Abstract
Oral leukoplakia and erythroleukoplakia are common oral potentially malignant disorders diagnosed in the oral cavity. The specific outcome of these lesions remains to be elucidated, as their malignant transformation rate exhibits great variation. The ability to predict which of those potentially malignant lesions are likely to progress to cancer would be vital to guide their future clinical management. The present study reported two patients with tongue squamous cell carcinoma: Case study 1 was diagnosed with a simultaneous leukoplakia and case study 2 developed an erythroleukoplakia following the primary tumor treatment. Whole genome copy number alterations were analyzed using array comparative genomic hybridization. The present study determined more genomic imbalances in the tissues from leukoplakia and erythroleukoplakia compared with their respective tumors. The present study also identified in tumor and potentially malignant lesions common alterations of chromosomal regions and genes, including FBXL5, UGT2B15, UGT2B28, KANSL1, GSTT1 and DUSP22, being some of these typical aberrations described in oral cancer and others are linked to chemoradioresistance. Several putative genes associated with hallmarks of malignancy that may have an important role in predicting the progression of leukoplakia and erythroleukoplakia to squamous cell carcinoma, namely gains in BNIPL, MCL1, STAG2, CSPP1 and ZNRF3 genes were also identified.
Keywords: oral leukoplakia, oral erythroleukoplakia, genomic imbalances, malignant transformation, oral carcinoma, biomarkers
Introduction
Oral potentially malignant lesions (OPMLs) are often clinically categorized as leukoplakia or erythroplakia, with being leukoplakia the most common, accounting for 85% of all these lesions (1,2). While oral leukoplakia is defined as a white plaque without immediate apparent cause, erythroplakia is a bright red patch, which is rarely characterized as another definitive disease (3). Erythroleukoplakia has a mixed red and white appearance.
The diagnosis of these lesions is frequently made excluding known diseases or disorders lacking increased risk for cancer (3). These lesions precede malignant development in 0.13–34% oral squamous cell carcinoma (OSCC) cases (4). Histologically, leukoplakia with dysplasia is often associated with a high risk of malignant transformation (5), dysplasia is currently the principal predictor of tumor development.
Oral leukoplakia is more frequent in males; however, the malignant transformation is significantly higher in females (4). In addition to the presence of OPMLs being a risk factor for OSCC, its malignant transformation may be dependent on clinical, demographic, etiologic, histological and/or molecular features (6). Co-incidence of leukoplakia at the time of diagnosis of OSCC was demonstrated in up to 60% of cases (7–9).
Patients with leukoplakia suffer frequently with recurrence and development of new leukoplakias after the primary treatment. The OPMLs may appear at any time, remaining stable for a considerable length of time or may progress into malignant tumors (10). The molecular mechanism underlying malignant transformation of OPMLs remains to be elucidated and biomarkers which may predict this risk have not been identified. OPMLs have revealed several genetic alterations associated with OSCC (11,12).
The present case report described one case of simultaneous OSCC and adjacent oral leukoplakia and another with erythroleukoplakia that evolved following treatment of primary OSCC. The patients were clinically followed for ~48 months. During this time, the patients developed local relapses of leukoplakia and erythroleukoplakia. The genomic analysis of the tumors and OPMLs allowed for the identification of some putative biomarkers of malignant transformation.
Case report
Case 1
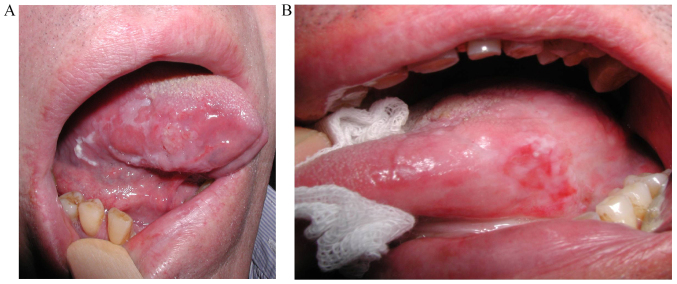
In March 2012, a Caucasian 59-year-old man, drinker, heavy smoker (≥20 cigarettes/day) and negative for human papillomavirus infection, was diagnosed at the Maxillofacial Surgery Unit, Coimbra Hospital and University Centre, EPE (Coimbra, Portugal) with a simultaneous primary squamous cell carcinoma in the right side of the tongue and a leukoplakia with severe dysplasia (Fig. 1A). The diagnosis was confirmed by a biopsy and the well differentiated tumor was classified as early stage (I), pT1, pN0, pMx, without compromised margins. The primary tumor and leukoplakia were simultaneously removed by surgery in April 2012 and the leukoplakia reached the surgical margins. Nine months after the initial diagnosis and surgery, the patient presented a leukoplakia, histopathologically classified as severe dysplasia. A local relapse of squamous carcinoma was diagnosed 28 months after the primary tumor diagnosis and total surgical excision was performed. The patient is alive and without signs of disease 48 months after the primary diagnosis.
Figure 1.
(A) Patient 1 presents tumor and simultaneous leukoplakia in the right side of the tongue. (B) Patient 2 was diagnosed with erythroleukoplakia after the treatment of primary tumor.
Case 2
In June 2012, a Caucasian 66-year-old man, drinker, smoker (<20 cigarettes/day) and negative for human papillomavirus infection, was diagnosed at the Maxillofacial Surgery Unit, Coimbra Hospital and University Centre, EPE with a primary squamous cell carcinoma in the left side of the tongue (photography unavailable). The diagnosis was confirmed by a biopsy and the well-differentiated tumor was classified as early stage (II), pT2, pN0, pMx, with close but non-compromised margins. In June 2012, the patient underwent glossectomy and ipsilateral neck dissection, followed by braquitherapy at 60 Gy between 27 August and 3 September 2012. The patient presented an erythroleukoplakia without dysplasia 15 months after the primary tumor diagnosis (Fig. 1B). A local relapse was suspected 46 months after the primary diagnosis; however, the patient refused biopsy and was lost to follow-up.
Genomic study
The present study was approved by the Ethics in Research Committee of the Faculty of Medicine of the University of Coimbra (ref: 030-CE-2015) and written informed consent from the patients was obtained. All the experiments were performed according to the regulations of Helsinki Declaration. From patient 1, samples of tumor tissue, macroscopically tumor-free tissue from surgical margins and from leukoplakia were simultaneously collected during surgery. From patient 2 samples of tumor tissue and macroscopically tumor-free tissue from surgical margins were simultaneously collected during surgery. Sample from the erythroleukoplakia that evolved after the treatment of primary tumor was obtained during the biopsy of patient 2. All samples were immediately stored in liquid nitrogen until use. DNA extraction was performed using a High Pure PCR Template Preparation kit (Roche GmbH, Mannheim, Germany) according to the manufacturer's protocol.
The genomic profile of all samples was analyzed through array Comparative Genomic Hybridization (aCGH) using Agilent SurePrint G3 Human Genome microarray 180K, (Agilent Technologies, Santa Clara, CA, USA) as previously described (13). Reference male DNA provided by Agilent was used as control.
Discussion
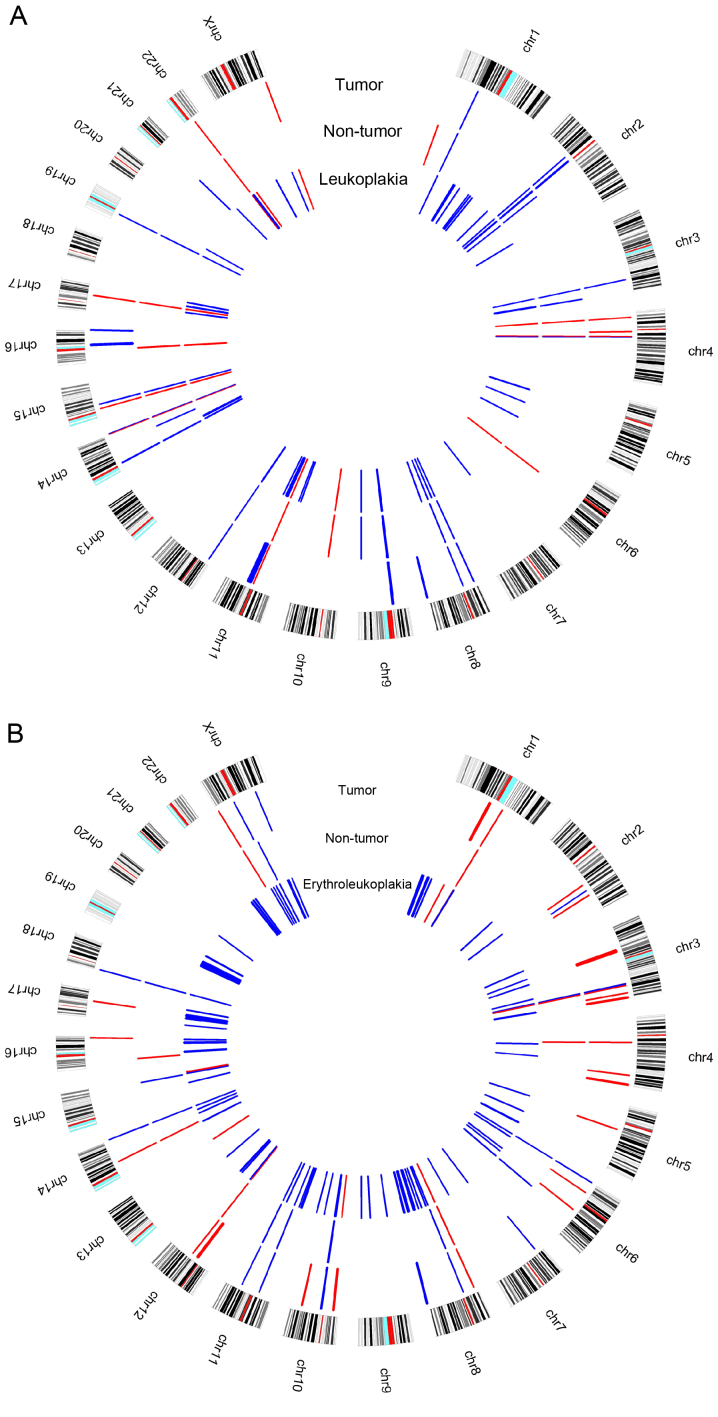
In case 1 it was observed that samples from tumor, non-tumor and simultaneous leukoplakia shared copy number alterations (CNAs) in some chromosomal regions, gains were identified at 1p31.1; 2p12, 2p11.2-p11.1; 3q26.1; 4q13.2; 8p23.1; 8p11.22; 9p13.1p11.2; 12p13.31; 14q11.2; 14q32.33; 15q14; 19p12, and losses at 4p15.32; 4q13.2; 11q11; 14q32.33; 15q11.2; 17q21.31 and 22q11.23 (Fig. 2A). In several of these regions the present study identified some relevant genes regarding their known function described in the University of California, Santa Cruz (UCSC) genome browser (https://genome.ucsc.edu/) and their relationship with oral carcinogenesis process, including FBXL5 (4p15.32), UGT2B15 and UGT2B28 (4q13.2), KANSL1 (17q21.31) and GSTT1 (22q11.23). Additionally, gains at 11q13.3-q13.4 were specifically observed in tumor and leukoplakia samples, which harbor the CCND1, ORAOV1, FGF19, FGF4, FGF3, FADD, CTTN, FOLR3, INPPL1, ARAP1 and P2RY2 genes. These chromosomal regions and genes have been associated with cancer, apoptosis, proliferation and OSCC (14–16). Regarding the clinical outcome, gains at CTTN and FADD were associated with lymph node metastasis (17,18) and reduced overall survival (19). Additionally, gain in the CCND1 was associated with poor prognosis, recurrence and metastasis (20). CCND1 amplification in OPMLs was associated with 8-fold increase in malignant transformation risk (21). In the tumor sample from case 1 a gain was observed at 16q24.3 harboring the FANCA gene, which is associated with locoregional progression-free survival (22). The presence of these CNAs in tumor or in leukoplakia may suggest poor prognosis and a recurrence/metastasis risk. Additionally, gain at the 3q29 that mapped the PAK2 gene and loss at 6q14.1, 10q11.22 and 16p11.2 were specifically identified in the non-tumor and leukoplakia samples. The present study specifically identified some CNAs in the non-tumor sample, namely loss at 1p36.33, mapped the CDK11B and CDK11A genes and gains at 2q13, mapped ZC3H8 gene, 14q24.3 and 21q11.2. The genomic imbalances observed in non-tumor tissue, similar to the ones observed in the tumors, may explain the local relapse observed, since these morphological normal cells presented already genomic aberrations characteristic of malignancy process; therefore, being able to suffer clonal expansion during the multistep process of malignization. This is particularly relevant for the oral carcinogenesis, since this patient has the traditional risk factors, tobacco and alcohol abuse, which may lead to local relapse disease through the field cancerization concept (23,24). It is on note that the present study observed more genomic imbalances in the leukoplakia compared with the tumor sample (Fig. 2A). Chromosomes 5, 7 and 10 did not present genomic imbalances in this tumor sample and chromosomes 13, 18 and 20 did not presented imbalances in both samples, tumor and leukoplakia.
Figure 2.
Circus plot with aberration pattern identified by array-CGH technique for (A) case report 1 and (B) case report 2. Blue color represents copy number gains and red copy number losses.
In this case, distinct chromosomal changes and genes have been described that may have a strong prognostic potential to predict patients' outcome and tumor transformation. However, further studies in larger cohorts are required in order to validate these findings.
In case 2 it was observed that tumor, non-tumor and the erythroleukoplakia that developed following primary tumor treatment shared CNAs in some chromosomal regions, particularly gains at 3q25.32, 8p11.22, 10q11.22, 11p12, 11q14.3, 14q32.33 and 18p11.32, and losses at 3q26.1, 6p25.3, 8p23.1, 12q14.2 and Yq11.223 (Fig. 2B). Taking the known function of the genes mapped in these regions into account DUP22 (6p25.3) may be the stronger putative candidate gene for the oral carcinogenesis process. Downregulation of DUP22 suggests its putative role as tumor suppressor gene; however, its function in cancer remains to be elucidated. This gene has been previously identified as a regulator of JNK signaling and with ability to dephosphorylate MAPKs (25). DUSP22 may also regulate metastasis, as its overexpression inhibited cell migration and reduced FAK phosphorylation (26). The present study observed losses at 1q21.2, 4q13.2, 14q21.1, Xp22.33 and Yp11.32 in tumor and non-tumor samples and in non-tumor and erythroleukoplakia samples gain was observed at 6q22.1 and Xq11.1. Tumor and erythroleukoplakia samples shared copy number gains at 8q24.3, which harbored GSDMD, SCRIB, PUF60, GRINA, SHARPIN and SCRT1. The 8q24.3 chromosomal band is frequently amplified in OSCC (15,16), which may suggest its importance for malignant transformation. These genes may be important predictors of tumor transformation risk, warranting further investigation. Copy number losses in Yq11.23 were also observed in both of these samples. MYC (8q24.21) and PTK2 (8q24.3) were detected only in the erythroleukoplakia sample and have already been associated with OSCC. A previous study revealed that overexpression of MYC was linked with malignant transformation and poor survival (27). PTK2 has been associated with resistance to radiotherapy (28). The presence of these genomic imbalances in the erythroleukoplakia, diagnosed 15 months following the primary tumor, may be the trigger for the development of relapse. There is no histological confirmation of relapse in this patient, as he refused to performed biopsy to verify the clinical suspicion. Additionally, these genes seem to be important candidates for the OSCC prognosis and especially for the prediction of the risk of relapse. The present study observed in the tumor, non-tumor and erythroleukoplakia collected from this patient, that was treated by surgery and braquitherapy several genomic alterations which were previously identified by Van den Broek et al (29) with chemoradioresistance and some with chemoradiosensitivity (Table I). Additionally, the present study highlighted some putative genes for these regions based in its known function described at the UCSC, including DUSP22 and JARID2 (Table I). Genomic imbalances in non-tumor tissues may indicate the presence of altered clones of cells even in the surrounding clinically and histologically normal oral mucosa, originating a progression to malignancy. Additionally, the present study detected more imbalances associated with chemoradiotherapy in erythroleukoplakia compared with the primary tumor sample, which may suggest that these alterations occurred following the treatment for the primary tumor or there was a selection of radioresistant cell populations due to the treatment.
Table I.
Chromosomal regions described in the study of Van den Broek et al (29), as associated with chemoradioresistance and chemoradiosensitivity and some putative candidate genes for these regions identified in the patient 2.
| Chromosomal region | Type of alteration | Clinical association | Patient 2-tumor | Patient 2-non-tumor | Patient 2-leukoplakia | Putative candidate genes |
|---|---|---|---|---|---|---|
| 3q21-q26.1 | Gain | Chemoradioresistance | 3q25.32 | |||
| 6p11-pter | 6p25.3 | 6p25.3; 6p23-p22.3 | DUSP22, JARID2 | |||
| 6q22-q27 | 6q22.1 | 6q22.1 | ||||
| Xq11-qter | Xq11.1 | Xq11.1 | ||||
| Xq25 | Xq25 | |||||
| 7p11.2–12 | Amplification | 7p11.2 | SEPT14, CHCHD2 | |||
| 8p11.1–12 | 8p11.22 | 8p11.22 | 8p11.22 | |||
| 12q15 | 12q15 | RAP1B, MDM2 | ||||
| 15q21 | 15q21.2 | |||||
| 18p11.3 | 18p11.32 | 18p11.32 | 18p11.32 | |||
| 3p11-pter | Loss | 3p14.2-p14.1 | FHIT, PTPR6, ADAMTS9 | |||
| 5q11-q12 | Gain | Chemoradiosensitivity | 5q11.2 | |||
| 10q11-q22 | 10q11.22 | 10q11.22 | 10q11.22; 10q11.21 | |||
| 14q distal | 14q32.33 | 14q32.33 | 14q32.33 | |||
| 14q13 | Amplification | 14q13.1-q13.2 | EAPP | |||
| 2q22-q25 | Loss | 2q23.3-q24.1 |
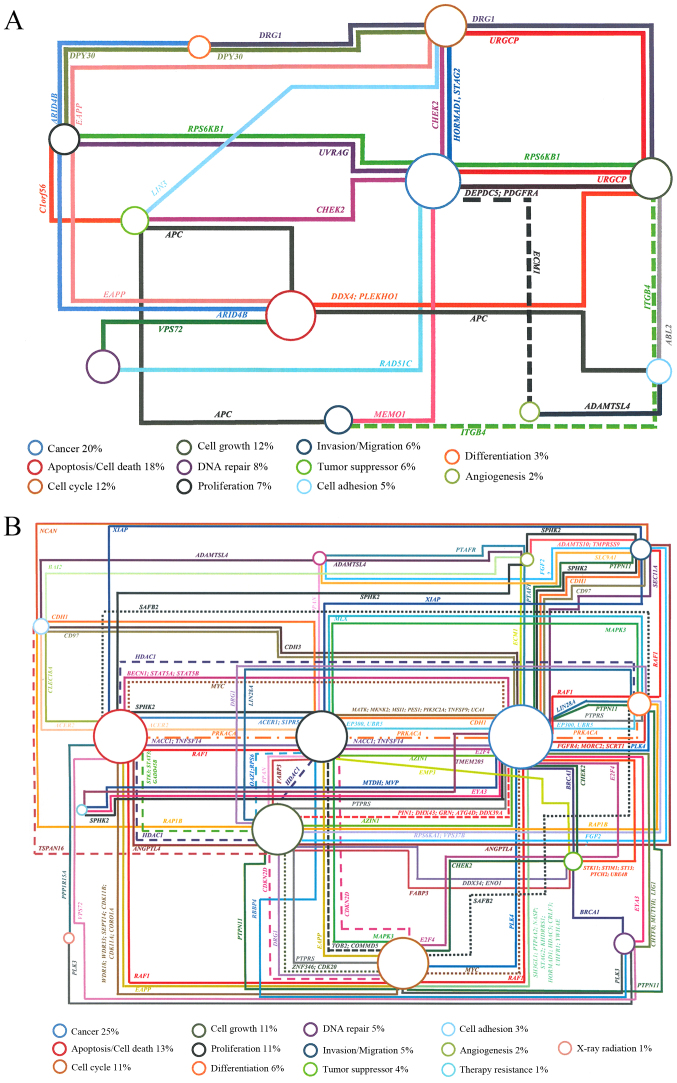
Overall, leukoplakia and erythroleukoplakia samples of the two patients presented more CNAs than the respective primary tumor. The erythroleukoplakia sample presented more CNAs than leukoplakia one (Fig. 1A and B), which may be due to the fact that erythroleukoplakia is associated with significantly higher rates of dysplasia, carcinoma in situ and invasive carcinoma compared with leukoplakia (30). As this patient presented an erythroleukoplakia without dysplasia, it is possible that erythroleukoplakia occurred following braquitherapy, which may induce several of the genomic alterations detected. CNAs were identified in several genes in these two OPMLs, which were associated with cancer and hallmarks of the carcinogenesis process, including cell cycle, cell growth, proliferation, differentiation, angiogenesis, apoptosis/cell death, DNA repair and invasion/migration (Fig. 3A and B). The present study observed that common genes altered in both cases, namely gains in BNIPL, MCL1, CSPP1 and ZNRF3 genes. These specific genes may represent important players in the malignant transformation of OPMLs into tumors, as these two patients relapsed. Further investigation is required to validate these results. Additionally, in the non-tumor sample of patient 1 and in the erythroleukoplakia sample of patient 2 a loss at 1p36.33 was identifies, where the CDK11B and CDK11A genes, which are associated with cell cycle and apoptosis, are mapped.
Figure 3.
Genes associated to diverse biological processes with impact on cancer development according to UCSC genome browser. The percentage represents the number of genes associated with each biological process. (A) Genes identified specifically in leukoplakia of patient 1. The following genes are associated only with one specific biological process: DNA repair: C11orf30, CHAF1B, GTF2H1, POLD3, RNF169. Apoptosis/cell death: ANO1, BIRC6, BNIPL, DAP3, MCL1, SAP30BP, SUMO2, PAK2, PTRH2. Cancer: SERPINH1, STAG2, TRIM37, WNT11, GSTT1, PPME1, PRUNE. Cell cycle: CCNO, COPS5, CSPP1. Invasion/migration: LLGL2, RAB25. Tumor suppressor: ZNRF3. X-ray radiation: XRRA1. (B) Genes identified specifically in erythroleukoplakia of patient 2. The following genes are associated only with one specific biological process: DNA repair: KIAA0146, MCM4, NSMCE2, PRKDC, RNF168, SF3B3, SIRT6, UIMC1, XRCC2, XRCC6. Apoptosis/cell death: ATAD5, BAX, BBC3, BCL2L13, BNIPL, CARD8, HCHD2, DAPK3, GRINA, IFI6, MAP15, MCL1, PYCARD, RDM25, RERE, TAOK2, TMEM161A, TRADD, TRIAP1. Angiogenesis: CCR10. Cancer: ALDH2, CREB3L3, CTCF, OX6C, DPP9, DUSP22, ELAVL1, EWSR1, FAM83A, GAL3ST1, HIGD1B, MDM2, MTSS1, NQO1, PGPEP1, PIK3CB, POUSF1B, PRDX1, PRDX2, PVT1, RFX1, RNF139, RPS15, RPS8, S100PBP, SAFB, SELENBP1, SPIN1, WWP2. Proliferation: ANGPTL6, CIB1, DLG1, HDGFRP3, FUT3, PLA2G1B, RASL10A, S1PR2. Cell cycle: ARID3A, BRD4, CDK2AP1, CSPP1, FZR1, GADD4SGIP1, NAE1, NPEPPS, PPP6C. Invasion/migration: ELMO3, MRI1, PARD6A, SCAI, ZRANB1. Cell growth: ARHGEF18, CD37, CDIPT, DDX19B, DDX20, DDX28, PPAN-P2RY11, PTK2, SESN2. Tumor suppressor: ADAM11, APC2, MAPKAPK5, ZNRF3. Tumor growth: ANXA13, KLF10, PDF. Differentiation: PUM1.
Identifying accurately and prospectively the OPMLs likely to progress to tumor is of paramount clinical significance. The present study identified the chromosomal regions and genes with CNAs in OSCCs and in OPMLs, such as FBXL5, UGT2B15, UGT2B28, KANSL1, GSTT1 and DUSP22 in the two patients presented. Leukoplakia and erythroleukoplakia had a high genomic heterogeneity with several genes altered; the genes that were altered were consistent across the two cases that were investigated specifically gains in BNIPL, MCL1, STAG2, CSPP1 and ZNRF3. Therefore, the current study identified several genes that may be associated with malignant transformation. The present study also highlighted several putative genes that may be associated with chemoradioresistance, particularly DUSP22 and JARID2.
Acknowledgements
The present study was in part supported by the Center of Investigation on Environment Genetics and Oncobiology, Faculty of Medicine, University of Coimbra. Ribeiro I.P. has a PhD fellowship (grant. no. SFRH/BD/52290/2013) supported by the Portuguese Foundation for Science and Technology.
References
- 1.Warnakulasuriya S. Lack of molecular markers to predict malignant potential of oral precancer. J Pathol. 2000;190:407–409. doi: 10.1002/(SICI)1096-9896(200003)190:4<407::AID-PATH546>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 2.Vigneswaran N, Williams MD. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26:123–141. doi: 10.1016/j.coms.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dionne KR, Warnakulasuriya S, Zain RB, Cheong SC. Potentially malignant disorders of the oral cavity: Current practice and future directions in the clinic and laboratory. Int J Cancer. 2015;136:503–515. doi: 10.1002/ijc.28754. [DOI] [PubMed] [Google Scholar]
- 4.Warnakulasuriya S, Ariyawardana A. Malignant transformation of oral leukoplakia: A systematic review of observational studies. J Oral Pathol Med. 2016;45:155–166. doi: 10.1111/jop.12339. [DOI] [PubMed] [Google Scholar]
- 5.Kobayashi T, Maruyama S, Abé T, Cheng J, Takagi R, Saito C, Saku T. Keratin 10-positive orthokeratotic dysplasia: A new leucoplakia-type precancerous entity of the oral mucosa. Histopathology. 2012;61:910–920. doi: 10.1111/j.1365-2559.2012.04283.x. [DOI] [PubMed] [Google Scholar]
- 6.William WN., Jr Oral premalignant lesions: Any progress with systemic therapies? Curr Opin Oncol. 2012;24:205–210. doi: 10.1097/CCO.0b013e32835091bd. [DOI] [PubMed] [Google Scholar]
- 7.Bouquot JE, Weiland LH, Kurland LT. Leukoplakia and carcinoma in situ synchronously associated with invasive oral/oropharyngeal carcinoma in Rochester, Minn., 1935–1984. Oral Surg Oral Med Oral Pathol. 1988;65:199–207. doi: 10.1016/0030-4220(88)90166-1. [DOI] [PubMed] [Google Scholar]
- 8.Gupta PC, Mehta FS, Daftary DK, Pindborg JJ, Bhonsle RB, Jalnawalla PN, Sinor PN, Pitkar VK, Murti PR, Irani RR, et al. Incidence rates of oral cancer and natural history of oral precancerous lesions in a 10-year follow-up study of Indian villagers. Community Dent Oral Epidemiol. 1980;8:283–333. doi: 10.1111/j.1600-0528.1980.tb01302.x. [DOI] [PubMed] [Google Scholar]
- 9.Reibel J. Prognosis of oral pre-malignant lesions: Significance of clinical, histopathological, and molecular biological characteristics. Crit Rev Oral Biol Med. 2003;14:47–62. doi: 10.1177/154411130301400105. [DOI] [PubMed] [Google Scholar]
- 10.Neville BW, Dann DD, Allen CM, Bouqout JE. Patologia Oral & Maxilofacial. Guanabara Koogan Brazil; 2009. Patologia epitelial; pp. 1–423. [Google Scholar]
- 11.Califano J, Westra WH, Meininger G, Corio R, Koch WM, Sidransky D. Genetic progression and clonal relationship of recurrent premalignant head and neck lesions. Clin Cancer Res. 2000;6:347–352. [PubMed] [Google Scholar]
- 12.Ha PK, Califano JA. The molecular biology of mucosal field cancerization of the head and neck. Crit Rev Oral Biol Med. 2003;14:363–369. doi: 10.1177/154411130301400506. [DOI] [PubMed] [Google Scholar]
- 13.Pinto-Leite R, Carreira I, Melo J, Ferreira SI, Ribeiro I, Ferreira J, Filipe M, Bernardo C, Arantes-Rodrigues R, Oliveira P, Santos L. Genomic characterization of three urinary bladder cancer cell lines: Understanding genomic types of urinary bladder cancer. Tumour Biol. 2014;35:4599–4617. doi: 10.1007/s13277-013-1604-3. [DOI] [PubMed] [Google Scholar]
- 14.Jin C, Jin Y, Gisselsson D, Wennerberg J, Wah TS, Strömbäck B, Kwong YL, Mertens F. Molecular cytogenetic characterization of the 11q13 amplicon in head and neck squamous cell carcinoma. Cytogenet Genome Res. 2006;115:99–106. doi: 10.1159/000095228. [DOI] [PubMed] [Google Scholar]
- 15.Ribeiro IP, Marques F, Caramelo F, Ferrão J, Prazeres H, Julião MJ, Rifi W, Savola S, de Melo JB, Baptista IP, Carreira IM. Genetic imbalances detected by multiplex ligation-dependent probe amplification in a cohort of patients with oral squamous cell carcinoma-the first step towards clinical personalized medicine. Tumour Biol. 2014;35:4687–4695. doi: 10.1007/s13277-014-1614-9. [DOI] [PubMed] [Google Scholar]
- 16.Ribeiro IP, Marques F, Caramelo F, Pereira J, Patrício M, Prazeres H, Ferrão J, Julião MJ, Castelo-Branco M, de Melo JB, et al. Genetic gains and losses in oral squamous cell carcinoma: Impact on clinical management. Cell Oncol (Dordr) 2014;37:29–39. doi: 10.1007/s13402-013-0161-5. [DOI] [PubMed] [Google Scholar]
- 17.Rothschild BL, Shim AH, Ammer AG, Kelley LC, Irby KB, Head JA, Chen L, Varella-Garcia M, Sacks PG, Frederick B, et al. Cortactin overexpression regulates actin-related protein 2/3 complex activity, motility, and invasion in carcinomas with chromosome 11q13 amplification. Cancer Res. 2006;66:8017–8025. doi: 10.1158/0008-5472.CAN-05-4490. [DOI] [PubMed] [Google Scholar]
- 18.Pattje WJ, Melchers LJ, Slagter-Menkema L, Mastik MF, Schrijvers ML, Gibcus JH, Kluin PM, Hoegen-Chouvalova O, Van Der Laan BF, Roodenburg JL, et al. FADD expression is associated with regional and distant metastasis in squamous cell carcinoma of the head and neck. Histopathology. 2013;63:263–270. doi: 10.1111/his.12174. [DOI] [PubMed] [Google Scholar]
- 19.Freier K, Hofele C, Knoepfle K, Gross M, Devens F, Dyckhoff G, Plinkert P, Lichter P, Herold-Mende C. Cytogenetic characterization of head and neck squamous cell carcinoma cell lines as model systems for the functional analyses of tumor-associated genes. J Oral Pathol Med. 2010;39:382–389. doi: 10.1111/j.1600-0714.2009.00864.x. [DOI] [PubMed] [Google Scholar]
- 20.Ruiz C, Martins JR, Rudin F, Schneider S, Dietsche T, Fischer CA, Tornillo L, Terracciano LM, Schreiber R, Bubendorf L, Kunzelmann K. Enhanced expression of ANO1 in head and neck squamous cell carcinoma causes cell migration and correlates with poor prognosis. PLoS One. 2012;7:e43265. doi: 10.1371/journal.pone.0043265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poh CF, Zhu Y, Chen E, Berean KW, Wu L, Zhang L, Rosin MP. Unique FISH patterns associated with cancer progression of oral dysplasia. J Dent Res. 2012;91:52–57. doi: 10.1177/0022034511425676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer VL, Braselmann H, Henke M, Mattern D, Walch A, Unger K, Baudis M, Lassmann S, Huber R, Wienberg J, et al. Chromosomal changes characterize head and neck cancer with poor prognosis. J Mol Med (Berl) 2008;86:1353–1365. doi: 10.1007/s00109-008-0397-0. [DOI] [PubMed] [Google Scholar]
- 23.Rettori MM, de Carvalho AC, Longo AL, de Oliveira CZ, Kowalski LP, Carvalho AL, Vettore AL. TIMP3 and CCNA1 hypermethylation in HNSCC is associated with an increased incidence of second primary tumors. J Transl Med. 2013;11:316. doi: 10.1186/1479-5876-11-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–968. doi: 10.1002/1097-0142(195309)6:5<963::AID-CNCR2820060515>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 25.Chen AJ, Zhou G, Juan T, Colicos SM, Cannon JP, Cabriera-Hansen M, Meyer CF, Jurecic R, Copeland NG, Gilbert DJ, et al. The dual specificity JKAP specifically activates the c-Jun N-terminal kinase pathway. J Biol Chem. 2002;277:36592–36601. doi: 10.1074/jbc.M200453200. [DOI] [PubMed] [Google Scholar]
- 26.Li JP, Fu YN, Chen YR, Tan TH. JNK pathway-associated phosphatase dephosphorylates focal adhesion kinase and suppresses cell migration. J Biol Chem. 2010;285:5472–5478. doi: 10.1074/jbc.M109.060186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gollin SM. Chromosomal alterations in squamous cell carcinomas of the head and neck: Window to the biology of disease. Head Neck. 2001;23:238–253. doi: 10.1002/1097-0347(200103)23:3<238::AID-HED1025>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 28.Skinner HD, Giri U, Yang L, Woo SH, Story MD, Pickering CR, Byers LA, Williams MD, El-Naggar A, Wang J, et al. Proteomic profiling identifies PTK2/FAK as a driver of radioresistance in HPV-negative head and neck cancer. Clin Cancer Res. 2016;22:4643–4650. doi: 10.1158/1078-0432.CCR-15-2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van den Broek GB, Wreesmann VB, van den Brekel MW, Rasch CR, Balm AJ, Rao PH. Genetic abnormalities associated with chemoradiation resistance of head and neck squamous cell carcinoma. Clin Cancer Res. 2007;13:4386–4391. doi: 10.1158/1078-0432.CCR-06-2817. [DOI] [PubMed] [Google Scholar]
- 30.Thomas G, Hashibe M, Jacob BJ, Ramadas K, Mathew B, Sankaranarayanan R, Zhang ZF. Risk factors for multiple oral premalignant lesions. Int J Cancer. 2003;107:285–291. doi: 10.1002/ijc.11383. [DOI] [PubMed] [Google Scholar]