Abstract
The current study presents a patient carrying a de novo ~6 Mb deletion of the isolated chromosome 8p23.2-pter that was identified with a single-nucleotide polymorphism array. The patient was characterized by developmental delay (DD)/intellectual disability (ID), microcephaly, autism spectrum disorder, attention-deficit/hyperactivity disorders and mildly dysmorphic features. The location, size and gene content of the deletion observed in this patient were compared with those in 7 patients with isolated 8p23.2 to 8pter deletions reported in previous studies (4 patients) or recorded in the Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources (DECIPHER) database (3 patients). The deletions reported in previous studies were assessed using a chromosomal microarray analysis. The 8p23.2-pter deletion was a distinct microdeletion syndrome, as similar phenotypes were observed in patients with this deletion. Furthermore, following a detailed review of the potential associations between the genes located from 8p23.2 to 8pter and their clinical significance, it was hypothesized that DLG associated protein 2, ceroid-lipofuscinosis neuronal 8, Rho guanine nucleotide exchange factor 10 and CUB and sushi multiple domains 1 may be candidate genes for DD/ID, microcephaly and neurobehavioral disorders. However, firm evidence should be accumulated from high-resolution studies of patients with small, isolated, overlapping and interstitial deletions involving the region from 8p23.2 to 8pter. These studies will allow determination of genotype-phenotype associations for the specific genes crucial to 8p23.2-pter.
Keywords: 8p23.2 terminal deletion, developmental delay, intellectual disability, microcephaly, neurobehavioral disorders, DLGAP2, CLN8, ARHGEF10, CSMD1
Introduction
The distal region of chromosome 8p is prone to various genomic rearrangements, mainly due to the presence of low copy repeats (LCRs). The LCRs flank an approximately 5 Mb region on 8p23.1 and contain two olfactory receptors gene clusters or defensin repeats, repeat-distal (REPD) in the distal 8p23.1 region and repeat-proximal (REPP) in the proximal 8p23.1 region (1,2). These LCRs are believed to mediate a U-type exchange, commonly resulting in a recurrent interstitial 8p23 deletion, a terminal 8p23 deletion and a variably sized interstitial inverted duplication in 8p with an 8p terminal deletion (1,3–6). The distal deletions of 8p23, including either the terminal or the sub-terminal 8p region, appear as an isolated finding or as a part of a complex chromosomal rearrangement. Both interstitial or terminal 8p23 deletions are relatively frequent among the structural variations associated with 8p, and patients with a deletion in this region are generally characterized by developmental delay (DD)/intellectual disability (ID), mildly dysmorphic features, microcephaly, congenital heart defects (CHD), congenital diaphragmatic hernia (CDH), seizure, and neuropsychiatric disorders (4,5,7,8). The deleted region often spans a large genomic segment containing numerous genes. Therefore, the genotype-phenotype correlations associated with the deletion are largely unknown. However, an increasing number of studies have contributed to narrowing the critical region (CR) to search for the candidate genes associated with the clinical features. A deletion at 8p23.1 was proposed as a CR for CHD, CDH and other common clinical features (4,5,8–10). Although genes in the deleted region may interact or participate in the same pathways, specific candidate genes are expected to play important roles in certain phenotypes. Notably, haploinsufficiency of GATA4, NEIL2 and/or SOX7 located at 8p23.1 were shown to be associated with CHD and CDH in several studies (8,11–13). Another gene in the region, MCPH1, has also been implicated as a candidate gene for primary microcephaly (14–16).
In addition to the common 8p23.1 deletion, a terminal deletion more distal to the REPD deletion involving 8p23.2 to 8pter has increasingly been reported as a CR associated with DD/ID, seizure and neurobehavioral disorders, such as autism spectrum disorder (ASD) and attention-deficit/hyperactivity disorders (ADHD) (7,17–19). Isolated 8p23.2-pter deletions are a rare chromosomal aberration. The recurrence of an isolated distal deletion involving the region from 8p23.2 to 8pter in patients with similar phenotypes is consistent with a distinctive microdeletion syndrome. To the best of our knowledge, cytogenetic and molecular characteristics have only been reported for four patients (7,17,19). The locations and sizes of the deletions detected in this region vary, similar to most of the sub-telomeric microdeletion syndromes. The majority of the examined patients with an 8p23 deletion were characterized using traditional techniques, including G-banded karyotyping, fluorescence in situ hybridization (FISH), microsatellite marker genotyping and multiplex ligation-dependent probe amplification (MLPA), with limited resolution and low efficiency. Therefore, an accurate definition of the locations, sizes and genes contained in the deletions has been difficult to obtain (5,9,20–23). Nevertheless, few microarray-based molecular studies have been performed to investigate these deletions. Current high-resolution chromosomal microarray analyses (CMA), including array comparative genome hybridization and single-nucleotide polymorphism (SNP) arrays, can now accurately determine the locations, sizes and genes contained in 8p23 deletions. This technique allows researchers to obtain more precise genotype-phenotype correlations and aids in identifying CRs and candidate genes. Here, we present a new case study of a patient with an isolated 8p23.2-pter deletion that was determined using a SNP array and compare the clinical features with patients with isolated deletions involving the region from 8p23.2 to 8pter that were previously identified using CMA or recorded in the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER; https://decipher.sanger.ac.uk/). The accumulation of patients with isolated 8p23.2-pter deletions at various locations and of different sizes will help in further defining the CRs and candidate genes for certain phenotypes.
Materials and methods
Case presentation
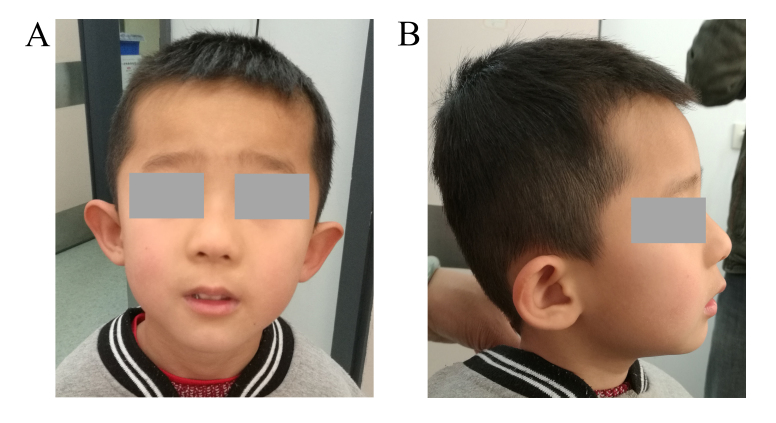
The patient was a 5-year-old Chinese male, who was the first child of healthy, unrelated parents that had no personal or family history of DD/ID, congenital malformations or psychiatric disorders. The patient was born at term via a natural delivery. His weight, length and head circumference at birth were 2.5 kg (<3rd percentile), 49 cm (<25th percentile) and 29 cm [≤3 standard deviations (SD)], respectively. The child progressed physiologically during the neonatal period. He sat alone at 7 months, crawled at 10 months, walked at 13 months and spoke his first words at 16 months. The parents noted that the child appeared to experience a growth delay, psychomotor delay and poor language/motor skills during preschool. He understood simple and blended verbal orders, but he had more difficulty in understanding complex verbal orders. He would sometimes talk to himself. He had deficiencies in his interactions with others, would not hold a direct gaze and often performed certain stereotyped or repetitive behaviours. He also had poor balance and coordination. At the age of 3 years and 11 months old, the child was diagnosed with ASD by qualified child psychiatrists based on both the childhood autism rating scale (CARS) and the autistic behaviour checklist (ABC). Additionally, attention deficits, impulsivity, unstable emotion and hyperactivity were also observed. The patient's intelligence quotient (IQ) was measured with the Wechsler Young Children Scale of Intelligence (C-WYCSI) at the age of 4 years and 4 months old. Psychomotor development was slightly delayed (IQ 50, verbal IQ 49, performance IQ 60). At the age of 5 years and 5 months, he weighed 15 kg (<3rd percentile) and had a height of 104 cm (<3rd percentile). He showed mild microcephaly, with a head circumference of 47.6 cm (≤2 SD). Mildly dysmorphic craniofacial abnormalities were noted, including low-set ears with a bilateral prominence of the antitragus, epicanthal folds and a long philtrum (Fig. 1). Electroencephalograms and cerebral magnetic resonance imaging (MRI) did not reveal any anomalies. The parents provided informed consent for clinical information and photographs of the child to be published.
Figure 1.
Facial features of the affected patient. (A) Frontal and (B) lateral views of the patient at the age of 5 years and 5 months. Note the mildly dysmorphic facial feature: Low-set ears with bilateral prominence of the antitragus, epicanthal folds and long philtrum.
Cytogenetic analysis
Using standard procedures, routine G-banded karyotyping was performed on cultured peripheral blood lymphocytes from the patient and his parents. A chromosome analysis was performed at the 400-band level in both the patient and his parents according to the International System for Human Cytogenomic Nomenclature (ISCN) 2013 (50 metaphases each).
High-resolution SNP array analysis
Genomic DNA was isolated from peripheral blood lymphocytes with the QIAamp DNA Blood Mini kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturer's instructions. The concentration of the extracted genomic DNA was determined using a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Berlin, Germany). The DNA sample (250 ng) was hybridized to CytoScan HD arrays on an Affymetrix SNP array platform (Affymetrix; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer's protocol. The CytoScan HD array contains more than 2.6 million markers for the copy number analysis. Of these markers, 1,950,000 are unique, non-polymorphic oligonucleotide probes, and 750,000 are SNP probes used for genotyping. The average marker spacing is one probe per 1.1 kb, with a mean spacing of one probe per 1.7 kb on non-gene backbones and one probe per 880 bp in intragenic regions. Using ChAS 3.0 software (Affymetrix; Thermo Fisher Scientific, Inc.), aberrations were filtered up to a minimum size of 100 kb, with at least 50 probe calls for deletions and duplications.
FISH analysis
A metaphase FISH analysis was performed on the patient and his parents using standard methods. Fluorescent probes specific for the sub-telomere region of chromosome 8p and the centromere of chromosome 8 (Cytocell, Inc., Cambridge, UK) were used for the FISH analysis and were labelled with SpectrumOrange and SpectrumGreen, respectively. FISH signals were observed using an Olympus BX-51 microscope (Olympus Corp., Tokyo, Japan).
Results
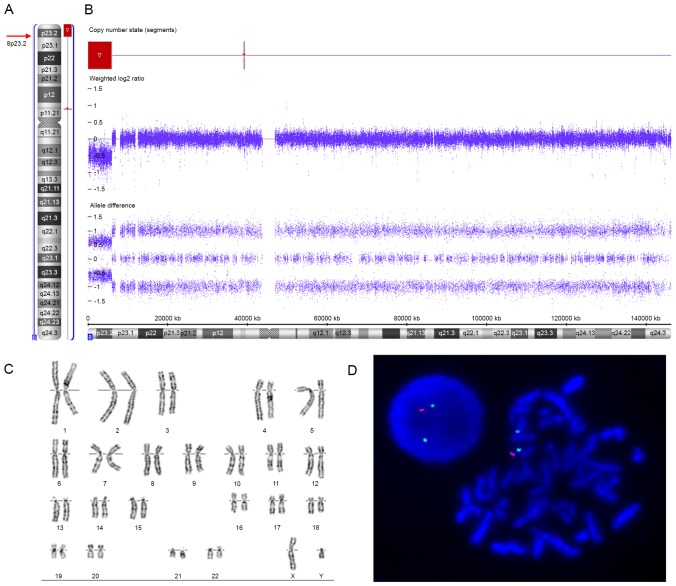
Routine G-banded karyotyping showed normal karyotypes in the patient (46, XY, Fig. 2C) and his parents (46, XX and 46, XY, respectively). The SNP array revealed an 8p23.2-pter terminal deletion in the patient, represented as arr[hg19] 8p23.3p23.2(158, 048 - 6, 004, 205)x1 (Fig. 2A and B). The deleted region was ~6.0 Mb in size and contained 23 RefSeq genes, including 11 protein-coding genes and 12 non-coding genes; 6 of the protein-coding genes were annotated in OMIM (FBXO25, DLGAP2, CLN8, ARHGEF10, MYOM2, and CSMD1) (Fig. 3A). Other copy number variants related to pathogenicity or of unknown significance were not identified. Furthermore, the two-colour FISH analysis of the patient's metaphase chromosomes using specific probes identified a terminal deletion of chromosome 8p (Fig. 2D), confirming the 8p terminal deletion identified by SNP array. However, FISH studies of the parents were normal, excluding cryptic rearrangements of the 8p terminal region in the parents and indicating the de novo deletion of 8p23.2-pter in the patient.
Figure 2.
Results of the SNP array, G-banded karyotyping and two-colour FISH analysis of the current patient. (A) Ideogram of chromosome 8 in the patient. Arrows indicate the location of the breakpoint. (B) A SNP array identified a deletion from 8p23.2 to 8pter in the patient. The SNP array data were analysed using ChAS 3.0 software. Both log2 ratios and SNP genotyping calls accurately identified the location and size of the deleted region. The red box outlines the deleted region. The deleted region spanned a ~6 Mb segment with a breakpoint at ~6,004,205 bp. (C) Routine G-banded karyotyping showed a normal karyotype of 46, XY. (D) The FISH analysis showed one orange signal (probe for the sub-telomere region of chromosome 8p labelled with spectrum orange) and two green signals (probe for the centromere of chromosome 8 labelled with spectrum green) for this patient, indicating a deletion of 8pter region. SNP, single nucleotide polymorphism; FISH, fluorescence in situ hybridization (FISH).
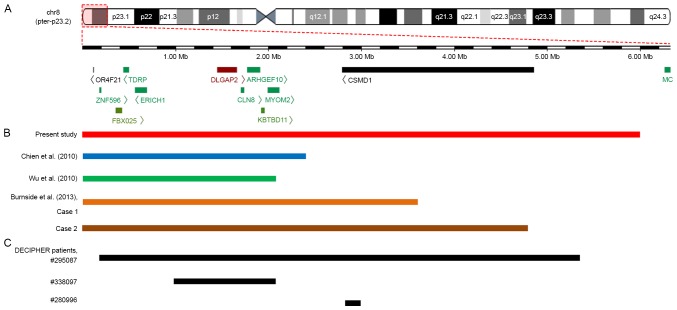
Figure 3.
An overlapping map of 8p23.2-pter deletions in the CMA-identified patients reported in the present study, previous studies and the DECIPHER database. (A) Schematic presentation of the 8p23.2-pter region and the genes located in this region, as determined by the University of California Santa Cruz Genome Browser. (B) An overview of CMA-identified patients with a deletion involving 8p23.2 to 8pter who were described in various reports. Among five patients, the deleted segment in the patient reported by Wu et al (19) was the smallest. Both deletions reported by Chien et al (17) and the deletion reported by Wu et al (19) encompassed DLGAP2, CLN8 and ARHGEF10 genes, whereas both deletions reported by Burnside et al (7) and the current study not only covered the DLGAP2, CLN8 and ARHGEF10 genes but also the CSMD1 gene. (C) An overview of patients from the DECIPHER database with deletions involving the 8p23.2 to the 8pter region that were regarded as definitely or likely pathogenic. A total of three patients presented with ID, and patient no. 338097 also exhibited behavioural problems (not specifically described). The deleted region in patient no. 338097 covered the DLGAP2, CLN8 and ARHGEF10 genes, whereas the deletion in patient no. 280996 only partially overlapped with the CSMD1 gene. CMA, chromosomal microarray analysis.
To date, only four patients with isolated deletions involving the region from 8p23.2 to 8pter that were identified using CMA have been reported in previous studies; all of them shared common clinical features. In addition, three patients were included in DECIPHER harbored deletions from 8p23.2 to 8pter that were recognized as definitely or likely pathogenic (Table I). The locations, sizes and genes contained in the deleted region in these patients were compared based on the genetic map (Fig. 3). In addition, we also performed a phenotypic comparison between the 8p23.2-pter deletion and 8p23.1 deletion (Table II).
Table I.
Summary of the clinical features of patients characterized by a CMA in the present study, previous studies and the DECIPHER database.
| Case study | Sex | Age, years | Deleted region | Genomic breakpoint (hg19), bp | Size, Mb | Intellectual disability | Developmental delay | Language delay | Neurobehavioral disorders | Craniofacial abnormalities | Other abnormalities | Other | Refs. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chien et al | M | 12 | 8p23.2-pter | ~2,410,000 | 2.4 | + | + | + | ASD and ADHD | − | Seizure | − | (17) |
| Wu et al | F | 1 | 8p23.3-pter | ~2,067,000 | 2.1 | + | + | ND | ND | Microcephaly, hypertelorism, long philtrum, malformed ears | − | − | (19) |
| Burnside et al | |||||||||||||
| Patient 1 | F | 2 | 8p23.2-pter | ~3,623,904 | 3.6 | − | + | + | − | Microcephaly, brachycephaly and mild maxillary, flattening medial epicanthal folds with upslanting palpebral a fissures, preauricular pit and bilateral prominence of the antitragus | Balance/coordination problems | − | (7) |
| Patient 2 | M | 4 | 8p23.2-pter | ~4,832,934 | 4.8 | − | + | − | − | Microcephaly, a palpable metopic ridge, upslanting palpebral fissures, nystagmus, aniridia, low-set and posteriorly rotated ears with overfolded helices, small upturned nose, and downturned corners of the mouth | Coarctation of the aorta and nail hypoplasia | − | |
| Present study | M | 5 | 8p23.2-pter | ~6,004,205 | 6.0 | + | + | + | ASD and ADHD | Microcephaly, low-set ears with bilateral prominence of the antitragus, epicanthal folds, and a long philtrum | Balance/coordination problems | − | |
| DECIPHER patients | |||||||||||||
| No. 295087 | F | 10 | 8p23.2-pter | 191,560–5,353,580 | 5.2 | + | unk | unk | unk | unk | − | Definitely pathogenica | |
| No. 338097 | M | unk | 8p23.3 | 974,820–2,067,679 | 1.1 | + | unk | unk | Behavioural abnormalities | unk | − | Likely pathogenica | |
| No. 280996 | M | 6 | 8p23.2 | 2,832,422–296,643 | 0.1 | + | unk | unk | unk | unk | unk | Likely pathogenica |
+, present; -, absent; M, male; F, female; ND, not determined; unk, unknown; ASD, autism spectrum disorder; ADHD; attention-deficit/hyperactivity disorders.
the contribution to pathogenicity was assigned by the DECIPHER database.
Table II.
Comparison of phenotypes in patients with the 8p23.2-pter deletion and 8p23.1 deletion.
| Clinical features | 8p23.2-pter deletion | Current patient | 8p23.1 deletion (terminal or interstitial) |
|---|---|---|---|
| DD/ID | + | + | + |
| Speech delay | + | + | + |
| Growth retardation | + | + | + |
| Neurobehavioral disorders | + | + | + |
| ASD | + | + | + |
| ADHD | + | + | + |
| Aggressiveness | + | − | + |
| Impulsiveness | + | − | + |
| Craniofacial features | + | + | + |
| Microcephaly | + | + | + |
| Brachycephaly | + | − | − |
| Palpable metopic ridge | + | − | − |
| Low-set ears | + | + | + |
| Malformed ears | + | + | + |
| Upslanting palpebral fissures | + | − | + |
| Epicanthal folds | + | + | + |
| Eye abnormalities | + | − | + |
| Bulbous nasal tip | − | − | + |
| Long philtrum | + | + | − |
| Widely spaced nipples | − | − | + |
| Thin upper lip | − | − | + |
| Micrognathia | − | − | + |
| Congenital heart disease | − | − | + |
| Congenital diaphragmatic hernia | − | − | + |
| Hypospadias | − | − | + |
| Cryptorchism | − | − | + |
| Short fingers | − | − | + |
| Seizure | + | − | + |
DD, developmental delay; ID, intellectual disability; ASD, autism spectrum disorder; ADHD; attention-deficit/hyperactivity disorders.
Discussion
Deletions of varying sizes at the distal region of the 8p chromosome are frequently reported, particularly the 8p23 deletion, but most have not been investigated using molecular cytogenetics. With the increasing use of high-resolution microarray technologies, a better understanding of the genotype-phenotype correlations of the distal 8p deletion have been obtained with the identification of CRs or candidate genes. Among distal 8p deletions, the 8p23.1 deletion is frequently observed, mainly due to the presence of LCRs. The recurrence of CHD and CDH in patients with various 8p23.1 deletions has led to the identification of a CR at 8p23.1 for CHD and CDH (4,5,8–10). Furthermore, the GATA4, NEIL2 and SOX7 genes in the deleted region have been recognized as candidate genes for CHD and CDH (8,11–13). In addition, a more distal deletion to 8p23.1 is the 8p23.2-pter deletion, a relatively rare aberration. Patients with an 8p23.2-pter deletion usually do not present CHD, CDH and urogenital abnormalities, but instead appear to have milder phenotypes than patients with an 8p23.1 deletion (Table II). The diversity in phenotypes suggests that the 8p23.2-pter deletion does not encompass the CR or candidate genes (such as the GATA4, NEIL2 and SOX7 genes) on 8p23.1 associated with these clinical features. Moreover, based on these data, the 8p23.1 region plays an important role in the manifestation of these clinical features.
However, common clinical features, including DD/ID, mildly dysmorphic features, microcephaly and neurobehavioral disorders, are usually shared by patients with either an 8p23.2-pter deletion or an 8p23.1 deletion (not only individuals with a large terminal 8p23.1 deletion but also in individuals with small interstitial 8p23.1 deletions) (Table II). Either an isolated 8p23.1 deletion or an isolated 8p23.2-pter deletion seems to contribute to these common clinical features, suggesting that the isolated 8p23.2-pter region may harbour other CRs and candidate genes associated with these features. The present study compared the phenotypic features between the present patient and four previously reported patients with isolated terminal deletions involving the region from 8p23.2 to 8pter that were characterized by CMA (7,17,19). Of these five patients, the most common phenotypes included growth retardation (5/5), microcephaly (4/5) and mildly dysmorphic features (4/5), followed by ID (3/5), language delay (3/5) and behavioural problem (3/5, including ASD and ADHD). In the DECIPHER database, three patients with isolated deletions involving the 8p23.2 to 8pter region were classified as definitely or likely pathogenic and all of them presented with ID. Taken together, although the phenotypes listed in DECIPHER might be incomplete and limited, ID was present in 6 out of the 8 patients with a deletion involving the region from 8p23.2 to 8pter reported in the literature and DECIPHER. A terminal 2.1 Mb deletion of 8p23.3 reported by Wu et al (19) was the smallest segment observed in the five patients from the present study and the literature. This deletion might be proposed as a CR for DD/ID, dysmorphic features and microcephaly (Fig. 2). The CR contains 5 OMIM genes, including FBXO25, DLGAP2, CLN8, ARHGEF10 and MYOM2. Most of these genes are known to be expressed in the brain.
The product encoded by FBXO25 is a member of the F-box protein family and is involved in phosphorylation-dependent ubiquitination. Fbxo25 may act as a ubiquitin E3 ligase to degrade cardiac transcription factors and control the transcriptional activity of the cardiac transcription factors Tbx5 and Nkx2-5, which play key roles in human cardiac development (24). The protein encoded by DLGAP2 is a membrane-associated protein that is biallelically expressed in the brain and plays roles in synapse organization and signalling in neuronal cells. It has been implicated as a candidate gene in neurobehavioral disorders, such as ASD and schizophrenia (25). Li et al (26) identified 5 rare, isolated variants and a DLGAP2 haplotype (CCACCAACT) in a cohort of patients with schizophrenia and showed that increased expression of the DLGAP2 gene is associated with schizophrenia. As shown in a study by Chien et al (17), an individual with DD/ID, ASD and seizures carried an 8p23.2-pter deletion. The DLGAP2 gene located in this region was suggested to be a candidate gene for ASD. Subsequently, the authors performed deep exon resequencing of DLGAP2 and detected two SNPs and some rare missense DLGAP2 mutations that might contribute to ASD (27). In a large autism case-control study, a de novo 817 kb duplication that partially overlapped with DLGAP2 was detected in a child with non-syndromic autism, and the authors also proposed DLGAP2 as a candidate gene involved in the pathogenesis of ASD (28). Notably, two SNPs in DLGAP2 (rs6558484 and rs7014992) were also revealed to be associated with alterations in brain volume (white matter volume of the prefrontal cortex) (29). CLN8 is recognized to have roles in lipid synthesis, transport, and sensing, thus contributing to cell proliferation during neuronal differentiation and defence against cell death stimuli (30). Mutations in this gene alter the levels of sphingolipids and phospholipids in the brain (30,31). A homozygous CLN8 mutation is one of the causes of autosomal recessive neuronal ceroid lipofuscinoses, which are mainly associated with progressive seizures and DD/ID (30). Allen et al (32) reported a patient with seizures, microcephaly, DD and behavioural problems, which were thought to result from the co-existence of a heterozygous CLN8 mutation and a de novo 8p23.3 terminal deletion. Notably, the 8p23.3 deletion was only identified by a FISH analysis using a probe (D8S504) located on the telomeric region of the chromosome (8:1017556-1017754) that did not cover the CLN8 gene. Therefore, the authors did not clearly determine whether this patient truly had a CLN8 gene deletion. The actual size and genes contained in the 8p23.3 deletion should be further investigated to obtain a better understanding of the genotype-phenotype correlation in this patient. ARHGEF10 encodes a Rho guanine nucleotide exchange factor and may play a role in neural morphogenesis (33). Mutations in the ARHGEF10 gene may cause demyelination and re-myelination of the peripheral nervous system and are associated with a slower nerve conduction velocity through an autosomal dominant pattern (33). Some SNPs in ARHGEF10 might confer a risk of developing schizophrenia (34). Notably, in DECIPHER, the 8p23.3-p23.2 and 8p23.3 deletions presented by two patients (nos. 295087 and 338097, respectively) with ID covered DLGAP2, CLN8 and ARHGEF10. Among all the patients carrying these genes (literature and DECIPHER), the deleted segment in patient no. 338097 was the smallest. Further studies of patients with similar phenotypes are required to determine whether the deleted region in patient no. 338097 is a potential CR for ID (only this phenotype was presented in DECIPHER). Although limited clinical features are listed in DECIPHER, the two patients also provided evidence for the potential role for either the deleted region or these genes in the pathogenesis of ID.
Furthermore, among the five patients with an 8p23.2-pter deletion from the present study and the literature, only the deletions reported by both Chien et al (17) and Wu et al (19) did not contain the CSMD1 gene. CSMD1 encodes a large multi-domain protein that mainly consists of CUB and Sushi domains. This protein may function as an important regulator of complement activation and inflammation in the developing central nervous system and is involved in brain mechanisms associated with memory and learning (35,36). Based on the evidence from previous studies, a SNP (rs10503253) located within the CSMD1 gene may be associated with the risk of multiple neurodevelopmental phenotypes, such as schizophrenia, seizure, speech delay, cognitive impairment and learning difficulties (37–39). Regarding cognitive ability, the ‘A’ allele of rs10503253 is considered to have deleterious impacts on verbal or performance intelligence, verbal episodic memory, and spatial working memory (35). In DECIPHER, patient no. 280996 had a maternally inherited 134 kb deletion that partially overlapped with CSMD1, and the deletion was recognized as a likely pathogenic variant. Both the patient and his mother showed moderate ID. Thus, CSMD1 might be a potential candidate gene for ID.
Microcephaly is caused by a congenital insufficiency during foetal brain (mainly cerebral cortex) development and is frequently observed in patients with either an 8p23.2-pter deletion or an 8p23.1 deletion (7,16,19). A candidate gene for microcephaly, the MCPH1 gene (16,40), is located on 8p23.1, but no candidate genes have been proposed for the region from 8p23.2 to 8pter. However, the DLGAP2, CLN8, ARHGEF10 and CSMD1 genes mentioned above are all expressed in the brain and implicated as being associated with multiple neurodevelopmental phenotypes. These genes should not be excluded as possibly having a role in microcephaly, particularly DLGAP2, which has been shown to be associated with alterations in brain volume alterations (29).
However, haploinsufficiency of each gene alone has not proven sufficient to explain the clinical phenotypes. These genes might function together, forming an interaction network in some pathway or may be modified by other genetic or environmental factors (27). Thus, the deletion of all these genes might conceivably have adverse effects on neurodevelopment. Compound inheritance of a rare null mutation and a common risk haplotype should also be considered in studies aiming to understand genotype-phenotype correlations. According to the study by Wu et al (41), compound inheritance of a heterozygous deletion and a common risk haplotype of the TBX6 allele are responsible for congenital scoliosis. Therefore, some genes in the deleted region, including DLGAP2, CLN8, ARHGEF10 and CSMD1, might contribute to DD/ID, microcephaly and neurobehavioral disorders, although the pathogenic mechanisms remain unclear.
In summary, the current study reported a new case study of a patient with a rarely isolated 8p23.2-pter deletion, and performed a review of patients with deletions in the region from 8p23.2 to 8pter that have been reported in the literature and the DECIPHER database. Similar clinical characteristics were observed among patients with the deletions, which were consistent with a distinctive microdeletion syndrome. Although a refined CR for DD/ID, microcephaly and neurobehavioral disorders was not conclusively determined due to the currently limited number of patients, the deleted region reported by Wu et al (19) as being responsible for these features is regarded as the smallest deletion among the five deletions reviewed in the present study. Furthermore, after a detailed review of the potential correlations between the genes located in 8p23.2 to 8pter region and their clinical significance, we hypothesized that DLGAP2, CLN8, ARHGEF10 and CSMD1 might be plausible candidate genes for DD/ID, microcephaly and neurobehavioral disorders. However, further high-resolution studies of patients with small isolated, overlapping and interstitial deletions involving the region from 8p23.2 to 8pter may allow us to obtain genotype-phenotype correlations for specific genes crucial to 8p23.2-pter.
Acknowledgements
No funding was applicable to this study. We are grateful to the patient and his parents for participating in the study.
References
- 1.Hollox EJ, Barber JC, Brookes AJ, Armour JA. Defensins and the dynamic genome: What we can learn from structural variation at human chromosome band 8p23.1. Genome Res. 2008;18:1686–1697. doi: 10.1101/gr.080945.108. [DOI] [PubMed] [Google Scholar]
- 2.Yu S, Fiedler S, Stegner A, Graf WD. Genomic profile of copy number variants on the short arm of human chromosome 8. Eur J Hum Genet. 2010;18:1114–1120. doi: 10.1038/ejhg.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rowe LR, Lee JY, Rector L, Kaminsky EB, Brothman AR, Martin CL, South ST. U-type exchange is the most frequent mechanism for inverted duplication with terminal deletion rearrangements. J Med Genet. 2009;46:694–702. doi: 10.1136/jmg.2008.065052. [DOI] [PubMed] [Google Scholar]
- 4.Páez MT, Yamamoto T, Hayashi K, Yasuda T, Harada N, Matsumoto N, Kurosawa K, Furutani Y, Asakawa S, Shimizu N, Matsuoka R. Two patients with atypical interstitial deletions of 8p23.1: Mapping of phenotypical traits. Am J Med Genet A. 2008;146A:1–1165. doi: 10.1002/ajmg.a.32205. [DOI] [PubMed] [Google Scholar]
- 5.Ballarati L, Cereda A, Caselli R, Selicorni A, Recalcati MP, Maitz S, Finelli P, Larizza L, Giardino D. Genotype-phenotype correlations in a new case of 8p23.1 deletion and review of the literature. Eur J Med Genet. 2011;54:55–59. doi: 10.1016/j.ejmg.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 6.García-Santiago FA, Martínez-Glez V, Santos F, García-Minañr S, Mansilla E, Meneses AG, Rosell J, Granero ÁP, Vallespín E, Fernández L, et al. Analysis of invdupdel(8p) rearrangement: Clinical, cytogenetic and molecular characterization. Am J Med Genet A. 2015;167A:1–1025. doi: 10.1002/ajmg.a.36879. [DOI] [PubMed] [Google Scholar]
- 7.Burnside RD, Pappas JG, Sacharow S, Applegate C, Hamosh A, Gadi IK, Jaswaney V, Keitges E, Phillips KK, Potluri VR, et al. Three cases of isolated terminal deletion of chromosome 8p without heart defects presenting with a mild phenotype. Am J Med Genet A. 2013;161A:1–828. doi: 10.1002/ajmg.a.35699. [DOI] [PubMed] [Google Scholar]
- 8.Wat MJ, Shchelochkov OA, Holder AM, Breman AM, Dagli A, Bacino C, Scaglia F, Zori RT, Cheung SW, Scott DA, Kang SH. Chromosome 8p23.1 deletions as a cause of complex congenital heart defects and diaphragmatic hernia. Am J Med Genet A. 2009;149A:1–1677. doi: 10.1002/ajmg.a.32896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devriendt K, Matthijs G, Van Dael R, Gewillig M, Eyskens B, Hjalgrim H, Dolmer B, McGaughran J, Bröndum-Nielsen K, Marynen P, et al. Delineation of the critical deletion region for congenital heart defects, on chromosome 8p23.1. Am J Hum Genet. 1999;64:1119–1126. doi: 10.1086/302330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Molck MC, Monteiro FP, Simioni M, Gil-da-Silva-Lopes VL. 8p23.1 interstitial deletion in a patient with congenital cardiopathy, neurobehavioral disorders and minor signs suggesting 22q11.2 deletion syndrome. J Dev Behav Pediatr. 2015;36:544–548. doi: 10.1097/DBP.0000000000000197. [DOI] [PubMed] [Google Scholar]
- 11.Longoni M, Lage K, Russell MK, Loscertales M, Abdul-Rahman OA, Baynam G, Bleyl SB, Brady PD, Breckpot J, Chen CP, et al. Congenital diaphragmatic hernia interval on chromosome 8p23.1 characterized by genetics and protein interaction networks. Am J Med Genet A. 2012;158A:1–3158. doi: 10.1002/ajmg.a.35665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wat MJ, Beck TF, Hernández-García A, Yu Z, Veenma D, Garcia M, Holder AM, Wat JJ, Chen Y, Mohila CA, et al. Mouse model reveals the role of SOX7 in the development of congenital diaphragmatic hernia associated with recurrent deletions of 8p23.1. Hum Mol Genet. 2012;21:4115–4125. doi: 10.1093/hmg/dds241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keitges EA, Pasion R, Burnside RD, Mason C, Gonzalez-Ruiz A, Dunn T, Masiello M, Gebbia JA, Fernandez CO, Risheg H. Prenatal diagnosis of two fetuses with deletions of 8p23.1, critical region for congenital diaphragmatic hernia and heart defects. Am J Med Genet A. 2013;161A:1–1758. doi: 10.1002/ajmg.a.35965. [DOI] [PubMed] [Google Scholar]
- 14.Garshasbi M, Motazacker MM, Kahrizi K, Behjati F, Abedini SS, Nieh SE, Firouzabadi SG, Becker C, Rüschendorf F, Nurnberg P, et al. SNP array-based homozygosity mapping reveals MCPH1 deletion in family with autosomal recessive mental retardation and mild microcephaly. Hum Genet. 2006;118:708–715. doi: 10.1007/s00439-005-0104-y. [DOI] [PubMed] [Google Scholar]
- 15.Jackson AP, Eastwood H, Bell SM, Adu J, Toomes C, Carr IM, Roberts E, Hampshire DJ, Crow YJ, Mighell AJ, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71:136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faheem M, Naseer MI, Rasool M, Chaudhary AG, Kumosani TA, Ilyas AM, Pushparaj P, Ahmed F, Algahtani HA, Al-Qahtani MH, Jamal H Saleh. Molecular genetics of human primary microcephaly: An overview. BMC Med Genomics. 2015;8(Suppl 1):S4. doi: 10.1186/1755-8794-8-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chien WH, Gau SS, Wu YY, Huang YS, Fang JS, Chen YJ, Soong WT, Chiu YN, Chen CH. Identification and molecular characterization of two novel chromosomal deletions associated with autism. Clin Genet. 2010;78:449–456. doi: 10.1111/j.1399-0004.2010.01395.x. [DOI] [PubMed] [Google Scholar]
- 18.Nucaro A, Pisano T, Chillotti I, Montaldo C, Pruna D. Chromosome 8p23. 2-pter: A critical region for mental retardation, autism and epilepsy? Clin Genet. 2011;79:394–396. doi: 10.1111/j.1399-0004.2010.01548.x. [DOI] [PubMed] [Google Scholar]
- 19.Wu Y, Ji T, Wang J, Xiao J, Wang H, Li J, Gao Z, Yang Y, Cai B, Wang L, et al. Submicroscopic subtelomeric aberrations in Chinese patients with unexplained developmental delay/mental retardation. BMC Med Genet. 2010;11:72. doi: 10.1186/1471-2350-11-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reddy KS. A paternally inherited terminal deletion, del(8)(p23.1)pat, detected prenatally in an amniotic fluid sample: A review of deletion 8p23.1 cases. Prenat Diagn. 1999;19:868–872. doi: 10.1002/(SICI)1097-0223(199909)19:9<868::AID-PD641>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 21.Giglio S, Graw SL, Gimelli G, Pirola B, Varone P, Voullaire L, Lerzo F, Rossi E, Dellavecchia C, Bonaglia MC, et al. Deletion of a 5-cM region at chromosome 8p23 is associated with a spectrum of congenital heart defects. Circulation. 2000;102:432–437. doi: 10.1161/01.CIR.102.4.432. [DOI] [PubMed] [Google Scholar]
- 22.Claeys I, Holvoet M, Eyskens B, Adriaensens P, Gewillig M, Fryns JP, Devriendt K. A recognisable behavioural phenotype associated with terminal deletions of the short arm of chromosome 8. Am J Med Genet. 1997;74:515–520. doi: 10.1002/(SICI)1096-8628(19970919)74:5<515::AID-AJMG12>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 23.Novo-Filho GM, Montenegro MM, Zanardo ÉA, Dutra RL, Dias AT, Piazzon FB, Costa TV, Nascimento AM, Honjo RS, Kim CA, et al. Subtelomeric copy number variations: The importance of 4p/4q deletions in patients with congenital anomalies and developmental disability. Cytogenet Genome Res. 2016;149:241–246. doi: 10.1159/000448905. [DOI] [PubMed] [Google Scholar]
- 24.Jeong HS, Jung ES, Sim YJ, Kim SJ, Jang JW, Hong KS, Lee WY, Chung HM, Park KT, Jung YS, et al. Fbxo25 controls Tbx5 and Nkx2-5 transcriptional activity to regulate cardiomyocyte development. Biochim Biophys Acta. 2015;1849:709–721. doi: 10.1016/j.bbagrm.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Li JM, Lu CL, Cheng MC, Luu SU, Hsu SH, Chen CH. Exonic resequencing of the DLGAP3 gene as a candidate gene for schizophrenia. Psychiatry Res. 2013;208:84–87. doi: 10.1016/j.psychres.2012.12.015. [DOI] [PubMed] [Google Scholar]
- 26.Li JM, Lu CL, Cheng MC, Luu SU, Hsu SH, Hu TM, Tsai HY, Chen CH. Role of the DLGAP2 gene encoding the SAP90/PSD-95-associated protein 2 in schizophrenia. PLoS One. 2014;9:e85373. doi: 10.1371/journal.pone.0085373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chien WH, Gau SS, Liao HM, Chiu YN, Wu YY, Huang YS, Tsai WC, Tsai HM, Chen CH. Deep exon resequencing of DLGAP2 as a candidate gene of autism spectrum disorders. Mol Autism. 2013;4:26. doi: 10.1186/2040-2392-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, et al. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466:368–372. doi: 10.1038/nature09146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu K, Hanna GL, Easter P, Kennedy JL, Rosenberg DR, Arnold PD. Glutamate system genes and brain volume alterations in pediatric obsessive-compulsive disorder: A preliminary study. Psychiatry Res. 2013;211:214–220. doi: 10.1016/j.pscychresns.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vantaggiato C, Redaelli F, Falcone S, Perrotta C, Tonelli A, Bondioni S, Morbin M, Riva D, Saletti V, Bonaglia MC, et al. A novel CLN8 mutation in late-infantile-onset neuronal ceroid lipofuscinosis (LINCL) reveals aspects of CLN8 neurobiological function. Hum Mutat. 2009;30:1104–1116. doi: 10.1002/humu.21012. [DOI] [PubMed] [Google Scholar]
- 31.Katata Y, Uematsu M, Sato H, Suzuki S, Nakayama T, Kubota Y, Kobayashi T, Hino-Fukuyo N, Saitsu H, Kure S. Novel missense mutation in CLN8 in late infantile neuronal ceroid lipofuscinosis: The first report of a CLN8 mutation in Japan. Brain Dev. 2016;38:341–345. doi: 10.1016/j.braindev.2015.09.008. [DOI] [PubMed] [Google Scholar]
- 32.Allen NM, O'HIci B, Anderson G, Nestor T, Lynch SA, King MD. Variant late-infantile neuronal ceroid lipofuscinosis due to a novel heterozygous CLN8 mutation and de novo 8p23.3 deletion. Clin Genet. 2012;81:602–604. doi: 10.1111/j.1399-0004.2011.01777.x. [DOI] [PubMed] [Google Scholar]
- 33.Verhoeven K, De Jonghe P, Van de Putte T, Nelis E, Zwijsen A, Verpoorten N, De Vriendt E, Jacobs A, Van Gerwen V, Francis A, et al. Slowed conduction and thin myelination of peripheral nerves associated with mutant rho Guanine-nucleotide exchange factor 10. Am J Hum Genet. 2003;73:926–932. doi: 10.1086/378159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jungerius BJ, Hoogendoorn ML, Bakker SC, Van't Slot R, Bardoel AF, Ophoff RA, Wijmenga C, Kahn RS, Sinke RJ. An association screen of myelin-related genes implicates the chromosome 22q11 PIK4CA gene in schizophrenia. Mol Psychiatry. 2008;13:1060–1068. doi: 10.1038/sj.mp.4002080. [DOI] [PubMed] [Google Scholar]
- 35.Donohoe G, Walters J, Hargreaves A, Rose EJ, Morris DW, Fahey C, Bellini S, Cummins E, Giegling I, Hartmann AM, et al. Neuropsychological effects of the CSMD1 genome-wide associated schizophrenia risk variant rs10503253. Genes Brain Behav. 2013;12:203–209. doi: 10.1111/gbb.12016. [DOI] [PubMed] [Google Scholar]
- 36.Kraus DM, Elliott GS, Chute H, Horan T, Pfenninger KH, Sanford SD, Foster S, Scully S, Welcher AA, Holers VM. CSMD1 is a novel multiple domain complement-regulatory protein highly expressed in the central nervous system and epithelial tissues. J Immunol. 2006;176:4419–4430. doi: 10.4049/jimmunol.176.7.4419. [DOI] [PubMed] [Google Scholar]
- 37.Glancy M, Barnicoat A, Vijeratnam R, de Souza S, Gilmore J, Huang S, Maloney VK, Thomas NS, Bunyan DJ, Jackson A, Barber JC. Transmitted duplication of 8p23.1–8p23.2 associated with speech delay, autism and learning difficulties. Eur J Hum Genet. 2009;17:37–43. doi: 10.1038/ejhg.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Havik B, Le Hellard S, Rietschel M, Lybaek H, Djurovic S, Mattheisen M, Muhleisen TW, Degenhardt F, Priebe L, Maier W, et al. The complement control-related genes CSMD1 and CSMD2 associate to schizophrenia. Biol Psychiatry. 2011;70:35–42. doi: 10.1016/j.biopsych.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 39.Shimizu A, Asakawa S, Sasaki T, Yamazaki S, Yamagata H, Kudoh J, Minoshima S, Kondo I, Shimizu N. A novel giant gene CSMD3 encoding a protein with CUB and sushi multiple domains: A candidate gene for benign adult familial myoclonic epilepsy on human chromosome 8q23.3-q24.1. Biochem Biophys Res Commun. 2003;309:143–154. doi: 10.1016/S0006-291X(03)01555-9. [DOI] [PubMed] [Google Scholar]
- 40.Trimborn M, Bell SM, Felix C, Rashid Y, Jafri H, Griffiths PD, Neumann LM, Krebs A, Reis A, Sperling K, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu N, Ming X, Xiao J, Wu Z, Chen X, Shinawi M, Shen Y, Yu G, Liu J, Xie H, et al. TBX6 null variants and a common hypomorphic allele in congenital scoliosis. N Engl J Med. 2015;372:341–350. doi: 10.1056/NEJMoa1406829. [DOI] [PMC free article] [PubMed] [Google Scholar]