Abstract
Anticholinergic agent, ipratropium bromide (IB) ameliorates symptoms of allergic rhinitis (AR) using neuroimmunologic mechanisms. However, the underlying molecular mechanism remains largely unclear. In the present study, 27 mice with AR induced by ovalbumin were randomly allocated to one of three groups: Model group, model group with IB treatment for 2 weeks, and model group with IB treatment for 4 weeks. Allergic symptoms were evaluated according to symptoms scores. Differentially expressed genes [microRNAs (miRNAs) and messenger RNAs (mRNAs)] of nasal mucosa were identified by microarray analysis. The expression levels of candidate genes were measured by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The data indicates that the symptoms scores in allergic mice were significantly reduced by IB treatment. In the nasal mucosa of allergic mice with IB treatment, 207 mRNAs and 87 miRNAs were differentially expressed, when compared with the sham group. IB treatment significantly downregulated the expression levels of interleukin-4Rα and prostaglandin D2 synthase, whereas the leukemia inhibitory factor, A20 and nuclear receptor subfamily 4, group A, member 1 expression levels were upregulated. Similarly, the expression levels of mmu-miR-124-3p/5p, −133b-5p, −133a-3p/5p, −384-3p, −181a-5p, −378a-5p and −3071-5p were significantly increased. RT-qPCR data further validated these mRNA and miRNA expression levels. Thus, IB treatment regulated expression of allergic immune-associated mRNAs and miRNAs of the nasal mucosa in allergic mice, which may be associated with ameliorated nasal allergic symptoms.
Keywords: allergic rhinitis, ipratropium bromide, miRNAs, allergic inflammation
Introduction
Allergic rhinitis (AR), an inflammation disorder of the nasal mucosa, is caused by various allergens, such as pollen. Vidian neurectomy or anticholinergic therapy with ipratropium bromide (IB) contributed to attenuation of nasal symptoms and downregulation of the Th2-polarized immune response of AR induced by ovalbumin (OVA) (1). MicroRNA (miRNA) profiles altered in AR and aberrant expression of miRNAs have been implicated in the modulation of Th2-type inflammatory reaction (2). However, the impact of anticholinergic treatment on the miRNA expression profile in AR following allergen exposure remains poorly understood, particularly the interaction between miRNAs and putative target messenger RNAs (mRNAs), which requires further investigation and may provide the missing link in the IB-mediated anti-allergic signaling pathway.
miRNAs are a class of small (18–24 nt) single-stranded, noncoding RNAs that, in the majority of situations, negatively regulate the expression of target genes. Previous studies have determined miRNA profiles in multiple allergic inflammatory disorders, including AR, bronchial asthma and atopic dermatitis (3). Furthermore, it was demonstrated that specific miRNAs were critical in regulating the pathogenesis in allergic inflammation, including polarization and activation of CD4+ T cells, regulation of airway eosinophilia and modulation of T helper (Th)2 cytokine-induced epithelial responses (3). Currently, to the best of our knowledge, the miRNA profiles, which are distinct from other allergic diseases, have not been specifically defined following an intervention with IB in AR induced by OVA. Thus, the present study focused on these areas to comprehensively elucidate the molecular mechanisms underlying the effects of IB (4).
The aim of the present study was to investigate the mRNA transcriptome and miRNA profiles in nasal mucosa of allergic mice using IB treatment, as well as the potential regulatory mechanism by which IB interferes with allergic inflammation. The study may elucidate the association between mRNA transcriptome alternation, particularly the miRNA expression and intranasal IB therapy, and the molecular mechanisms intimately involved in the development, maintenance and exacerbation or attenuation of AR. The findings may facilitate with the screening and development of novel miRNA-targeting compounds for AR treatment.
Materials and methods
Experimental animals
Eight-week-old healthy male BALB/c mice, free of murine-specific pathogens, were used in the present study. A total of 66 mice (weight, 24.76±2.26 g) were purchased from and maintained at the Experimental Animal Center of the Third Xiangya Hospital, Central South University (Changsha, China). The mice were housed in a controlled environment under a 12-h light/dark cycle with free access to food and water [Experimental Animal License no. SCXK-0020003; Environmental Facilities Certificate of Conformity SYXK-(Hunan) 200,200.20]. All animals used in the current study were handled according to a protocol approved by the institutional animal care and use committee of Central South University.
Establishment of the AR model and treatment protocol
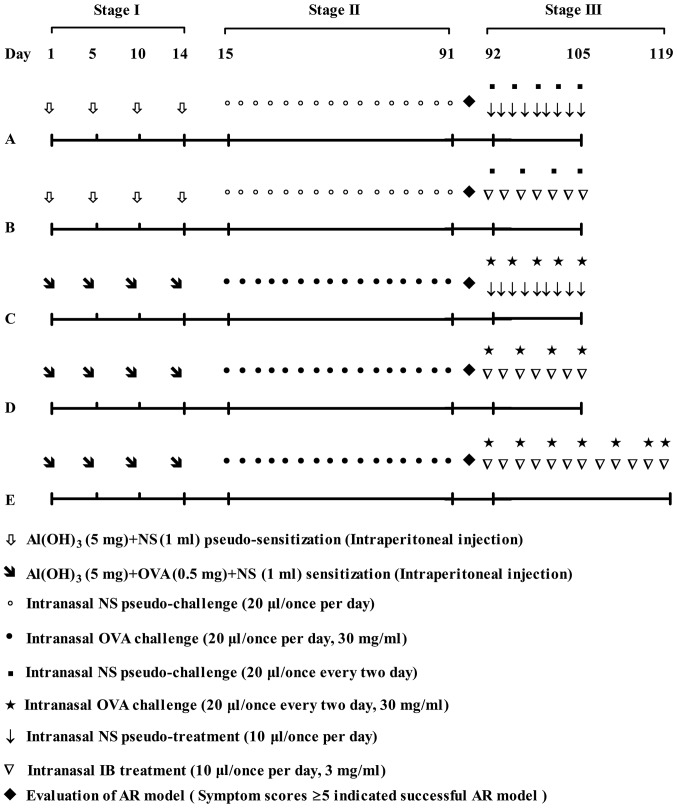
The mouse model of AR was produced as described previously, but with certain modifications (1). Briefly, 46 mice were randomly assigned to two groups: Control group (n=16) and model group (n=30). The model group mice were sensitized with a mixture of OVA (0.5 mg) (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany), normal saline (NS; 1 ml) and aluminum hydroxide gel (5 mg) (Sigma-Aldrich; Merck KGaA) four times (on days 1, 5, 10 and 14) via intra-peritoneal injection, which was followed by intranasal 3% (w/v) OVA challenge (20 µl) once per day from week 3 to week 13 (11 weeks in total). In the control group, OVA was substituted with an equal volume of NS. The allergic mouse model was assessed using symptom scores on the last day of week 13 (1). Subsequently, mice in the control group (n=15) were randomly allocated to two groups: Group A (n=7) and group B (n=8), which received intranasal NS and 0.3% IB (China Pharmaceutical and Biological Products, Beijing, China) treatment daily, respectively from week 14 to week 15. Mice in the model group (n=27) were randomly allocated to three groups: Group C (n=8; sham group) received NS sham-treatment (10 µl per day) for 2 weeks, group D (n=10) received intranasal 0.3% IB pre-treatment (10 µl per day) for 2 weeks and group E (n=9) received intranasal 0.3% IB pre-treatment (10 µl per day) for 4 weeks. Allergic mice received the intranasal 3% OVA challenge (20 µl) every 2 days and non-allergic mice received intranasal NS pseudo-challenge (20 µl) every 2 days during IB treatment. All mice were sacrificed within 30 min of the final intranasal OVA challenge. Evaluation of symptom scores was performed prior to sacrifice. Nasal mucosa tissue samples were harvested within a few min of sacrifice for further investigation (Fig. 1).
Figure 1.
Flowchart demonstrating the establishment of the AR model and treatment protocol. Stage I, systemic sensitization by OVA; stage II, nasal mucosa local challenge with OVA; Stage III, IB treatment with a concomitant OVA challenge. Control: Groups A and B. Model: Groups C-E. AR, allergic rhinitis; OVA, ovalbumin; IB, ipratropium bromide; NS, normal saline.
Evaluation of induced allergic symptom
Allergic symptoms, such as sneezing, nasal itching and rhinorrhea were observed and counted within 10 min of the last challenge with OVA or IB treatment. Symptom scores were calculated as described previously, but with modifications (1). Briefly, sneezing, nasal itching and rhinorrhea were graded from one to three based on the degree of severity. Sneezing: The frequency of sneezing <3 within 10 min after the final OVA challenge were scored 1; 4–10 within 10 min were scored 2; >11 within 10 min were scored 3. Nasal itching: The occurrence of mild and occasional nasal scratching within 10 min of the final OVA challenge were scored 1; more severe and persistent nasal scratching within 10 min were scored 3; nasal itching in-between these frequencies were scored 2. Rhinorrhea: Observable watery discharges within the nasal cavity, but without spilling from the anterior naris within 10 min of the final OVA challenge were scored 1; watery discharge spilling from the anterior naris within 10 min were scored 2, the face covered with abundant watery discharges within 10 min were scored 3. The symptom scores refer to the accumulative scores of sneezing, nasal itching and rhinorrhea. The mouse model of AR was regarded as successful case when the symptom scores were >5.
Total RNA isolation and detection
The RNA sequencing procedure was performed by Beijing Novogene Bioinformatics Technology Co., Ltd (Beijing, China). Total RNA was extracted from the mixed nasal mucosa tissue samples (three samples per group) using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's instructions. RNA purity was assessed using a NanoPhotometer® spectrophotometer (Implen, Inc., Westlake Village, CA, USA) and the RNA concentration was measured using a Qubit® RNA Assay kit in a Qubit® 2.0 Flurometer (Thermo Fisher Scientific, Inc.). RNA integrity was assessed using the RNA Nano 6000 Assay kit of the Bioanalyzer 2100 system (Agilent Technologies, Inc., Santa Clara, CA, USA).
mRNA library preparation for transcriptome sequencing and data analysis
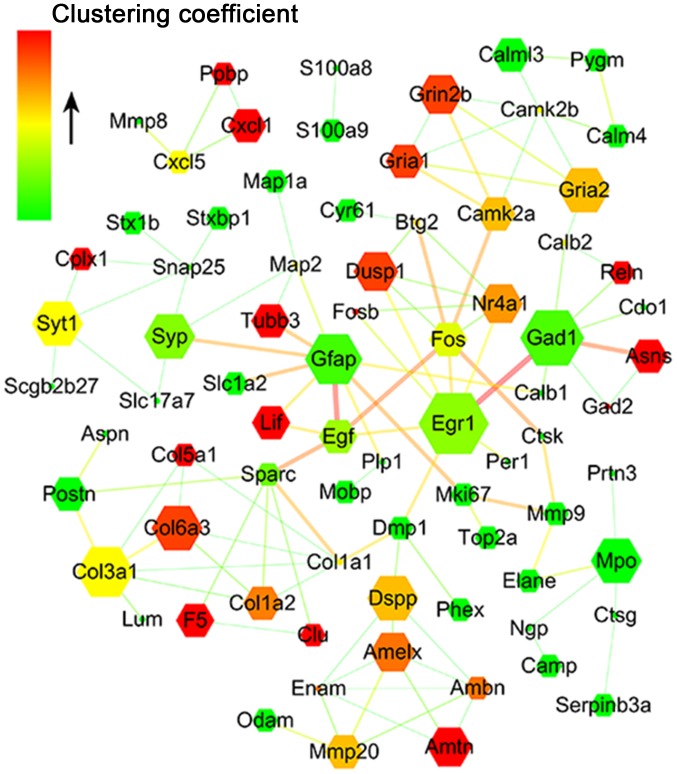
The mRNA libraries were generated using NEBNext® Ultra™ RNA Library Prep kit for Illumina® (New England BioLabs, Inc., Ipswich, MA, USA) according to the manufacturer's instructions. The mRNA library for groups A-E were designated as Am, Bm, Cm, Dm and Em, respectively. The five libraries were sequenced on an Illumina Hiseq 2000 platform for raw data. Clean reads were obtained by removing unqualified data and were aligned to the reference genome using TopHat v2.0.9 (https://ccb.jhu.edu/software/tophat/index.shtml). The mapped gene expression level was evaluated by HTSeq software v0.6.1 with Reads Per Kilobase of exon model per Million mapped reads (RPKM) (5). RPKM>1 was considered as the threshold for gene expression. Subsequently, the DEGSeq R package (1.12.0) was used to identify differentially expressed genes (DEGs), and a corrected P<0.005 value and |log2(Fold change)|>1 were considered statistically significant (6). A protein-protein interaction (PPI) network was constructed using Cytoscape software v3.2.1.0 to investigate the PPI of DEGs and a confidence score of PPI >700 was selected (7). The node represents a single DEG, and the edges between nodes indicate the confidence score. The size of the node reflects the node degree; genes that exhibit a greater number of interconnections are represented with a larger node. The width of the edge is positively associated with the confidence score of the PPI. The color of the nodes increases from green to red and represents the clustering coefficient. The clustering coefficient is an indicator of the network connectivity of the node; a higher clustering coefficient is representative of a more central status of the gene in the network.
Small RNA library preparation, sequencing and data analysis
The small RNA libraries were generated using NEBNext® Multiplex Small RNA Library Prep Set for Illumina® (New England BioLabs, Inc.) according to the manufacturer's instructions. The sRNA library for groups A-E were designated as Ami, Bmi, Cmi, Dmi and Emi, respectively. The five sRNA libraries were sequenced on Illumina Hiseq 2500/2000 platform for raw data. Clean reads, length 18–35 nt, were obtained by removing unqualified data and mapped to the reference sequence using Bowtie software v0.12.9 (8). Subsequently, mapped small RNAs (sRNAs) were aligned to miRBase20.0 (http://www.mirbase.org/) to obtain known miRNAs with software mirdeep2 v0.0.5 and srna-tools-cli (http://Srna-tools.cmp.uea.ac.uk/) (9–11). The level of identified miRNAs was accessed by ‘transcripts per million’ according to the following normalization formula: miRNA expression level = mapped read counts × 1,000,000 /total read counts (12). The DEGSeq R package was used to identify the differentially expressed miRNAs, and q-value (adjusted P-value) <0.01 and log2(fold change)>1 were considered to be statistically significant. Target genes for differentially expressed miRNAs were predicted using miRanda (http://www.microrna.org/) (13). Finally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of DEGs was implemented with KOBAS software v2.0, and corrected P<0.05 values were considered to indicate statistically significant differences (14).
Determination of nasal mucosa candidate mRNAs and miRNAs by reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA and sRNA within nasal mucosa were extracted using a total RNA/sRNA kit (Omega Bio-Tek, Inc., Norcross, GA, USA) according to the manufacturer's protocol. RT-qPCR for sRNA was performed using the All-in-One™ qRT-PCR miRNA Detection kit (GeneCopoeia, Inc., Rockville, MD, USA) according to the manufacturer's protocol, and U6 served as the reference gene. RT-qPCR for total RNA was performed using a commercial kit (Toyobo Co., Ltd., Osaka, Japan) according to the manufacturer's protocol and β-actin served as the reference gene. A melting curve was performed to determine potential non-specific amplifications. The relative expression level of candidate miRNAs or mRNAs was calculated using the 2−ΔΔCq method (15). The primers used are presented in Table I.
Table I.
Primers used for reverse transcription-quantitative polymerase chain reaction.
| mRNA | Sense | Sequence (5′-3′) |
|---|---|---|
| IL-4 receptor-α | Forward | 5′-GTGCCCTTATTTACTTTCGG-3′ |
| Reverse | 5′-CTGGCTGTGGTGTTGCTT-3′ | |
| Prostaglandin D2 synthase | Forward | 5′-TGCTGTGGATGGGTTTGG-3′ |
| Reverse | 5′-TTGAGTTGGAGGCGAGGC-3′ | |
| Leukemia inhibitory factor | Forward | 5′-AAGTTGGTGGAGCTGTATAGG-3′ |
| Reverse | 5′-AAGGCTTCTTTGTCAGAGTGGT-3′ | |
| A20 | Forward | 5′-GCCTCACTTCCAGTATCCC-3′ |
| Reverse | 5′-CTTGCTTGTCCCTGCTCT-3′ | |
| Nuclear receptor subfamily 4, group A, member 1 | Forward | 5′-AAAATCCCTGGCTTCATTG-3′ |
| Reverse | 5′-AGGACCATGAACCCAAGT-3′ | |
| Eotaxin-2 | Forward | 5′-GCTGCACGTCCTTTATTTCC-3′ |
| Reverse | 5′-TCTTATGGCCCTTCTTGGTG-3′ | |
| IL-4 | Forward | 5′-CATCGGCATTTTGAACGAG-3′ |
| Reverse | 5′-TGGAAGCCCTACAGACAAGC-3′ | |
| Interferon-γ | Forward | 5′-CTGCTGATGGGAGGAGATGT-3′ |
| Reverse | 5′-ATTTGTCATTCGGGTGTAGTCA-3′ | |
| IL-5 | Forward | 5′-GGGCTTCCTGCTCCTATCTAA-3′ |
| Reverse | 5′-CAACCTTCTCTCTCCCCAAG-3′ | |
| IL-13 | Forward | 5′-GAGGAGGGTTGAGGAGGAAG-3′ |
| Reverse | 5′-TTTCTGTAGGGATGGGATGG-3′ | |
| IL-17 | Forward | 5′-CGGCTGACCCCTAAGAAAC-3′ |
| Reverse | 5′-CAGAAAAACAAACACGAAGCAG-3′ | |
| Forkhead box P3 | Forward | 5′-CCAGTGCCCATCCAATAAAC-3′ |
| Reverse | 5′-GTATCCGCTTTCTCCTGCTG-3′ | |
| IL-10 | Forward | 5′-GGACAACATACTGCTAACCGACT-3′ |
| Reverse | 5′-TGGGGCATCACTTCTACCA-3′ |
IL, interleukin.
Statistical analysis
Statistical comparisons of symptoms scores were performed using one-way analysis of variance followed by the Student-Newman-Keuls post hoc test, and RT-qPCR data were analyzed using the Kruskal-Wallis test followed by pairwise multiple comparisons. PASW Statistics 19.0 (IBM Corp., Armonk, NY, USA) was used to perform all analyzes and P<0.05 was considered to indicate a statistically significant difference. The data are presented as means ± standard deviations.
Results
Animal model of AR evaluated by symptom scores
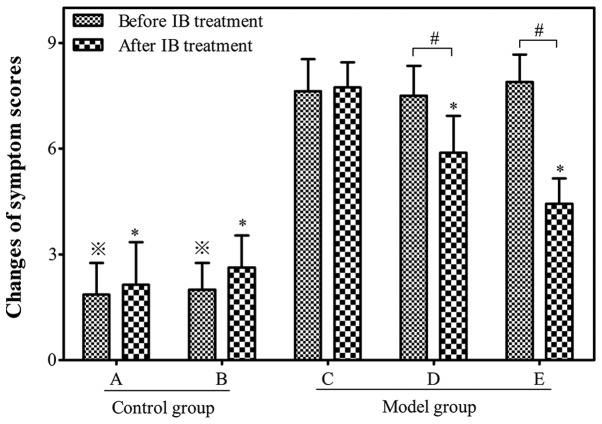
The number of sneezes, nasal itching and rhinorrhea were observed in order to calculate the symptom scores of the allergic mouse models (Fig. 2). A successful mouse model of AR was determined by symptom scores >5. By week 14, 27 of the 30 OVA-sensitized mice in the model group had symptom scores >5. The average score was significantly higher in the allergen-challenged model group than those in the non-allergic control group prior to IB treatment (7.72±1.03 vs. 1.93±0.35; P<0.05).
Figure 2.
Symptom scores in different groups before and after IB treatment. Control: Group A (n=7) and group B (n=8). Model: Group C (n=8), group D (n=10) and group E (n=9). Data are expressed as the mean ± standard deviation. ※P<0.05 vs. group C before IB treatment; *P<0.05 vs. group C after IB treatment; #P<0.05. IB, ipratropium bromide.
Attenuation of allergic symptoms of AR using anticholinergic drug IB
The symptom scores decreased significantly to 5.89±1.05 and 4.44±0.72 in the allergic mice of group D and E, respectively following IB treatment when compared with group C (7.63±0.92; P<0.05). The symptom scores of allergic mice in group D were higher than those of group E following IB treatment, indicating that IB pre-treatment may reduce the symptom scores in a time-dependent manner (Fig. 2).
DEGs in nasal mucosa following IB treatment
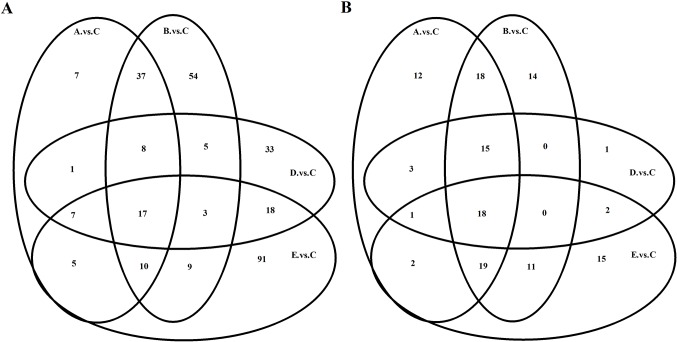
Compared with group C, 92 transcripts were identified to be differentially expressed in the nasal mucosa of allergic mice in group D, and 92, 143, and 160 transcripts in group A, B, and E, respectively. Of which 81, 112, 67 and 116 transcripts were upregulated, whereas 11, 31, 25 and 44 transcripts were downregulated in the group A, B, D and E, respectively (data not shown). The transcripts of the different groups overlap and the Venn diagram for mRNA demonstrates the similarity in their responses to IB treatment among the different groups (Fig. 3A). Notably, 207 mRNAs were differentially expressed in the nasal mucosa of the allergic mice that received IB treatment compared with the NS sham-treatment (data not shown).
Figure 3.
Venn diagram of differentially expressed (A) mRNAs and (B) miRNAs. Control: Group A (n=7) and group B (n=8). Model: Group C (n=8), group D (n=10) and group E (n=9). When mRNAs or miRNAs exhibit similar characteristics, they appear in the overlapping boxes. mRNA, messenger RNA; miRNA, microRNA.
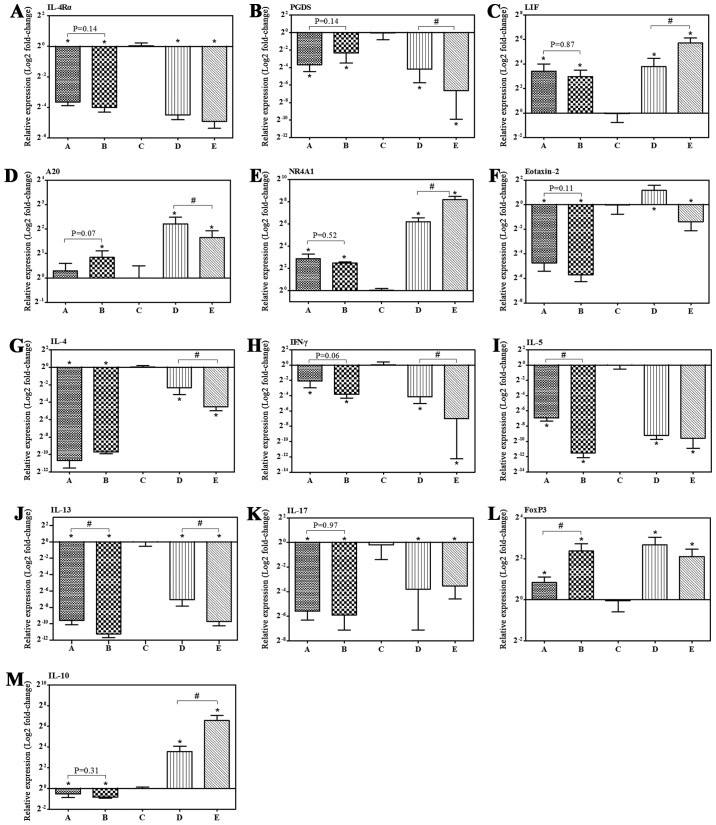
Compared with group C, various allergic-associated genes changed their expression response to IB, which was validated by RT-qPCR. IB treatment induced significant 23- and 30-fold decreases in interleukin (IL)-4 receptor-α (IL-4Rα) mRNA expression levels in groups D and E, respectively. Furthermore, 30- and 100-fold decreases in prostaglandin D2 synthase (PGDS) were observed in groups D and E, respectively. Additionally, leukemia inhibitory factor (LIF), tumor necrosis factor α-induced protein 3/A20 and nuclear receptor subfamily 4, group A, member 1 (NR4A1) in group D exhibited significant 14-, 5- and 74-fold increases in mRNA expression, respectively, and a 53-, 3- and 290-fold increase in group E, respectively. The expression levels of IL-4Rα and PGDS exhibited a decreasing tendency, whereas LIF and NR4A1 demonstrated a tendency to increase following IB treatment (Fig. 4 and Table II).
Figure 4.
IB treatment regulated mRNA expression levels of allergic-associated genes. (A-M) The mRNA expression level in nasal mucosa was determined by reverse transcription-quantitative polymerase chain reaction with β-actin as the reference gene. Control: Group A (n=7) and group B (n=8). Model: Group C (n=8), group D (n=10) and group E (n=9). Data are presented as the mean ± standard deviation. *P<0.05 vs. group C following IB treatment; #P<0.05. IB, ipratropium bromide; mRNA, messenger RNA; IL, interleukin; PGDS, prostaglandin D2 synthase; LIF, leukemia inhibitory factor; A20, tumor necrosis factor α-induced protein 3; NR4A1, nuclear receptor subfamily 4, group A, member 1; IFN-γ, interferon-gamma; FoxP3, forkhead box P3.
Table II.
Fold change of differentially expressed genes in the nasal mucosa of allergic mice post-treatment with ipratropium bromide.
| Expression level (RPKMa) | log2 (Fold change)a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene description | A | B | C | D | E | A/C | B/C | D/C | E/C |
| Interleukin 4 receptor-α | 66.08 | 62.76 | 64.87 | 37.37 | 28.08 | 0 | −0.03 | −0.9 | −1.34 |
| Prostaglandin D2 synthase | 112.15 | 161.23 | 101.91 | 62.51 | 34.47 | 0.01 | 0.47 | −0.66 | −1.69 |
| Leukemia inhibitory factor | 10.42 | 10.28 | 10.16 | 18.05 | 26.86 | −0.1 | −0.18 | 0.73 | 1.27 |
| Tumor necrosis factor α-induced protein 3 | 39.35 | 49.74 | 35.47 | 62.61 | 97.08 | 0.02 | 0.29 | 0.72 | 1.32 |
| Nuclear receptor subfamily 4, group A, member 1 | 15.62 | 16.09 | 22.38 | 25.38 | 51.83 | −0.65 | −0.67 | 0.08 | 1.08 |
Fold change, RPKM of group D (or E or A or B)/RPKM of group C. RPKM, reads per kilo-base of exon model per million mapped reads (reflection of gene relative expression level).
Interaction network analysis for DEGs following IB treatment
The interaction network for differentially expressed transcripts was established based on the string score to investigate the role and function of the DEG response to IB treatment. Finally, a total of 162 transcripts were identified to participate in the formation of the interaction network (Fig. 5). According to the network, a series of the above-mentioned genes were associated with allergic inflammation responses, and interacted in the network directly or indirectly (data not shown). These gene products are located in the cytoplasm (A20), nucleus [early growth response protein 1 (EGR-1), dual-specificity phosphatase 1 (DUSP-1), serine (or cysteine) peptidase inhibitor, clade B (ovalbumin), member 3A, Fos/FosB and NR4A] or secreted into the extracellular space [LIF and chemokine (C-X-C motif) ligand 1)], which may locally co-regulate the nasal mucosa allergic immune response.
Figure 5.
PPI network for differentially expressed proteins. The gene network was constructed using Cytoscape software based on the STRING database. The circular node represents a single differentially expressed gene, and edges between nodes indicate the confidence score between them. The size of the node is correlated with the positivity degree, namely the number of genes interacting with it, and the width of the edge is positively associated with the confidence score of PPI. The colors of the nodes gradually change from green to red, reflecting the clustering coefficient depicting the positive connectivity of the nodes. PPI, protein-protein interaction.
Differentially expressed miRNAs in the nasal mucosa following IB treatment
The specific miRNA responses in nasal mucosa with AR following treatment with IB were investigated. Compared with group C, 40 miRNAs were identified to be differentially expressed in the nasal mucosa of the allergic mice in group D, and 88, 95 and 68 miRNAs in group A, B, and E, respectively. Of which 60, 77, 31 and 40 miRNAs, respectively, were upregulated, and 28, 18, 9 and 28 miRNAs were downregulated in groups A, B, D and E, respectively (data not shown). Similarly, there were overlaps in the miRNAs of the different groups and the Venn diagram of the miRNAs demonstrates the similarity of responses to IB treatment between the different groups (Fig. 3B). Notably, 87 miRNAs were differentially expressed in the nasal mucosa of the allergic mice with IB treatment compared with the NS sham-treatment (data not shown).
Prediction of potential target genes of differentially expressed miRNAs and function analysis of putative target genes
Based on the differentially expressed miRNAs induced by IB treatment, the potential targets of these dysfunctional miRNAs were identified, and the interaction between miRNAs and corresponding predicted genes associated with allergic inflammation were analyzed. IL-4Rα was identified to be potentially regulated by mmu-miR-124-3p, −181a/b/c-5p, −128-3p, −129-5p, 129b-3p. In addition, PGDS was predicted as the target gene of mmu-miR-378a-3p. A20 was likely to be negatively regulated by mmu-miR-384-3p, −7b-5p and −592-5p. mmu-miR-133a-3p, −133b-3p, −181a/b/c-5p, −138-5p and −128-3p may be negative regulators of LIF, and mmu-miR-124-3p, −138-5p and −200a/b-5p were expected to target NR4A1 (data not shown). In order to further understand the role of these miRNAs, their candidate targets underwent KEGG analysis. T/B cell receptor, high-affinity IgE receptor (FcεRI) and neurotrophin signaling pathways, as well as leukocyte transendothelial migration were perceived as the most enriched pathways (data not shown).
Modulation of allergic inflammation-associated genes expression by IB treatment
The microarray analysis identified a substantial number of differentially expressed mRNAs and miRNAs with various functions. The genes associated with allergic inflammation were subsequently selected for further confirmation by RT-qPCR. Nasal mucosa IL-4Rα, PGDS, LIF, A20 and NR4A1 mRNA expression responses to IB were presented above, and are consistent with the microarray analysis. Furthermore, no significant differences in IL-4Rα, PGDS, LIF, A20 and NR4A1 mRNA expression levels were identified between group A and B (Fig. 4). In addition, we have determined the mRNA expression of Th2 response-promoting or -restraining genes, which were not in included in the aforementioned DEGs. The mRNA expression levels in response to IB treatment in the nasal mucosa of allergic mice for forkhead box P3 (FOXP3), IL-10 and eotaxin-2 (termed C-C motif chemokine ligand 24; CCL24) increased, whereas those for interferon (IFN)-γ, IL-4, IL-5, IL-13 and IL-17 decreased (Fig. 4).
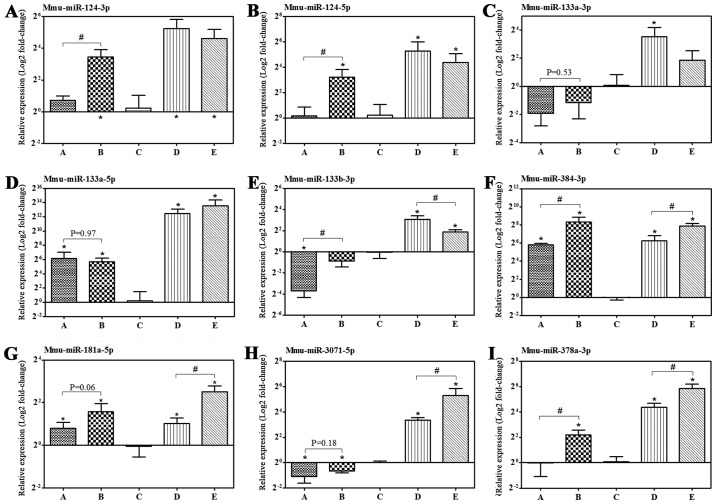
Furthermore, the expression levels of miRNAs targeting the above-mentioned corresponding putative genes were confirmed by RT-qPCR. The expression levels of mmu-miR-124-3p/5p, −133a-5p, −384-3p, −181a-5p, −378a-3p and −3071-5p in the nasal mucosa of allergic mice increased by IB treatment compared with the NS sham-treatment group, which are consistent with the observations of microarray analysis, apart from mmu-miR-378a-3p (Fig. 6 and Table III).
Figure 6.
IB treatment regulated allergic-associated miRNA expression levels. (A-I) The miRNA expression level in nasal mucosa was determined by reverse transcription-quantitative polymerase chain reaction with U6 as the reference gene. Control: Group A (n=7) and group B (n=8); model: Group C (n=8), group D (n=10) and group E (n=9). Data are presented as the mean ± standard deviation. *P<0.05 vs. group C after IB treatment; #P<0.05. IB, ipratropium bromide; miRNA, microRNA; Mmu: Mus musculus.
Table III.
Differentially expressed nasal mucosa miRNAs in allergic mice post-treatment with ipratropium bromide.
| Expression level (TPM) | log2 (Fold change)a | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| miRNA | A | B | C | D | E | A/C | B/C | D/C | E/C |
| mmu-miR-124-3p | 293.5 | 377.3 | 4.4 | 101.4 | 14.4 | 5.6 | 5.9 | 4.5 | 1.2 |
| mmu-miR-124-5p | 26.8 | 32.6 | 0.3 | 10 | 1.2 | 5.9 | 6.1 | 4.9 | 1.3 |
| mmu-miR-133a-3p | 755.3 | 964.6 | 16.3 | 72.6 | 1,272.6 | 5.1 | 5.4 | 2.2 | 5.8 |
| mmu-miR-133a-5p | 48.9 | 55.9 | 0.7 | 2.4 | 60.1 | 5.7 | 5.8 | 1.8 | 5.9 |
| mmu-miR-133b-3p | 46.5 | 47.9 | 1.7 | 5.3 | 74.9 | 4.4 | 4.3 | 1.7 | 4.9 |
| mmu-miR-138-5p | 69 | 93.6 | 6.1 | 23.8 | 22.8 | 3.1 | 3.4 | 2 | 1.4 |
| mmu-miR-181a-5p | 2,114.3 | 2,962.1 | 967.2 | 1,279.7 | 2,957.1 | 0.7 | 1.1 | 0.4 | 1.1 |
| mmu-miR-181b-5p | 364.9 | 453.5 | 139.3 | 223.3 | 493.2 | 1 | 1.2 | 0.7 | 1.3 |
| mmu-miR-181c-5p | 28.7 | 32.2 | 4.1 | 4.4 | 35.7 | 2.4 | 2.5 | 0.1 | 2.6 |
| mmu-miR-181d-5p | 102.7 | 87.2 | 33.2 | 45.2 | 95 | 1.2 | 0.9 | 0.5 | 1 |
| mmu-miR-1a-3p | 18,773.7 | 21,540.8 | 728.6 | 4,349.9 | 21,603.1 | 4.3 | 4.4 | 2.6 | 4.4 |
| mmu-miR-1b-5p | 18,757.4 | 21,523.9 | 727.9 | 4,348.7 | 21,586.7 | 4.3 | 4.4 | 2.6 | 4.4 |
| mmu-miR-206-3p | 79.1 | 99.1 | 2.4 | 19.7 | 165.9 | 4.6 | 4.9 | 3.1 | 5.6 |
| mmu-miR-3071-3p | 39.2 | 83.8 | 6.1 | 24.4 | 41.7 | 2.3 | 3.3 | 2 | 2.2 |
| mmu-miR-3071-5p | 178 | 160.1 | 15.2 | 74.6 | 82.5 | 3.1 | 2.9 | 2.3 | 1.9 |
| mmu-miR-378a-3p | 3,206.9 | 4,143.5 | 6,531.4 | 4,011.7 | 3,798.9 | −1.5 | −1.2 | −0.7 | −1.3 |
| mmu-miR-378c | 529.6 | 558.1 | 900.1 | 408.4 | 618.3 | −1.2 | −1.2 | −1.1 | −1.1 |
| mmu-miR-384-3p | 14.3 | 17.8 | 0.3 | 9.1 | 2.4 | 5 | 5.2 | 4.8 | 2.3 |
| mmu-miR-384-5p | 12.8 | 12.7 | 0.3 | 6.2 | 1.2 | 4.8 | 4.7 | 4.2 | 1.3 |
Fold change, TPM of group D (or E or A or B)/TPM of group C. TPM, transcripts per million (evaluates the relative expression level of differentially expressed miRNAs); miRNA, micro RNA.
Discussion
AR is an inflammatory disorder within the nasal mucosa. Currently, correlation between the immune and nervous systems in AR have gained substantial attention. IB is a nonselective anticholinergic agent and its anti-inflammatory effect may function via muscarinic acetylcholine receptors (mAChRs) (16). In the current study, the effects of mAChR antagonist, IB on nasal mucosa allergic-associated inflammation were investigated by microarray analysis of the transcriptome and miRNAome in a mouse model of AR induced by OVA. Treatment with IB mitigated the nasal mucosa allergic symptoms induced by OVA challenge. IB treatment significantly modulated the expression levels of multiple mRNAs and miRNAs. IL-4Rα and PGDS mRNA expression levels were downregulated, and LIF, A20 and NR4A1 mRNA expression levels were upregulated. mmu-miR-124-3p/5p, −133a-5p, −384-3p, −181a-5p, −138-5p and −3071-5p expression levels increased, while the mmu-miR-378a-5p expression level decreased. Furthermore, IB treatment promoted or inhibited the cytokine mRNA level of particular CD4+ T cell subpopulations, and enhanced FOXP3 and CCL24 mRNA expression levels. These observations indicate the involvement of IB in modulating upper airway allergic inflammation, influencing regulatory T cell (Treg) differentiation and eosinophil chemotaxis.
The immunoregulatory effect of IB was previously demonstrated in a mouse model of allergic nasal mucosa inflammation, which highlighted the inhibition of IL-4 production, the increase in FOXP3 expression level and reduction of eosinophil infiltration (1). However, the present study profiled allergic nasal mucosa mRNA and miRNA expression levels in response to IB treatment, which indicated the beneficial effect of IB on nasal mucosa allergic inflammation partially via a complex immune mechanism involved in various allergic reaction-associated candidate genes.
IL-4/-13 were pivotal in allergic inflammation (17). In the present study, IB treatment significantly decreased the OVA-triggered upper airway IL-4 and IL-13 mRNA expression levels, which was consistent with the previous study (1). IL-4Rα, a common subunit shared by IL-4R and IL-13R, which is responsible for IL-4/IL-13 pathway signal transduction (18,19), exhibited a decreased mRNA expression level in the nasal mucosa following IB treatment compared with the NS-sham treatment. Therefore, IB treatment appeared to inhibit nasal allergic inflammation via a dual mechanism with decreased IL-4/IL-13 expression levels and reduced IL-4/IL-4Rα and IL-13/IL-4Rα signaling. The percentage of IFN-γ-expressing cells within the nasal mucosa of allergic mice remained unchanged following IB treatment for two weeks (1), whereas the IFN-γ mRNA expression level decreased following IB treatment for two or four weeks. The discrepancy may partially be attributed to the prolonged OVA intranasal challenge in the present study. Repetitive OVA exposure induced eosinophil accumulation within the nasal mucosa (20). Furthermore, IL-5 acted synergistically with eotaxins leading to airway eosinophilia, and IB indeed inhibited eosinophil infiltration in the nasal mucosa via an undefined mechanism (1). The present study demonstrated that the IL-5 mRNA expression level decreased, although the CCL24 mRNA expression level increased significantly following IB treatment. Considering that eotaxins primarily account for eosinophil tissue recruitment (21), the current observations indicate the potential mechanism for the decreased nasal mucosa eosinophil count (22).
Th17/Treg cell imbalance has been associated with airway allergic inflammation (23,24). A previous study demonstrated that the FOXP3+ Treg cell count in the nasal mucosa of allergic mice increased significantly following IB treatment (1), which was consistent with the current results of enhanced nasal mucosa FOXP3 mRNA expression following IB treatment. Furthermore, IB inhibited the nasal mucosa IL-17 mRNA expression level and increased the IL-10 mRNA expression level, which are predominantly secreted by IL-10-producing Treg1 cells (25). The LIF/IL-6 axis formed opposing feed-forward loops at the Th17/Treg axis. LIF directly promoted FOXP3 expression levels and simultaneously repressed retinoic acid-related orphan receptor-γt expression levels, resulting in the increased expansion of inducible FOXP3+ Treg (iTreg) cells; however, IL-6 demonstrated the opposite effect, leading to impaired LIF signaling and enhanced Th17 cell development (26,27). Allergic mice with IB treatment exhibited significantly elevated LIF mRNA expression levels and concomitant decreasing IL-17 mRNA expression levels compared with the NS sham treatment group, which indicated the role of IB in modulation of the LIF/IL-6 axis and the subsequent Th17/Treg axis. Th17/Treg axis imbalance was observed in the present study and regarded as a promising therapeutic target for AR (23). NR4A receptors promoted Treg differentiation by directly enhancing the transcription of FOXP3, and NR4A deficiency within Tregs induced an augmented conversion to cells with a Th2-like phenotype due to impaired depression of IL-4 expression levels (28–30). The current study demonstrated that nasal mucosa NR4A1 mRNA expression levels were significantly upregulated by IB treatment.
Communication between barrier epithelial cells (ECs) and dendritic cells determined the outcome of local immune responses to allergens, and A20 expressed in ECs markedly ameliorated allergic reactions (31). A20, as an inhibitor of nuclear factor-κB signaling, attenuated IL-5 and IL-13 expression levels, as well as the eotaxin level within bronchoalveolar fluid induced by OVA challenge (32). The present study demonstrated that nasal mucosa A20 levels significantly increased, whereas IL-5 and IL-13 mRNA expression levels significantly decreased, subsequent to IB treatment in allergic mice. Thus, IB treatment may modulate various allergic immune-associated candidate genes leading to resolution of allergic inflammation.
In the present study IB significantly attenuated the nasal allergic symptoms of sneezing, watery rhinorrhea and nasal itching. However, the symptom scores of allergic mice were >5 despite IB treatment. Nasal hyper-responsiveness, such as enhanced sneezing response to allergens in allergic mice was dependent on allergen-specific CD4+ T cells (33). Presumably, IB partially inhibited, rather than completely abrogated, the allergen-specific CD4+ T cell response. Nsal congestion was not evaluated in the current study. However, the PGDS, associated with induction of late-phase nasal blockage (34,35), exhibited markedly decreased mRNA expression levels in the nasal mucosa following IB treatment. Whether the IB-induced decrease in PGDS, and even subsequent PGD2 production, improved nasal obstruction requires further investigation. Furthermore, A20 attenuates OVA-induced airway hyper-responsiveness and mucus production (32), which may account for the IB-mediated improved nasal symptoms.
To investigate the potential function of DEGs by IB, PPI networks were constructed in the present study. Multiple allergic inflammation response-associated genes coexisted within the PPI regulatory network. Among them, NR4A1, dual specificity phosphatase 1 (DUSP-1), Fos, FosB, early growth response 1 (EGR-1), epidermal growth factor (EGF) and LIF interconnected in a direct or indirect manner. Reduced EGR-1 and/or DUSP-1 expression levels in the nasal epithelium resulted in prolonged activated response to allergens (36). Increased DUSP-1 expression levels in airway ECs enhanced the anti-inflammatory effect of glucocorticoids (37) and DUSP-1 knockout mast cells demonstrated an enhanced degranulation response (38). EGR-1 expressed in Th2 cells directly increased T cell receptor-induced IL-4 transcription (39). However, EGR-1 deficiency impaired IL-13-induced inflammatory responses and EGR-1-deficient mast cells demonstrated reduced stem cell factor-induced IL-13 expression levels (40). Indeed, IB treatment increased nasal mucosa NR4A1, LIF, DUSP-1 and EGR-1 expression levels. Although the current study does not clarify the mechanism by which IB modulates the expression levels of the above-mentioned genes, the network implied that the expression levels of these allergic response-associated candidate genes changed in response to IB in the nasal mucosa in a specific manner, which may jointly affect the local immune status; however, this requires further investigation.
Dysregulation of nasal mucosa miRNAs is associated with an allergen-induced Th2-polarized immune response (41). However, which miRNAs are directly implicated in the upper airway allergic inflammation was poorly understood. In the current study, the expression levels of various miRNAs in the nasal mucosa of allergic mice were demonstrated to change significantly in response to IB treatment.
miR-124 positively modulated neuropilin-1-expressing cell responsiveness to semaphorin-3A, which alleviated allergic nasal symptoms (42,43). Methyl CpG binding protein 2-induced miR-124 promoted the Th1 response via signal transducer and activator of transcription (STAT)1/STAT3 derepression due to miR-124-mediated reduced expression of suppressor of cytokine signaling 5 (44). The nasal mucosa miR-124 expression level in allergic mice was significantly upregulated with intranasal IB. A decreased miR-384 expression level was observed in allergic inflammation and miR-384 mitigated allergic inflammation by inhibiting the level of histone deacetylase 3 (HDAC3) expression (45,46). In the present study, the nasal mucosa miR-384 expression level in allergic mice was undetectable following OVA challenge, but increased significantly following IB treatment, which is consistent with a previous study (45). Notably, HDAC3 downregulated miR-384 expression forming the negative feedback loop of miR-384/HDAC3, which appeared to be a candidate target for allergic inflammation resolution (47).
The canonical myomiRs (miR-1, −133a/b and −206) are expressed in the nasal mucosa. These miRNA levels in the nasal mucosa of allergic mice were significantly increased by IB treatment, and OVA challenge decreased the level of nasal mucosa miR-133a expression. Circulating miR-206 and miR-133b expression levels were regarded as biomarkers for allergic and asthmatic status (48), whereas miR-133b and miR-206 co-expression in CD4+ T cells were deemed to be specific for Th17 cells (49). The role and function of miR-133b and miR-206 expression requires further investigation. Furthermore, the allergic mice in the present study exhibited a declined nasal mucosa miR-181a expression level, which was consistent with a previous study (50). IB treatment markedly increased the local level of miR-181a, which may negatively regulate allergic inflammation by targeting osteopontin and transglutaminase II (51). Taken together, IB treatment may modulate the expression levels of various immune-associated candidate miRNAs in the nasal mucosa, which downregulate the allergic inflammation directly or indirectly. However, further studies are required to elucidate the molecular mechanism underlying altered miRNA expression levels. Subsequently, the putative target gene of these differentially expressed miRNAs were predicted using bioinformatics. Unexpectedly, allergic response-associated genes, such as IL-4Rα, PGDS, A20, LIF and NR4A1 were identified as putative target gene of these miRNAs. Furthermore, the miRNA putative target gene-KEGG pathway analysis indicated IB treatment-induced changes in the miRNA transcriptome influenced a broad range of cellular signaling pathways. Among them, T/B cell receptor, FcεRI and neurotrophins, and leukocyte transendothelial migration signaling pathways are perceived as the most enriched pathways. These miRNAs may present as potential novel targets for anti-allergic therapy, which require further investigation focused on their effects on allergic inflammation.
In conclusion, the present study demonstrated the effect of IB treatment on the nasal mucosa mRNA transcriptome and miRNA profile in allergic mice induced by OVA. A unique panel of differentially expressed miRNAs and mRNAs implicated in the Th2 immune response were identified. Furthermore, IB treatment was shown to downregulate the expression levels of Th2 cytokines, and upregulate the expression levels of FOXP3 and IL-10 locally, which may account for the IB-elicited ameliorated nasal allergic symptoms. In addition, IB or other mAChR antagonists may be beneficial for the treatment of AR.
Acknowledgements
The present study was supported by Funds from the Hunan Scientific Plan in China (grant no. S2013F1023). The authors would like to thank the staff at Beijing Novogene Bioinformatics Technology Co., Ltd for their help with high-throughput RNA sequencing and their technical assistance during the bioinformatics analysis.
References
- 1.Daoud A, Xie Z, Ma Y, Wang T, Tan G. Changes of T-helper type 1/2 cell balance by anticholinergic treatment in allergic mice. Ann Allergy Asthma Immunol. 2014;112:249–255. doi: 10.1016/j.anai.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 2.Teng Y, Zhang R, Liu C, Zhou L, Wang H, Zhuang W, Huang Y, Hong Z. miR-143 inhibits interleukin-13-induced inflammatory cytokine and mucus production in nasal epithelial cells from allergic rhinitis patients by targeting IL13Rα1. Biochem Biophys Res Commun. 2015;457:58–64. doi: 10.1016/j.bbrc.2014.12.058. [DOI] [PubMed] [Google Scholar]
- 3.Lu TX, Rothenberg ME. Diagnostic, functional, and therapeutic roles of microRNA in allergic diseases. J Allergy Clin Immunol. 2013;132:3–14. doi: 10.1016/j.jaci.2013.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogoda M, Niiya R, Koshika T, Yamada S. Comparative characterization of lung muscarinic receptor binding after intratracheal administration of tiotropium, ipratropium, and glycopyrrolate. J Pharmacol Sci. 2011;115:374–382. doi: 10.1254/jphs.10311FP. [DOI] [PubMed] [Google Scholar]
- 5.Anders S, Pyl PT, Huber W. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang L, Feng Z, Wang X, Zhang X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26:136–138. doi: 10.1093/bioinformatics/btp612. [DOI] [PubMed] [Google Scholar]
- 7.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: Tools for microRNA genomics. Nucleic Acids Res. 2008;36(Database issue):D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedländer MR, Chen W, Adamidi C, Maaskola J, Einspanier R, Knespel S, Rajewsky N. Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol. 2008;26:407–415. doi: 10.1038/nbt1394. [DOI] [PubMed] [Google Scholar]
- 11.Moxon S, Schwach F, Dalmay T, Maclean D, Studholme DJ, Moulton V. A toolkit for analysing large-scale plant small RNA datasets. Bioinformatics. 2008;24:2252–2253. doi: 10.1093/bioinformatics/btn428. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L, Chen J, Li Z, Li X, Hu X, Huang Y, Zhao X, Liang C, Wang Y, Sun L, et al. Integrated profiling of microRNAs and mRNAs: microRNAs located on Xq27.3 associate with clear cell renal cell carcinoma. PLoS One. 2010;5:e15224. doi: 10.1371/journal.pone.0015224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human MicroRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: A web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34(Web Server issue):W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arocho A, Chen B, Ladanyi M, Pan Q. Validation of the 2-DeltaDeltaCt calculation as an alternate method of data analysis for quantitative PCR of BCR-ABL P210 transcripts. Diagn Mol Pathol. 2006;15:56–61. doi: 10.1097/00019606-200603000-00009. [DOI] [PubMed] [Google Scholar]
- 16.Kistemaker LE, Bos ST, Mudde WM, Hylkema MN, Hiemstra PS, Wess J, Meurs H, Kerstjens HA, Gosens R. Muscarinic M3 receptors contribute to allergen-induced airway remodeling in mice. Am J Respir Cell Mol Biol. 2014;50:690–698. doi: 10.1165/rcmb.2013-0220OC. [DOI] [PubMed] [Google Scholar]
- 17.Oeser K, Maxeiner J, Symowski C, Stassen M, Voehringer D. T cells are the critical source of IL-4/IL-13 in a mouse model of allergic asthma. Allergy. 2015;70:1440–1449. doi: 10.1111/all.12705. [DOI] [PubMed] [Google Scholar]
- 18.Wei Q, Sha Y, Bhattacharya A, Fattah E Abdel, Bonilla D, Jyothula SS, Pandit L, Hershey GK Khurana, Eissa NT. Regulation of IL-4 receptor signaling by STUB1 in lung inflammation. Am J Respir Crit Care Med. 2014;189:16–29. doi: 10.1164/rccm.201305-0874OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moshkovits I, Karo-Atar D, Itan M, Reichman H, Rozenberg P, Morgenstern-Ben-Baruch N, Shik D, Ejarque-Ortiz A, Hershko AY, Tian L, et al. CD300f associates with IL-4 receptor α and amplifies IL-4-induced immune cell responses; Proc Natl Acad Sci USA; 2015; pp. 8708–8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang MJ, Fu QL, Jiang HY, Chen FH, Chen D, Chen DH, Lin ZB, Xu R. Immune responses to different patterns of exposure to ovalbumin in a mouse model of allergic rhinitis. Eur Arch Otorhinolaryngol. 2016;273:3783–3788. doi: 10.1007/s00405-016-4128-9. [DOI] [PubMed] [Google Scholar]
- 21.Nussbaum JC, Van Dyken SJ, von Moltke J, Cheng LE, Mohapatra A, Molofsky AB, Thornton EE, Krummel MF, Chawla A, Liang HE, Locksley RM. Type 2 innate lymphoid cells control eosinophil homeostasis. Nature. 2013;502:245–248. doi: 10.1038/nature12526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pope SM, Brandt EB, Mishra A, Hogan SP, Zimmermann N, Matthaei KI, Foster PS, Rothenberg ME. IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J Allergy Clin Immunol. 2001;108:594–601. doi: 10.1067/mai.2001.118600. [DOI] [PubMed] [Google Scholar]
- 23.Zhao J, Lloyd CM, Noble A. Th17 responses in chronic allergic airway inflammation abrogate regulatory T-cell-mediated tolerance and contribute to airway remodeling. Mucosal Immunol. 2013;6:335–346. doi: 10.1038/mi.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang X, Chen Y, Zhang F, Yang Q, Zhang G. Peripheral Th17/Treg cell-mediated immunity imbalance in allergic rhinitis patients. Braz J Otorhinolaryngol. 2014;80:152–155. doi: 10.5935/1808-8694.20140031. (In English, Portuguese) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coomes SM, Kannan Y, Pelly VS, Entwistle LJ, Guidi R, Perez-Lloret J, Nikolov N, Müller W, Wilson MS. CD4+ Th2 cells are directly regulated by IL-10 during allergic airway inflammation. Mucosal Immunol. 2017;10:150–161. doi: 10.1038/mi.2016.47. [DOI] [PubMed] [Google Scholar]
- 26.Janssens K, van den Haute C, Baekelandt V, Lucas S, van Horssen J, Somers V, Van Wijmeersch B, Stinissen P, Hendriks JJ, Slaets H, Hellings N. Leukemia inhibitory factor tips the immune balance towards regulatory T cells in multiple sclerosis. Brain Behav Immun. 2015;45:180–188. doi: 10.1016/j.bbi.2014.11.010. [DOI] [PubMed] [Google Scholar]
- 27.Metcalfe SM. LIF in the regulation of T-cell fate and as a potential therapeutic. Genes Immun. 2011;12:157–168. doi: 10.1038/gene.2011.9. [DOI] [PubMed] [Google Scholar]
- 28.Sekiya T, Kashiwagi I, Inoue N, Morita R, Hori S, Waldmann H, Rudensky AY, Ichinose H, Metzger D, Chambon P, Yoshimura A. The nuclear orphan receptor Nr4a2 induces Foxp3 and regulates differentiation of CD4+ T cells. Nat Commun. 2011;2:269. doi: 10.1038/ncomms1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sekiya T, Kondo T, Shichita T, Morita R, Ichinose H, Yoshimura A. Suppression of Th2 and Tfh immune reactions by Nr4a receptors in mature T reg cells. J Exp Med. 2015;212:1623–1640. doi: 10.1084/jem.20142088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fassett MS, Jiang W, D'Alise AM, Mathis D, Benoist C. Nuclear receptor Nr4a1 modulates both regulatory T-cell (Treg) differentiation and clonal deletion; Proc Natl Acad Sci USA; 2012; pp. 3891–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schuijs MJ, Willart MA, Vergote K, Gras D, Deswarte K, Ege MJ, Madeira FB, Beyaert R, van Loo G, Bracher F, et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science. 2015;349:1106–1110. doi: 10.1126/science.aac6623. [DOI] [PubMed] [Google Scholar]
- 32.Kang NI, Yoon HY, Lee YR, Won M, Chung MJ, Park JW, Hur GM, Lee HK, Park BH. A20 attenuates allergic airway inflammation in mice. J Immunol. 2009;183:1488–1495. doi: 10.4049/jimmunol.0900163. [DOI] [PubMed] [Google Scholar]
- 33.Nishimura T, Kaminuma O, Saeki M, Kitamura N, Matsuoka K, Yonekawa H, Mori A, Hiroi T. Essential contribution of CD4+ T cells to antigen-induced nasal hyperresponsiveness in experimental allergic rhinitis. PLoS One. 2016;11:e0146686. doi: 10.1371/journal.pone.0146686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nabe T, Kuriyama Y, Mizutani N, Shibayama S, Hiromoto A, Fujii M, Tanaka K, Kohno S. Inhibition of hematopoietic prostaglandin D synthase improves allergic nasal blockage in guinea pigs. Prostaglandins Other Lipid Mediat. 2011;95:27–34. doi: 10.1016/j.prostaglandins.2011.05.001. [DOI] [PubMed] [Google Scholar]
- 35.Kajiwara D, Aoyagi H, Shigeno K, Togawa M, Tanaka K, Inagaki N, Miyoshi K. Role of hematopoietic prostaglandin D synthase in biphasic nasal obstruction in guinea pig model of experimental allergic rhinitis. Eur J Pharmacol. 2011;667:389–395. doi: 10.1016/j.ejphar.2011.05.041. [DOI] [PubMed] [Google Scholar]
- 36.Golebski K, van Egmond D, de Groot EJ, Roschmann KI, Fokkens WJ, van Drunen CM. EGR-1 and DUSP-1 are important negative regulators of pro-allergic responses in airway epithelium. Mol Immunol. 2015;65:43–50. doi: 10.1016/j.molimm.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 37.Luo X, Ma R, Wu X, Xian D, Li J, Mou Z, Li H. Azelastine enhances the clinical efficacy of glucocorticoid by modulating MKP-1 expression in allergic rhinitis. Eur Arch Otorhinolaryngol. 2015;272:1165–1173. doi: 10.1007/s00405-014-3191-3. [DOI] [PubMed] [Google Scholar]
- 38.Maier JV, Brema S, Tuckermann J, Herzer U, Klein M, Stassen M, Moorthy A, Cato AC. Dual specificity phosphatase 1 knockout mice show enhanced susceptibility to anaphylaxis but are sensitive to glucocorticoids. Mol Endocrinol. 2007;21:2663–2671. doi: 10.1210/me.2007-0067. [DOI] [PubMed] [Google Scholar]
- 39.Lohoff M, Giaisi M, Köhler R, Casper B, Krammer PH, Li-Weber M. Early growth response protein-1 (Egr-1) is preferentially expressed in T helper type 2 (Th2) cells and is involved in acute transcription of the Th2 cytokine interleukin-4. J Biol Chem. 2010;285:1643–1652. doi: 10.1074/jbc.M109.011585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li B, Berman J, Wu P, Liu F, Tang JT, Lin TJ. The early growth response factor-1 contributes to interleukin-13 production by mast cells in response to stem cell factor stimulation. J Immunotoxicol. 2008;5:163–171. doi: 10.1080/15476910802129612. [DOI] [PubMed] [Google Scholar]
- 41.Liu Z, Zhang XH, Callejas-Diaz B, Mullol J. MicroRNA in united airway diseases. Int J Mol Sci. 2016;17(pii):E716. doi: 10.3390/ijms17050716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sawaki H, Nakamura F, Aihara M, Nagashima Y, Komori-Yamaguchi J, Yamashita N, Nakazawa M, Goshima Y, Ikezawa Z. Intranasal administration of semaphorin-3A alleviates sneezing and nasal rubbing in a murine model of allergic rhinitis. J Pharmacol Sci. 2011;117:34–44. doi: 10.1254/jphs.11005FP. [DOI] [PubMed] [Google Scholar]
- 43.Baudet ML, Zivraj KH, Abreu-Goodger C, Muldal A, Armisen J, Blenkiron C, Goldstein LD, Miska EA, Holt CE. miR-124 acts through CoREST to control onset of Sema3A sensitivity in navigating retinal growth cones. Nat Neurosci. 2011;15:29–38. doi: 10.1038/nn.2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jiang S, Li C, McRae G, Lykken E, Sevilla J, Liu SQ, Wan Y, Li QJ. MeCP2 reinforces STAT3 signaling and the generation of effector CD4+ T cells by promoting miR-124-mediated suppression of SOCS5. Sci Signal. 2014;7:ra25. doi: 10.1126/scisignal.2004824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eom S, Kim Y, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. Histone deacetylase-3 mediates positive feedback relationship between anaphylaxis and tumor metastasis. J Biol Chem. 2014;289:12126–12144. doi: 10.1074/jbc.M113.521245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kim Y, Kim K, Park D, Lee E, Lee H, Lee YS, Choe J, Jeoung D. Histone deacetylase 3 mediates allergic skin inflammation by regulating expression of MCP1 protein. J Biol Chem. 2012;287:25844–25859. doi: 10.1074/jbc.M112.348284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim Y, Eom S, Park D, Kim H, Jeoung D. The hyaluronic acid-HDAC3-miRNA network in allergic inflammation. Front Immunol. 2015;6:210. doi: 10.3389/fimmu.2015.00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Panganiban RP, Wang Y, Howrylak J, Chinchilli VM, Craig TJ, August A, Ishmael FT. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. J Allergy Clin Immunol. 2016;137:1423–1432. doi: 10.1016/j.jaci.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 49.Haas JD, Nistala K, Petermann F, Saran N, Chennupati V, Schmitz S, Korn T, Wedderburn LR, Förster R, Krueger A, Prinz I. Expression of miRNAs miR-133b and miR-206 in the Il17a/f locus is co-regulated with IL-17 production in αβ and γδ T cells. PLoS One. 2011;6:e20171. doi: 10.1371/journal.pone.0020171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eom S, Kim Y, Kim M, Park D, Lee H, Lee YS, Choe J, Kim YM, Jeoung D. Transglutaminase II/microRNA-218/-181a loop regulates positive feedback relationship between allergic inflammation and tumor metastasis. J Biol Chem. 2014;289:29483–29505. doi: 10.1074/jbc.M114.603480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu W, Zeng Q, Luo R. Correlation between serum osteopontin and miR-181a levels in allergic rhinitis children. Mediators Inflamm. 2016;2016:9471215. doi: 10.1155/2016/9471215. [DOI] [PMC free article] [PubMed] [Google Scholar]