc., San Diego, CA, USA). These exome arrays include putative functional exonic variants selected from >12,000 individual exome or whole-genome sequences. The exonic content consists of ~244,000 SNPs representing diverse populations, including European, African, Chinese and Hispanic individuals (12). SNPs contained in only one of the exome arrays (~3.6%) were excluded from analysis. Quality control (13) was performed as follows: i) Genotyping data with a call rate of <97% were discarded, with the mean call rate for the remaining data at 99.9%; ii) gender specification was checked for all samples, and those for which gender in the clinical records was inconsistent with genetic sex were discarded; iii) duplicate samples and cryptic relatedness were checked by calculation of identity by descent, all pairs of DNA samples with identity by descent of >0.1875 were inspected and one sample from each pair was excluded; iv) the frequency of SNP heterozygosity was calculated for all samples, and those with extremely low or high heterozygosity (>3 standard deviations from the mean) were discarded; v) SNPs in sex chromosomes or mitochondrial DNA were excluded from the analysis, as were nonpolymorphic SNPs or SNPs with a MAF of <0.1%; vi) SNPs whose genotype distributions deviated significantly (P<0.001) from Hardy-Weinberg equilibrium in control individuals were discarded; vii) the genotype data for the EWAS were examined for population stratification by principal components analysis (14), and population outliers were excluded from the analysis. A total of 41,243 SNPs passed quality control and was subjected to analysis.
Statistical analysis
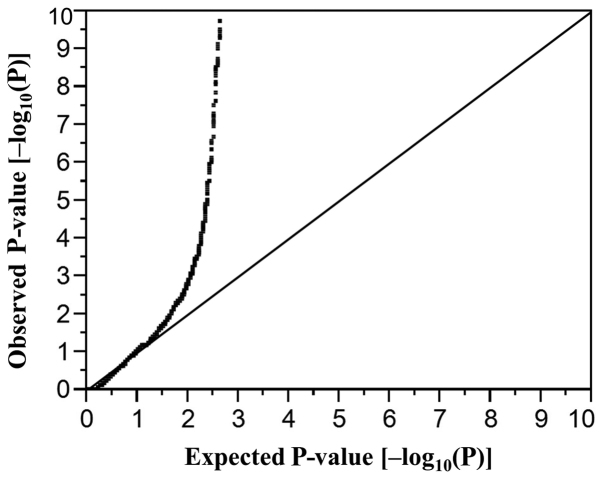
For analysis of characteristics of the study subjects, quantitative data were compared between patients with AF and control individuals with the unpaired Student's t test. Categorical data were compared between the two groups with Fisher's exact test. Allele frequencies were estimated by the gene counting method, and Fisher's exact test was applied to identify departure from Hardy-Weinberg equilibrium. Allele frequencies of SNPs were compared between subjects with AF and controls with Fisher's exact test in the EWAS. Multivariable logistic regression analysis was performed with AF as a dependent variable and independent variables including age, sex (0, woman; 1, man), the prevalence of hypertension (0, no history of this condition; 1, positive history), and genotype of each SNP. Genotypes of SNPs were assessed according to dominant [0, AA; 1, AB+BB (A, major allele; B, minor allele)], recessive (0, AA+AB; 1, BB), and additive genetic models, and the P-value, odds ratio, and 95% confidence interval were calculated. Additive models comprised additive 1 (0, AA; 1, AB; 0, BB) and additive 2 (0, AA; 0, AB; 1, BB) scenarios, which were analyzed simultaneously with a single statistical model. The relation of SNPs to the prevalence of intermediate phenotypes of AF was compared among genotypes with Fisher's exact test (2×2) or Pearson's chi-square test (2×3). To compensate for multiple comparisons of genotypes with AF, we applied Bonferroni's correction for statistical significance of association. Given that 41,243 SNPs were analyzed, the significance level was set at P<1.21×10−6 (0.05/41,243) for the EWAS. A quantile-quantile plot for P-values of allele frequencies in the EWAS for AF is shown in Fig. 1. The inflation factor (λ) was 1.52. Bonferroni's correction was also applied to other statistical comparisons as indicated. Statistical tests were performed with JMP Genomics version 6.0 software (SAS Institute, Inc., Cary, NC, USA).
Figure 1.
Quantile-quantile plot for P-values of allele frequencies in the exome-wide association study of atrial fibrillation. The observed P-values (y-axis) were compared with the expected P-values (x-axis) under the null hypothesis, with points plotted actually corresponding to the negative log-transformed P-values [-log10 (P-values)].
Results
Characteristics of the study subjects
The characteristics of the subjects enrolled in the study are presented in Table I. Age, the number of men, and the prevalence of hypertension, diabetes mellitus, chronic kidney disease and hyperuricemia were significantly greater in patients with AF than in control subjects.
Table I.
Characteristics of the 13,166 study subjects.
| Characteristic | Atrial fibrillation | Control | P-value |
|---|---|---|---|
| No. of subjects | 884 | 12,282 | |
| Age (years) | 75.8±12.3 | 59.4±13.2 | <0.0001 |
| Sex (male/female, %) | 67.8/32.2 | 56.8/43.2 | <0.0001 |
| Body mass index (kg/m2) | 23.4±3.7 | 23.3±3.5 | 0.7412 |
| Current or former smoker (%) | 33.6 | 36.3 | 0.2576 |
| Hypertension (%) | 83.9 | 51.1 | <0.0001 |
| Diabetes mellitus (%) | 51 | 23.2 | <0.0001 |
| Dyslipidemia (%) | 65.3 | 62 | 0.188 |
| Chronic kidney disease (%) | 40.3 | 23 | <0.0001 |
| Hyperuricemia (%) | 31.9 | 17 | <0.0001 |
Quantitative data are presented as the mean ± standard deviation and were compared between patients with atrial fibrillation and control individuals using the unpaired Student's t test. Categorical data were compared between the two groups using Fisher's exact test. Based on Bonferroni's correction, P<0.0056 (0.05/9) was considered to indicate a statistically significant difference.
EWAS of AF
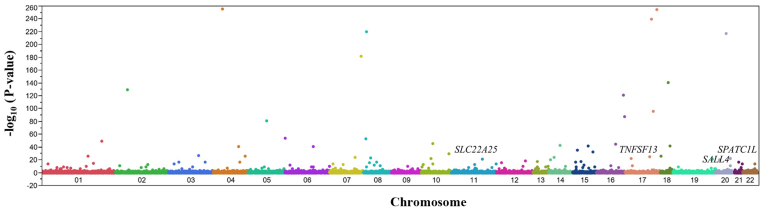
The association of AF with allele frequencies of 41,243 SNPs that passed quality control was determined using Fisher's exact test. A Manhattan plot for the EWAS of AF is presented in Fig. 2. Following Bonferroni's correction, 122 SNPs were significantly [P<1.21×10−6 (0.05/41,243)] associated with AF (Table II). The genotype distributions of these SNPs were in Hardy-Weinberg equilibrium (P>0.001) both among subjects with AF and among control individuals (data not shown).
Figure 2.
Manhattan plot for P-values of allele frequencies in the exome-wide association study of AF. P-values [-log10 (P-values)] on the y-axis are shown according to the physical chromosomal position of the corresponding single nucleotide polymorphism on the x-axis. The SNPs ultimately identified as being associated with AF are indicated. AF, atrial fibrillation; SLC22A25, solute carrier family 22 member 25; TNFSF13, TNF superfamily member 13; SALL4, spalt like transcription factor 4; SPATC1L, spermatogenesis and centriole associated 1 like.
Table II.
The 122 SNPs significantly associated with atrial fibrillation by exome-wide association analysis.
| Gene | SNP | Nucleotide substitution (amino acid)a | Chromosome: Position | MAF (%) | P-value (allele) | Allele OR |
|---|---|---|---|---|---|---|
| N4BP2 | rs61748749 | T/G (S1353R) | 4: 40122170 | 2.3 | <1.0×10−255 | 0.74 |
| DNAH17 | rs690844 | A/C (I1742M) | 17: 78501838 | 2.5 | 8.94×10−255 | 1.31 |
| HELZ | rs184499441 | C/T (G1288R) | 17: 67114380 | 1.6 | 4.40×10−240 | 1.08 |
| rs7828656 | A/C | 8: 11645425 | 44.8 | 1.87×10−220 | 1.1 | |
| SLA2 | rs221308 | T/C | 20: 36647995 | 46.7 | 4.11×10−218 | 0.99 |
| SSPO | rs55976638 | G/T | 7: 149832881 | 4.3 | 2.99×10−182 | 1.31 |
| TCEB3B | rs2010834 | A/C (F254C) | 18: 47034504 | 24.5 | 2.60×10−141 | 1.1 |
| FANCL | rs149731356 | T/C (T224A) | 2: 58165745 | 0.7 | 7.35×10−130 | 0.75 |
| PIEZO1 | rs143004911 | G/A (R333C) | 16: 88737957 | 2.7 | 5.07×10−122 | 0.87 |
| TTYH2 | rs9899862 | C/A (D423E) | 17: 74253090 | 6.3 | 1.31×10−96 | 0.86 |
| TUBB3 | rs2302898 | A/G | 16: 89932386 | 23.5 | 1.13×10−87 | 0.87 |
| SLCO6A1 | rs17150488 | T/C (K381R) | 5: 102438751 | 0.6 | 3.30×10−81 | 1.05 |
| GMDS | rs9378305 | C/T | 6: 1703056 | 41.4 | 1.20×10−54 | 0.99 |
| RP1L1 | rs79329877 | T/C | 8: 10622701 | 9.2 | 2.05×10−53 | 1.05 |
| FCMR | rs150080259 | T/G (S61R) | 1: 206902976 | 1.5 | 1.07×10−49 | 1.02 |
| RTKN2 | rs7090884 | A/G | 10: 62202267 | 33.2 | 4.97×10−46 | 1.05 |
| UTP4 | rs193164904 | A/G (I534V) | 16: 69163131 | 0.2 | 1.90×10−45 | 1.58 |
| SNAPC1 | rs74810099 | T/G (M36R) | 14: 61762567 | 2.8 | 1.99×10−43 | 1.02 |
| ALPK2 | rs3809977 | G/T (P1174H) | 18: 58536666 | 17.2 | 3.05×10−42 | 0.93 |
| CSPG4 | rs137981794 | T/C (D1936G) | 15: 75676712 | 0.4 | 4.05×10−42 | ND |
| MDN1 | rs9294445 | A/G (Y3423H) | 6: 89692763 | 7.5 | 1.15×10−41 | 0.99 |
| SETD7 | rs6814310 | C/A | 4: 139534374 | 48.4 | 1.64×10−41 | 0.97 |
| PLA2G4E | rs4924595 | T/C (N400S) | 15: 41995408 | 18.2 | 1.87×10−35 | 1.01 |
| KIF7 | rs117123311 | C/G (S788R) | 15: 89642233 | 3.1 | 1.03×10−32 | 1 |
| CTBP2 | rs3781411 | C/T (R298Q) | 10: 125026867 | 7 | 1.46×10−30 | 0.9 |
| GATA2 | rs78245253 | G/C (A250P) | 3: 128485850 | 4.6 | 1.37×10−27 | 0.79 |
| DLGAP1 | rs3745051 | C/T | 18: 3729175 | 34.4 | 1.64×10−26 | 0.86 |
| rs1711393 | T/C | 4: 178074088 | 31 | 1.70×10−26 | 1.14 | |
| SLAMF7 | rs117009784 | A/C (R96S) | 1: 160750053 | 5.8 | 3.86×10−26 | 1.07 |
| USP32 | rs8079220 | C/T | 17: 60239372 | 20.5 | 3.56×10−25 | 0.91 |
| rs8011192 | T/G | 14: 28319412 | 33.1 | 8.46×10−25 | 0.99 | |
| IMPDH1 | rs201001000 | G/A (T369M) | 7: 128395003 | 0.1 | 3.85×10−24 | 0.79 |
| ADRA1A | rs151273238 | G/A (T391M) | 8: 26770378 | 0.2 | 2.12×10−23 | 0.71 |
| TNFSF13 | rs11552708 | G/A (G67R) | 17: 7559238 | 36.6 | 1.32×10−22 | 0.54 |
| SLC18A3 | rs118107581 | A/G (I426V) | 10: 49612016 | 2 | 1.62×10−22 | 1.11 |
| NFATC2 | rs12479626 | T/C (H446R) | 20: 51475656 | 4.7 | 2.79×10−22 | 1.06 |
| TENM4 | rs3812723 | C/T (V396I) | 11: 78863031 | 1.7 | 1.03×10−21 | 0.89 |
| EPN1 | rs200478642 | C/T (P203L) | 19: 55689301 | 0.1 | 1.52×10−21 | 0.35 |
| HNRNPC | rs17197037 | A/G | 14: 21257495 | 29.1 | 3.26×10−21 | 0.99 |
| TMX4 | rs2076015 | T/C (R303G) | 20: 7982394 | 41.1 | 4.49×10−21 | 1.16 |
| FOXN4 | rs140167217 | G/A (S308F) | 12: 109281778 | 0.4 | 4.51×10−19 | 1.12 |
| CEP152 | rs145138194 | G/A (S894F) | 15: 48760148 | 1.2 | 6.92×10−18 | 0.73 |
| FREM2 | rs114864077 | C/T (P128L) | 13: 38687727 | 0.1 | 1.04×10−17 | 1.81 |
| CPA6 | rs4737845 | T/C | 8: 67542707 | 43.3 | 1.87×10−17 | 1.04 |
| KIF15 | rs146292440 | G/A (R1199H) | 3: 44843135 | 0.6 | 3.33×10−17 | 0.57 |
| MFSD1 | rs3765083 | A/G (I230V) | 3: 158819654 | 30.3 | 4.19×10−17 | 1.05 |
| BAHD1 | rs3743143 | A/G (E26G) | 15: 40458541 | 10.5 | 5.85×10−17 | 0.99 |
| rs1395821 | A/G | 4: 147126398 | 39.6 | 6.48×10−17 | 1.05 | |
| BRWD1 | rs2183573 | G/A (P1511S) | 21: 39202379 | 46 | 1.11×10−16 | 1.06 |
| CD69 | rs199676648 | G/A (R32C) | 12: 9756390 | 0.3 | 2.93×10−16 | 0.9 |
| HR | rs12675375 | C/T (G337D) | 8: 22127432 | 43.4 | 8.47×10−16 | 1.02 |
| SOAT1 | rs143616084 | G/A (R292Q) | 1: 179345008 | 0.1 | 6.19×10−15 | ND |
| JMJD1C | rs149833441 | T/C (K878E) | 10: 63208491 | 1.2 | 9.49×10−15 | 1.44 |
| VWDE | rs848016 | A/G (F142S) | 7: 12389177 | 36.7 | 1.41×10−14 | 1.05 |
| VPS13D | rs143833298 | G/A (R830Q) | 1: 12276077 | 0.8 | 1.59×10−14 | 0.86 |
| SPATC1L | rs113710653 | C/T (E231K) | 21: 46161921 | 1.9 | 1.60×10−14 | 5.36 |
| SNX19 | rs117834100 | C/A (G416C) | 11: 130914694 | 6.9 | 1.83×10−14 | 0.89 |
| rs9854207 | A/C | 3: 27572825 | 40.3 | 3.41×10−14 | 1.04 | |
| ARHGAP8 | rs5766113 | A/G | 22: 44855543 | 36.8 | 7.18×10−14 | 1.03 |
| rs4407763 | G/A | 7: 67553705 | 35.8 | 1.14×10−13 | 1.08 | |
| SLC22A25 | rs11231397 | G/C (R300T) | 11: 63183749 | 43.7 | 1.53×10−13 | 1.55 |
| XIRP2 | rs77219745 | G/A (G1839D) | 2: 167246908 | 7 | 2.89×10−13 | 0.81 |
| MCM10 | rs7905784 | A/T (T541S) | 10: 13192356 | 2.2 | 5.31×10−13 | 1.05 |
| HIST1H2AC | rs198823 | G/T | 6: 26122705 | 22.9 | 1.60×10−12 | 0.86 |
| rs10102598 | G/A | 8: 47178128 | 37.7 | 1.61×10−12 | 0.96 | |
| VPS13C | rs77555508 | G/A (S1798F) | 15: 61940726 | 0.6 | 2.47×10−12 | 1.44 |
| ADCY3 | rs7586879 | C/T | 2: 24894108 | 44.7 | 1.18×10−11 | 0.99 |
| CTC1 | rs183966301 | G/A (A1025V) | 17: 8229384 | 0.2 | 1.84×10−11 | 0.56 |
| SALL4 | rs77538589 | C/T (G117R) | 20: 51792134 | 4.3 | 2.24×10−11 | 2.61 |
| ADCY7 | rs201661947 | G/A (A475T) | 16: 50304414 | 0.2 | 3.37×10−11 | ND |
| TP53INP1 | rs896854 | G/A | 8: 94948283 | 30.7 | 3.76×10−11 | 0.96 |
| TMEM245 | rs2271877 | C/T (A314T) | 9: 109091132 | 11.5 | 8.13×10−11 | 0.98 |
| FCRL1 | rs149740001 | A/T (K103I) | 1: 157803856 | 1 | 8.38×10−11 | 1.22 |
| SCYL2 | rs200554353 | T/C (M256T) | 12: 100312568 | 1.6 | 8.73×10−11 | 0.36 |
| TMCO3 | rs185071949 | C/T (P14L) | 13: 113495622 | 0.4 | 1.97×10−10 | 1.02 |
| WDR27 | rs3734905 | C/T | 6: 169558886 | 19.5 | 3.00×10−10 | 0.99 |
| NGB | rs117207261 | C/G (Q60E) | 14: 77269238 | 0.5 | 3.59×10−10 | 1.24 |
| rs6695567 | A/G | 1: 53163913 | 43.6 | 4.89×10−10 | 1.05 | |
| FAP | rs151314911 | C/T | 2: 162173786 | 2.9 | 5.26×10−10 | 1.02 |
| rs13277113 | A/G | 8: 11491677 | 31.9 | 5.36×10−10 | 1.01 | |
| ACER1 | rs72981971 | T/C (M74V) | 19: 6312279 | 36.5 | 5.41×10−10 | 1 |
| FREM2 | rs2496425 | T/C (F1070S) | 13: 38690553 | 38.3 | 7.29×10−10 | 1.01 |
| ASB13 | rs138695721 | A/C (V139G) | 10: 5649071 | 0.6 | 7.67×10−10 | 1.09 |
| rs10943716 | T/C | 6: 80543356 | 40.2 | 1.05×10−9 | 1.05 | |
| CCDC168 | rs1449707 | A/G (I3015T) | 13: 102741653 | 44.4 | 1.82×10−9 | 0.98 |
| ADGRV1 | rs2366928 | A/G (K3471E) | 5: 90728918 | 17.9 | 2.17×10−9 | 0.97 |
| MDN1 | rs115931523 | G/A (T3130M) | 6: 89695987 | 0.2 | 2.44×10−9 | 1.58 |
| CD96 | rs140727933 | A/G (Y11C) | 3: 111542280 | 0.2 | 2.63×10−9 | 1.16 |
| rs4965121 | G/C | 15: 97975562 | 8.5 | 2.69×10−9 | 0.97 | |
| KNL1 | rs11858113 | T/C (M598T) | 15: 40621979 | 27.4 | 3.29×10−9 | 0.99 |
| OR4X2 | rs7120775 | C/G (Y27*) | 11: 48245184 | 16.9 | 3.65×10−9 | 1.05 |
| TRPM2 | rs144412484 | A/G (E450G) | 21: 44390934 | 0.4 | 3.68×10−9 | 0.82 |
| MGAT5 | rs66523341 | C/T | 2: 134364804 | 37 | 4.17×10−9 | 0.93 |
| GCOM1 | rs4774980 | G/A | 15: 57691677 | 6 | 5.40×10−9 | 0.98 |
| CSMD2 | rs1874045 | T/C (K2096R) | 1: 33605925 | 45.1 | 7.56×10−9 | 1.08 |
| ADAT1 | rs200524721 | G/C (Q167H) | 16: 75612785 | 0.3 | 8.75×10−9 | 1.12 |
| rs4420065 | T/C | 1: 65695778 | 13.1 | 1.52×10−8 | 1.02 | |
| NLRX1 | rs149129258 | C/A (P262Q) | 11: 119174034 | 0.5 | 2.31×10−8 | 0.99 |
| DNAAF3 | rs890871 | A/G (L280P) | 19: 55160687 | 25.2 | 3.01×10−8 | 0.98 |
| ZNF25 | rs150582814 | T/C (Y202C) | 10: 37952893 | 0.1 | 3.01×10−8 | ND |
| CMYA5 | rs62621915 | C/T (L1038F) | 5: 79731877 | 49.3 | 5.62×10−8 | 0.93 |
| SYDE2 | rs141587551 | C/A (D173Y) | 1: 85200480 | 6.6 | 6.20×10−8 | 1.03 |
| SLC15A5 | rs3915247 | C/T | 12: 16247679 | 47.6 | 7.01×10−8 | 0.97 |
| CDC42BPG | rs3741395 | T/C (Q1135R) | 11: 64830034 | 5.6 | 8.31×10−8 | 1.11 |
| rs8030485 | G/A | 15: 79116590 | 35.2 | 9.03×10−8 | 0.98 | |
| rs2564486 | G/T | 18: 59579061 | 12.9 | 9.24×10−8 | 1.05 | |
| SLC4A4 | rs1062677 | A/C (I1074L) | 4: 71567828 | 6.1 | 1.20×10−7 | 0.91 |
| STEAP1B | rs17364464 | A/G | 7: 22474434 | 12.2 | 2.03×10−7 | 0.99 |
| KLF17 | rs11210969 | T/A (I35N) | 1: 44129375 | 22.5 | 2.83×10−7 | 1.11 |
| ADAMTS13 | rs78977446 | C/T (S903L) | 9: 133445796 | 4.5 | 3.10×10−7 | 1.24 |
| ZNF879 | rs17078988 | A/G (T112A) | 5: 179032282 | 33.8 | 4.42×10−7 | 1.01 |
| rs1464833 | T/C | 7: 125475608 | 10.2 | 4.58×10−7 | 1.01 | |
| PKD1L1 | rs10951936 | A/T | 7: 47882084 | 21 | 7.76×10−7 | 1.01 |
| SNX32 | rs200684568 | G/A (G179R) | 11: 65850787 | 0.5 | 9.00×10−7 | 0.56 |
| NTF3 | rs6332 | G/A | 12: 5494466 | 41.8 | 9.12×10−7 | 1.09 |
| EFHD1 | rs4072149 | T/C | 2: 232643967 | 36.7 | 9.18×10−7 | 1.01 |
| URB2 | rs3811473 | G/T (G778V) | 1: 229636946 | 15.8 | 9.48×10−7 | 1.05 |
| CCDC71 | rs4955419 | A/T (Q317L) | 3: 49163259 | 4.6 | 9.91×10−7 | 1.13 |
| rs543588 | T/G | 13: 30096848 | 31.4 | 1.01×10−6 | 0.9 | |
| TRIM40 | rs757259 | G/A (E244K) | 6: 30147765 | 16 | 1.06×10−6 | 1.12 |
| rs3129264 | T/C | 6: 33133825 | 8.3 | 1.07×10−6 | 0.8 | |
| SEMA6A | rs12516652 | G/T (D567E) | 5: 116475552 | 1.9 | 1.14×10−6 | 1.2 |
Allele frequencies were analyzed with Fisher's exact test. P<1.21×10−6 was considered to indicate a significant association.
Major allele/minor allele. SNP, single nucleotide polymorphism; MAF, minor allele frequency; OR, odds ratio; ND, not determined. SNPs without gene names are not located within the genes.
Multivariable logistic regression analysis of the relation of SNPs to AF
The association of AF with 122 SNPs identified by the EWAS was further examined by multivariable logistic regression analysis with adjustment for age, sex and the prevalence of hypertension. Eight SNPs were identified to be associated with AF (P<0.01 in at least one genetic model; Table III). Among these eight SNPs, rs11552708 [G/A (G67R)] of TNF superfamily member 13 (TNFSF13), rs113710653 [C/T (E231 K)] of spermatogenesis and centriole associated 1 like (SPATC1L), and rs11231397 [G/C (R300T)] of solute carrier family 22 member 25 (SLC22A25) were significantly [P<1.02×10−4 (0.05/488)] associated with AF. The minor T allele of rs113710653 and the minor C allele of rs11231397 were risk factors for AF, whereas the minor A allele of rs11552708 was protective against this condition (Table III). In addition, the association of rs77538589 [C/T (G117R)] of spalt like transcription factor 4 (SALL4) with AF almost achieved significance, with the minor T allele representing a risk factor for this condition.
Table III.
Association of SNPs to atrial fibrillation as determined by multivariable logistic regression analysis.
| Dominant | Recessive | Additive 1 | Additive 2 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| SNP | Nucleotide substitution (amino acid) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) | P-value | OR (95% CI) |
| rs11552708 | G/A (G67R) | 9.36×10−9 | 0.58 (0.49–0.70) | 3.44×10−5 | 0.54 (0.39–0.73) | 3.31×10−6 | 0.63 (0.52–0.77) | 3.66×10−8 | 0.43 (0.30–0.59) |
| rs2076015 | T/C (R303G) | 0.1339 | 0.0028 | 1.38 (1.12–1.69) | 0.6253 | 0.0038 | 1.41 (1.12–1.78) | ||
| rs113710653 | C/T (E231K) | 1.09×10−5 | 3.27 (2.00–5.14) | 0.6731 | 9.01×10−6 | 3.32 (2.02–5.21) | 0.6802 | ||
| rs11231397 | G/C (R300T) | 0.0016 | 1.40 (1.13–1.73) | 0.0004 | 1.51 (1.20–1.88) | 0.03 | 1.28 (1.02–1.60) | 3.71×10−5 | 1.77 (1.35–2.31) |
| rs77219745 | G/A (G1839D) | 0.2458 | 0.0076 | <0.01 (ND) | 0.4205 | 0.0072 | <0.01 (ND) | ||
| rs77538589 | C/T (G117R) | 0.0002 | 1.88 (1.36–2.56) | 0.4697 | 0.0002 | 1.90 (1.37–2.58) | 0.4825 | ||
| rs141587551 | C/A (D173Y) | 0.7965 | 0.0046 | 3.39 (1.50–7.07) | 0.6564 | 0.0048 | 3.37 (1.49–7.02) | ||
| rs3129264 | T/C | 0.0422 | 0.78 (0.61–0.99) | 0.0016 | <0.01 (0.00–0.36) | 0.1138 | 0.0014 | <0.01 (ND) | |
Multivariable logistic regression analysis was performed with adjustment for age, sex, and the prevalence of hypertension. Based on Bonferroni's correction, P<1.02×10−4 (0.05/488) were considered statistically significant and are shown in bold. SNP, single nucleotide polymorphism; OR, odds ratio; CI, confidence interval; ND, not determined.
Association of SNPs with intermediate phenotypes of AF
The association of the four SNPs, rs11552708, rs113710653, rs11231397 and rs77538589, with intermediate phenotypes of AF was examined. The rs11552708 SNP was significantly associated with hyperuricemia [P<0.0016 (0.05/32); Table IV].
Table IV.
P-values of the association between SNPs and intermediate phenotypes of atrial fibrillation.
| SNP | Nucleotide substitution (amino acid) | Hypertension | DM | Hyper-TG | Hypo-HDL | Hyper-LDL | CKD | Obesity | HU |
|---|---|---|---|---|---|---|---|---|---|
| rs11552708 | G/A (G67R) | 0.9943 | 0.7439 | 0.8296 | 0.0622 | 0.8902 | 0.0754 | 0.472 | 0.0006 |
| rs113710653 | C/T (E231K) | 0.1278 | 0.5273 | 0.1465 | 0.6862 | 0.2732 | 0.4215 | 0.9986 | 0.1295 |
| rs11231397 | G/C (R300T) | 0.2471 | 0.9445 | 0.12 | 0.1683 | 0.9415 | 0.9571 | 0.1993 | 0.9519 |
| rs77538589 | C/T (G117R) | 0.1179 | 0.8113 | 0.4675 | 0.6656 | 0.5844 | 0.8815 | 0.8741 | 0.6006 |
The prevalence of each phenotype was compared among genotypes with Fisher's exact test (2×2) or Pearson's chi-square test (2×3). Based on Bonferroni's correction, P<0.0016 (0.05/32) were considered statistically significant and are shown in bold. SNP, single nucleotide polymorphism; DM, diabetes mellitus; hyper-TG, hypertriglyceridemia; hypo-HDL, hypo-high density lipoprotein-cholesterolemia; hyper-LDL, hyper-low density lipoprotein-cholesterolemia; CKD, chronic kidney disease; HU, hyperuricemia.
Association of genes and SNPs identified in the present study with phenotypes previously examined in GWASs
The association of the four genes and four SNPs identified in the present study with phenotypes previously examined in GWASs available in public databases [GWAS Catalog (http://www.ebi.ac.uk/gwas) and GWAS Central (http://www.gwascentral.org/browser)] was determined. None of the genes or SNPs were identified to be associated with AF in previous studies (data not shown).
Discussion
The development of AF has been reported to be associated with endothelial dysfunction, including the upregulation of adhesion molecules, and increased inflammation and oxidative stress (15). Such endothelial dysfunction also promotes the electrophysiological remodeling observed in AF (16) and accelerates atrial ectopy through electrical excitation of cells near the pulmonary vein (17). In addition, coronary atherosclerosis may injure atrial tissue as result of myocardial ischemia and give rise to histological changes, including myocyte hypertrophy, necrosis and apoptosis (18,19). These processes may promote atrial remodeling with structural, functional, electrical, and metabolic consequences that underlie the development of AF (18,19).
The current study demonstrated that rs11552708 [G/A (G67R)] of TNFSF13, rs113710653 [C/T (E231K)] of SPATC1L and rs11231397 [G/C (R300T)] of SLC22A25 were significantly associated with AF in Japanese individuals. The minor T allele of rs113710653 and the minor C allele of rs11231397 were risk factors for AF, whereas the minor A allele of rs11552708 was protective against this condition. The rs77538589 [C/T (G117R)] SNP of SALL4 also showed a tendency to be associated to AF, although the association did not achieve statistical significance, with the minor T allele representing a risk factor for this condition.
The TNFSF13 gene is located at chromosomal region 17p13.1 (http://www.ncbi.nlm.nih.gov/gene/8741) and is expressed in various tissues and organs including heart muscle (The Human Protein Atlas, www.proteinatlas.org/ENSG00000161955-TNFSF13/tissue). TNFSF13 is a regulator of the development and function of B cells (20). It has been suggested that TNFSF13 has important roles in normal immune responses, and also in the development of autoimmune and inflammatory diseases. Plasma levels of TNFSF13 were reported to be increased in patients with multiple sclerosis (21), Sjögren's syndrome (22), systemic lupus erythematosus (23) and rheumatoid arthritis (24). The levels of TNFSF13 were also demonstrated to be significantly higher in the synovial fluid of patients with rheumatoid arthritis than in the serum, suggesting that this protein may be produced locally at the site of inflammation (25). Previous GWASs identified TNFSF13 as a susceptibility locus for immunoglobulin A nephropathy (26). Chronic vascular inflammation is an important feature of atherosclerosis (27), and atherosclerosis was previously reported to be associated with increased expression of TNFSF13. The plasma levels of TNFSF13 were demonstrated to be greater in patients with coronary artery disease than in healthy controls, and TNFSF13 immunoreactivity was detected in aggregated platelets within the ruptured plaque of patients with myocardial infarction (28). Pronounced TNFSF13 immunostaining was also observed in macrophages from carotid plaques (28). The current study demonstrated that rs11552708 [G/A (G67R)] of TNFSF13 was significantly associated with AF, with the minor A allele being protective against this condition. Chronic vascular inflammation, atherosclerosis, endothelial dysfunction and subsequent atrial remodeling are important in the pathogenesis of AF. Given that TNFSF13 is implicated in vascular inflammation and atherosclerosis (28), the association of TNFSF13 rs11552708 [G/A (G67R)] with AF may be attributable to the effect of this gene on atrial remodeling.
The SPATC1L gene is located at chromosomal region 21q22.3 (http://www.ncbi.nlm.nih.gov/gene/84221) and is expressed in various tissues and organs including heart muscle (The Human Protein Atlas, www.proteinatlas.org/ENSG00000160284-SPATC1L/tissue). SPATC1L is distributed in the cytoplasm, nucleus and perinuclear region of cells, and it translocates to sites of cell-cell junctions in response to cell stimulation with neurokinin A (29). Expression of SPATC1L modulates the response of human cells to N-methyl-N'-nitro-N-nitrosoguanidine, suggesting that the encoded protein may have a role in protecting cells from cell death induced by this DNA-damaging agent (30). The current study demonstrated that rs113710653 [C/T (E231K)] of SPATC1L was significantly associated with AF, with the minor T allele representing a risk factor for this condition, although the underlying molecular mechanism of this association remains unclear.
The SLC22A25 gene is located at chromosomal region 11q12.3 (http://www.ncbi.nlm.nih.gov/gene/387601) and is abundantly expressed in the liver (The Human Protein Atlas, www.proteinatlas.org/ENSG00000196600-SLC22A25/tissue). SLC22 is a large family of transmembrane organic cation and anion transporter proteins expressed predominantly in kidney and liver that mediate the uptake and excretion of environmental toxins, endogenous substances and drugs in the body (31). The results of the present study indicated that rs11231397 [G/C (R300T)] of SLC22A25 was significantly associated with AF, with the minor C allele representing a risk factor for this condition, although the functional relevance of this association remains unknown.
The SALL4 gene is located at chromosomal region 20q13.2 (http://www.ncbi.nlm.nih.gov/gene/57167) and is expressed in various tissues and organs including heart muscle (The Human Protein Atlas, www.proteinatlas.org/ENSG00000101115-SALL4/tissue). Mutations in SALL4 have been identified as a cause of Okihiro syndrome, which is characterized by developmental defects in limbs and multiple organs including the heart (32,33). Expression of SALL4 was detected in the left ventricular myocardium and interventricular septum of the mouse heart at embryonic day 10.5 (34). The current study demonstrated that rs77538589 [C/T (G117R)] of SALL4 tended to be associated with AF, with the minor T allele representing a risk factor for this condition. Given that SALL4 may have an important role in heart development, the association of rs77538589 of SALL4 with AF may reflect an effect of this gene on atrial structure or electrical function.
A meta-analysis of GWASs for AF (10) identified 10 SNPs (rs6666258 of potassium calcium-activated channel subfamily N member 3, rs3903239 of paired related homeobox 1, rs6817105 of paired like homeodomain 2, rs2040862 of Wnt family member 8A, rs3807989 of caveolin 1, rs10821415 of chromosome 9 open reading frame 3, rs10824026 of synaptopodin 2 like, rs1152591 of spectrin repeat containing nuclear envelope protein 2, rs7164883 of hyperpolarization activated cyclic nucleotide gated potassium channel 4 and rs2106261 of zinc finger homeobox 3) as susceptibility loci for this condition. The MAF of these SNPs ranged from 13.1–44.7% and the odds ratio from 0.87–1.64 (10). In the current study, four novel loci that may confer susceptibility to AF were identified, with the allele odds ratio (MAF, %) of rs11552708 of TNFSF13, rs113710653 of SPATC1L, rs11231397 of SLC22A25 and rs77538589 of SALL4 being 0.54 (36.6%), 5.36 (1.9%), 1.55 (43.7%) and 2.61 (4.3%), respectively. Both rs11552708 of TNFSF13 and rs11231397 of SLC22A25 were thus common variants with a low effect size, whereas rs113710653 of SPATC1L and rs77538589 of SALL4 were low-frequency variants with a moderate to high effect size.
There are several limitations to the present study: i) Given that the results of the study were not replicated, the findings will require validation in other independent subject panels or other ethnic groups; ii) it is possible that rs11552708 of TNFSF13, rs113710653 of SPATC1L, rs11231397 of SLC22A25 or rs77538589 of SALL4 is in linkage disequilibrium with other polymorphisms in the same gene or other nearby genes that are actually responsible for the development of AF; and iii) the functional relevance of these SNPs to the pathogenesis of AF remains to be elucidated.
In conclusion, rs11552708 of TNFSF13, rs113710653 of SPATC1L, rs11231397 of SLC22A25, and rs77538589 of SALL4 may be susceptibility loci for AF in Japanese individuals. Determination of genotypes of these SNPs may prove informative for assessment of the genetic risk for AF in Japanese individuals.
Acknowledgements
This work was supported by CREST (grant no. JPMJCR1302), Japan Science and Technology Agency (to Y.Y, J.S. and I.T.) and by Japan Society for the Promotion of Science KAKENHI grants no. JP15H04772 (to Y.Y.), no. JP25242062 (to M.T.) and no. JP16H01872 (to M.T.).
References
- 1.Chugh SS, Havmoeller R, Narayanan K, Singh D, Rienstra M, Benjamin EJ, Gillum RF, Kim YH, McAnulty JH, Jr, Zheng ZJ, et al. Worldwide epidemiology of atrial fibrillation: A global burden of disease 2010 study. Circulation. 2014;129:837–847. doi: 10.1161/CIRCULATIONAHA.113.005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Go AS, Hylek EM, Phillips KA, Chang Y, Henault LE, Selby JV, Singer DE. Prevalence of diagnosed atrial fibrillation in adults: National implications for rhythm management and stroke prevention: The An ticoagulation and risk factors in atrial fibrillation (ATRIA) study. JAMA. 2001;285:2370–2375. doi: 10.1001/jama.285.18.2370. [DOI] [PubMed] [Google Scholar]
- 3.Lip GY, Tse HF, Lane DA. Atrial fibrillation. Lancet. 2012;379:648–661. doi: 10.1016/S0140-6736(11)61514-6. [DOI] [PubMed] [Google Scholar]
- 4.Schnabel RB, Sullivan LM, Levy D, Pencina MJ, Massaro JM, D'Agostion RB, Sr, Newton-Cheh C, Yamamoto JF, Magnani JW, Tadros TM, et al. Development of a risk score for atrial fibrillation (Framingham Heart Study): A community-based cohort study. Lancet. 2009;373:739–745. doi: 10.1016/S0140-6736(09)60443-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lubitz SA, Ozcan C, Magnani JW, Kääb S, Benjamin EJ, Ellinor PT. Genetics of atrial fibrillation: Implications for future research directions and personalized medicine. Circ Arrhythm Electrophysiol. 2010;3:291–299. doi: 10.1161/CIRCEP.110.942441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, Gretarsdottir S, Holm H, Sigurdsson A, Jonasdottir A, Baker A, Thorleifsson G, Kristjansson K, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–357. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 7.Gudbjartsson DF, Holm H, Gretarsdottir S, Thorleifsson G, Walters GB, Thorgeirsson G, Gulcher J, Mathiesen EB, Njølstad I, Nyrnes A, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–878. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benjamin EJ, Rice KM, Arking DE, Pfeufer A, van Noord C, Smith AV, Schnabel RB, Bis JC, Boerwinkle E, Sinner MF, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–881. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinor PT, Lunetta KL, Glazer NL, Pfeufer A, Alonso A, Chung MK, Sinner MF, de Bakker PI, Mueller M, Lubitz SA, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–244. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ellinor PT, Lunetta KL, Albert CM, Glazer NL, Ritchie MD, Smith AV, Arking DE, Müller-Nurasyid M, Krijthe BP, Lubitz SA, et al. Meta-analysis identifies six new susceptibility loci for atrial fibrillation. Nat Genet. 2012;44:670–675. doi: 10.1038/ng.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grove ML, Yu B, Cochran BJ, Haritunians T, Bis JC, Taylor KD, Hansen M, Borecki IB, Cupples LA, Fornage M, et al. Best practices and joint calling of the HumanExome BeadChip: The CHARGE consortium. PLoS One. 2013;8:e68095. doi: 10.1371/journal.pone.0068095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Anderson CA, Pettersson FH, Clarke GM, Cardon LR, Morris AP, Zondervan KT. Data quality control in genetic case-control association studies. Nat Protoc. 2010;5:1564–1573. doi: 10.1038/nprot.2010.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.O'Neal WT, Efird JT, Yeboah J, Nazarian S, Alonso A, Heckbert SR, Soliman EZ. Brachial flow-mediated dilation and incident atrial fibrillation: The multi-ethnic study of atherosclerosis. Arterioscler Thromb Vasc Biol. 2014;34:2717–2720. doi: 10.1161/ATVBAHA.114.304560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim YM, Guzik TJ, Zhang YH, Zhang MH, Kattach H, Ratnatunga C, Pillai R, Channon KM, Casadei B. A myocardial Nox2 containing NAD (P)H oxidase contributes to oxidative stress in human atrial fibrillation. Circ Res. 2005;97:629–636. doi: 10.1161/01.RES.0000183735.09871.61. [DOI] [PubMed] [Google Scholar]
- 17.Haïssaguerre M, Jaïs P, Shah DC, Takahashi A, Hocini M, Quiniou G, Garrigue S, Le Mouroux A, Le Métayer P, Clémenty J. Spontaneous initiation of atrial fibrillation by ectopic beats origination in the pulmonary veins. N Engl J Med. 1998;339:659–666. doi: 10.1056/NEJM199809033391003. [DOI] [PubMed] [Google Scholar]
- 18.Herringa J, Van Der Kuip DA, Hofman A, Kors JA, van Rooij FJ, Lip GY, Witteman JC. Subclinical atherosclerosis and risk of atrial fibrillation: The rotterdam study. Arch Intern Med. 2007;167:382–387. doi: 10.1001/archinte.167.4.382. [DOI] [PubMed] [Google Scholar]
- 19.Casaclang-Verzosa G, Gresh BJ, Tsang TS. Structural and functional remodeling of the left atrium: Clinical and therapeutic implications for atrial fibrillation. J Am Coll Cardiol. 2008;51:1–11. doi: 10.1016/j.jacc.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 20.Dillon SR, Gross JA, Ansell SM, Novak AJ. An APRIL to remember: Novel TNF ligands as therapeutic targets. Nat Rev Drug Discov. 2006;5:235–246. doi: 10.1038/nrd1982. [DOI] [PubMed] [Google Scholar]
- 21.Thangarajh M, Masterman T, Rot U, Duvefelt K, Brynedal B, Karrenbauer VD, Hillert J. Increased levels of APRIL (a proliferation-inducing ligand) mRNA in multiple sclerosis. J Neuroimmunol. 2005;167:210–214. doi: 10.1016/j.jneuroim.2005.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Jonsson MV, Szodoray P, Jellestad S, Jonsson R, Skarstein K. Association between circulating levels of the novel TNF family members APRIL and BAFF and lymphoid organization in primary Sjögren's syndrome. J Clin Immunol. 2005;25:189–201. doi: 10.1007/s10875-005-4091-5. [DOI] [PubMed] [Google Scholar]
- 23.Koyama T, Tsukamoto H, Miyagi Y, Himeji D, Otsuka J, Miyagawa H, Harada M, Horiuchi T. Raised serum APRIL levels in patients with systemic lupus erythematosus. Ann Rheum Dis. 2005;64:1065–1067. doi: 10.1136/ard.2004.022491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheema GS, Roschke V, Hilbert DM, Stohl W. Elevated serum B lymphocyte stimulator levels in patients with systemic immune-based rheumatic diseases. Arthritis Rheum. 2001;44:1313–1319. doi: 10.1002/1529-0131(200106)44:6<1313::AID-ART223>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 25.Tan SM, Xu D, Roschke V, Perry JW, Arkfeld DG, Ehresmann GR, Migone TS, Hilbert DM, Stohl W. Local production of B lymphocyte stimulator protein and APRIL in arthritic joints of patients with inflammatory arthritis. Arthritis Rheum. 2003;48:982–992. doi: 10.1002/art.10860. [DOI] [PubMed] [Google Scholar]
- 26.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Libby P. Inflammation in atherosclerosis. Nature. 2002;420:868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 28.Sandberg WJ, Otterdal K, Gullestad L, Halvorsen B, Ragnarsson A, Frøland SS, Damås JK, Oie E, Aukrust P, Hansson GK, Yndestad A. The tumour necrosis factor superfamily ligand APRIL (TNFSF13) is released upon platelet activation and expressed in atherosclerosis. Thromb Haemost. 2009;102:704–710. doi: 10.1160/TH08-10-0665. [DOI] [PubMed] [Google Scholar]
- 29.Lecat S, Matthes HW, Pepperkok R, Simpson JC, Galzi JL. A fluorescent live imaging screening assay based on translocation criteria identifies novel cytoplasmic proteins implicated in G protein-coupled receptor signaling pathways. Mol Cell Proteomics. 2015;14:1385–1399. doi: 10.1074/mcp.M114.046698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fry RC, Svensson JP, Valiathan C, Wang E, Hogan BJ, Bhattacharya S, Bugni JM, Whittaker CA, Samson LD. Genomic predictors of interindividual differences in response to DNA damaging agents. Genes Dev. 2008;22:2621–2626. doi: 10.1101/gad.1688508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacobsson JA, Haitina T, Lindblom J, Fredriksson R. Identification of six putative human transporters with structural similarity to the drug transporter SLC22 family. Genomics. 2007;90:595–609. doi: 10.1016/j.ygeno.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 32.Kohlhase J, Heinrich M, Schubert L, Liebers M, Kispert A, Laccone F, Turnpenny P, Winter RM, Reardon W. Okihiro syndrome is caused by SALL4 mutations. Hum Mol Genet. 2002;11:2979–2987. doi: 10.1093/hmg/11.23.2979. [DOI] [PubMed] [Google Scholar]
- 33.Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- 34.Koshiba-Takeuchi K, Takeuchi JK, Arruda EP, Kathiriya IS, Mo R, Hui CC, Srivastava D, Bruneau BG. Cooperative and antagonistic interactions between Sall4 and Tbx5 pattern the mouse limb and heart. Nat Genet. 2006;38:175–183. doi: 10.1038/ng1707. [DOI] [PubMed] [Google Scholar]