Abstract
The transcription factor, interferon regulatory factor 4 (IRF4), serves an essential role in the regulation of immune responses, and has been reported to act as a diagnostic and prognostic marker for various hematological malignancies. The present study aimed to investigate whether IRF4 could exert effects on human non-small cell lung cancer (NSCLC) and to explore the underlying mechanism. The mRNA and protein expression of IRF4 was detected in NSCLC tissues using reverse-transcription quantitative polymerase chain reaction and western blotting, respectively. In the in vitro experiment, IRF4 expression was knocked down or overexpressed using lentivirus in human lung adenocarcinoma A549 and lung squamous cell carcinoma LC-AI cell lines. Cell proliferation and colony number were analyzed using MTT and colony formation assays, respectively. The expression levels of IRF4 mRNA and protein were significantly higher in NSCLC tissues (n=54) compared with that in adjacent non-tumor tissues. Similarly, the expression levels of Notch1 and Notch2 mRNA were significantly higher in NSCLC tissues. Furthermore, the expression level of IRF4 mRNA was positively correlated with the levels of Notch1 and Notch2 mRNA in NSCLC tissues. Consequently, using NSCLC cell lines, it was demonstrated that the knockdown of IRF4 expression significantly reduced the cell proliferation rate and colony formation, whereas IRF4-overexpression significantly increased them. Notably, the IRF4 knockdown significantly decreased the expression levels of Notch1 and Notch2 mRNA, and phosphorylated protein kinase B (AKT), whereas IRF4 overexpression resulted in the opposite. The results of the present study indicate that IRF4 is overexpressed and serves as a tumor promoter in human NSCLC, at least partially, through activating the Notch-Akt signaling pathway.
Keywords: cell growth, interferon regulatory factor 4, MK-0752, Non-small cell lung cancer, Notch/Akt signaling pathway
Introduction
Lung cancer is a serious public health problem and remains the most common cause of cancer-related mortality (1). Non-small cell lung cancer (NSCLC) accounts for approximately 80% of all lung cancers (2). Although many advances have achieved in cancer biology, such as the development of specific therapies for distinct subtypes of NSCLC, appropriate treatment options for the majority of patients are unsatisfactory (3). To identify new therapeutic targets and options, it is of great necessity and importance to have a better understanding of the pathogenesis of NSCLC.
Accumulating evidence suggests that, in many types of malignancies, inflammatory signals could be regulated by cytokines and chemokines, and thus exerting vital effects on tumor progression including cancer cell proliferation, survival, metastasis and angiogenesis (4). The nine members of the interferon regulatory factor (IRF) family are transcription factors that play diverse roles in innate and adaptive immune responses, cell growth regulation, cell apoptosis, and hematopoietic development (5–7).
Particularly, IRF4 is a diagnostic and prognostic marker for various hematological malignancies (8). IRF4 is lymphocyte specific and is overexpressed in Epstein-Barr virus transformed cells, multiple myeloma, and human T cell leukemia virus 1 (HTLV1)-infected cell lines and associated adult T cell lymphoma/leukemia (9–12). There are several reports suggest a possible role of IRF4 in the pathogenesis of chronic myeloid leukemia (13,14). Besides, IRF4 upregulation has been proved to induce the growth of lymphomas or multiple myeloma (10,15). Yang et al (16) reported that IRF 4 binding protein is a novel p53 target gene and suppresses cisplatin-induced apoptosis of breast cancer cells. In a previous study by Chen et al (17), IRF4 was shown to predict poorer survival of NSCLC patients. Based on these investigations, it raises the possibility that IRF4 may be involved in NSCLC tumor progression, however, the underlying mechanism was not clear yet. In the present study, we investigated whether IRF4 could exert effects on human NSCLC and to explore the underlying mechanism in the in vitro experiment. IRF4 expression was upregulated in NSCLC tissues as compared to the corresponding adjacent non-tumor tissue. In vitro experiments showed that IRF4 knockdown by shRNA significantly reduced cell proliferation and colony number of NSCLC cells; whereas IRF4 overexpression showed the absolutely opposite results. Further investigations were performed to explore the underlying mechanism in human NSCLC cells. The present study suggests that IRF4 may be a new potential target for NSCLC treatment.
Materials and methods
Human samples
Paired NSCLC (34 females and 20 males; with an average age of 54; 22 patients with adenocarcinoma and 32 patients with squamous cell carcinoma) and non-tumor adjacent lung tissues (more than 5 cm from the edge of tumor) were obtained, with informed consent, from 54 patients who underwent primary surgical resection of NSCLC at the Second Affiliated Hospital of Soochow University (SuZhou, China). Among them, 22 patients had negative lymph nodes metastasis and 32 patients had positive lymph nodes metastasis. 19 patients were at the I–II TNM stages and 35 were at the III–IV TNM stages. None of the patients had received preoperative radiotherapy or chemotherapy and chronic obstructive pulmonary disease was excluded in the present study. Tissues were obtained and frozen immediately with liquid nitrogen and stored in a freezer at −70°C. The present study was approved by the Ethics Committee of the Second Affiliated Hospital of Soochow University. Written informed consent was obtained from all the participants.
Quantitative real-time PCR
Total RNA was extracted from paired NSCLC and non-tumor adjacent lung tissues with Trizol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. First-strand cDNA was synthesized from total RNA using Super Script II first-strand synthesis system (Invitrogen). Real-time PCR was performed using an Applied Biosystems 7300 Sequence Detection system (Applied Biosystems, Foster City, CA, USA). The primers for IRF4: forward, 5′-CTACACCATGACAACGCCTTACC-3′ and reverse, 5′-GGCTGATCCGGGACGTAGT-3′. Notch1: forward, 5′-CTTAGATGTGCTGAGCGCGTCAATGTGTC-3′ and reverse, 5′-GCGCGATCCTTGATAACCTGCGGAT-3′. Notch 2: forward, 5′-CATAGAATGATTAGCAGAGAG-3′ and reverse, 5′-CAACATCAGAGCTAGCAAGAG-3′. GAPGH, forward 5′-GGTGGAGGTCGGGAGTCAACGGA-3′, reverse 5′-GAGGGATCTCGCTCCTGGAGGA-3′. The comparative threshold cycle (Ct) method was used to analyze the results.
Western blot analysis
For western blot analysis, cell lysates were prepared from cell lines with RIPA lysis buffer kit (Santa Cruz Biotechnology, Santa Cruz, CA), and the protein concentrations were quantified using a Bio-Rad protein assay (Bio-Rad, Hercules, CA). Whole-cell proteins (30 µg) were separated on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride membranes (Amersham Corp., Arlington Heights, IL). The membranes were incubated with primary antibodies anti-IRF4 (1:500, cat. sc-6059; Santa Cruz Biotechnology), anti-Notch1 (1:100, cat. Val1744, Cell Signaling, Danvers, MA), anti-Notch2 (1:500, cat. D67C8; Cell Signaling), anti-AKT (1:500, cat. 9272; Cell Signaling), anti-AKT phosphorylation (1:500, cat. 9271; Cell Signaling) or anti-GAPDH (1:1,000, cat. 2111; Cell Signaling) overnight at 4°C. The horseradish peroxidase-conjugated secondary antibodies (1:1,000, cat. A50-106P; Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd., Beijing, China;) were subsequently used at room temperature for 1 h. Signals were detected using enhanced chemiluminescence kit (Wuhan Booute Biotechnology Co., Ltd, Wuhan, China; cat. no. orb90504) and exposed to Kodak X-OMAT film (Eastman Kodak, Rochester, NY, USA).
Cell lines and cell culture
Human lung adenocarcinoma cell line (A549) and lung squamous cell carcinoma cell line (RERF-LC-AI, LC-AI) were obtained from Shanghai Cell Bank (Chinese Academy of Sciences, China) and maintained in −80°C freezer or cultured in RPMI (Roswell Park Memorial Institute) 1640 medium containing 10% fetal bovine serum (FBS) before experiments. Cells were maintained in 5% CO2 incubator at 37°C and used for experiments in the exponential phase of their growth. To detect the effect of Notch on NSCLC cells, A549 and LC-AI were treated with 20 µM MK-0752, a Notch pathway inhibitor (18), for 48 h.
Lentivirus packaging
For IRF4 knockdown, the lentivirus expressing short hairpin RNA (shRNA) targeting the sequence of IRF4 (5′-GCCGTACAAAGTTCAGGATCC-3′) and the control sequence (5′-TTCTCCGAACGTGTCACGT-3′) was purchased from Genechem Co. Ltd (Shanghai, China). Lentivirus expressing scrambled shRNA was used as a negative control (sh-Ctrl; Genechem Co. Ltd.). For IRF4 overexpression, the targeted IRF4 gene was amplified in human sample by PCR with the following primers: forward: 5′-CACCATGACAACGCCTTACC-3′ and reverse 5′-CATTTTCACAAGCTGGGCCT-3′. The PCR product containing PmeI and BstBI site introduced by primer was digested, and then cloned into lentivirus vector to construct a lentiviral vector carrying IRF4. The empty lentivirus vector was used as a negative control (Ctrl). The shRNA-expressing, overexpressing lentivirus and the corresponding controls were transfected into 293 T cells, which are always used in producing lentiviruses (19), together with the lentivirus helper plasmids to generate respective lentivirus. After transfection, the lentivirus supernatant was collected to transfect A549 and LC-AI cells. Infectious lentivirus was harvested 48 h post-transfection, centrifuged to remove cell debris, and then filtered through 0.45 µm cellulose acetate filters.
Cell proliferation assay
Cells were seeded into a 96-well plate (1×104 cells/well) and cultured in 200 µl RPMI-1640. At 4 h before terminating culture, 20 µl MTT (5 mg/ml) was added into each well. After cell culture, the supernatant was discarded and 100 µl dimethylsulfoxide (DMSO) was added into each well. After the formazan granules were completely dissolved, a value of each well was measured at 490 nm, and the corresponding cell proliferation number was calculated.
Colony formation assay
NSCLC cells were suspended in 1.5 ml complete medium supplemented with 0.45% low melting point agarose. The cells were placed in 35 mm tissue culture plates containing 1.5 ml complete medium and agarose (0.75%) on the bottom layer. The plates were incubated at 37°C with 5% CO2 for 2 weeks. Cell colonies were stained with 0.005% crystal violet and analyzed using a microscope.
Statistical analysis
Data were presented as means ± SEM. The statistical analysis was performed with SPSS 10.0 (SPSS, Inc., Chicago, IL, USA). For two groups, Student's t test was performed to analysis the data, and one-way ANOVA analysis was applied when three or more groups exist. The associations between Notch1, Notch2 mRNA expression levels and IRF4 mRNA level in NSCLC tissues were tested with the linear regression analysis. P<0.05 was considered statistically significant.
Results
IRF4 is upregulated in human NSCLC tissues
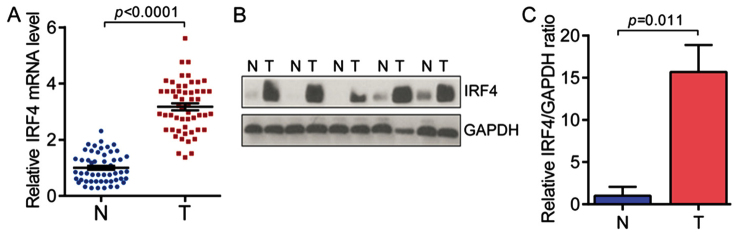
To investigate the role of IRF4 in NSCLC development, we first analyzed the expression of IRF4 in NSCLC tissues (n=54, 22 cases with adenocarcinoma and 32 cases with squamous cell carcinoma). As shown in Fig. 1A, the expression levels of IRF4 mRNA were significantly highe rin NSCLC tissues as compared with non-tumor adjacent normal tissues (n=54 for each group). Moreover, the expression levels of IRF4 protein were significantly higher in NSCLC tissues (Fig. 1B and C). These findings indicate the potential role of IRF4 in NSCLC development and progression.
Figure 1.
IRF4 is up-regulated in human NSCLC (A) IRF4 mRNA level is overexpressed in human non-small cell lung cancer (NSCLC) tissues (T) compared with non-tumor adjacent normal tissues (N) (n=54 in each group). (B) IRF4 protein level is up-regulated in NSCLC tissues (T) compared with non-tumor adjacent normal tissues (N) (n=54 in each group). Representative western blot images were shown (n=5, 1case (lane 1) with adenocarcinoma and 4 cases (lane 2–5) with squamous cell carcinoma). (C) Quantitative results of IRF4 protein level in (B). The protein level of IRF4 is normalized by GAPDH protein level. N=54 in each group. 54 NSCLC cases consist of 22 cases with adenocarcinoma and 32 cases with squamous cell carcinoma. IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer; T, tumor; N, non-tumor.
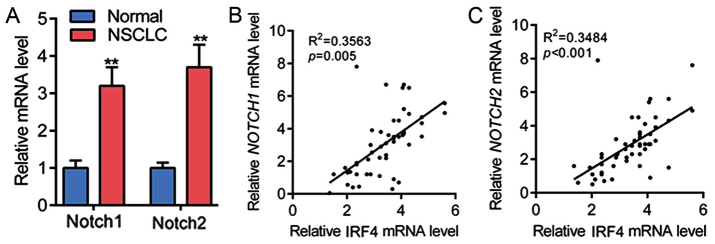
IRF4 level is positively correlated with the expression of Notch in NSCLC tissues
Then, we want to further explore the underlying mechanism of the tumorigenic role of IRF4 in NSCLC. Notch, firstly identified in human T-cell neoplasia, is a potent regulator for cellular differentiation, development, proliferation and survival (20–22). Besides, previous study indicated that IRF4 acts as a critical regulator of Notch signaling during chronic lymphocytic leukemia (CLL) development (23). Therefore, the mRNA levels of Notch1 and Notch2 in NSCLC tissues were determined, as shown in Fig. 2A, Notch1 and Notch2 mRNA expression were significantly upregulated in NSCLC tissues as compared with non-tumor adjacent normal tissues. Subsequently, linear regression analysis showed that IRF4 mRNA expression was positively correlated with the mRNA level of Notch1 in 54 cases of NSCLC tissues (Fig. 2B). The linear regression analysis also showed that IRF4 mRNA expression was positively correlated with the mRNA level of Notch2 in NSCLC tissues in 54 cases of NSCLC tissues (Fig. 2C). These results showed that IRF4 level was positively correlated with the expression of Notch in NSCLC tissues.
Figure 2.
IRF4 level was positively correlated with the expression of Notch in NSCLC tissues (A) Notch1 and Notch2 are overexpressed in NSCLC tissues (N=54 in each group). **P<0.01 vs. normal. (B) Linear regression analysis was performed to analyze the correlation between IRF4 and Notch1 (N=54 in each group). (C) Linear regression analysis was performed to analyze the correlation between IRF4 and Notch2 (N=54 in each group). IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer.
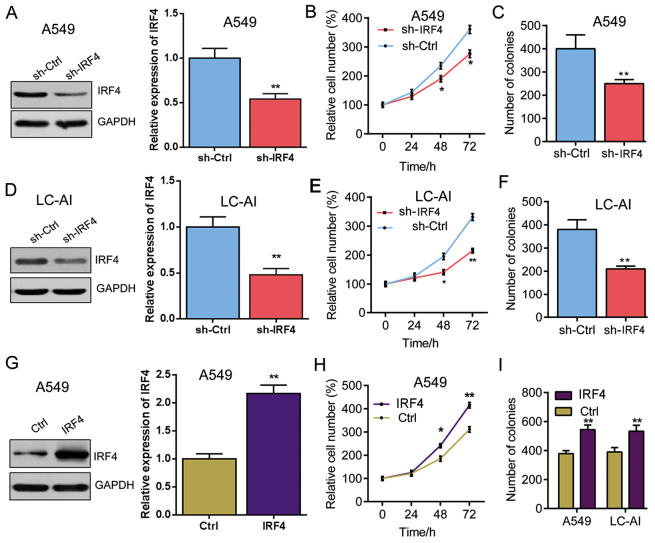
IRF4 promotes cell growth of human NSCLC cells
To understand the tumorigenic role of IRF4, we knock-downed the expression of IRF4 with lentivirus expressing sh-IRF4 in A549 and LC-AI cells, respectively. The IRF4 expression was significantly decreased by IRF4 knockdown in A549, which is determined by western blot analysis (Fig. 3A). As shown in Fig. 3B, cell proliferation was significantly suppressed in A549 that with IRF4 knockdown in a time-dependent manner. And IRF4 knockdown also markedly reduced the colony number in A549 cells (Fig. 3C). Similarly, IRF4 expression was also significantly inhibited in the LC-AI cells (Fig. 3D) and the cell proliferation rate and colony number of LC-AI cells were markedly reduced by IRF4 knockdown (Fig. 3E and F).
Figure 3.
IRF4 promotes NSCLC cell proliferation and increases colony number (A) IRF4 knockdown in A549 cells. A549 cells were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors for 48 h. Western blotting was performed to analyze protein level. **P<0.01 vs. sh-Ctrl. (B) IRF4 knockdown inhibits proliferation of A549 cells. A549 cell were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors. Relative cell numbers were evaluated at the indicated time points using MTT method. *P<0.05 vs. sh-Ctrl. (C) IRF4 knockdown decreases colony number of A549 cells. A549 cells infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors were subjected to soft sugar colony formation assay. The colony number in each well was evaluated two weeks later. **P<0.01 vs. sh-Ctrl. (D) IRF4 knockdown in LC-AI cells. LC-AI cells were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors for 48 h. Western blotting was performed to analyze protein level. **P<0.01 vs. sh-Ctrl. (E) IRF4 knockdown inhibits proliferation of LC-AI cells. LC-AI cell were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors. Relative cell numbers were evaluated at the indicated time points using MTT method. *P<0.05 and **P<0.01 vs. sh-Ctrl. (F) IRF4 knockdown decreases colony number of LC-AI cell. LC-AI cells infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors were subjected to soft sugar colony formation assay. The colony number in each well was evaluated two weeks later. **P<0.01 vs. sh-Ctrl. (G) IRF4 overexpression in A549 cells. A549 cells were infected with lentivirus expressing Ctrl or IRF4 vectors for 48 h. Western blotting was performed to analyze protein level. **P<0.01 vs. Ctrl. (H) IRF4 overexpression facilitates proliferation of A549 cells. A549 cell were infected with lentivirus expressing Ctrl or IRF4 vectors. Relative cell numbers were evaluated at the indicated time points using MTT method. *P<0.05 and **P<0.01 vs. Ctrl. (I) IRF4 overexpression increases colony number of A549 and LC-AI cells. A549 and LC-AI cells infected with lentivirus expressing Ctrl or IRF4 vectors were subjected to soft sugar colony formation assay. The colony number in each well was evaluated two weeks later. **P<0.01 vs. Ctrl. IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer. IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer; sh-Ctrl, negative control; Ctrl, control.
Additionally, IRF4-overexpressing lentivirus (IRF4) was also generated and infectedA549 cells, which was confirmed by western blotting analysis (Fig. 3G). In contrast to the results of IRF4 knockdown, IRF4 overexpression significantly promoted the proliferation and increased colony number of A549 cells (Fig. 3H and I). Besides, similar results were found in LC-AI cells (Fig. 3I). These results indicate that IRF4 promotes NSCLC cell proliferation and increases colony number.
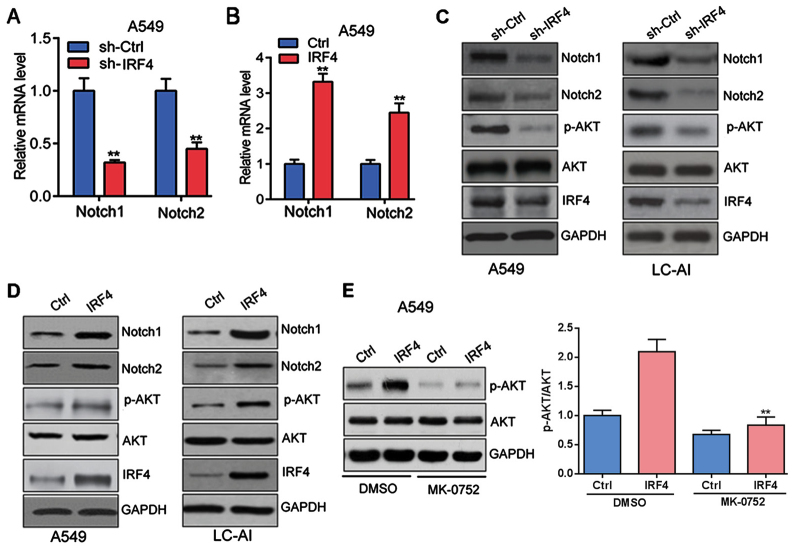
IRF4 activates notch-Akt signaling in NSCLC cells
Furthermore, as shown in Fig. 4A and B, IRF4 knockdown significantly decreased both the mRNA expression of Notch1 and Notch2 in A549 cells; whereas IRF4 overexpression markedly increased the mRNA expression of Notch1 and Notch2 in A549 cells.
Figure 4.
IRF4 activates Notch-Akt signaling in NSCLC cells (A) IRF4 knockdown inhibits Notch1 and Notch2 expression in A549 cells. A549 cells were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors for 48 h. qRT-PCR was performed to analyzed mRNA level. **P<0.01 vs. sh-Ctrl. (B) IRF4overexpression promotes Notch1 and Notch2 expression in A549 cells. A549 cells were infected with lentivirus expressing Ctrl or IRF4 vectors for 48 h. qRT-PCR was performed to analyzed mRNA level. **P<0.01 vs. Ctrl. (C) IRF4 knockdown inhibits Notch-Akt signaling in A549 and LC-AI cells. A549 and LC-AI cells were infected with lentivirus expressing sh-Ctrl or sh-IRF4 vectors for 48 h. Western blotting was performed to analyze protein level. (D) IRF4 overexpression promotes Notch-Akt signaling in A549 and LC-AI cells. A549 and LC-AI cells were infected with lentivirus expressing Ctrl or IRF4 vectors for 48 h. Western blotting was performed to analyze protein level. (E) Notch pathway inhibitor MK-0752 reverses the effect of IRF4 on activation of Akt signaling in A549 cells. A549 cells were infected with lentivirus expressing Ctrl or IRF4 vectors with/without MK-0752 (20 µM) treatment for 48 h. **P<0.01 vs. IRF+DMSO group. IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer, sh-Ctrl, negative control; Ctrl, control; qRT-PCR, quantitative reverse transcription-polymerase chain reaction; p-, phosphorylated; AKT, protein kinase B.
There is growing evidence that Notch regulates the AKT pathway in several normal and cancer cell types (24), we then investigated whether Notch/AKT pathway was activated by IRF4. Indeed, IRF4 knockdown significantly suppressed the protein expression of Notch1, Notch2 and phosphorylation of AKT (p-AKT) in A549 and LC-AI cells (Fig. 4C); whereas IRF4 overexpression markedly facilitated the protein expressionof Notch1, Notch2 and p-AKT in A549 and LC-AI cells (Fig. 4D). Furthermore, A549 cells with IRF4 overexpression (IRF4) were treated with Notch pathway inhibitor MK-0752 (20 µM) or DMSO for 48 h. Western blotting was performed and the results showed that MK-0752 could significantly reverse the effect of IRF4 overexpressionon activation of Akt signaling in A549 cells (Fig. 4E). Totally, those investigations indicated that IRF4 activates Notch-Akt signaling in NSCLC cells.
Notch inhibition reverses the effects of IRF4 overexpression on cell growth
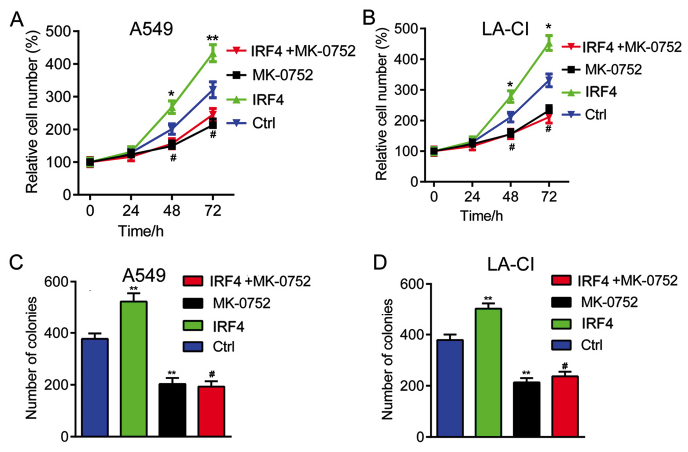
Finally, we explored whether Notch-Akt pathway was involved in the effects of IRF4 on human NSCLC cell growth. As shown in Fig. 5, IRF4 overexpression (IRF4) could significantly increased cell proliferation and colony number. However, when treated with Notch pathway inhibitor MK-0752 (IRF4+MK-0752), the enhancement of cell proliferation and colony number induced by IRF4 overexpression were significantlyreversed. It seems that Notch inhibition significantly reverses the effects of IRF4 overexpression on human NSCLC cell growth.
Figure 5.
Notch pathway inhibitor MK-0752 reverses the effects of IRF4 on NSCLC cell growth (A) Notch pathway inhibitor MK-0752 reverses the effect of IRF4 on proliferation of A549 cells. A549 cell were infected with lentivirus expressing Ctrl or IRF4 vectors with/without MK-0752 (20 µM) treatment. Relative cell numbers were evaluated at the indicated time points using MTT method. *P<0.05 and **P<0.01 vs. Ctrl. #P<0.05 indicates IRF4+MK-0752 vs. IRF4. (B) Notch pathway inhibitor MK-0752 reverses the effect of IRF4 on proliferation of LC-AI cells. LC-AI cells were infected with lentivirus expressing Ctrl or IRF4 vectors with/without MK-0752 (20 µM) treatment. Relative cell numbers were evaluated at the indicated time points using MTT method. *P<0.05 and **P<0.01 vs. Ctrl. #P<0.05 indicates IRF4+MK-0752 vs. IRF4. (C) Notch pathway inhibitor MK-0752 reverses the effect of IRF4 on colony formation of A549 cells. A549 cells were infected with lentivirus expressing Ctrl or IRF4 lentivirus vectors with/without MK-0752 (20 µM) treatment. The colony number in each well was evaluated two weeks later by soft sugar colony formation assay. **P<0.01 vs. Ctrl. #P<0.05 indicates IRF4+MK-0752 vs. IRF4. (D) Notch pathway inhibitor MK-0752 reverses the effect of IRF4 on colony formation of LC-AI cells. A549 cells were infected with lentivirus expressing Ctrl or IRF4 lentivirus vectors with/without MK-0752 (20 µM) treatment. The colony number in each well was evaluated two weeks later by soft sugar colony formation assay. **P<0.01 vs. Ctrl. #P<0.05 indicates IRF4+MK-0752 vs. IRF4. IRF4, interferon regulatory factor 4; NSCLC, non-small cell lung cancer, sh-Ctrl, negative control; Ctrl, control.
Discussion
In the present study, the function of IRF4 in human NSCLC was investigated. IRF4 mRNA and protein were found highly expressed in NSCLC tissues as compared to the corresponding adjacent non-tumor tissues, indicating its potential role in NSCLC development and progression. The cell proliferation rate and colony number of A549 and LC-AI were significantly declined when IRF4 expression was knocked down. However, the cell proliferation rate and colony number of A549 and LC-AI cells were markedly increased when IRF4 expression was overexpressed. These investigations validated the important role of IRF4 in NSCLC cell growth. This is the first evidence demonstrating the oncogenic role of IRF4 in NSCLC. IRF4 is overexpressed in many hematological malignancies, serves as a diagnostic and prognostic marker of hematological malignancies and is a promising therapeutic target for treatment of hematological malignancies (25). In a previous study by Chen et al (17), IRF4 was shown to predict poorer survival of NSCLC patients. To some extent, te present study is consistent with our study. By contrast, Wu et al (26) demonstrated that IRF4 was a protective prognostic factor in NSCLC patients. These contradictory data may suggest that IRF4 may reflect tumor-infiltrating lymphocyte activity in tissue section.
Besides, in the present study, Notch1 and Notch2 expression were significantly upregulated in NSCLC tissues as compared to corresponding adjacent non-tumor tissues, also indicating their potential role in NSCLC development and progression. Notch1 and Notch2 expression were found positively correlated with IRF4 level in NSCLC tissues. The expression of Notch1, Notch2 and p-AKT were found regulated by IRF4 in NSCLC cells. In the cells with IRF4 overexpressed that treated with MK-0752, it showed the significant decreased cell proliferation rate and colony number as compared with cells treated with DMSO. Mechanically, our study suggests that the oncogenic role of IRF4 in NSCLC was, at least partially, through regulating Notch expression and activating Notch-Akt signaling pathway. Previous study indicated that low level of IRF4 is a common feature of CLL, and the deregulated IRF4-Notch axis may represent a major pathway in the molecular pathogenesis of CLL (23). It suggests the positive relation between IRF4 and Notch signaling pathway, to some extent, this is consistent with our study. Besides, Previous study also identified Nedd4 as a key IRF4 target gene involved in impeding the responses of CLL cells and their precursors to Notch signaling (23), whether Nedd4 acts as a linkage between IRF4 and Notch signaling in our study remain further exploration.
It is well-known that Notch receptors (Notch1-4) are critical for tumorigenesis (27). Upon binding to its ligands, Notch receptors are proteolytically processed bymetal loproteases and the g-secretase complex. This process releases the intracellular domain of Notch, which thentranslocates into the nucleus to modulate expression of its target genes (28). Our study is consistent with a serious of previous studies that indicating the tumorgenic role of Notch1 (29–31). Particularly, a previous work has shown that Notch1 signaling promotes tumorigenicity and survival in NSCLC cells in vitro (32). The present study also showed that Notch2 mRNA was upregulated in NSCLC tissues, consistently, the transcription factors Notch2 was upregulated in invasive cancer cells in all 11 minimally invasive adenocarcinomas (33). In embryonal brain tumors, Notch2 also functions as an oncogene (34). However, a study reported that Notch2 mediates differentiation and has tumor suppressor functions during lung carcinogenesis (3). Notch2 also functions as a tumor suppressor in bronchial epithelial cells modulating tumor initiation and progression (35).
There is growing evidence that Notch regulates the AKT pathway in several normal and cancer cell types (24). The present study showed IRF4 upregulated the expression of Notch-1, Notch-2 and p-Akt, it seems that Notch-1 and Notch-2 have consistently regulative effects on p-Akt levels. Differently, aprevious study indicated that Notch-1 led to Akt phosphorylation and promoted cell survival, whereas Notch-2 signaling led to Akt dephosporylation and suppressed cell survival of malignant mesothelioma cells (35).
In summary, our results indicate that IRF4 acts as a tumor promoter in human NSCLC, at least partially, through activating Notch-Akt signaling pathway.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Yuan X, Wu H, Han N, Xu H, Chu Q, Yu S, Chen Y, Wu K. Notch signaling and EMT in non-small cell lung cancer: Biological significance and therapeutic application. J Hematol Oncol. 2014;7:87. doi: 10.1186/s13045-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumgart A, Mazur P, Anton M, Rudelius M, Schwamborn K, Feuchtinger A, Behnke K, Walch A, Braren R, Peschel C, et al. Opposing role of Notch1 and Notch2 in a Kras(G12D)-driven murine non-small cell lung cancer model. Oncogene. 2015;34:578–588. doi: 10.1038/onc.2013.592. [DOI] [PubMed] [Google Scholar]
- 4.Coffelt SB, Hughes R, Lewis CE. Tumor-associated macrophages: Effectors of angiogenesis and tumor progression. Biochimica et Biophysica Acta (BBA)-Reviews on Cancer. 2009;1796:11–18. doi: 10.1016/j.bbcan.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Honda K, Taniguchi T. IRFs: Master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- 6.Mamane Y, Heylbroeck C, Génin P, Algarté M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: The next generation. Gene. 1999;237:1–14. doi: 10.1016/S0378-1119(99)00262-0. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annual Rev immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, Ning S. Interferon regulatory factor 4 is activated through c-Src-mediated tyrosine phosphorylation in virus-transformed cells. J Virol. 2013;87:9672–9679. doi: 10.1128/JVI.01435-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spender LC, Lucchesi W, Bodelon G, Bilancio A, Karstegl CE, Asano T, Dittrich-Breiholz O, Kracht M, Vanhaesebroeck B, Farrell PJ. Cell target genes of Epstein-Barr virus transcription factor EBNA-2: Induction of the p55alpha regulatory subunit of PI3-kinase and its role in survival of EREB2. 5 cells. J Gen Virol. 2006;87:2859–2867. doi: 10.1099/vir.0.82128-0. [DOI] [PubMed] [Google Scholar]
- 10.Iida S, Rao PH, Butler M, Corradini P, Boccadoro M, Klein B, Chaganti RS, Dalla-Favera R. Deregulation of MUM1/IRF4 by chromosomal translocation in multiple myeloma. Nat Genet. 1997;17:226–230. doi: 10.1038/ng1097-226. [DOI] [PubMed] [Google Scholar]
- 11.Mamane Y, Grandvaux N, Hernandez E, Sharma S, Innocente SA, Lee JM, Azimi N, Lin R, Hiscott J. Repression of IRF-4 target genes in human T cell leukemia virus-1 infection. Oncogene. 2002;21:6751–6765. doi: 10.1038/sj.onc.1205843. [DOI] [PubMed] [Google Scholar]
- 12.Sharma S, Mamane Y, Grandvaux N, Bartlett J, Petropoulos L, Lin R, Hiscott J. Activation and regulation of interferon regulatory factor 4 in HTLV type 1-infected T lymphocytes. AIDS Res Hum Retroviruses. 2000;16:1613–1622. doi: 10.1089/08892220050193047. [DOI] [PubMed] [Google Scholar]
- 13.Ortmann CA, Burchert A, Hölzle K, Nitsche A, Wittig B, Neubauer A, Schmidt M. Down-regulation of interferon regulatory factor 4 gene expression in leukemic cells due to hypermethylation of CpG motifs in the promoter region. Nucleic Acids Res. 2005;33:6895–6905. doi: 10.1093/nar/gki1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schmidt M, Hochhaus A, König-Merediz SA, Brendel C, Proba J, Hoppe GJ, Wittig B, Ehninger G, Hehlmann R, Neubauer A. Expression of interferon regulatory factor 4 in chronic myeloid leukemia: Correlation with response to interferon alfa therapy. J Clin Oncol. 2000;18:3331–3338. doi: 10.1200/JCO.2000.18.19.3331. [DOI] [PubMed] [Google Scholar]
- 15.Tsuboi K, Iida S, Inagaki H, Kato M, Hayami Y, Hanamura I, Miura K, Harada S, Kikuchi M, Komatsu H, et al. MUM 1/IRF4 expression as a frequent event in mature lymphoid malignancies. Leukemia. 2000;14:449–456. doi: 10.1038/sj.leu.2401696. [DOI] [PubMed] [Google Scholar]
- 16.Yang M, Yuan F, Li P, Chen Z, Chen A, Li S, Hu C. Interferon regulatory factor 4 binding protein is a novel p53 target gene and suppresses cisplatin-induced apoptosis of breast cancer cells. Mol Cancer. 2012;11:54. doi: 10.1186/1476-4598-11-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen HY, Yu SL, Chen CH, Chang GC, Chen CY, Yuan A, Cheng CL, Wang CH, Terng HJ, Kao SF, et al. A five-gene signature and clinical outcome in non-small-cell lung cancer. N Engl J Med. 2007;356:11–20. doi: 10.1056/NEJMoa060096. [DOI] [PubMed] [Google Scholar]
- 18.Blackman SC, Podtelezhnikov A, Railkar RA, Loboda A, Tanis K, Klappenbach JA, Watters J, Iannone R, Herman G, Bergtrom DA. Abstract 26: Notch pathway inhibition with MK-0752 leads to dose- and time-dependent transcriptional alterations in proliferation, PI3K, and Wnt pathway genes in plucked human hair follicles. Cancer Res. 2010;70:17–21. doi: 10.1158/1538-7445.AM10-26. [DOI] [Google Scholar]
- 19.Zhao J, Lever AM. Lentivirus-mediated gene expression. Methods Mol Biol. 2007;366:343–355. doi: 10.1007/978-1-59745-030-0_20. [DOI] [PubMed] [Google Scholar]
- 20.Harper JA, Yuan JS, Tan JB, Visan I, Guidos CJ. Notch signaling in development and disease. Clin Genet. 2003;64:461–472. doi: 10.1046/j.1399-0004.2003.00194.x. [DOI] [PubMed] [Google Scholar]
- 21.Kadesch T. Notch signaling: The demise of elegant simplicity. Curr Opin Genet Dev. 2004;14:506–512. doi: 10.1016/j.gde.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-B. [DOI] [PubMed] [Google Scholar]
- 23.Shukla V, Shukla A, Joshi SS, Lu R. Interferon regulatory factor 4 attenuates Notch signaling to suppress the development of chronic lymphocytic leukemia. Oncotarget. 2016;7:41081–41094. doi: 10.18632/oncotarget.9596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H, Bhaijee F, Ishaq N, Pepper DJ, Backus K, Brown AS, Zhou X, Miele L. Correlation of Notch1, pAKT and nuclear NF-κB expression in triple negative breast cancer. Am J Cancer Res. 2013;3:230–239. [PMC free article] [PubMed] [Google Scholar]
- 25.Gualco G, Weiss LM, Bacchi CE. MUM1/IRF4: A Review. Appl Immunohistochem Mol Morphol. 2010;18:301–310. doi: 10.1097/PAI.0b013e3181cf1126. [DOI] [PubMed] [Google Scholar]
- 26.Wu YY, Hwang YT, Perng WC, Chian CF, Ho CL, Lee SC, Chang H, Terng HJ, Chao TY. CPEB4 and IRF4 expression in peripheral mononuclear cells are potential prognostic factors for advanced lung cancer. J Formos Med Assoc. 2017;116:114–122. doi: 10.1016/j.jfma.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 27.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 28.Bray SJ. Notch signalling: A simple pathway becomes complex. Nat Rev Mol Cell Biol. 2006;7:678–689. doi: 10.1038/nrm2009. [DOI] [PubMed] [Google Scholar]
- 29.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, et al. Activation of Notch-1 signaling maintains the neoplastic phenotype in human Ras-transformed cells. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 30.Allen TD, Rodriguez EM, Jones KD, Bishop JM. Activated Notch1 induces lung adenomas in mice and cooperates with Myc in the generation of lung adenocarcinoma. Cancer Res. 2011;71:6010–6018. doi: 10.1158/0008-5472.CAN-11-0595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osanyingbemi-Obidi J, Dobromilskaya I, Illei PB, Hann CL, Rudin CM. Notch signaling contributes to lung cancer clonogenic capacity in vitro but may be circumvented in tumorigenesis in vivo. Mol Cancer Res. 2011;9:1746–1754. doi: 10.1158/1541-7786.MCR-11-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baumgart A, Seidl S, Vlachou P, Michel L, Mitova N, Schatz N, Specht K, Koch I, Schuster T, Grundler R, et al. ADAM17 regulates epidermal growth factor receptor expression through the activation of Notch1 in non-small cell lung cancer. Cancer Res. 2010;70:5368–5378. doi: 10.1158/0008-5472.CAN-09-3763. [DOI] [PubMed] [Google Scholar]
- 33.Mimae T, Okada M, Hagiyama M, Miyata Y, Tsutani Y, Inoue T, Murakami Y, Ito A. Upregulation of notch2 and six1 is associated with progression of early-stage lung adenocarcinoma and a more aggressive phenotype at advanced stages. Clin Cancer Res. 2012;18:945–955. doi: 10.1158/1078-0432.CCR-11-1946. [DOI] [PubMed] [Google Scholar]
- 34.Fan X, Mikolaenko I, Elhassan I, Ni X, Wang Y, Ball D, Brat DJ, Perry A, Eberhart CG. Notch1 and notch2 have opposite effects on embryonal brain tumor growth. Cancer Res. 2004;64:7787–7793. doi: 10.1158/0008-5472.CAN-04-1446. [DOI] [PubMed] [Google Scholar]
- 35.Graziani I, Eliasz S, De Marco MA, Chen Y, Pass HI, De May RM, Strack PR, Miele L, Bocchetta M. Opposite effects of Notch-1 and Notch-2 on mesothelioma cell survival under hypoxia are exerted through the Akt pathway. Cancer Res. 2008;68:9678–9685. doi: 10.1158/0008-5472.CAN-08-0969. [DOI] [PubMed] [Google Scholar]