Abstract
Insulin resistance (IR) is known to be an important factor, which can lead to the onset of type 2 diabetes. Autophagy is a cellular process, which sequesters senescent or damaged proteins in autophagosomes for recycling of their products. Insulin and intracellular molecules, including mammalian target of rapamycin (mTOR), are well-known inhibitors of autophagy. In patients with type 2 diabetes, the expression levels of glucose transporter 4 (GLUT-4) in skeletal muscles are significantly decreased, indicating decreased glucose-processing ability. Geniposide is an iridoid compound isolated from Gardenia jasminoides Ellis. Previously, it was reported that geniposide significantly promoted glucose uptake. In the present study, a HepG2 cell model of IR was constructed to determine whether geniposide can promote autophagy to inhibit insulin resistance in HepG2 cells via P62/nuclear factor (NF)-κB/GLUT-4. Cell proliferation was analyzed by performing an MTT assay, and the mRNA expression levels of NF-κB and GLUT-4 were assessed using semi-quantitative polymerase chain reaction and immunohistochemical staining. In addition, the protein levels of GLUT-4, P62 and phosphorylated-P65 were assessed by western blotting. The expression of GLUT-4 was initially increased following geniposide treatment, decreasing in time to its lowest level at 8 h. The expression levels of NF-κB and GLUT-4 in the IR cells treated with and without geniposide were significantly different, compared with those in the control group. Geniposide promoted autophagy in the IR HepG2 cells and significantly improved IR in the HepG2 cells, which may be associated with the dynamic regulation of the P62/NF-κB/GLUT-4 pathway.
Keywords: geniposide, autophagy, insulin resistance, P62, nuclear factor-κB, glucose transporter 4
Introduction
Insulin resistance (IR) is known to be an important factor, which can lead to the onset of type 2 diabetes (1). The mechanism by which IR develops is complex; during the insulin signal transduction process, alterations to the structure or activity of certain molecules, including insulin receptor, insulin receptor substrate, phosphoinositide 3-kinase, glucose transporter 4 (GLUT-4), and glucokinase, can result in IR. Additionally, the abnormal expression of nuclear factor κB (NF-κB) and its associated genes in different regions of an organism can affect the occurrence of IR directly or indirectly (2).
Autophagy is a cellular process, which sequesters senescent or damaged proteins in autophagosomes for recycling of their products (3). Autophagy is also involved in the removal of cells, which have undergone classical type 1 programmed cell death or apoptosis. Therefore, autophagy can be generally considered as a protector of cells against various stressors and a cellular response to stress (4). Paradoxically, autophagy can also lead to a form of non-apoptotic cell death, which is termed type 2 programmed cell death. Thus, autophagy can either protect cells or promote cell death, depending on the cellular and environmental context.
Dysfunctional autophagy may be involved in the pathophysiology of several diseases, including neurodegenerative disorders, cardiomyopathy and cancer (5). This may involve the accumulation of damaged molecules and organelles. Autophagy also appears to be involved in the cellular changes associated with aging (6). Insulin and intracellular molecules, including mammalian target of rapamycin (mTOR) are well-known inhibitors of autophagy, whereas glucagon, a counter-regulatory hormone of insulin, induces autophagy (7,8). These observations support the hypothesis that autophagy is involved in the natural history of diabetes via its involvement in hormone activity and organelle function (9).
As a key element in extracellular glucose transport to insulin-sensitive cells, the translocation of GLUT-4 to the cell membrane is regulated by insulin (10). In patients with type 2 diabetes, the expression levels of GLUT-4 in skeletal muscles are significantly decreased, which indicates decreased glucose-processing ability (11). Autophagy is a cellular degrading process, which can promote cell survival and cell death. Microtubule-associated protein 1 light chain 3 (LC3) and p62/SQSTM1 (P62) are associated with autophagosomal membranes, which engulf cytoplasmic content for subsequent degradation (12). Hyperglycemia stimulates the p62/protein kinase Cζ interaction, which mediates the activation of NF-κB, increases the expression of NADPH oxidase 4, and activates inflammatory cytokines in the vascular smooth muscle (13).
At present, there are no effective treatments specific for IR. Geniposide is an iridoid compound isolated from Gardenia jasminoides Elli, which has been previously reported to significantly promote glucose uptake (14). In the present study, a HepG2 cell model of IR was constructed to determine whether geniposide can promote autophagy to inhibit IR in HepG2 cells via the P62/NF-κB/GLUT-4 pathway.
Materials and methods
Reagents
RPMI-1640 medium and foetal bovine serum (FBS) were from Gibco; Thermo Fisher Scientific, Inc. (Waltham, MA, USA). Geniposide was from Xi'an Hao-xuan Bio-tech Co., Ltd. (Xi'an, China). Tetrazolium (MTT), recombinant human insulin and trypsin were from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany). Rabbit anti-human NF-κB (P65), phosphorylated (p-)NF-κB (p-P65), GLUT-4, LC3, P62 and β-actin were from Cell Signalling Technology, Inc. (Danvers, MA, USA). Rapamycin and 3-methyladenine (3-MA) were purchased from Selleck Chemicals (Houston, TX, USA). The streptavidin-peroxidase immunostaining kit was from Zymed; Thermo Fisher Scientific, Inc. Diaminobenzidine was from Beijing Dingguo Changsheng Biotechnology Co., Ltd. (Beijing, China) and the glucose detection kit was from Shanghai Rongsheng Biotechnology Co., Ltd. (Shanghai, China).
Cell culture
The HepG2 cells were obtained from the Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in RPMI-1640 medium containing 10% FBS at 37°C in 5% CO2. The medium was replaced every 2 days and passaged every 3–4 days following trypsinisation.
IR cell model construction
The IR cell model was constructed as previously described (15). Briefly, log-phase HepG2 cells were harvested following trypsinisation. The cells (5×104 /ml) were seeded into 96-well plates and cultured for 4 h. Following attachment of the cells to the bottom of the plate, the medium was replaced with RPMI-1640 medium supplemented with 1% FBS and containing 0, 50, 100, 200 or 500 nmol/l insulin. The control cells were treated with RPMI-1640 medium supplemented with 1% FBS. The cells were cultured at 37°C in 5% CO2 for 12, 24 and 36 h. The glucose levels in the supernatants were detected using a glucose detection kit according to the manufacturer's protocol, and were used as an index for determining IR. In the rapamycin group, the cells were treated with 40 nmol/l rapamycin for 2 h, followed by IR model establishment, and (7) in the 3-MA group, the cells were treated with 60 µM 3-MA for 2 h, followed by IR model establishment.
MTT assay
The present study evaluated cell survival using an MTT assay. The cells (~5,000 cells/well) were plated in a 96-well plate and incubated overnight in 100 µl culture medium. The cells were then treated with 15.63–125 mg/l geniposide. Following treatment for 20 h at 37°C, 20 µl MTT (5 mg/ml) was added to each well. Following incubation for 4 h at 37°C in 5% CO2, the MTT was removed, replaced with 200 µl dimethyl sulfoxide, and incubated for 20 min at 37°C until the crystals had dissolved. The optical density (OD) of each well was measured using an ELx800 microculture plate reader (BioTek Instruments, Winooski, VT, USA) with a test wavelength of 490 nm. The cell survival rate (SR) was determined as follows: SR = [(OD drug well - OD treated cell well)/(OD control well - OD cell well)] × 100%. Each experiment was repeated at least five times.
Fluorescence microscopy for detection of autophagy
The cells (~10,000 cells/well) were cultured in 6-well plates to 60–70% confluence. Lipofectamine 3000® was mixed with Opti-minimum essential media containing 2 µg/l green fluorescent protein (GFP)-LC3 plasmid (Hanbio Biotechnology Co., Ltd., Shanghai, China) according to the manufacturer's protocol. The medium was then replaced with RPMI-1640 containing 10% FBS for 12 h, following which the cells were treated as described above. The cells then were washed with PBS and mounted using Vectashield containing 1 µg/ml DAPI (Vector Laboratories, Inc., Burlingame, CA, USA). Images of the cells were captured using a fluorescence microscope.
Transmission electron microscopy (TEM)
The cell samples were placed in 1% glutaraldehyde and post-fixed with 2% osmium tetroxide. The cells were centrifuged at 1,000 × g for 15 min at 4°C and then the cell pellets were embedded in epon resin. The data were quantified by counting the number of autophagosomes per cross-sectioned cell by transmission electron microscopy.
Immunostaining
The log-phase HepG2 cells were harvested and 5×104/ml cells were seeded on glass coverslips in 24-well plates. When the cells had attached to the coverslips, the medium was replaced with RPMI-1640 medium supplemented with 1% FBS containing 100 nmol/l insulin. The cells were divided into three groups comprising a control group of untreated HepG2 cells, an untreated group of IR cells without geniposide treatment, and a treated group of IR cells treated with 62.5 mg/l geniposide. All groups were cultured for 4, 8, 12 and 24 h prior to fixing of the cells 4% paraformaldehyde for 30 min at room temperature. Immunostaining was performed using GLUT-4, P62 and p-P65 detection kits according to the manufacturer's protocol. Staining was visualised in five visual fields and the differences in staining intensity were measured using ImageJ software v6.0 (National Institutes of Health, Bethesda, MD, USA).
Semi-quantitative polymerase chain reaction (sqPCR) analysis
The mRNA expression levels of NF-κB and GLUT-4 in the cells were detected using sqPCR analysis. The RNA was isolated using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) Briefly, cDNA was synthesized from 1 µg RNA in the presence of a ribonuclease inhibitor (Sigma-Aldrich; Merck KGaA), dNTPs, Oligo(dT) 18 primers, and RevertAid™ M-MuLV reverse transcriptase (Fermentas; Thermo Fisher Scientific, Inc.) in a total volume of 25 µl. The primers used were as follows: NF-κB, sense 5′-TAGCCCAGCATCGCCTCT-3′ and antisense 5′-TTTGACACGGCAGTCCTCCATGGGA-3′ (target product of 641 bp); GLUT-4, sense 5′-CCCCGCTACCTCTACATCATCCA-3′ and antisense 5′-CCACCAACAACACCGAGACCAA-3′ (355 bp); internal control β-actin, sense 5′-TGGCATCCACGAAACTAC-3′ and antisense 5′-GCATCCTGTCGGCAAT-3′ (125 bp).
sqPCR was performed using a Takara mRNA Selective PCR kit (Takara Bio, Inc., Otsu, Japan) according to the manufacturer's instructions, with a total reaction volume of 25 µl, under the following thermocycling conditions: Initial denaturation at 94°C for 10 min, denaturation at 94°C for 1 min, annealing for 1.5 min (NF-κB and GLUT-4 at 65°C, β-actin at 51°C), and extension at 72°C for 2 min for 35 cycles, followed by a final extension at 72°C for 10 min. The PCR products were separated by 1.5% agarose gel electrophoresis and ethidium bromide staining was used for visualization. The target bands were analysed densitometrically using a GS-800 calibrated densitometer (Bio-Rad Laboratories, Inc., Hercules, CA, USA) and with Gel-Pro Analyser 4.0 software (Media Cybernetics, Inc., Rockville, MD, USA). The results were calculated as the ratio of the OD value relative to that of β-actin.
Western blot analysis
The cells were lysed in radioimmunoprecipitation assay buffer containing phosphatase inhibitor cocktail I (Sigma-Aldrich; Merck KGaA) and a protease inhibitor cocktail mini-tablet (Roche Diagnostics GmbH, Mannheim, Germany), followed by centrifugation at 8,000 × g at 4°C for 15 min. Protein concentration was determined by performing a bicinchoninic acid assay. An equal quantity of protein (50 µg) from each sample was separated on a 10, 12 or 15% SDS-polyacrylamide gel and then transferred onto nitrocellulose membranes. The membranes were probed with antibodies against P65 (rabbit IgG; cat. no. 4767; 1:1,000), p-P65 (rabbit IgG; cat. no. 8214; 1:1,000), GLUT-4 (rabbit IgG; cat. no. 2213; 1:1,000), LC3 (rabbit IgG; cat. no. 12513; 1:1,000), P62 (rabbit IgG; cat. no. 5114; 1:1,000) and β-actin (rabbit IgG, cat. no. 4970, 1:1,000; all Cell Signaling Technology, Inc.) at 4°C overnight. Following washing with PBS, a horseradish peroxidase-conjugated secondary antibody (anti-rabbit IgG; cat. no. 14708S; 1:2,000; Cell Signaling Technology, Inc.) was added to the membranes for 2 h at room temperature. The bound antibody was visualized using an enhanced chemiluminescence system (EMD Millipore, Billerica, MA, USA).
Statistical analysis
Statistical analysis was performed using SPSS 16.0 (SPSS, Inc., Chicago, IL, USA). All values are presented as the mean ± standard deviation. Data were analyzed by one-way analysis of variance followed by Tukey's post hoc test (equal variances) or Dunnett's T3 post hoc test (unequal variances). P<0.05 was considered to indicate a statistically significant difference.
Results
Geniposide decreases supernatant glucose levels
As shown in Table I, the glucose levels in the supernatant of all groups treated with 100 nmol/l recombinant human insulin were significantly higher, compared with those in the blank groups at the indicated time points (P<0.01). However, when the IR HepG2 cells were treated with 62.5 mg/l geniposide, the glucose levels in the supernatant decreased in a time-dependent manner (P<0.01). These results indicated that geniposide was able to ameliorate IR in HepG2 cells following insulin treatment, improve the utilization of insulin, and decrease the glucose levels in the supernatant.
Table I.
Glucose content in culture supernatant at different insulin concentrations and treatment times.
| Glucose content (mmol/l) | |||
|---|---|---|---|
| Duration (h) | Control | IR (100 nmol/l insulin) | Geniposide (62.5 mg/l) |
| 12 | 17.93±0.83b | 22.95±0.47 | 21.17±0.65a |
| 24 | 16.16±0.46b | 20.91±1.04 | 17.38±0.82b |
| 36 | 13.97±0.87b | 18.21±0.64 | 14.95±0.93b |
P<0.05
P<0.01 vs. the IR group. IR, insulin resistance.
Geniposide has no significant effect on IR cell survival
As shown in Table II, treatment with 15.63–125 µg/ml geniposide had no significant effect on the proliferation of the IR cells (P>0.05), compared with cells in the blank groups. Therefore, 62.50 µg/ml was selected as an effective concentration of geniposide in the present study.
Table II.
Effects of geniposide on the survival rate of insulin-resistant HepG2 cells.
| Geniposide (µg/ml) | Cell survival rate (100%) | P-value |
|---|---|---|
| 15.63 | 0.99±0.14 | >0.05 |
| 31.25 | 1.12±0.13 | >0.05 |
| 62.50 | 1.08±0.21 | >0.05 |
| 125.0 | 1.03±0.19 | >0.05 |
Cells were exposed to various concentrations of geniposide for 20 h. The assay was performed in quadruplet and the results presented as the mean ± standard error of the mean.
Effect of geniposide on the mRNA expression of GLUT-4
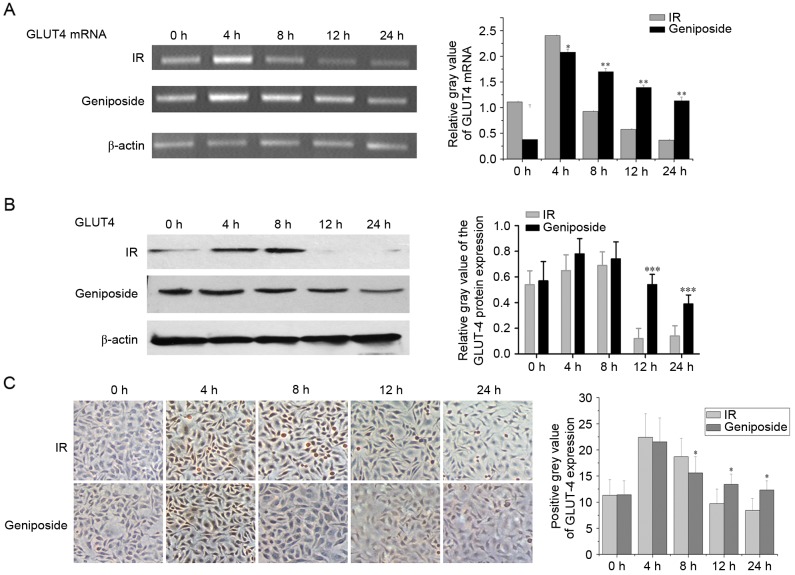
As shown in Fig. 1A, in the IR cells, the mRNA expression of GLUT-4 peaked at 4 h (P<0.01) and began to decline at 8 h, but remained higher than that at 0 h (P<0.05). The mRNA expression of GLUT-4 decreased at 12 h to below the level at 0 h, reaching a nadir at 24 h (P<0.05). In the geniposide-treated cells, the mRNA expression of GLUT-4 peaked at 4 h (P<0.01), although the increase was less than that observed in the control group. Subsequently, the mRNA expression of GLUT-4 began to decline at 8 and 12 h, but remained higher than that at 0 h (P<0.01), reaching a nadir at 24 h. The mRNA expression of GLUT-4 was higher, compared with that in the control group (P<0.05).
Figure 1.
Expression levels of GLUT-4 in IR HepG2 cells. Data are presented as the mean ± standard deviation (n=3). (A) mRNA expression of GLUT-4 in IR cells and geniposide-treated cells were detected using semi-quantitative polymerase chain reaction analysis. (B) Protein expression levels of GLUT-4 in HepG2 cells. (C) Expression of GLUT-4 in IR and geniposide-treated cells were detected using immunostaining (magnification, ×200). *P<0.05, **P<0.01 and ***P<0.001, geniposide-treated vs. IR group. GLUT-4, glucose transporter 4; IR, insulin resistance.
Effect of geniposide on the expression of NF-κB and GLUT-4
GLUT-4 is present on the cell membrane and in the cytoplasm; positive staining signals are brown in colour. In the control group, the protein expression levels of GLUT-4 were highest at 4 and 8 h, and staining intensity was higher, compared with that at 0 h (P<0.05; Fig. 1B). The staining intensity began to decline at 12 h, with levels below that at 0 h and reaching a nadir at 24 h. Following geniposide treatment, the expression levels of GLUT-4 peaked at 4 h, although the increase was less than that observed in the control group. The expression levels began to decline at 8 and 12 h, but remained higher than the level at 0 h. The expression of GLUT-4 reached a nadir at 24 h and was lower than that in the control group at the same time point (Fig. 1B and C).
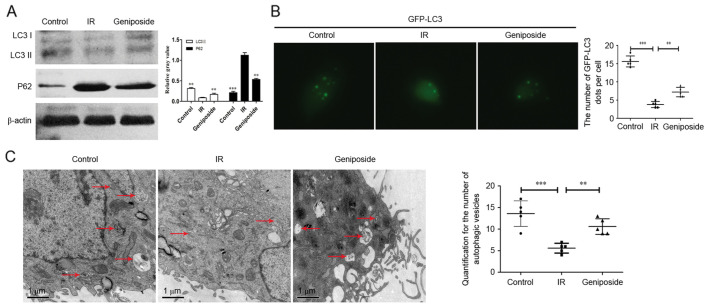
Geniposide promotes autophagy in IR HepG2 cells
As shown in Fig. 2A, compared with the control group, the protein expression levels of LC3II were downregulated in the IR HepG2 cells. There was also a significant increase in the expression of LC3II in the geniposide-treated HepG2 cells, compared with the cells in the IR group (P<0.01). By contrast, the expression levels of P62 were increased in the IR HepG2 cells, whereas geniposide decreased the expression of P62 in the IR cells.
Figure 2.
Geniposide promotes autophagy to inhibit IR in HepG2 cells. (A) Western blot analysis for LC3 and P62 in HepG2 cells. (B) Immunofluorescence (magnification, ×400) of HepG2 cells shows numbers of GFP-LC3 dots in the control, IR and geniposide groups. The data were quantified by counting the number of GFP-LC3 dots per cross-sectioned cell. (C) TEM images for HepG2 cells. TEM images show representative examples of autophagosomes (red arrows). Scale bar, 1.0 µm. Data were quantified by counting the number of autophagosomes per cross-sectioned cell. **P<0.01 and ***P<0.001 vs. IR group. IR, insulin resistance; LC3, microtubule-associated protein 1 light chain 3; GFP, green fluorescent protein; TEM, transmission electron microscopy.
The appearance of GFP-LC3 within the cytoplasm reflects the recruitment of LC3 protein to autophagosomes. As shown in Fig. 2B, IR significantly decreased the number of GFP-LC3 dots, compared with those in the control group (P<0.001). There was also a significant increase in the number of GFP-LC3 dots in the geniposide-treated HepG2 cells, compared with the number in the IR group (P<0.01). In addition, autophagosomes, containing partially degraded cytoplasmic material, were observed using TEM (Fig. 2C). The number of autophagosomes within the IR group was significantly decreased, compared with that in the control group (P<0.001), however, geniposide decreased the number of autophagosomes (P<0.01).
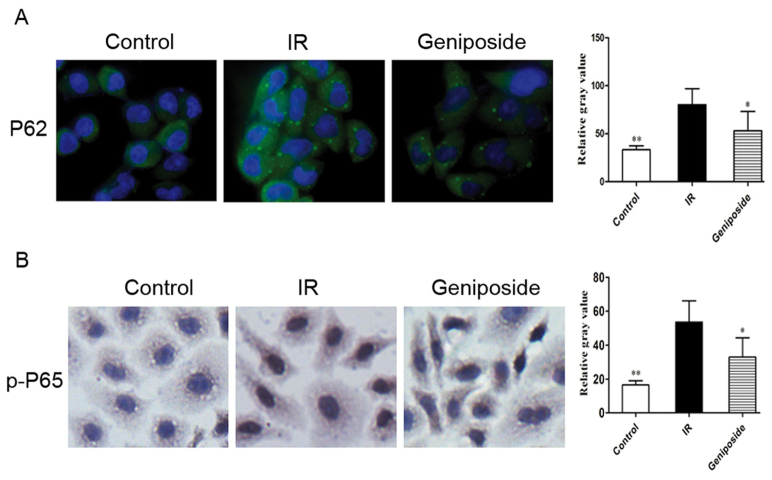
Effect of geniposide on the expression of P62 and p-P65
P62 is present in the cytoplasm; positive staining signals are green. In the control group, marked positive staining of P62 was apparent in the IR cells, with a higher intensity, compared with that in control (P<0.05). Following geniposide treatment, the P62 staining intensity decreased, compared with that in the control group (Fig. 3A).
Figure 3.
Effect of geniposide on expression levels of P62 and p-P65 in IR HepG2 cells. (A) Immunofluorescence (magnification, ×400) staining demonstrating intranuclear P62 in HepG2 cells. (B) Immunocytochemistry (magnification, ×400) demonstrating intranuclear p-P65 in HepG2 cells. *P<0.05 and **P<0.01 vs. IR group. IR, insulin resistance; p-, phosphorylated.
As shown in Fig. 3B, p-P65 was present in the nucleus and cytoplasm. p-P65 is translocated from the cytoplasm into the nucleus upon cell activation, increasing its presence in the nucleus; positive staining signals are brown in colour. Compared with the control group, the relative gray value of positive cytoplasmic p-P65 staining was the highest, and there was an increase in positive nuclear staining in the IR cells (P<0.05). Following geniposide treatment, the relative gray value of p-P65 positive staining decreased, compared with that in IR group.
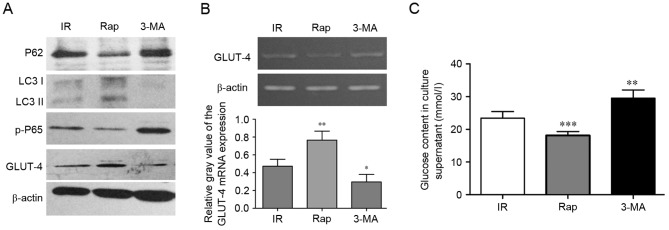
Activating autophagy can inhibit IR in HepG2 cells via P62/NF-κB/GLUT-4
To assess whether activating autophagy can inhibit IR in the cell model, the effects of activating or inhibiting autophagy were examined using rapamycin and 3-MA. As shown in Fig. 4A, compared with the IR group, the protein expression levels of LC3II and GLUT-4 were upregulated in the rapamycin treated HepG2 cells, whereas the protein expression levels of P62 were decreased. However, 3-MA decreased the expression levels of LC3II and GLUT-4, and increased the expression of P62 in the IR HepG2 cells, whereas the protein expression levels of P62 decreased. Rapamycin also promoted the mRNA expression of GLUT-4 (P<0.01; Fig. 4B). These histopathological changes were accompanied by increased and decreased glucose content in the culture supernatant. As shown in Fig. 4C, rapamycin decreased glucose content, whereas 3-MA increased glucose content in the culture supernatant, respectively (P<0.001, rapamycin vs. IR; P<0.01, 3-MA vs. IR).
Figure 4.
Activating autophagy can inhibit IR in HepG2 cells via P62/NF-κB/GLUT-4. (A) Western blot analysis for P62, LC3, p-P65 and GLUT-4 in HepG2 cells. (B) mRNA expression of GLUT-4 in HepG2 cells treated with IR, Rap and 3-MA were detected using semi-quantitative polymerase chain reaction analysis. (C) Measurements of glucose content in culture supernatants of HepG2 cells treated with IR, Rap and 3-MA. *P<0.05, **P<0.01 and ***P<0.001 vs. IR group. IR, insulin resistance; LC3, GLUT-4, glucose transporter 4; Rap, rapamycin; 3-MA, 3-methyladenine; p-, phosphorylated.
Discussion
IR is a condition, which can lead to type 2 diabetes (1), however, no specific treatment has been identified. The present study investigated the effect of geniposide, an iridoid compound derived from the fruit of G. jasminoides Ellis, which can be hepatotoxic, in a HepG2 cell model of IR. Geniposide significantly improved IR, which may have been associated with its dynamic regulation of the expression of NF-κB and GLUT-4. The peroxisome proliferator-activated receptor-γ (PPARγ) receptor is an important element responsible for regulating blood sugar by mediating the effects of insulin on glucose uptake (16). The downregulation of PPARγ contributes to IR (17). Using PPARγ to investigate the inducible expression of signalling pathway reporter genes, the hypoglycaemic effect of geniposide in vivo and in vitro has been suggested to be associated with PPARγ receptor activation (18). However, Sriwijitkamol et al (19) reported that the NF-κB pathway was overactivated in the muscular tissue of subjects with type 2 diabetes. In animal studies, inhibition of NF-κB β/NF-κB pathway inhibition significantly improved insulin sensitivity (20). In the present study, the expression of NF-κB was downregulated following geniposide treatment. It has been reported that NF-κB downregulates the expression of GLUT-4 (21) and that genipin, a geniposide metabolite, inhibits NF-κB (22). Therefore, it is possible that geniposide treatment reversed the inhibitory effect of NF-κB on the expression of GLUT-4. Noguchi et al (23) reported that IR is caused partly by the downregulation of GLUT-4, although it is also associated with impaired GLUT-4 translocation to the plasma membrane under insulin stimulation. The results of the present study suggested that geniposide improved the expression of GLUT-4, an important downstream insulin receptor site.
A product of the G. jasminoides Ellis fruit, geniposide, has been reported to cause hepatotoxicity in rats (24). However, in the present study, geniposide did not affect HepG2 cell viability, indicating that the concentration used (62.50 µg/ml) may be safe for use in animals and subsequently in humans. Liu et al (25) also reported that geniposide had a protective effect on insulin secretion in rat INS-1 cells, which indicated that it may also have a protective effect against IR in humans.
In the present study, the expression levels of GLUT-4 decreased over time following a peak at 4 h. At its lowest point, 24 h following geniposide treatment, the mRNA expression of GLUT-4 remained higher than that in the control at the same time point. Of note, the peak protein expression of GLUT-4 did not exceed that of the control at the same time point, whereas the nadir of the protein expression of GLUT-4 was lower, compared with that of the control. Similarly, Leguisamo et al (17) reported lower protein levels of GLUT-4 in induced IR in spontaneously hypertensive neonate rats, although the mRNA levels of GLUT-4 were not investigated.
Autophagy is a highly conserved intracellular lysosomal catabolic process, which degrades aged, damaged or dysfunctional proteins, intracellular organelles and cytoplasmic components to maintain cellular homeostasis (26). The basal level of autophagy has a unique housekeeping role in the regulation of cardiac geometry and function (27). Impaired autophagy may contribute to various end organ complications in IR and diabetes, including cardiomyopathy and nephropathy (28).
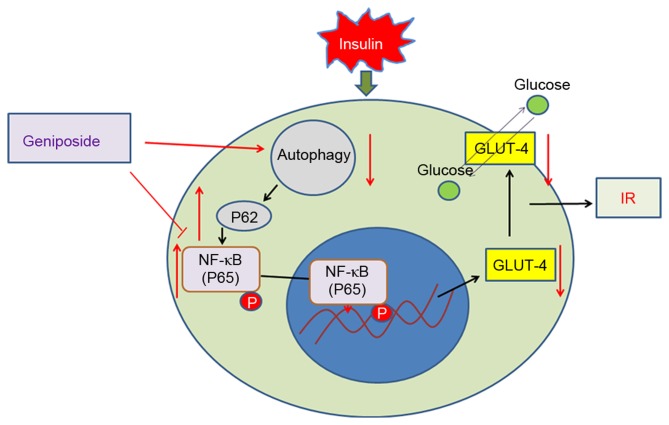
In the present study, it was shown that the protein expression levels of LC3II were downregulated in IR HepG2 cells. IR decreased autophagic activity, as evidenced by the decrease in the expression of LC3 and the accumulation of P62 protein. These results suggested that the level of autophagy can be altered by the duration of exposure to high insulin, which also indicates the putative role of autophagy under IR conditions. There was also a significant increase in the expression of LC3II in the geniposide-treated HepG2 cells, compared with that in the IR group. By contrast, the expression levels of P62 increased in the IR HepG2 cells, whereas geniposide decreased the expression of P62 in the IR cells. Geniposide promoted autophagy in the IR HepG2 cells, and activating autophagy inhibited IR in HepG2 cells via P62/NF-κB/GLUT-4 (Fig. 5).
Figure 5.
Model of geniposide promotion of autophagy to inhibit IR in HepG2 cells via P62/NF-κB/GLUT-4. IR, insulin resistance; NF-κB, nuclear factor-κB; GLUT-4, glucose transporter.
Taken together, the findings of the present study indicated that geniposide may be a viable method for treating IR, a condition, which can escalate to type 2 diabetes but for which no specific treatment has been identified. This requires further attention, and future investigations are required with a focus on characterising geniposide.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (grant nos. 81141059 and U1404805).
References
- 1.Lillioja S, Mott DM, Spraul M, Ferraro R, Foley JE, Ravussin E, Knowler WC, Bennett PH, Bogardus C. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of pima indians. N Engl J Med. 1993;329:1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 2.Kadowaki T. Insights into insulin resistance and type 2 diabetes from knockout mouse models. J Clin Invest. 2000;106:459–465. doi: 10.1172/JCI10830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yin XM, Ding WX, Gao W. Autophagy in the liver. Hepatology. 2008;47:1773–1785. doi: 10.1002/hep.22146. [DOI] [PubMed] [Google Scholar]
- 4.Thapalia BA, Zhou Z, Lin X. Autophagy, a process within reperfusion injury: An update. Int J Clin Exp Pathol. 2014;7:8322–8341. [PMC free article] [PubMed] [Google Scholar]
- 5.Hashimoto S. Autophagy in the respiratory diseases. Respir Investig. 2016;54:383–384. doi: 10.1016/j.resinv.2016.10.002. [DOI] [PubMed] [Google Scholar]
- 6.Vucicevic L, Misirkic-Marjanovic M, Paunovic V, Kravic-Stevovic T, Martinovic T, Ciric D, Maric N, Petricevic S, Harhaji-Trajkovic L, Bumbasirevic V, Trajkovic V. Autophagy inhibition uncovers the neurotoxic action of the antipsychotic drug olanzapine. Autophagy. 2014;10:2362–2378. doi: 10.4161/15548627.2014.984270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rovira J, Ramirez-Bajo MJ, Banon-Maneus E, Moya-Rull D, Ventura-Aguiar P, Hierro-Garcia N, Lazo-Rodriguez M, Revuelta I, Torres A, Oppenheimer F, et al. mTOR inhibition: Reduced insulin secretion and sensitivity in a rat model of metabolic syndrome. Transplant Direct. 2016;2:e65. doi: 10.1097/TXD.0000000000000576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Q, Ren J. mTOR-Independent autophagy inducer trehalose rescues against insulin resistance-induced myocardial contractile anomalies: Role of p38 MAPK and Foxo1. Pharmacol Res. 2016;111:357–373. doi: 10.1016/j.phrs.2016.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sarparanta J, Garcia-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev. 2016 doi: 10.2174/1573399812666160217122530. [DOI] [PubMed] [Google Scholar]
- 10.Roccisana J, Sadler JB, Bryant NJ, Gould GW. Sorting of GLUT4 into its insulin-sensitive store requires the Sec1/Munc18 protein mVps45. Mol Biol Cell. 2013;24:2389–2397. doi: 10.1091/mbc.E13-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaster M, Staehr P, Beck-Nielsen H, Schrøder HD, Handberg A. GLUT4 is reduced in slow muscle fibers of type 2 diabetic patients: Is insulin resistance in type 2 diabetes a slow, type 1 fiber disease? Diabetes. 2001;50:1324–1329. doi: 10.2337/diabetes.50.6.1324. [DOI] [PubMed] [Google Scholar]
- 12.Schläfli AM, Adams O, Galván JA, Gugger M, Savic S, Bubendorf L, Schmid RA, Becker KF, Tschan MP, Langer R, Berezowska S. Prognostic value of the autophagy markers LC3 and p62/SQSTM1 in early-stage non-small cell lung cancer. Oncotarget. 2016;7:39544–39555. doi: 10.18632/oncotarget.9647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xi G, Shen X, Wai C, Vilas CK, Clemmons DR. Hyperglycemia stimulates p62/PKCζ interaction, which mediates NF-κB activation, increased Nox4 expression, and inflammatory cytokine activation in vascular smooth muscle. FASEB J. 2015;29:4772–4782. doi: 10.1096/fj.15-275453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo LX, Xia ZN, Gao X, Yin F, Liu JH. Glucagon-like peptide 1 receptor plays a critical role in geniposide-regulated insulin secretion in INS-1 cells. Acta Pharmacol Sin. 2012;33:237–241. doi: 10.1038/aps.2011.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang WY, Lee JJ, Kim Y, Kim IS, Park JS, Myung CS. Amelioration of insulin resistance by scopoletin in high-glucose-induced, insulin-resistant HepG2 cells. Horm Metab Res. 2010;42:930–935. doi: 10.1055/s-0030-1265219. [DOI] [PubMed] [Google Scholar]
- 16.Kim HI, Ahn YH. Role of peroxisome proliferator-activated receptor-gamma in the glucose-sensing apparatus of liver and beta-cells. Diabetes. 2004;53(Suppl 1):S60–S65. doi: 10.2337/diabetes.53.2007.S60. [DOI] [PubMed] [Google Scholar]
- 17.Leguisamo NM, Lehnen AM, Machado UF, Okamoto MM, Markoski MM, Pinto GH, Schaan BD. GLUT4 content decreases along with insulin resistance and high levels of inflammatory markers in rats with metabolic syndrome. Cardiovasc Diabetol. 2012;11:100. doi: 10.1186/1475-2840-11-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y, Xia Z, Liu J, Yin F. Cell signaling mechanisms by which geniposide regulates insulin-degrading enzyme expression in primary cortical neurons. CNS Neurol Disord Drug Targets. 2015;14:370–377. doi: 10.2174/1871527314666141229110156. [DOI] [PubMed] [Google Scholar]
- 19.Sriwijitkamol A, Christ-Roberts C, Berria R, Eagan P, Pratipanawatr T, DeFronzo RA, Mandarino LJ, Musi N. Reduced skeletal muscle inhibitor of kappaB beta content is associated with insulin resistance in subjects with type 2 diabetes: Reversal by exercise training. Diabetes. 2006;55:760–767. doi: 10.2337/diabetes.55.03.06.db05-0677. [DOI] [PubMed] [Google Scholar]
- 20.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med. 2005;11:183–190. doi: 10.1038/nm1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hommelberg PP, Plat J, Remels AH, van Essen AL, Kelders MC, Mensink RP, Schols AM, Langen RC. Trans-10, cis-12 conjugated linoleic acid inhibits skeletal muscle differentiation and GLUT4 expression independently from NF-κB activation. Mol Nutr Food Res. 2010;54:1763–1772. doi: 10.1002/mnfr.201000103. [DOI] [PubMed] [Google Scholar]
- 22.Koo HJ, Song YS, Kim HJ, Lee YH, Hong SM, Kim SJ, Kim BC, Jin C, Lim CJ, Park EH. Antiinflammatory effects of genipin, an active principle of gardenia. Eur J Pharmacol. 2004;495:201–208. doi: 10.1016/j.ejphar.2004.05.031. [DOI] [PubMed] [Google Scholar]
- 23.Noguchi Y, Yoshikawa T, Marat D, Doi C, Makino T, Fukuzawa K, Tsuburaya A, Satoh S, Ito T, Mitsuse S. Insulin resistance in cancer patients is associated with enhanced tumor necrosis factor-alpha expression in skeletal muscle. Biochem Biophys Res Commun. 1998;253:887–892. doi: 10.1006/bbrc.1998.9794. [DOI] [PubMed] [Google Scholar]
- 24.Ding Y, Zhang T, Tao JS, Zhang LY, Shi JR, Ji G. Potential hepatotoxicity of geniposide, the major iridoid glycoside in dried ripe fruits of Gardenia jasminoides (Zhi-zi) Nat Prod Res. 2013;27:929–933. doi: 10.1080/14786419.2012.673604. [DOI] [PubMed] [Google Scholar]
- 25.Liu J, Guo L, Yin F, Zhang Y, Liu Z, Wang Y. Geniposide regulates glucose-stimulated insulin secretion possibly through controlling glucose metabolism in INS-1 cells. PLoS One. 2013;8:e78315. doi: 10.1371/journal.pone.0078315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cicchini M, Chakrabarti R, Kongara S, Price S, Nahar R, Lozy F, Zhong H, Vazquez A, Kang Y, Karantza V. Autophagy regulator BECN1 suppresses mammary tumorigenesis driven by WNT1 activation and following parity. Autophagy. 2014;10:2036–2052. doi: 10.4161/auto.34398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, Omiya S, Mizote I, Matsumura Y, Asahi M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007;13:619–624. doi: 10.1038/nm1574. [DOI] [PubMed] [Google Scholar]
- 28.Nemchenko A, Chiong M, Turer A, Lavandero S, Hill JA. Autophagy as a therapeutic target in cardiovascular disease. J Mol Cell Cardiol. 2011;51:584–593. doi: 10.1016/j.yjmcc.2011.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]