Abstract
Malignant gliomas are high-grade gliomas, which are derived from glial cells in the spine or brain. To examine the mechanisms underlying malignant gliomas in the present study, the expression profile of GSE54004, which included 12 grade II astrocytomas, 33 grade III astrocytomas and 98 grade IV astrocytomas, was downloaded from the Gene Expression Omnibus. Using the Limma package in R, the differentially expressed genes (DEGs) in grade III, vs. grade II astrocytoma, grade IV, vs. grade II astrocytoma, and grade IV, vs. grade III astrocytoma were analyzed. Venn diagram analysis and enrichment analyses were performed separately for the DEGs using VennPlex software and the Database for Annotation, Visualization and Integrated Discovery. Protein-protein interaction (PPI) networks were visualized using Cytoscape software, and subsequent module analysis of the PPI networks was performed using the ClusterONE tool. Finally, glioma-associated genes and glioma marker genes among the DEGs were identified using the CTD database. A total of 27, 1,446 and 776 DEGs were screened for the grade III, vs. grade II, grade IV, vs. grade II, and grade IV, vs. grade III astrocytoma comparison groups, respectively. Functional enrichment analyses showed that matrix metalloproteinase 9 (MMP9) and chitinase 3-like 1 (CHI3L1) were enriched in the extracellular matrix and extracellular matrix structural constituent, respectively. In the PPI networks, annexin A1 (ANXA1) had a higher degree and MMP9 had interactions with vascular endothelial growth factor A (VEGFA). There were 10 common glioma marker genes between the grade IV, vs. grade II and the grade IV, vs. grade III comparison groups, including MMP9, CHI3L1, VEGFA and S100 calcium binding protein A4 (S100A4). This suggested that MMP9, CHI3L1, VEGFA, S100A4 and ANXA1 may be involved in the progression of malignant gliomas.
Keywords: malignant gliomas, differentially expressed genes, common genes, glioma marker genes, protein-protein interaction network
Introduction
As a type of tumor derived from glial cells in the spine or brain (1), gliomas account for ~80% of malignant brain tumors, and 30% of central nervous system and brain tumors (2). According to their histological features, gliomas are classified into astrocytomas, ependymomas, oligodendrogliomas, optic nerve gliomas, mixed gliomas and brainstem gliomas, among which astrocytomas are the common type of primary brain tumor among adults (3). Gliomas are also divided into low-grade gliomas (grade I and II) and high-grade gliomas (grade III and IV) according to the World Health Organization classification criteria (4). High grade gliomas are malignant gliomas comprising glioblastoma multiforme and anaplastic astrocytomas, and the median overall survival rates of patients with glioblastoma multiforme and anaplastic astrocytomas are ~15 months and 3 years, respectively (5). Therefore, elucidation of the mechanisms underlying malignant gliomas and the development of novel therapeutic strategies are urgently required.
Several studies have investigated the genes in involved in gliomas. In children with malignant gliomas, the overexpression of p53 is closely associated with adverse outcomes, independently of histological findings and clinical prognostic factors (6,7). As a critical mediator of the unfolded protein response, 78-kDa glucose-regulated protein (GRP78) is significantly upregulated in human malignant glioma and associated with its proliferation rate, suggesting that drugs capable of inhibiting GRP78 may be applied in the treatment of malignant glioma (8). Promoter methylation-induced silencing of the O6-methylguanine-DNA methyltransferase DNA-repair gene contributes to longer survival rates in patients with glioblastoma who are treated with alkylating agents (9). A previous study demonstrated that the increased level of hypoxia-inducible factor-1α (HIF-1α) is critical for the activation of glioma cell motility through affecting molecules associated with invasion (10,11). The Decoy receptor 3 (DcR3) soluble protein may be implicated in the immune evasion and progression of malignant gliomas through inhibiting CD95 ligand (CD95 L) (12). It has also been reported that the overexpression of Neuropilin 1 (NRP1) promotes tumor progression and is associated with poor prognosis in glioma (13). Spy1, which belongs to the Speedy/RINGO family, is correlated with the poor prognosis in patients with glioma and may serve as an independent prognostic predictor for patients with the disease (14). However, a comprehensive understanding of the mechanisms underlying gliomas is required.
In 2014, Guan et al used newly sequenced glioma datasets and downloaded glioma gene expression profiles to investigate the association between known molecular subtypes of grade IV glioblastoma (GBM) with grade II/III gliomas (GII/III), and found shared patterns between the GBMs and GII/IIIs (15). Using the data deposited by Guan et al (15), the present study further identified the differentially expressed genes (DEGs) in three comparison groups (grade III, vs. grade II, grade IV, vs. grade II, and grade IV, vs. grade III), and their functions were predicted using enrichment analysis. Subsequently, protein-protein interaction (PPI) networks were constructed and module analysis was performed to analyze the interactions among the DEGs. In addition, glioma-associated genes and glioma marker genes among the DEGs were screened to identify the key genes implicated in malignant glioma.
Materials and methods
Microarray data
The gene expression profile of GSE54004, which was sequenced on the platform of GPL18281 Illumina HumanHT-12 WG-DASL V4.0 R2 expression beadchip (gene symbol version), was downloaded from the Gene Expression Omnibus database (http://www.ncbi.nih.gov/geo). To identify the key genes involved in the progression of malignant glioma, a total 143 samples were selected from the GSE54004 dataset, including 12 grade II astrocytomas, 33 grade III astrocytomas and 98 grade IV astrocytomas. The astrocytomas were collected from patients with glioma, following which the tissues were fixed in formalin and embedded in paraffin at MD Anderson Cancer Center (Houston, TX, USA) (15). The GSE54004 dataset was deposited by Guan et al (15), and this study by Guan et al (15) was approved by the Institutional Review Board of MD Anderson Cancer Center, with informed consent provided by all participants.
DEG screening
The downloaded gene expression files were merged to obtain a gene expression matrix. Using the Limma package (16) in R (version 3.22.7, http://www.bioconductor.org/packages/release/bioc/html/limma.html), the DEGs in three comparison groups (grade III, vs. grade II, grade IV, vs. grade II, and grade IV, vs. grade III) were analyzed. The thresholds of P<0.05 and |log2fold change (FC)|≥1 were used. VennPlex software (version 1.0.0.2; http://www.irp.nia.nih.gov/bioinformatics/vennplex.html) enables the screening of upregulated, downregulated or contraregulated individual factors between complex data sets (17). Venn diagram analysis was performed for the DEGs screened from different groups using VennPlex software (17) (version 1.0.0.2; http://www.irp.nia.nih.gov/bioinformatics/vennplex.html).
Functional and pathway enrichment analysis
Gene Ontology (GO; http://www.geneontology.org/) can be utilized to predict the potential functions for genes and their products in terms of molecular function (MF), cellular component (CC) and biological process (BP) (18). The Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/) database contains integrated knowledge of information regarding biochemical reactions and compounds, information on proteins and genes, and information on molecular interaction networks (19). GO functional and KEGG pathway enrichment analyses were performed separately for the upregulated and downregulated genes in each comparison group, using Database for Annotation, Visualization and Integrated Discovery software (DAVID version 6.7; http://www.david.niaid.nih.gov). Those terms involving at least two genes and with a P-value of P<0.05 were selected.
PPI network and module analyses
The Search Tool for the Retrieval of Interacting Genes (STRING version 10.0; http://string-db.org/) web resource and database contributes to the identification of PPIs, involving functional and physical associations (20). The PPI associations among the DEGs in each comparison group were searched using the STRING database (21), with a combined score (required confidence) >0.9. Following this, the PPI networks for the DEGs in each comparison group were visualized separately using Cytoscape software (version 3.2.0; http://www.cytoscape.org/) (22). In the PPI network, the degrees of nodes were determined by the number of edges involved, and nodes with higher degrees were determined as key nodes. Module analysis was also performed for the PPI networks using the ClusterONE tool (23).
Identification of glioma-associated genes and glioma marker genes
The Comparative Toxicogenomics Database (CTD; http://ctdbase.org/) (24) collects gene-disease, chemical-disease and chemical-gene interactions, which are manually searched from scientific literature through strict text mining using structured notation, ontologies and controlled vocabularies. Combined with the CTD database, glioma-associated genes and glioma marker genes among the DEGs screened for each group were analyzed.
Results
DEG analysis
With the thresholds of P<0.05 and |log2FC| ≥1, the DEGs in the three comparison groups were analyzed. Compared with grade II samples, a total of 27 DEGs (grade III, vs. grade II), including nine upregulated and 18 downregulated genes, were screened in the grade III samples. A total of 1,446 DEGs (grade IV, vs. grade II), including 643 upregulated and 803 downregulated genes, were identified in the grade IV samples. A total of 776 DEGs (grade IV, vs. grade III), including 410 upregulated and 366 downregulated genes, were identified in the grade IV samples relative to the grade III samples. There were more DEGs in the grade III and grade IV samples.
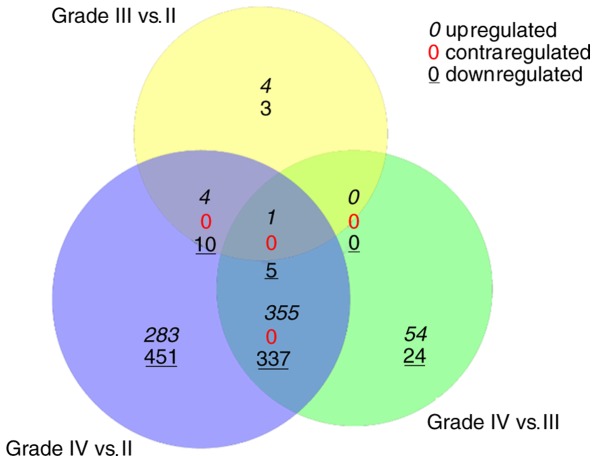
Venn diagram analysis showed that 20 DEGs, including five upregulated and 15 downregulated genes, were common genes between the grade III, vs. grade II and grade IV, vs. grade II comparison groups. In addition, 698 DEGs, including 356 upregulated and 342 downregulated genes, were common genes between the grade IV, vs. grade II, and grade IV, vs. grade III comparison groups (Fig. 1).
Figure 1.
Results of Venn diagram analysis for the differentially expressed genes identified from different groups.
Functional and pathway enrichment analysis
No functions were enriched for the upregulated genes in the grade III, vs. grade II group. However, the downregulated genes in the grade III, vs. grade II group were significantly enriched in functions including synaptic transmission (GO_BP; P=8.06E-04) and plasma membrane (GO_CC; P=1.98E-02; Table I). No pathways were enriched for the DEGs in the grade III, vs. grade II group.
Table I.
Functions enriched for the downregulated genes in the grade III, vs. grade II group.
| Category | Term | P-value | Genes (n) | Gene symbol |
|---|---|---|---|---|
| BP | GO:0007268~synaptic transmission | 8.06E-04 | 4 | NOS1, PCDHB4, UNC13C, HTR2A |
| GO:0019226~transmission of nerve impulse | 1.28E-03 | 4 | NOS1, PCDHB4, UNC13C, HTR2A | |
| GO:0050877~neurological system process | 5.56E-03 | 5 | NOS1, PCDHB4, POU4F1, UNC13C, HTR2A | |
| GO:0007267~cell-cell signaling | 5.97E-03 | 4 | NOS1, PCDHB4, UNC13C, HTR2A | |
| GO:0043271~negative regulation of ion transport | 1.52E-02 | 2 | NOS1, HTR2A | |
| GO:0007416~synaptogenesis | 2.11E-02 | 2 | PCDHB4, POU4F1 | |
| GO:0009408~response to heat | 3.67E-02 | 2 | NOS1, XYLT1 | |
| GO:0050808~synapse organization | 3.99E-02 | 2 | PCDHB4, POU4F1 | |
| CC | GO:0005886~plasma membrane | 1.98E-02 | 8 | CAMKV, EPHB6, NOS1, PCDHB4, UNC13C, TMEM25, ABCC8, HTR2A |
GO, Gene Ontology; BP, biological process; CC, cellular component.
For the upregulated genes in the grade IV, vs. grade II group, functions including extracellular matrix organization (GO_BP; P=2.64E-17), extracellular matrix (GO_CC; P=2.90E-25), which involves matrix metallopeptidase 9 (MMP9) and extracellular matrix structural constituent (GO_MF; P=4.83E-10), in addition to the systemic lupus erythematosus pathway (P=4.70E-16), were significantly enriched. For the downregulated genes in the grade IV, vs. grade II group, functions including transmission of nerve impulse (GO_BP; P=1.12E-20), synapse (GO_CC; P=1.96E-21) and ion channel activity (GO_MF; P=1.35E-18), in addition to the neuroactive ligand-receptor interaction pathway (P=6.62E-07), were significantly enriched (Table II).
Table II.
Functions and pathways separately enriched for the upregulated and downregulated genes in the grade IV, vs. grade II group.
| Category | Term | P-value | Genes (n) | Gene symbol |
|---|---|---|---|---|
| Upregulated | ||||
| BP | GO:0030198~extracellular matrix organization | 2.64E-17 | 29 | ADAMTS14, MMP9, LUM, COL3A1, ELN, POSTN, DCN, SERPINH1, TGFB2, TNFRSF11B… |
| GO:0009611~response to wounding | 2.31E-15 | 61 | F2RL2, NRP1, S100A8, S100A9, C1QC, CXCL10, TGFB2, CD97, S1PR3, SAA2… | |
| GO:0022403~cell cycle phase | 2.89E-15 | 53 | KIF23, KIFC1, PRC1, PTTG1, GTSE1, CDKN2C, CDCA2, DNAJC2, CCNA2, SNHG3-RCC1… | |
| CC | GO:0031012~extracellular matrix | 2.90E-25 | 62 | CTHRC1, LTBP1, NPNT, MMP9, MMP7, POSTN, MMP2, TGFB2, TNFRSF11B, TGFBI… |
| GO:0005576~extracellular region | 1.42E-23 | 161 | F2RL2, CTHRC1, LTBP1, C9ORF47, FAM20A, MMP9, FAM20C, MMP7, TNFSF14, POSTN… | |
| GO:0005578~proteinaceous extracellular matrix | 5.13E-23 | 57 | CTHRC1, LTBP1, NPNT, MMP9, MMP7, POSTN, MMP2, TNFRSF11B, TGFBI, LOX… | |
| MF | GO:0005201~extracellular matrix structural constituent | 4.83E-10 | 19 | COL4A2, COL4A1, LUM, ELN, COL3A1, CHI3L1, MGP, COL5A3, EMILIN2, COL5A2… |
| GO:0050840~extracellular matrix binding | 5.02E-10 | 12 | BGN, TGFBI, C6ORF15, VEGFA, ELN, OLFML2A, NID1, DCN, THBS1, ADAM9… | |
| GO:0030246~carbohydrate binding | 1.18E-09 | 38 | CCL2, C21ORF63, CD248, SUSD2, HEXB, POSTN, DCN, MDK, SIGLEC9, HMMR… | |
| Pathway | hsa05322: Systemic lupus erythematosus | 4.70E-16 | 28 | HIST1H2AB, HIST4H4, HIST1H4L, HIST2H2AA4, HLA-DRB1, C1R, C1S, C1QC, HIST1H2BO, HIST1H2BM… |
| hsa04512: ECM-receptor interaction | 6.20E-11 | 21 | COL4A2, COL4A1, TNC, COL3A1, ITGA1, HSPG2, ITGA3, COL5A3, COL5A2, COL5A1… | |
| hsa04510: Focal adhesion | 5.34E-09 | 29 | CAV2, CAV1, TNC, COL3A1, COL6A3, COL6A1, ZYX, PDGFD, LAMB1, THBS1… | |
| Downregulated | ||||
| BP | GO:0019226~transmission of nerve impulse | 1.12E-20 | 58 | SYT1, GABRB3, SYT4, GLRA3, CNP, GABBR2, KCNIP2, VIPR1, SLC1A4, KCNQ5… |
| GO:0007268~synaptic transmission | 1.47E-19 | 52 | SYT1, GABRB3, SYT4, GLRA3, CNP, GABBR2, KCNIP2, VIPR1, SLC1A4, KCNQ5… | |
| GO:0006811~ion transport | 2.91E-15 | 79 | KCNC2, KCNH1, SLC22A17, SLC8A3, JPH4, JPH3, GABRB3, SCN3A, KCNAB1, SCN3B… | |
| CC | GO:0045202~synapse | 1.96E-21 | 63 | SYT1, SEPT3, ENAH, CLSTN2, GABRB3, SYT4, GRIP1, GLRA3, GABRB1, SYT9… |
| GO:0044456~synapse part | 7.63E-19 | 49 | SYT1, SYT4, GABRB3, CLSTN2, GRIP1, GABRB1, GLRA3, SYT9, GABBR2, GRIN3A… | |
| GO:0043005~neuron projection | 2.24E-12 | 48 | SNCG, SYT1, CDK5R1, CCK, ADCY2, SYT4, GABRB3, SNCB, ALDOC, GRIN3A… | |
| MF | GO:0005216~ion channel activity | 1.35E-18 | 59 | KCNC2, KCNH1, SCN3A, GABRB3, KCNAB1, SCN3B, GLRA3, GABRB1, GRIN3A, KCNIP2… |
| GO:0015267~channel activity | 1.56E-18 | 61 | KCNC2, KCNH1, SCN3A, GABRB3, KCNAB1, SCN3B, GLRA3, GABRB1, GRIN3A, KCNIP2… | |
| GO:0022803~passive transmembrane transporter activity | 1.76E-18 | 61 | KCNC2, KCNH1, SCN3A, GABRB3, KCNAB1, SCN3B, GLRA3, GABRB1, GRIN3A, KCNIP2… | |
| Pathway | hsa04080: Neuroactive ligand-receptor interaction | 6.62E-07 | 28 | GPR83, THRA, PRLHR, GABRB3, GABRB1, DRD5, GLRA3, GRIN3A, GABBR2, VIPR1… |
| hsa04020: Calcium signaling pathway | 6.83E-06 | 21 | SLC8A3, ADCY2, SLC8A2, NOS1, ADCY8, DRD5, GRIN1, CACNA1I, GRM1, ATP2B2… | |
| hsa04360: Axon guidance | 2.96E-03 | 13 | NGEF, PLXNB1, ABLIM3, NTN4, SLIT1, EPHB1, PAK7, EPHB6, SEMA6B, EPHA6… |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
For the upregulated genes in the grade IV, vs. grade III group, terms including extracellular matrix organization (GO_BP; P=9.69E-20), extracellular region part (GO_CC; P=5.11E-28), extracellular matrix structural constituent (GO_MF; P=3.47E-13), which involves chitinase 3-like 1 (CHI3L1), and ECM-receptor interaction (P=1.35E-10) were significantly enriched. For the downregulated genes in the grade IV, vs. grade III group, terms including transmission of nerve impulse (GO_BP; P=1.02E-14), synapse (GO_CC; P=4.37E-22), calcium ion binding (GO_MF; P=2.79E-13) and calcium signaling pathway (P=2.15E-05) were significantly enriched (Table III).
Table III.
Functions and pathways separately enriched for the upregulated and downregulated genes in the grade IV, vs. grade III group.
| Category | Term | P-value | Genes (n) | Gene symbol |
|---|---|---|---|---|
| Upregulated | ||||
| BP | GO:0030198~extracellular matrix organization | 9.69E-20 | 27 | IBSP, ADAMTS14, LUM, MMP9, COL3A1, POSTN, SERPINH1, TGFBI, BCL3, LOX… |
| GO:0001944~vasculature development | 1.06E-15 | 34 | CAV1, NRP1, TNFRSF12A, COL3A1, ENPEP, TYMP, ACE, HOXA3, ANG, HMOX1… | |
| GO:0043062~extracellular structure organization | 1.42E-15 | 28 | IBSP, ADAMTS14, MMP9, LUM, COL3A1, POSTN, SERPINH1, TGFBI, BCL3, LOX… | |
| CC | GO:0044421~extracellular region part | 5.11E-28 | 87 | CTHRC1, MMP9, IGFBP6, MMP7, TNFSF14, POSTN, HP, CXCL10, ISG15, SAA2… |
| GO:0031012~extracellular matrix | 3.40E-26 | 52 | CTHRC1, MMP9, MMP7, POSTN, ANG, TGFBI, LOX, SPON2, VWA1, LOXL1… | |
| GO:0005578~proteinaceous extracellular matrix | 6.96E-26 | 50 | ADAMTS18, CTHRC1, ADAMTS14, CD248, MMP9, LUM, COL3A1, MMP7, POSTN, TIMP1… | |
| MF | GO:0005201~extracellular matrix structural constituent | 3.47E-13 | 19 | COL18A1, COL4A2, COL4A1, LUM, EFEMP2, COL3A1, CHI3L1, MGP, EMILIN2, COL5A2… |
| GO:0019838~growth factor binding | 1.25E-11 | 19 | COL4A1, IL2RA, OSMR, IGFBP6, COL3A1, ESM1, COL5A1, CD36, IL1RAP, COL1A2… | |
| GO:0030247~polysaccharide binding | 7.81E-09 | 19 | FGFR1, SUSD2, C6ORF15, POSTN, CXCL6, COL5A1, PCOLCE2, TNFAIP6, BGN, ANG… | |
| Pathway | hsa04512: ECM-receptor interaction | 1.35E-10 | 17 | IBSP, COL4A2, COL4A1, COL3A1, ITGA1, ITGA3, COL5A2, COL5A1, CD36, ITGA5… |
| hsa04510: Focal adhesion | 2.82E-09 | 23 | IBSP, CAV2, CAV1, COL4A2, COL4A1, VAV3, COL3A1, MET, ITGA1, ACTN1… | |
| hsa05322: Systemic lupus erythematosus | 2.98E-05 | 12 | HIST1H2AB, HIST2H3A, HIST2H2AA4, HIST1H4L, HIST1H2BH, ACTN1, C1R, HIST2H3C, HIST2H3D… | |
| Downregulated | ||||
| BP | GO:0019226~transmission of nerve impulse | 1.02E-14 | 34 | SYT1, SYT4, GABRB3, GLRA3, GABBR2, KCNIP2, SLC1A4, NPTX1, GAD2, S1PR1… |
| GO:0007268~synaptic transmission | 2.14E-13 | 30 | SYT1, SYT4, GABRB3, GLRA3, GABBR2, KCNIP2, SLC1A4, NPTX1, GAD2, SYN1… | |
| GO:0007267~cell-cell signaling | 5.65E-12 | 40 | SYT1, EDN3, CCL3, SYT4, GABRB3, FGF17, CAMK2G, GLRA3, FGF12, GABBR2… | |
| CC | GO:0045202~synapse | 4.37E-22 | 45 | SYT1, SEPT3, CDK5R1, ENAH, SNAP91, CLSTN2, GABRB3, SYT4, GLRA3, GABRB1… |
| GO:0044456~synapse part | 1.31E-20 | 37 | SYT1, SNAP91, CLSTN2, GABRB3, SYT4, GABRB1, GLRA3, SYT9, BCAN, ATP6V1G2… | |
| GO:0043005~neuron projection | 2.66E-11 | 31 | SNCG, SYT1, CDK5R1, CCK, ADCY2, SNCB, SYT4, GABRB3, GRIN3A, GABBR2… | |
| MF | GO:0005509~calcium ion binding | 2.79E-13 | 53 | SYT1, CLSTN2, SYT4, MASP1, SYT9, GRIN3A, KCNIP2, KCNIP3, SYP, ATP2B2…… |
| GO:0022836~gated channel activity | 4.21E-12 | 29 | KCNC2, GABRB3, GABRB1, GLRA3, GRIN3A, KCNIP2, KCNK12, KCNJ3, KCNIP3, GRIN2C…… | |
| GO:0005216~ion channel activity | 3.09E-11 | 31 | KCNC2, GABRB3, GABRB1, GLRA3, GRIN3A, KCNIP2, KCNK12, KCNJ3, KCNIP3, SLC1A4…… | |
| Pathway | hsa04080: Neuroactive ligand-receptor interaction | 9.42E-07 | 19 | GABRG2, GABRA1, THRA, PRLHR, GABRB3, GLRA3, GABRB1, ADCYAP1R1, GRIN1, GRIN3A…… |
| hsa04020: Calcium signaling pathway | 2.15E-05 | 14 | ADCY2, SLC8A2, ADCY8, CAMK2G, CACNA1I, GRIN1, GRM1, ATP2B2, CD38, GRIN2C…… | |
| hsa04720: Long-term potentiation | 1.65E-03 | 7 | GRIA2, ADCY8, GRIN2C, CAMK2G, PPP1R1A, GRIN1, GRM1 |
GO, Gene Ontology; BP, biological process; CC, cellular component; MF, molecular function.
PPI network and module analyses
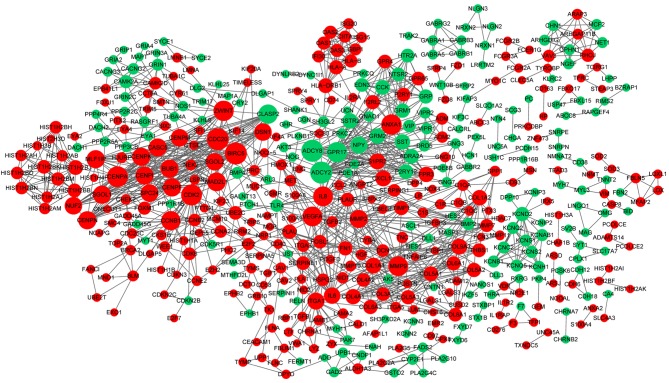
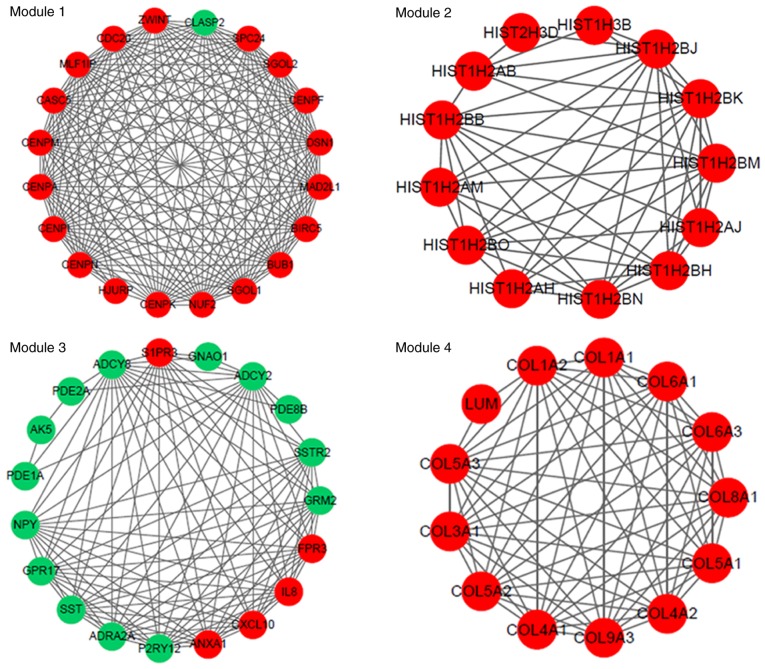
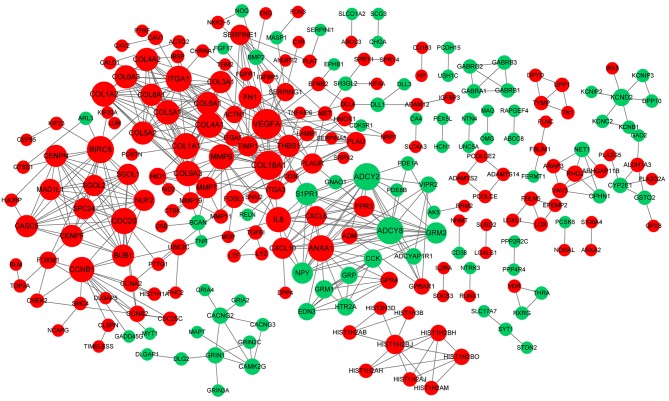
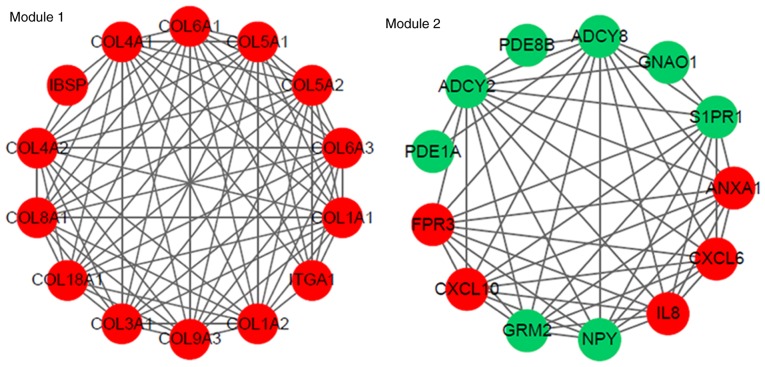
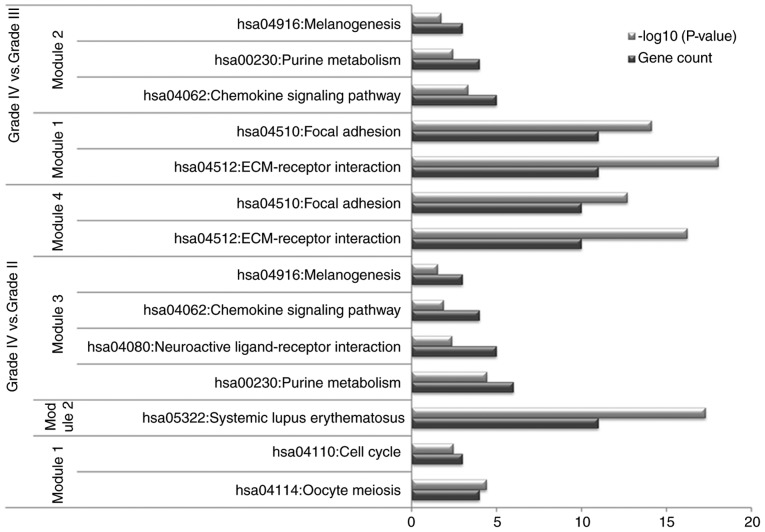
The PPI network constructed for the DEGs in the grade IV, vs. grade II group had 489 nodes and 1,244 interactions (Fig. 2), in which four significant modules were identified (Fig. 3). The PPI network constructed for the DEGs in the grade IV, vs. grade III group had 243 nodes and 500 interactions (Fig. 4), from which two significant modules were identified (Fig. 5). The nodes with higher degrees, including MMP9, vascular endothelial growth factor A (VEGFA) and annexin A1 (ANXA1), in the two networks are listed in Table IV. MMP9 interacted with VEGFA in these two PPI networks. No PPI network was constructed for the DEGs in grade III, vs. grade II group. The pathways enriched for the genes involved in the modules are shown in Fig. 6.
Figure 2.
Protein-protein interaction network constructed for the differentially expressed genes in the grade IV, vs. grade II group. The green nodes indicate the downregulated genes, and the red nodes indicate the upregulated genes. The higher the degree, the larger the node. The lines represent the interaction of the protein and the other proteins in the network.
Figure 3.
Four modules (module 1, 2, 3 and 4) identified from the protein-protein interaction network constructed for the differentially expressed genes in the grade IV, vs. grade II group. The green nodes indicate the downregulated genes, and the red nodes indicate the upregulated genes. The lines represent the interaction of the protein and the other proteins in the modules.
Figure 4.
Protein-protein interaction network constructed for the differentially expressed genes in the grade IV, vs. grade III group. The green nodes indicate the downregulated genes, and the red nodes indicate the upregulated genes. The higher the degree, the larger the node. The lines represent the interaction of the protein and the other proteins in the network.
Figure 5.
Two modules (module 1 and 2) identified from the protein-protein interaction network constructed for the differentially expressed genes in the grade IV, vs. grade III group. The green nodes indicate the downregulated genes, and the red nodes indicate the upregulated genes. The lines represent the interaction of the protein and the other proteins in the modules.
Table IV.
Top 15 nodes with highest degrees in the grade IV, vs. grade II protein-protein interaction networks.
| Node | Log fold change | Degree |
|---|---|---|
| ADCY2 | −1.98963 | 26 |
| ADCY8 | −1.17088 | 26 |
| BIRC5 | 1.359254 | 26 |
| CDC20 | 1.816218 | 26 |
| CENPA | 1.236634 | 24 |
| CDK2 | 1.239547 | 24 |
| GPR17 | −1.2297 | 23 |
| ANXA1 | 2.129899 | 23 |
| ZWINT | 1.196125 | 23 |
| IL8 | 1.634318 | 22 |
| MMP9 | 3.458891 | 22 |
| VEGFA | 2.236317 | 22 |
| CLASP2 | −1.04519 | 21 |
| CCNB1 | 1.290933 | 21 |
| MAD2L1 | 1.046268 | 20 |
| VEGFA | 2.225246 | 19 |
| COL18A1 | 1.107411 | 18 |
| BIRC5 | 1.006404 | 16 |
| ADCY8 | −1.09838 | 16 |
| ADCY2 | −1.40787 | 16 |
| CDC20 | 1.497869 | 15 |
| COL4A1 | 1.877445 | 15 |
| COL1A1 | 2.614414 | 15 |
| ANXA1 | 1.793973 | 15 |
| ITGA1 | 1.174656 | 15 |
| MMP9 | 3.32357 | 14 |
| CCNB1 | 1.097028 | 14 |
| IL8 | 1.420499 | 14 |
| COL1A2 | 2.180694 | 14 |
| COL9A3 | 1.231098 | 14 |
Figure 6.
Pathways enriched for the genes involved in modules.
Identification of glioma-associated genes and glioma marker genes
The glioma-associated genes and glioma marker genes among the DEGs screened for each group were further analyzed using the CTD database. In general, 81.48, 86.93 and 89.95% of the DEGs in the grade III, vs. grade II, grade IV, vs. grade II, and grade IV, vs. grade III comparison groups, respectively, were glioma-associated genes. Of note, there were 10 common glioma marker genes, including MMP9, CHI3L1, VEGFA and S100 calcium binding protein A4 (S100A4) between the grade IV, vs. grade II, and grade IV, vs. grade III comparison groups (Table V).
Table V.
Numbers of glioma-associated genes and glioma marker genes among the differentially expressed genes screened for each group.
| Group | Glioma-associated genes, n (%) | Glioma marker genes (n) |
|---|---|---|
| Grade III vs. grade II | 22 (81.48) | 1 |
| Grade IV vs. grade II | 1,257 (86.93) | 24 |
| Grade IV vs. grade III | 698 (89.95) | 13 |
Discussion
In the present study, a total of 27 (nine upregulated and 18 downregulated), 1,446 (643 upregulated and 803 downregulated) and 776 (410 upregulated and 366 downregulated) DEGs were identified in the grade III, vs. grade II, grade IV, vs. grade II, and grade IV, vs. grade III comparison groups, respectively. Venn diagram analysis showed that 20 DEGs, including five upregulated and 15 downregulated genes, were common genes between the grade III, vs. grade II and grade IV, vs. grade II comparison groups. A total of 698 DEGs, including 356 upregulated and 342 downregulated genes) were common genes between the grade IV, vs. grade II and grade IV, vs. grade III comparison groups. Four significantly modules were identified from the PPI network constructed for the DEGs in the grade IV, vs. grade II group, and two significantly modules were identified from the PPI network constructed for the DEGs in the grade IV, vs. grade III group. No PPI network was constructed for the DEGs in the grade III, vs. grade II group. It was found that 81.48, 86.93 and 89.95% of the DEGs in the grade III, vs. grade II, grade IV vs. grade II, and grade IV vs. grade III comparison groups, respectively, were glioma-associated genes. In addition, there were 10 common glioma marker genes, including MMP9, CHI3L1, VEGFA and S100A4, between the grade IV, vs. grade II and grade IV, vs. grade III comparison groups.
Inhibiting the expression of MMP9 through RNA interference represses the malignancy of glioma cells, indicating that it can be applied in the treatment of malignant gliomas (25–27). MMP2 and MMP9 have significant effects on the degradation of extracellular matrix (ECM) and angiogenesis, and on the invasiveness of gliomas, therefore, they can be utilized in targeted therapy of malignant glioma (28). CHI3L1 and MMP-9 are overexpressed in malignant gliomas, and can serve as a predictors of survival rates in patients with the disease (29). In addition, CHI3L1 is important in regulating local invasiveness and malignant transformation in gliomas, therefore, CHI3L1 may be a used as a molecular target in the treatment of gliomas (30). In patients with glioma, the serum level of CHI3L1, which encodes a secreted glycoprotein, is associated with tumor grade and possibly tumor burden in glioblastoma multiforme (31). Functional enrichment analyses have shown that MMP9 and CHI3L1 were separately enriched in the ECM and its structural constituent, respectively. ECM rigidity can mediate the invasion of glioblastoma multiforme cells through actomyosin contractility (32,33). These findings indicate that MMP9 and CHI3L1 may function in the progression of malignant gliomas through the ECM.
VEGF is an effective mediator of vascular permeability, and its inhibition can decrease tumor burden and edema production in malignant glioma (34). The growth and progression of astrocytoma is dependent on neovascularization, and the angiogenesis factor VEGFA may be essential for the infiltrative and aggressive growth of astrocytomas (35). VEGFA affects the neovascularization and invasion of glioblastoma, not only by promoting endothelial mitogenesis and permeability, but also by regulating MMP2 (36). In the two PPI networks in the present study, VEGFA interacted with MMP9, indicating that VEGFA may also affect malignant gliomas via interacting with MMP9.
The expression of S100A4 is promoted by neutrophil infiltration, and targeting S100A4 may be promising in reducing antiangiogenic therapy resistance and inhibiting the glioma malignant phenotype (37). S100A4/Mts1 has a higher expression in high-grade glioblastomas, compared with low-grade astrocytic tumors, indicating that it has an effect on brain tumor progression (38). ANXA1, targeted by forkhead box M1 (FOXM1) has a high expression in gliomas and can function as a predictor of poor prognosis in patients with the disease (39). A previous study demonstrated that ANXA1 may contribute to maintaining brain homeostasis and may be used as chemotherapeutic target in the treatment of glioblastoma multiforme (40). Therefore, S100A4 and ANXA1 may be involved in the development of malignant gliomas.
In conclusion, the present study identified 27, 1,446 and 776 DEGs in the grade III, vs. grade II, grade IV, vs. grade II, and grade IV vs. grade III comparison groups respectively. It was found that MMP9, CHI3L1, VEGFA, S100A4 and ANXA1 may act in the progression of malignant gliomas. However, these findings were obtained from bioinformatics analysis and require further validation.
Acknowledgements
This study was financially supported by the Heilongjiang Province Postdoctoral Scientific Research Developmental Fund (grant no. LBH-Q14113).
References
- 1.Mamelak AN, Jacoby DB. Targeted delivery of antitumoral therapy to glioma and other malignancies with synthetic chlorotoxin (TM-601) Expert Opin Drug Deliv. 2007;4:175–186. doi: 10.1517/17425247.4.2.175. [DOI] [PubMed] [Google Scholar]
- 2.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Kleihues P, Soylemezoglu F, Schäuble B, Scheithauer BW, Burger PC. Histopathology, classification, and grading of gliomas. Glia. 1995;15:211–221. doi: 10.1002/glia.440150303. [DOI] [PubMed] [Google Scholar]
- 4.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0278-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleeker FE, Molenaar RJ, Leenstra S. Recent advances in the molecular understanding of glioblastoma. J Neurooncol. 2012;108:11–27. doi: 10.1007/s11060-011-0793-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pollack IF, Finkelstein SD, Woods J, Burnham J, Holmes EJ, Hamilton RL, Yates AJ, Boyett JM, Finlay JL, Sposto R, Children's Cancer Group Expression of p53 and prognosis in children with malignant gliomas. N Engl J Med. 2002;346:420–427. doi: 10.1056/NEJMoa012224. [DOI] [PubMed] [Google Scholar]
- 7.Hirose Y, Berger MS, Pieper RO. p53 effects both the duration of G2/M arrest and the fate of temozolomide-treated human glioblastoma cells. Cancer Res. 2001;61:1957–1963. [PubMed] [Google Scholar]
- 8.Pyrko P, Schönthal AH, Hofman FM, Chen TC, Lee AS. The unfolded protein response regulator GRP78/BiP as a novel target for increasing chemosensitivity in malignant gliomas. Cancer Res. 2007;67:9809–9816. doi: 10.1158/0008-5472.CAN-07-0625. [DOI] [PubMed] [Google Scholar]
- 9.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara S, Nakagawa K, Harada H, Nagato S, Furukawa K, Teraoka M, Seno T, Oka K, Iwata S, Ohnishi T. Silencing hypoxia-inducible factor-1alpha inhibits cell migration and invasion under hypoxic environment in malignant gliomas. Int J Oncol. 2007;30:793–802. [PubMed] [Google Scholar]
- 11.Zagzag D, Lukyanov Y, Lan L, Ali MA, Esencay M, Mendez O, Yee H, Voura EB, Newcomb EW. Hypoxia-inducible factor 1 and VEGF upregulate CXCR4 in glioblastoma: Implications for angiogenesis and glioma cell invasion. Lab Invest. 2006;86:1221–1232. doi: 10.1038/labinvest.3700482. [DOI] [PubMed] [Google Scholar]
- 12.Roth W, Isenmann S, Nakamura M, Platten M, Wick W, Kleihues P, Bähr M, Ohgaki H, Ashkenazi A, Weller M. Soluble decoy receptor 3 is expressed by malignant gliomas and suppresses CD95 ligand-induced apoptosis and chemotaxis. Cancer Res. 2001;61:2759–2765. [PubMed] [Google Scholar]
- 13.Osada H, Tokunaga T, Nishi M, Hatanaka H, Abe Y, Tsugu A, Kijima H, Yamazaki H, Ueyama Y, Nakamura M. Overexpression of the neuropilin 1 (NRP1) gene correlated with poor prognosis in human glioma. Anticancer Res. 2004;24:547–552. [PubMed] [Google Scholar]
- 14.Zhang L, Shen A, Ke Q, Zhao W, Yan M, Cheng C. Spy1 is frequently overexpressed in malignant gliomas and critically regulates the proliferation of glioma cells. J Mol Neurosci. 2012;47:485–494. doi: 10.1007/s12031-012-9709-5. [DOI] [PubMed] [Google Scholar]
- 15.Guan X, Vengoechea J, Zheng S, Sloan AE, Chen Y, Brat DJ, O'Neill BP, de Groot J, Yust-Katz S, Yung WK, et al. Molecular subtypes of glioblastoma are relevant to lower grade glioma. PLoS One. 2014;9:e91216. doi: 10.1371/journal.pone.0091216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai H, Chen H, Yi T, Daimon CM, Boyle JP, Peers C, Maudsley S, Martin B. VennPlex-a novel Venn diagram program for comparing and visualizing datasets with differentially regulated datapoints. PLoS One. 2013;8:e53388. doi: 10.1371/journal.pone.0053388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harris MA, Clark J, Ireland A, Lomax J, Ashburner M, Foulger R, Eilbeck K, Lewis S, Marshall B, Mungall C, et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 2004;32(Database Issue):D258–D261. doi: 10.1093/nar/gkh036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database Issue):D277–D280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37(Database Issue):D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2014;43(Database Issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saito R, Smoot ME, Ono K, Ruscheinski J, Wang PL, Lotia S, Pico AR, Bader GD, Ideker T. A travel guide to Cytoscape plugins. Nat Methods. 2012;9:1069–1076. doi: 10.1038/nmeth.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ. The Comparative Toxicogenomics Database's 10th year anniversary: Update 2015. Nucleic Acids Res. 2015;43(Database Issue):D914–D920. doi: 10.1093/nar/gku935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lakka SS, Gondi CS, Yanamandra N, Olivero WC, Dinh DH, Gujrati M, Rao JS. Inhibition of cathepsin B and MMP-9 gene expression in glioblastoma cell line via RNA interference reduces tumor cell invasion, tumor growth and angiogenesis. Oncogene. 2004;23:4681–4689. doi: 10.1038/sj.onc.1207616. [DOI] [PubMed] [Google Scholar]
- 26.Sun C, Wang Q, Zhou H, Yu S, Simard AR, Kang C, Li Y, Kong Y, An T, Wen Y, et al. Antisense MMP-9 RNA inhibits malignant glioma cell growth in vitro and in vivo. Neurosci Bull. 2013;29:83–93. doi: 10.1007/s12264-012-1296-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gondi CS, Lakka SS, Dinh DH, Olivero WC, Gujrati M, Rao JS. Downregulation of uPA, uPAR and MMP-9 using small, interfering, hairpin RNA (siRNA) inhibits glioma cell invasion, angiogenesis and tumor growth. Neuron Glia Biol. 2004;1:165–176. doi: 10.1017/S1740925X04000237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang M, Wang T, Liu S, Yoshida D, Teramoto A. The expression of matrix metalloproteinase-2 and-9 in human gliomas of different pathological grades. Brain Tumor Pathol. 2003;20:65–72. doi: 10.1007/BF02483449. [DOI] [PubMed] [Google Scholar]
- 29.Hormigo A, Gu B, Karimi S, Riedel E, Panageas KS, Edgar MA, Tanwar MK, Rao JS, Fleisher M, DeAngelis LM, Holland EC. YKL-40 and matrix metalloproteinase-9 as potential serum biomarkers for patients with high-grade gliomas. Clin Cancer Res. 2006;12:5698–5704. doi: 10.1158/1078-0432.CCR-06-0181. [DOI] [PubMed] [Google Scholar]
- 30.Ku BM, Lee YK, Ryu J, Jeong JY, Choi J, Eun KM, Shin HY, Kim DG, Hwang EM, Yoo JC, et al. CHI3L1 (YKL-40) is expressed in human gliomas and regulates the invasion, growth and survival of glioma cells. Int J Cancer. 2011;128:1316–1326. doi: 10.1002/ijc.25466. [DOI] [PubMed] [Google Scholar]
- 31.Tanwar MK, Gilbert MR, Holland EC. Gene expression microarray analysis reveals YKL-40 to be a potential serum marker for malignant character in human glioma. Cancer Res. 2002;62:4364–4368. [PubMed] [Google Scholar]
- 32.Ulrich TA, de Juan Pardo EM, Kumar S. The mechanical rigidity of the extracellular matrix regulates the structure, motility, and proliferation of glioma cells. Cancer Res. 2009;69:4167–4174. doi: 10.1158/0008-5472.CAN-08-4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bellail AC, Hunter SB, Brat DJ, Tan C, Van Meir EG. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int J Biochem Cell Biol. 2004;36:1046–1069. doi: 10.1016/j.biocel.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 34.Carlson MR, Pope WB, Horvath S, Braunstein JG, Nghiemphu P, Tso CL, Mellinghoff I, Lai A, Liau LM, Mischel PS, et al. Relationship between survival and edema in malignant gliomas: Role of vascular endothelial growth factor and neuronal pentraxin 2. Clin Cancer Res. 2007;13:2592–2598. doi: 10.1158/1078-0432.CCR-06-2772. [DOI] [PubMed] [Google Scholar]
- 35.Johansson M, Brännström T, Bergenheim AT, Henriksson R. Spatial expression of VEGF-A in human glioma. J Neurooncol. 2002;59:1–6. doi: 10.1023/A:1016317407043. [DOI] [PubMed] [Google Scholar]
- 36.Gong J, Zhu S, Zhang Y, Wang J. Interplay of VEGFa and MMP2 regulates invasion of glioblastoma. Tumor Biol. 2014;35:11879–11885. doi: 10.1007/s13277-014-2438-3. [DOI] [PubMed] [Google Scholar]
- 37.Liang J, Piao Y, Holmes L, Fuller GN, Henry V, Tiao N, de Groot JF. Neutrophils promote the malignant glioma phenotype through S100A4. Clin Cancer Res. 2014;20:187–198. doi: 10.1158/1078-0432.CCR-13-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takenaga K, Nygren J, Zelenina M, Ohira M, Iuchi T, Lukanidin E, Sjöquist M, Kozlova EN. Modified expression of Mts1/S100A4 protein in C6 glioma cells or surrounding astrocytes affects migration of tumor cells in vitro and in vivo. Neurobiol Dis. 2007;25:455–463. doi: 10.1016/j.nbd.2006.10.021. [DOI] [PubMed] [Google Scholar]
- 39.Cheng SX, Tu Y, Zhang S. FoxM1 promotes glioma cells progression by up-regulating Anxa1 expression. PLoS One. 2013;8:e72376. doi: 10.1371/journal.pone.0072376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Festa M, Caputo M, Cipolla C, D'Acunto C, Rossi A, Tecce M, Capasso A. The involvement of xanthohumol in the expression of annexin in human malignant glioblastoma cells. Open Biochem J. 2013;7:1–10. doi: 10.2174/1874091X01307010001. [DOI] [PMC free article] [PubMed] [Google Scholar]