Abstract
Ulcerative colitis (UC) is a chronic inflammatory bowel disease that is associated with both genetic and environmental factors; however, the underlying pathogenesis of UC remains unclear. The present study aimed to further explore 12 microarray datasets from patients with UC obtained from the Gene Expression Omnibus repository, for potential genetic pathogenesis of UC through a global bioinformatics view, which included identification of differentially expressed genes (DEGs), functional enrichments, protein-protein interactions, transcriptional and post-transcriptional regulation and drug-gene associations. This integrated analysis screened 233 DEGs that were compared between UC and normal control tissue samples; these included 173 upregulated and 60 downregulated DEGs. Subsequently, transcription factors, such as TATA-binding protein 1 (TBP1; hsa_TATAAA_V$TATA_01) and nuclear factor-κB (NF-κB; hsa_V$NFKAPPAB_01) and microRNAs (miRNAs; such as miR-516-3p and miR-23a) were revealed to be associated with 233 DEGs. Notably, further analysis indicated that these DEGs were enriched in certain diseases, including inflammation, fibrosis and immune system diseases, and were also associated with some drugs, including prednisone, collagenase and mycophenolate mofetil, which may provide choice for treatment of UC. In conclusion, this study may provide novel insights into discovering potential molecular targets involved in the pathogenesis and treatment of UC.
Keywords: ulcerative colitis, integrated analysis, bioinformatics, differentially expressed genes, pathogenesis
Introduction
Ulcerative colitis (UC) is one of 2 major phenotypes of inflammatory bowel disease (IBD), whose etiology is associated with a complex interplay between genetics, the immune system and environmental factors such as enteric microflora (1,2). UC is associated with chronic colonic inflammation and damage to the mucosal surface, which commonly induces hematochezia and abdominal pain (1,2). The incidence of UC has been recorded as 9 to 20 cases per 100,000 people each year, and the prevalence rate as 156 to 291 cases per 100,000 people in both North America and Northern Europe (2). UC is clinically characterized by alternating periods of remission and relapse. Colectomy rates associated with postoperative morbidity and mortality, and the increasing incidence of colorectal cancer in the chronic intermittent course of UC must be taken into account when considering treatments (3). To date, the best treatment strategies for UC includes the early use of biological agents such as infliximab, more frequent monitoring when therapy is initiated and maintenance care; these have increased the number of patients with UC who have achieved clinical remission and even endoscopic remission (4). In addition, important candidate genes and causal genetic variants of UC, including ring finger protein 186, caspase recruitment domain family member 9 and PR/SET domain 1, have been identified by a genome-wide association study (5). However, the underlying pathogenesis and pivotal molecular interactions of UC require further exploration.
Recently, the roles of several key molecules, such as prostaglandin-endoperoxide synthase 2 (PTGS2; also known as COX-2), interleukin-23 receptor, mucin-2 and C-X-C chemokine receptor type 3, have been examined in UC to some extent (6–9). However, the pathogenesis of UC involves a number of biological processes, such as gene expression alteration, and transcriptional and post-transcriptional dysregulation. Studies of the molecular interactions based on differentially expressed genes (DEGs) by hypergeometric method are popular and provide a certain level of confidence for exploration of UC pathogenesis. Therefore, the present study used an integrated analysis approach that combined gene expression profiling, functional enrichments, protein-protein interactions (PPIs), transcriptional and post-transcriptional regulation, and drug-gene associations to identify biological molecular interactions in UC. This study may provide novel insight into the molecular targets involved in the pathogenesis and treatment of UC.
Materials and methods
Data resources
A total of 12 microarray data sets were downloaded from the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo), which included GSE3365 (10), GSE36807 (11), GSE37283 (12), GSE38713 (13), GSE42911, GSE47908 (14), GSE48634 (15), GSE48959 (16), GSE59071 (17), GSE6731 (18), GSE71730 (19), and GSE9452 (20) (Table I). The microarray data sets were selected according to following criteria: i) Expression profiling by array contain that compare samples from patients with UC and normal group; ii) samples in the UC group were obtained from patients whom did not receive any interventions or treatments; and iii) DEGs were statistically significantly different between the UC group and the normal control group, with a threshold of P<0.05 indicating a statistically significant difference and |log2(FC)|>1, where FC is fold change.
Table I.
Characteristics of the expression profiling microarray data sets analyzed in this study.
| Ulcerative colitis | Normal controls | |||||||
|---|---|---|---|---|---|---|---|---|
| GEO accession | Country | Sample source | Age (year, mean) | Sex (M/F) | n | Age (year, mean) | Sex (M/F) | n |
| GSE3365 | USA | PBMCs | 46.7 | 8/18 | 26 | 44.1 | 24/18 | 42 |
| GSE36807 | UK | Colon mucosa | N/A | 7/8 | 15 | N/A | N/A | 7 |
| GSE37283 | USA | Colon mucosa | N/A | N/A | 15 | N/A | N/A | 5 |
| GSE38713 | Spain | Colon mucosa | 42.4 | 7/16 | 23 | 41.6 | 5/8 | 13 |
| GSE42911 | USA | Colon mucosa | 11.3 | 2/2 | 4 | 13.8 | 2/4 | 6 |
| GSE47908 | Denmark | Colon mucosa | N/A | N/A | 45 | N/A | N/A | 15 |
| GSE48634 | UK | Colon mucosa | 48.3 | 16/8 | 24 | 45.3 | 15/11 | 26 |
| GSE48959 | Belgium | Colon mucosa | 55.3 | 13/14 | 27 | 57.7 | 5/5 | 10 |
| GSE59071 | Belgium | Colon mucosa | 44.9 | 55/42 | 97 | 69.6 | 5/6 | 11 |
| GSE6731 | USA | Colon mucosa | N/A | N/A | 8 | N/A | N/A | 4 |
| GSE71730 | USA | PBMCs | 12 | 11/4 | 15 | 12 | 1/9 | 10 |
| GSE9452 | Denmark | Colon mucosa | 46 | 2/6 | 8 | 35 | 4/14 | 18 |
PBMC, peripheral blood mononuclear cell; F, female; M, male; N/A, not available online; GEO, Gene Expression Omnibus.
Data set preprocessing and DEG identification
Expression profiling microarrays were preprocessed by background correction and normalization, followed by statistical analysis to identify the DEGs between the UC group and the control group, which was conducted using the limma package in R (http://www.bioconductor.org/packages/release/bioc/html/limma.html) (21), a commonly used bioinformatics method to identify DEGs. Each DEG had to meet both P<0.05 and |log2(FC)| >1. The identified DEGs from each data set were overlapped for further bioinformatics analysis.
Functional enrichment and analysis of DEGs
Functional enrichment was classified into two groups, Gene ontology (GO) function annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis. The Database for Annotation, Visualization and Integrated Discovery (DAVID; version 6.7; www.david.ncifcrf.gov) provides a comprehensive set of GO function annotation and pathway enrichment analysis to understand the biological meanings of large lists of genes (22). In this study, DAVID was used to explore the functional enrichments of DEGs including GO and KEGG. P<0.05 indicated significant functional annotations and pathways.
Construction and visualization of PPI networks among DEGs
PPIs are reflected as interactions between DEGs, which indicate upstream and/or downstream associations, including direct (physical) and indirect (functional) ones. In the present study, the online server Search Tool for the Retrieval of Interacting Genes (STRING; version 10; www.string-db.org/) was used to explore these interactions (23). DEG pairs were identified by a combined score of >0.4, which was used to evaluate the level of interactions between DEGs with the following scoring system: i) <0.4, low confidence; ii) 0.4 to 0.7, medium; iii) >0.7, high (24). These DEGs were subsequently chosen to construct PPI networks, which were further visualized with Cytoscape v3.4.0 software used for visualizing bimolecular interaction networks (25).
Regulation and drug-gene associations of DEGs
The biological function of a gene is regulated at the transcriptional and post-transcriptional level by transcription factors (TFs) and micro RNAs (miRNAs), respectively. Genes associated with disease, drug-gene associations, TF targets and miRNA targets of DEGs were predicted using the Web-based Gene Set Analysis Toolkit (WebGestalt 2013; www.webgestalt.org/webgestalt_2013/) (26). The present study explored these enrichment analyses using a hypergeometric statistical method and the top 10 results are presented based on significance level (P<0.05). The regulatory networks were constructed and visualized by Cytoscape v3.4.0 software (25).
Results
Microarray data sets and identification of DEGs
A total of 12 expression data sets were used in the present study, which comprised 307 samples from patients with UC and 167 samples from healthy control patients; all patients were from Western populations (Table I). Following screening and overlapping of data sets, 233 DEGs were identified, including 173 upregulated and 60 downregulated DEGs. The DEGs were revealed to be associated with a number of diseases, such as inflammation, infection, fibrosis, chronic disease, immune system diseases, necrosis, respiratory tract infections, virus diseases, rupture and bronchiolitis (Table II).
Table II.
Top 10 diseases associated with the identified differentially expressed genes.
| Disease | Gene count | Genes | P-value |
|---|---|---|---|
| Inflammation | 48 | MMP9, LCN2, CXCL3, SELL, S100A8, CXCL2, CD274, PECAM1, IL1B, TNC, ICAM1, NAMPT, CXCL9, IDO1, IL33, SPP1, PLAU, TIMP1, IL1RN, S100A9, CASP1, MMP12, CXCL13, CCL20, FPR1, CXCL6, MMP1, PI3, ANXA1, PLA2G2A, SOCS3, ENTPD1, CXCL10, PTGS2, CXCL11, SERPINA1, MMP3, GBP1, STAT1, CXCL1, OSMR, NFKBIZ, LY96, TNIP3, IL13RA2, NOS2, CCL11 and CHI3L1 | 1.58×10−48 |
| Infection | 39 | CCL19, IFI44, SELL, CXCL2, TRIM22, CD274, IL1B, GZMK, ICAM1, IFITM1, CXCL9, IDO1, DMBT1, DEFA5, IL1RN, S100A9, CASP1, APOL1, CD55, CXCL13, IFITM3, CCL20, DEFA6, CR2, IFIT3, PSMB9, FCGR3B, PI3, FCGR3A, SOCS3, PTPRC, HLA-DMA, PTGS2, CXCL10, STAT1, CXCL1, LY96, NOS2 and CCL11 | 5.52×10−33 |
| Fibrosis | 27 | MMP9, GREM1, SERPINB3, COL1A1, MMP1, S100A8, LOXL2, TNC, PI3, CXCL9, THY1, SLC26A3, CXCL10, IL33, SPP1, MMP7, SERPINA1, MMP3, TIMP1, SPARC, TGM2, COL1A2, S100A9, ENPP2, IL13RA2, NOS2 and CHI3L1 | 5.67×10−31 |
| Chronic disease | 31 | MMP9, UGT1A1, MMP12, SERPINB3, LCN2, MGP, MMP1, CD274, RHOH, FCGR3B, IL1B, NCF2, ICAM1, MS4A1, FCGR3A, ADH1C, NAMPT, CXCL9, ENTPD1, CXCL10, PTGS2, SPP1, MMP7, SERPINA1, MMP3, TIMP1, IL1RN, S100A9, NOS2, APOL1 and CHI3L1 | 3.37×10−30 |
| Immune system diseases | 37 | CDKN2B, CCL19, SELL, CD274, IL1B, NCF2, ICAM1, IGHM, CXCL9, IDO1, IL33, CD79A, SPP1, IL1RN, DPP10, POU2AF1, CXCL13, BIRC3, CCL20, DEFA6, PADI2, CR2, PRDM1, RHOH, FCGR3B, FCGR3A, MS4A1, PTPRC, HLA-DMA, CXCL10, CXCL11, BCL2A1, ABCB1, CCL11, NOS2, CHI3L1 and TRIM29 | 3.22×10−26 |
| Necrosis | 28 | MMP9, BIRC3, CCL20, CXCL3, MMP1, CXCL2, IL1B, ICAM1, FCGR3A, PI3, SOCS3, NAMPT, IDO1, CXCL10, IL33, UBD, MMP3, BCL2A1, STAT1, CXCL1, NFKBIZ, LY96, IL1RN, STEAP4, CASP1, TNIP3, NOS2 and CCL11 | 1.85×10−25 |
| Respiratory tract infections | 24 | MMP9, DPP10, CXCL13, CCL19, IGKC, IFITM3, CXCL3, CXCL6, CD274, PECAM1, IL1B, NCF2, ICAM1, CXCL9, PTGS2, CXCL10, SERPINA1, CXCL1, IL1RN, S100A9, CASP1, NOS2, IL13RA2 and CCL11 | 2.64×10−23 |
| Virus diseases | 30 | CXCL13, CD55, CCL19, CCL20, CXCL3, DEFA6, IFI44, SELL, CR2, TRIM22, CD274, PSMB9, IL1B, ICAM1, FCGR3A, IFITM1, SOCS3, PTPRC, CXCL9, HLA-DMA, IDO1, CXCL10, CXCL11, DEFA5, GBP1, ABCB1, STAT1, CXCL1, CASP1 and APOL1 | 6.81×10−23 |
| Rupture | 20 | MMP9, MMP12, MMP10, COL1A1, MGP, MMP1, IL1B, TNC, PI3, COL12A1, MMP7, DEFA5, MMP3, TIMP1, PLAU, VWF, IL1RN, COL1A2, IL13RA2 and CHI3L1 | 2.04×10−22 |
| Bronchiolitis | 22 | DPP10, CXCL13, CCL19, CXCL3, CXCL6, SELL, CXCL2, PECAM1, NCF2, ICAM1, SOCS3, CXCL9, IDO1, PTGS2, MMP7, STAT1, CXCL1, IL1RN, CASP1, IL13RA2, NOS2 and CCL11 | 9.64×10−22 |
Genes presented in bold and italics indicate a gene of interest used as an example in text: COL1A2, collagen type I α1.
Functional enrichment and analysis of DEGs
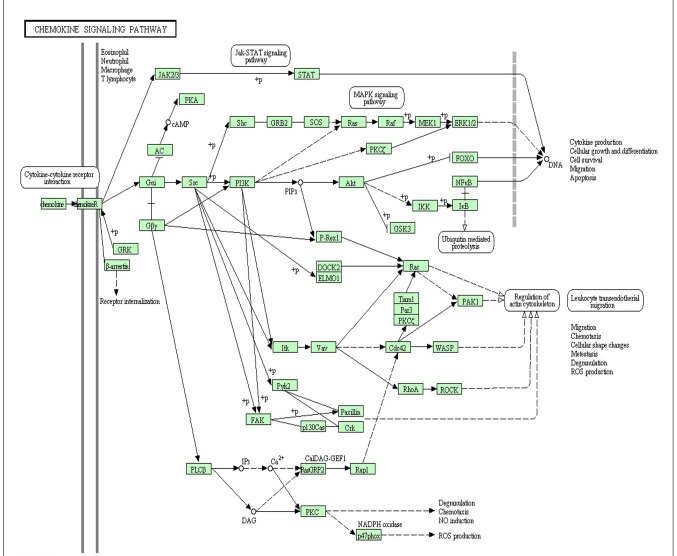
Functional analyses demonstrated that 185 GO functions (P<0.05) and 11 KEGG pathways (P<0.05) were significantly enriched by the upregulated DEGs, whereas 12 GO functions (P<0.05) and 6 KEGG pathways (P<0.05) were significantly enriched by the downregulated DEGs. Top 5 GO terms and top 5 KEGG pathways of the upregulated and downregulated DEGS are provided in Tables III and IV, respectively. Notably, the chemokine signaling pathway was enriched by 13 DEGs (Fig. 1), which has been previously verified as important pathway in UC (9,27,28).
Table III.
Top 5 GO functions of upregulated and downregulated differentially expressed genes.
| Category | GO ID | Term | Gene count | P-value |
|---|---|---|---|---|
| Up | GO:0006955 | Immune response | 49 | 1.18×10−25 |
| Up | GO:0006952 | Defense response | 41 | 3.55×10−20 |
| Up | GO:0006954 | Inflammatory response | 31 | 2.28×10−19 |
| Up | GO:0009611 | Response to wounding | 37 | 9.35×10−19 |
| Up | GO:0006935 | Chemotaxis | 17 | 2.95×10−11 |
| Down | GO:0055085 | Transmembrane transport | 8 | 9.09×10−04 |
| Down | GO:0042493 | Response to drug | 5 | 3.05×10−03 |
| Down | GO:0007584 | Response to nutrient | 4 | 7.02×10−03 |
| Down | GO:0031667 | Response to nutrient levels | 4 | 1.765×10−02 |
| Down | GO:0009725 | Response to hormone stimulus | 5 | 1.902×10−02 |
GO, gene ontology.
Table IV.
Top 5 Kyoto Encyclopedia of Genes and Genomes pathways of upregulated and downregulated differentially expressed genes.
| Category | Term | Gene count | P-value |
|---|---|---|---|
| Up | hsa04610:Complement and coagulation cascades | 9 | 1.59×10−05 |
| Up | hsa04062:Chemokine signaling pathway | 13 | 5.37×10−05 |
| Up | hsa04512:ECM-receptor interaction | 8 | 4.55×10−04 |
| Up | hsa04060:Cytokine-cytokine receptor interaction | 13 | 1.231×10−03 |
| Up | hsa04621:NOD-like receptor signaling pathway | 6 | 3.553×10−03 |
| Down | hsa00910:Nitrogen metabolism | 3 | 2.866×10−03 |
| Down | hsa00830:Retinol metabolism | 3 | 1.519×10−02 |
| Down | hsa00980:Metabolism of xenobiotics by cytochrome P450 | 3 | 1.8555×10−02 |
| Down | hsa00982:Drug metabolism | 3 | 1.9741×10−02 |
| Down | hsa03320:PPAR signaling pathway | 3 | 2.4136×10−02 |
ECM, extracellular matrix; NOD, nucleotide-binding oligomerization domain; PPAR, peroxisome proliferator-activated receptor.
Figure 1.
Diagram of the chemokine signaling pathway significantly enriched in differentially expressed genes.
Construction of PPI networks associated with DEGs
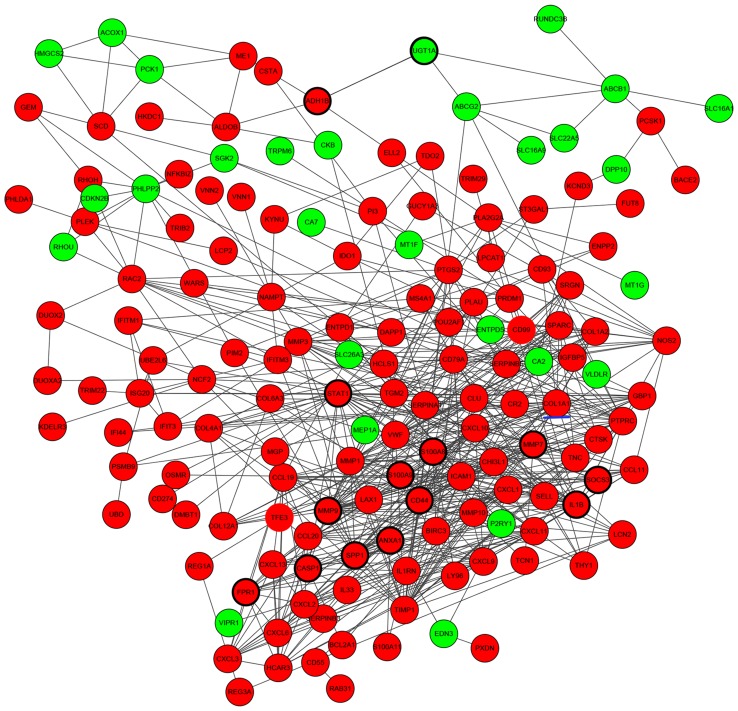
PPIs between DEGs were investigated by STRING, and a total of 608 protein-protein interactions were identified. Based on these protein-protein pairs, PPI networks were constructed involving 173 upregulated DEGs and 60 downregulated DEGs (Fig. 2). In this network, S100 calcium-binding protein A9 (S100A9), UDP-glucuronosyltransferase 1A (UGT1A), CD44, caspase 1 (CASP1), signal transducer and activator of transcription 1 (STAT1), secreted phosphoprotein 1 (SPP1), N-formyl peptide receptor 1 (FPR1), S100A8, interleukin 1β (IL1B), suppressor of cytokine signaling 3 (SOCS3), matrix metalloproteinase (MMP) 9, MMP7, annexin A1 (ANXA1) and alcohol dehydrogenase 1B (class I), β-polypeptide had higher node degrees and were defined as hub genes of PPI networks (Fig. 2).
Figure 2.
Predicted interaction network for DEGs in ulcerative colitis. Red nodes indicate upregulated genes and green nodes indicate downregulated genes. Lines indicate interactions between DEGs, genes circled by a black ellipse are the hub genes and the gene underlined in blue is a gene of interest. DEG, differentially expressed gene.
Regulatory networks of DEGs
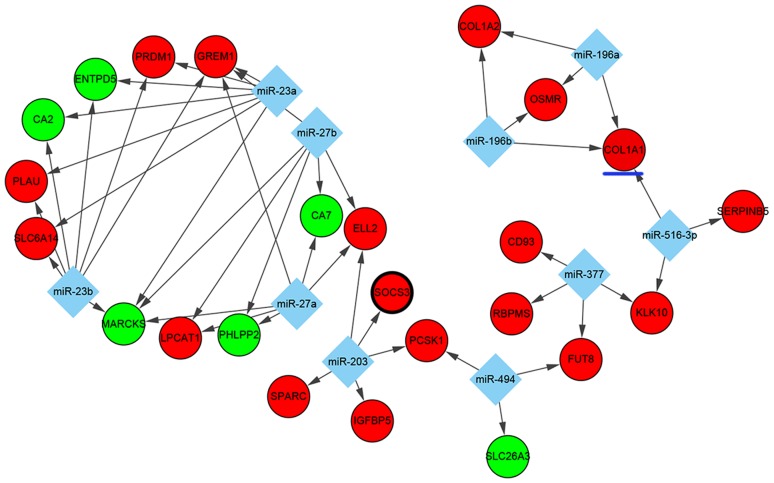
As for TF-targeted and miRNA-targeted DEGs, which serve pivotal roles in transcriptional and post-transcriptional regulation, a number of significant regulators of DEGs were identified in UC. Based on the WebGestalt data, the top 10 miRNAs (miR) including miR-516-3p, miR-203, miR-27a, miR-27b, miR-494, miR-196a, miR-196b, miR-23a, miR-23b and miR-377 were predicted to regulate some DEGs. In this interaction network, 10 miRNAs involving 47 miRNA-DEG regulatory pairs were constructed, which included 6 downregulated DEGs and 18 upregulated DEGs (Fig. 3). Among the DEGs, the expression of MARCKS and GREM1 were predicted to be co-regulated by miR-27a, miR-27b, miR-23a and miR-23b, whereas the overexpression of KLK10 was predicted to be co-regulated by miR-377 and miR-516-3p.
Figure 3.
Interaction network of the top 10 miRNAs-DEGs pairs. Red nodes indicate upregulated genes and green nodes indicate downregulated genes. Diamonds indicate miRNAs and lines represent interactions between miRNAs and DEGs. Genes circled by a black ellipse are the hub genes and the gene underlined in blue is of interest. DEG, differentially expressed gene; miRNA, microRNA.
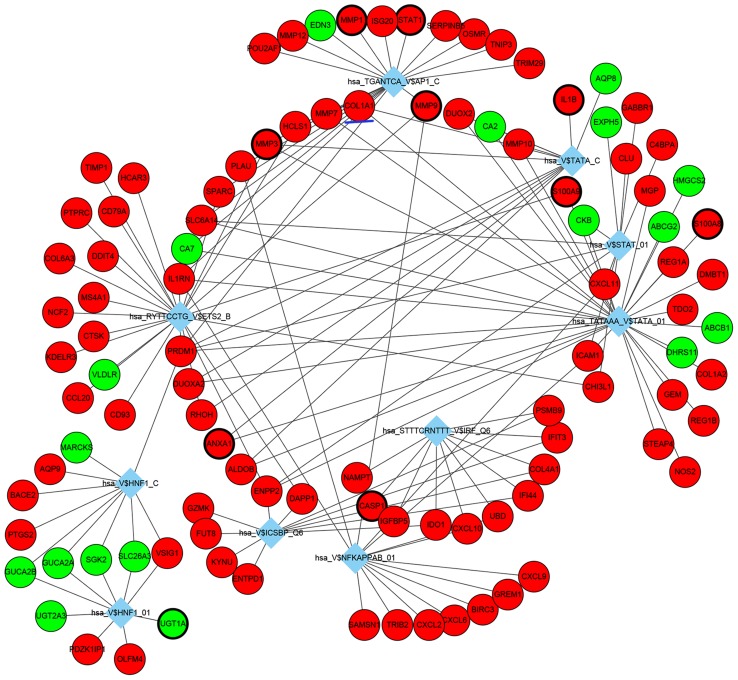
TF-DEG regulatory networks were also analyzed, the top 10 identified TF-DEG interactions are presented in Fig. 4. Among these identified TFs, TATA-binding protein 1 (TBP1; hsa_TATAAA_V$TATA_01), nuclear factor-κB (hsa_V$NFKAPPAB_01) and ETS proto-oncogene 2 (ETS2; hsa_RYTTCCTG_V$ETS2_B) exhibited the highest degree and were considered as important TFs in the present study, which may be potentially important in the underlying pathogenesis of UC.
Figure 4.
Interaction network of the top 10 TFs-DEGs pairs. Red nodes indicate upregulated genes and green nodes indicate downregulated genes. Diamonds indicate TFs and lines represent interactions between TFs and DEGs. Genes circled by a black ellipse are the hub genes and the gene underlined in blue is of interest. TFs, transcription factors; DEG, differentially expressed gene.
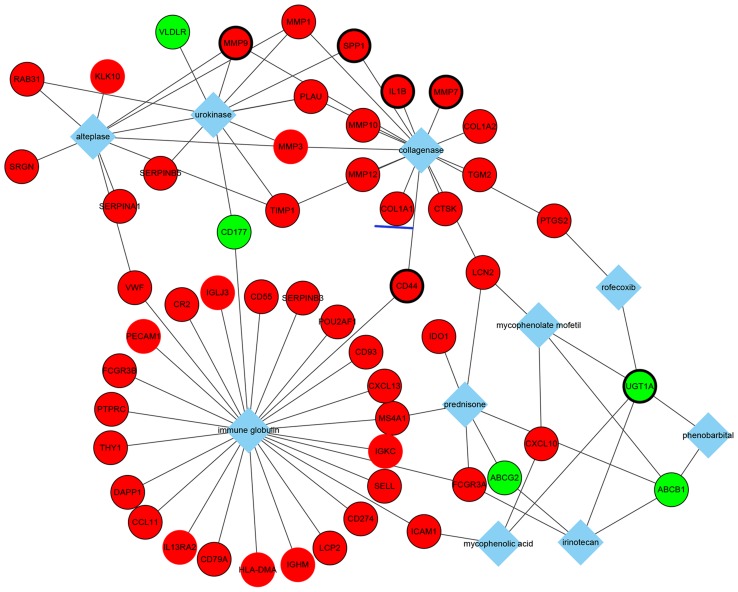
In addition, the top 10 drugs associated with DEGs were identified (Fig. 5). Notably, these drugs included mycophenolate mofetil and prednisone, which are immunosuppressive drugs used for UC treatment in clinical practice. These results may provide some considerations for choosing medicine therapy for patients with UC. Moreover, the results suggested that these two immunosuppressive drugs may be associated with corresponding DEGs, such as the common genes, lipocalin 2 and ATP-binding cassette, sub-family B (MDR/TAP), member 1 (Fig. 5).
Figure 5.
Interaction network of the top 10 drugs-DEGs pairs. Red nodes indicate upregulated genes and green nodes indicate downregulated genes. Diamonds indicate drugs and lines represent interactions between drugs and DEGs. Genes circled by a black ellipse are the hub genes and the gene underlined in blue is of interest. DEG, differentially expressed gene.
Overall, a number of molecular interactions were identified and predicted from DEGs. For example, collagen type I α1 (COL1A1) is an upregulated gene in patients with UC, and is associated with fibrosis and collagenase, which suggested that patients with UC with increased COL1A1 expression may be more likely to develop fibrosis and collagenase therapy may be effective in this group of patients (Fig. 5). COL1A1 is a profibrotic gene that was demonstrated to be co-regulated by three transcription factors (Fig. 4), including TBP1 (hsa_TATAAA_V$TATA_01), TBPC (hsa_V$TATA_C) and activator protein 1 (AP1; hsa_TGANTCA_V$AP1_C), and three miRNAs (Fig. 3), such as miR-196a, miR-196b and miR-516-3p. GO analysis revealed that COL1A1 may serve a role in responses to nutrient levels, extracellular stimuli, organic substances and hormone stimuli (Table V). Additionally, COL1A1 was significantly associated with the ECM-receptor interaction pathway and the focal adhesion pathway (Table VI).
Table V.
GO analysis of collagen type 1 α1.
| GO ID | Term | P-value | Fold enrichment | FDR |
|---|---|---|---|---|
| GO:0031667 | Response to nutrient levels | 1.25×10−07 | 5.723 | 2.10×10−04 |
| GO:0010033 | Response to organic substance | 3.12×10−07 | 2.932 | 5.25×10−04 |
| GO:0009991 | Response to extracellular stimulus | 5.22×10−07 | 5.124 | 8.78×10−04 |
| GO:0007584 | Response to nutrient | 6.70×10−07 | 6.543 | 1.13×10−03 |
| GO:0009719 | Response to endogenous stimulus | 1.74×10−05 | 3.305 | 2.93×10−02 |
| GO:0009725 | Response to hormone stimulus | 1.77×10−05 | 3.456 | 2.98×10−02 |
FDR, false discovery rate; GO, gene ontology.
Table VI.
Kyoto Encyclopedia of Genes and Genomes pathway analysis of collagen type 1 α1.
| Term | P-value | Fold enrichment | FDR |
|---|---|---|---|
| hsa04512: ECM-receptor interaction | 1.52×10−03 | 4.612 | 1.71×10−02 |
| hsa04510: Focal adhesion | 4.39×10−02 | 5.168 | 4.36×10−02 |
ECM, extracellular matrix; FDR, false discovery rate.
Discussion
Data from the present study indicated that numerous genes are differently expressed in UC tissue compared with normal control tissue. For a better understanding of these DEGs, the mechanisms of UC need to be identified via integrated bioinformatics methods. Hence, the present study established an integrated analysis based on a combination of gene expression profiling, PPIs, transcriptional and post-transcriptional regulation data and drug-gene associations. For example, compared with non-UC controls, S100A9 was demonstrated to have a significantly increased expression in UC tissue; increased S100A9 expression has been previously linked to colonic mucosal immune and defense responses (12,29–32). In addition, S100A9 expression was able to distinguish UC from diarrhea-predominant irritable bowel syndrome and correlated with the UC disease activity index (31). S100A9 was also reported to be upregulated in Crohn's disease (29). Similarly, the matrix metallopeptidase 9 (MMP9) gene was also upregulated in UC tissue in the present study. Notably, in a previous study of experimental UC (33), the increased activity of MMP9 was demonstrated to diminish significantly by treatment with mesalamine, which is an effective drug in patients with UC. ANXA1, a phospholipid-binding protein, exhibited increased expression when patients with UC were under antitumor necrosis factor (TNF)-α therapy (34). Furthermore, following administration of ANXA1 receptor antagonists, experimental colitis in TNF receptor 1−/− mice was exacerbated (35). ANXA1 may be able to stimulate epithelial cell migration for restitution and wound healing process (36). Another study revealed that ANXA1 expression was upregulated in intestinal epithelial cells of biopsies from patients with active UC when received medically induced remission (34). The increase of ANXA1 expression was diffuse and was associated with the degree of mucosal inflammation and the rate of cell turnover at any given time. These data suggested that ANXA1 expression serves a crucial role in the early recovery of colitis (34–37). In agreement with the results of previous studies (38–40), aquaporin 9 (AQP9), C-X-C motif chemokine ligand (CXCL) 2 and nicotinamide phosphoribosyltransferase (NAMPT) were previously revealed to be differently expressed and discriminated between patients with UC and healthy controls. In addition, the expressions of CXCL9 and S100A8 have been demonstrated to be upregulated in UC, which involved cytokine-cytokine receptor pathway dysregulation (39). High SOCS3 expression in intestinal epithelial cells of patients with UC in remission was reported to increase the risk of relapse by regulating IL-22-induced expression of deleted in malignant brain tumors 1 (DMBT1) and c-Myc, as well as proliferation and migration of intestinal epithelial cells (38).
Many of the upregulated genes were enriched in the GO subcategories of immune response and inflammatory response. Immune system homeostasis maintains the appropriate reactions to commensal bacteria and against pathogenic invasion; the pathogenesis of UC has been closely associated with immune disorders, which manifest as chronic nonspecific intestinal inflammation (41). A previous study identified AQP9, CXCL2 and NAMP expressions were associated with the inflammatory response (40). Hair and enhancer of split 1 was previously reported to promote the IL-22-mediated antimicrobial response and encoding antimicrobial peptides, such as regenerating islet-derived protein (REG) 1A, REG3A and REG3G which are involved in STAT3-dependent transcription in UC (42). These peptides were observed in the GO subcategory of ‘inflammatory response’ in the present study. The nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway and the Toll-like receptor (TLR) signaling pathway have both been previously reported to be associated with UC (43), and both were enriched in upregulated genes in this study. Numerous clinical and experimental studies have shown that the intestinal microbes are important for the initiation and progression of UC, and are involved in the activation of pattern recognition receptor signaling through members of the TLR-like and NOD-like families (44,45).
TFs and miRNAs are important regulators of the accuracy of gene expression at the stage of transcription and post-transcription, respectively. Therefore, TFs and miRNAs with their targets were used to construct an integrated regulation network in a cooperative manner in the present study. For example, COL1A1 was demonstrated to be associated with three TFs and three miRNAs. In addition, COL1A1 expression was previously reported to be increased in inflammation-driven intestinal fibrosis of UC (46), which was consistent with the predictions by the present study of the genes associated with the disease. Intestinal fibrosis is a common complication of IBDs, occurring in 5% of UC patients (47). Notably, COL1A1 expression in the present study also exhibited a significant relationship with collagenase in the analysis of genes associated with drug.
PTGS2 serves a key role in regulating the inflammatory response upon luminal pathogen stimulation through the Toll-like receptors. Carriers of PTGS2 polymorphisms were reported to have an increased risk of UC (48–50), which suggested that PTGS2 may be a better biomarker to diagnosis UC; and a previous study revealed that PTGS2 expression was significantly different between patients with UC-related neoplasm and UC controls (51). Data from the present study indicated that PTGS2 was regulated by miR-101 and TF hepatocyte nuclear factor 1 (HNF1; hsa_V$HNF1_C); however, further experimentation is required to confirm these relationships.
In conclusion, the present study used a novel integrated analysis method to predict molecular interactions that regulate DEGs in patients with UC. The integrated analysis identified a series of molecules such as TFs and miRNAs, which may serve an important role in the pathogenesis of UC. In addition, these findings may provide potential biomarkers for diagnosing UC, and target drugs were identified for providing more precise therapy according to relevant DEGs.
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Ordas I, Eckmann L, Talamini M, Baumgart DC, Sandborn WJ. Ulcerative colitis. Lancet. 2012;380:1606–1619. doi: 10.1016/S0140-6736(12)60150-0. [DOI] [PubMed] [Google Scholar]
- 3.Jess T, Loftus EV, Jr, Velayos FS, Harmsen WS, Zinsmeister AR, Smyrk TC, Schleck CD, Tremaine WJ, Melton LJ, III, Munkholm P, Sandborn WJ. Risk of intestinal cancer in inflammatory bowel disease: A population-based study from olmsted county, Minnesota. Gastroenterology. 2006;130:1039–1046. doi: 10.1053/j.gastro.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 4.Colombel JF, Narula N, Peyrin-Biroulet L. Management strategies to improve outcomes of patients with inflammatory bowel diseases. Gastroenterology. 2017;152:351–361.e5. doi: 10.1053/j.gastro.2016.09.046. [DOI] [PubMed] [Google Scholar]
- 5.Venema WT Uniken, Voskuil MD, Dijkstra G, Weersma RK, Festen EA. The genetic background of inflammatory bowel disease: From correlation to causality. J Pathol. 2017;241:146–158. doi: 10.1002/path.4817. [DOI] [PubMed] [Google Scholar]
- 6.Sivanesan D, Beauchamp C, Quinou C, Lee J, Lesage S, Chemtob S, Rioux JD, Michnick SW. IL23R (interleukin 23 receptor) variants protective against inflammatory bowel diseases (IBD) display loss of function due to impaired protein stability and intracellular trafficking. J Biol Chem. 2016;291:8673–8685. doi: 10.1074/jbc.M116.715870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alipour M, Zaidi D, Valcheva R, Jovel J, Martínez I, Sergi C, Walter J, Mason AL, Wong GK, Dieleman LA, et al. Mucosal barrier depletion and loss of bacterial diversity are primary abnormalities in paediatric ulcerative colitis. J Crohns Colitis. 2016;10:462–471. doi: 10.1093/ecco-jcc/jjv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spisni E, Valerii MC, De Fazio L, Cavazza E, Borsetti F, Sgromo A, Candela M, Centanni M, Rizello F, Strillacci A. Cyclooxygenase-2 silencing for the treatment of colitis: A combined in vivo strategy based on RNA interference and engineered Escherichia coli. Mol Ther. 2015;23:278–289. doi: 10.1038/mt.2014.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ostvik AE, Granlund AV, Bugge M, Nilsen NJ, Torp SH, Waldum HL, Damås JK, Espevik T, Sandvik AK. Enhanced expression of CXCL10 in inflammatory bowel disease: Potential role of mucosal Toll-like receptor 3 stimulation. Inflamm Bowel Dis. 2013;19:265–274. doi: 10.1002/ibd.23034. [DOI] [PubMed] [Google Scholar]
- 10.Burczynski ME, Peterson RL, Twine NC, Zuberek KA, Brodeur BJ, Casciotti L, Maganti V, Reddy PS, Strahs A, Immermann F, et al. Molecular classification of Crohn's disease and ulcerative colitis patients using transcriptional profiles in peripheral blood mononuclear cells. J Mol Diagn. 2006;8:51–61. doi: 10.2353/jmoldx.2006.050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montero-Meléndez T, Llor X, Garcia-Planella E, Perretti M, Suárez A. Identification of novel predictor classifiers for inflammatory bowel disease by gene expression profiling. PLoS One. 2013;8:e76235. doi: 10.1371/journal.pone.0076235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pekow J, Dougherty U, Huang Y, Gometz E, Nathanson J, Cohen G, Levy S, Kocherginsky M, Venu N, Westerhoff M, et al. Gene signature distinguishes patients with chronic ulcerative colitis harboring remote neoplastic lesions. Inflamm Bowel Dis. 2013;19:461–470. doi: 10.1097/MIB.0b013e3182802bac. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Planell N, Lozano JJ, Mora-Buch R, Masamunt MC, Jimeno M, Ordás I, Esteller M, Ricart E, Piqué JM, Panés J, Salas A. Transcriptional analysis of the intestinal mucosa of patients with ulcerative colitis in remission reveals lasting epithelial cell alterations. Gut. 2013;62:967–976. doi: 10.1136/gutjnl-2012-303333. [DOI] [PubMed] [Google Scholar]
- 14.Bjerrum JT, Nielsen OH, Riis LB, Pittet V, Mueller C, Rogler G, Olsen J. Transcriptional analysis of left-sided colitis, pancolitis, and ulcerative colitis-associated dysplasia. Inflamm Bowel Dis. 2014;20:2340–2352. doi: 10.1097/MIB.0000000000000235. [DOI] [PubMed] [Google Scholar]
- 15.Smith PJ, Levine AP, Dunne J, Guilhamon P, Turmaine M, Sewell GW, O'Shea NR, Vega R, Paterson JC, Oukrif D, et al. Mucosal transcriptomics implicates under expression of BRINP3 in the pathogenesis of ulcerative colitis. Inflamm Bowel Dis. 2014;20:1802–1812. doi: 10.1097/MIB.0000000000000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Goten J, Vanhove W, Lemaire K, Van Lommel L, Machiels K, Wollants WJ, De Preter V, De Hertogh G, Ferrante M, Van Assche G, et al. Integrated miRNA and mRNA expression profiling in inflamed colon of patients with ulcerative colitis. PLoS One. 2014;9:e116117. doi: 10.1371/journal.pone.0116117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanhove W, Peeters PM, Staelens D, Schraenen A, Van Der Goten J, Cleynen I, De Schepper S, Van Lommel L, Reynaert NL, Schuit F, et al. Strong upregulation of AIM2 and IFI16 inflammasomes in the Mucosa of patients with active inflammatory bowel disease. Inflamm Bowel Dis. 2015;21:2673–2682. doi: 10.1097/MIB.0000000000000535. [DOI] [PubMed] [Google Scholar]
- 18.Wu F, Dassopoulos T, Cope L, Maitra A, Brant SR, Harris ML, Bayless TM, Parmigiani G, Chakravarti S. Genome-wide gene expression differences in Crohn's disease and ulcerative colitis from endoscopic pinch biopsies: Insights into distinctive pathogenesis. Inflamm Bowel Dis. 2007;13:807–821. doi: 10.1002/ibd.20110. [DOI] [PubMed] [Google Scholar]
- 19.Gurram B, Salzman NH, Kaldunski ML, Jia S, Li BU, Stephens M, Sood MR, Hessner MJ. Plasma-induced signatures reveal an extracellular milieu possessing an immunoregulatory bias in treatment-naive paediatric inflammatory bowel disease. Clin Exp Immunol. 2016;184:36–49. doi: 10.1111/cei.12753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen J, Gerds TA, Seidelin JB, Csillag C, Bjerrum JT, Troelsen JT, Nielsen OH. Diagnosis of ulcerative colitis before onset of inflammation by multivariate modeling of genome-wide gene expression data. Inflamm Bowel Dis. 2009;15:1032–1038. doi: 10.1002/ibd.20879. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.da W Huang, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.von Mering C, Jensen LJ, Snel B, Hooper SD, Krupp M, Foglierini M, Jouffre N, Huynen MA, Bork P. STRING: Known and predicted protein-protein associations, integrated and transferred across organisms. Nucleic Acids Res. 2005;33(Database issue):D433–D437. doi: 10.1093/nar/gki005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): Update 2013. Nucleic Acids Res. 2013;41:W77–W83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Deuring J, Peppelenbosch MP, Kuipers EJ, de Haar C, van der Woude CJ. STAT1, STAT6 and adenosine 3′,5′-cyclic monophosphate (cAMP) signaling drive SOCS3 expression in inactive ulcerative colitis. Mol Med. 2012;18:1412–1419. doi: 10.2119/molmed.2012.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Christophi GP, Rong R, Holtzapple PG, Massa PT, Landas SK. Immune markers and differential signaling networks in ulcerative colitis and Crohn's disease. Inflamm Bowel Dis. 2012;18:2342–2356. doi: 10.1002/ibd.22957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holgersen K, Kutlu B, Fox B, Serikawa K, Lord J, Hansen AK, Holm TL. High-resolution gene expression profiling using RNA sequencing in patients with inflammatory bowel disease and in mouse models of colitis. J Crohn's Colitis. 2015;9:492–506. doi: 10.1093/ecco-jcc/jjv050. [DOI] [PubMed] [Google Scholar]
- 30.Harris RA, Nagy-Szakal D, Mir SA, Frank E, Szigeti R, Kaplan JL, Bronsky J, Opekun A, Ferry GD, Winter H, et al. DNA methylation-associated colonic mucosal immune and defense responses in treatment-naïve pediatric ulcerative colitis. Epigenetics. 2014;9:1131–1137. doi: 10.4161/epi.29446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shiotani A, Kusunoki H, Kimura Y, Ishii M, Imamura H, Tarumi K, Manabe N, Kamada T, Hata J, Haruma K. S100A expression and interleukin-10 polymorphisms are associated with ulcerative colitis and diarrhea predominant irritable bowel syndrome. Dig Dis Sci. 2013;58:2314–2323. doi: 10.1007/s10620-013-2677-y. [DOI] [PubMed] [Google Scholar]
- 32.Miao YL, Xiao YL, Du Y, Duan LP. Gene expression profiles in peripheral blood mononuclear cells of ulcerative colitis patients. World J Gastroenterol. 2013;19:3339–3346. doi: 10.3748/wjg.v19.i21.3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deng X, Tolstanova G, Khomenko T, Chen L, Tarnawski A, Szabo S, Sandor Z. Mesalamine restores angiogenic balance in experimental ulcerative colitis by reducing expression of endostatin and angiostatin: Novel molecular mechanism for therapeutic action of mesalamine. J Pharmacol Exp Ther. 2009;331:1071–1078. doi: 10.1124/jpet.109.158022. [DOI] [PubMed] [Google Scholar]
- 34.Vong L, Ferraz JG, Dufton N, Panaccione R, Beck PL, Sherman PM, Perretti M, Wallace JL. Up-regulation of Annexin-A1 and lipoxin A(4) in individuals with ulcerative colitis may promote mucosal homeostasis. PLoS One. 2012;7:e39244. doi: 10.1371/journal.pone.0039244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sena AA, Pedrotti LP, Barrios BE, Cejas H, Balderramo D, Diller A, Correa SG. Lack of TNFRI signaling enhances annexin A1 biological activity in intestinal inflammation. Biochem Pharmacol. 2015;98:422–431. doi: 10.1016/j.bcp.2015.09.009. [DOI] [PubMed] [Google Scholar]
- 36.Babbin BA, Lee WY, Parkos CA, Winfree LM, Akyildiz A, Perretti M, Nusrat A. Annexin I regulates SKCO-15 cell invasion by signaling through formyl peptide receptors. J Biol Chem. 2006;281:19588–19599. doi: 10.1074/jbc.M513025200. [DOI] [PubMed] [Google Scholar]
- 37.Leoni G, Alam A, Neumann PA, Lambeth JD, Cheng G, McCoy J, Hilgarth RS, Kundu K, Murthy N, Kusters D, et al. Annexin A1, formyl peptide receptor, and NOX1 orchestrate epithelial repair. J Clin Invest. 2013;123:443–454. doi: 10.1172/JCI65831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu AT, Li Y, Zhao D, Shen J, Xu XT, Qiao YQ, Zhu MM, Wang TR, Cui Y, Ai LY, et al. High suppressor of cytokine signaling-3 expression impairs STAT3-dependent protective effects of interleukin-22 in ulcerative colitis in remission. Inflamm Bowel Dis. 2015;21:241–250. doi: 10.1097/MIB.0000000000000267. [DOI] [PubMed] [Google Scholar]
- 39.Fang K, Grisham MB, Kevil CG. Application of comparative transcriptional genomics to identify molecular targets for pediatric IBD. Front Immunol. 2015;6:165. doi: 10.3389/fimmu.2015.00165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mesko B, Poliska S, Szegedi A, Szekanecz Z, Palatka K, Papp M, Nagy L. Peripheral blood gene expression patterns discriminate among chronic inflammatory diseases and healthy controls and identify novel targets. BMC Med Genomics. 2010;3:15. doi: 10.1186/1755-8794-3-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang Y, Chen Z. Inflammatory bowel disease related innate immunity and adaptive immunity. Am J Transl Res. 2016;8:2490–2497. [PMC free article] [PubMed] [Google Scholar]
- 42.Murano T, Okamoto R, Ito G, Nakata T, Hibiya S, Shimizu H, Fujii S, Kano Y, Mizutani T, Yui S, et al. Hes1 promotes the IL-22-mediated antimicrobial response by enhancing STAT3-dependent transcription in human intestinal epithelial cells. Biochem Biophys Res Commun. 2014;443:840–846. doi: 10.1016/j.bbrc.2013.12.061. [DOI] [PubMed] [Google Scholar]
- 43.Clavel T, Haller D. Bacteria- and host-derived mechanisms to control intestinal epithelial cell homeostasis: Implications for chronic inflammation. Inflamm Bowel Dis. 2007;13:1153–1164. doi: 10.1002/ibd.20174. [DOI] [PubMed] [Google Scholar]
- 44.Yadav V, Varum F, Bravo R, Furrer E, Bojic D, Basit AW. Inflammatory bowel disease: Exploring gut pathophysiology for novel therapeutic targets. Transl Res. 2016;176:38–68. doi: 10.1016/j.trsl.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 45.Geremia A, Biancheri P, Allan P, Corazza GR, Di Sabatino A. Innate and adaptive immunity in inflammatory bowel disease. Autoimmun Rev. 2014;13:3–10. doi: 10.1016/j.autrev.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 46.Speca S, Rousseaux C, Dubuquoy C, Rieder F, Vetuschi A, Sferra R, Giusti I, Bertin B, Dubuquoy L, Gaudio E, et al. Novel PPARγ modulator GED-0507-34 levo ameliorates inflammation-driven intestinal fibrosis. Inflamm Bowel Dis. 2016;22:279–292. doi: 10.1097/MIB.0000000000000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rieder F, Fiocchi C. Intestinal fibrosis in IBD - a dynamic, multifactorial process. Nat Rev Gastroenterol Hepatol. 2009;6:228–235. doi: 10.1038/nrgastro.2009.31. [DOI] [PubMed] [Google Scholar]
- 48.Andersen V, Nimmo E, Krarup HB, Drummond H, Christensen J, Ho GT, Ostergaard M, Ernst A, Lees C, Jacobsen BA, et al. Cyclooxygenase-2 (COX-2) polymorphisms and risk of inflammatory bowel disease in a Scottish and Danish case-control study. Inflamm Bowel Dis. 2011;17:937–946. doi: 10.1002/ibd.21440. [DOI] [PubMed] [Google Scholar]
- 49.Østergaard M, Ernst A, Labouriau R, Dagiliené E, Krarup HB, Christensen M, Thorsgaard N, Jacobsen BA, Tage-Jensen U, Overvad K, et al. Cyclooxygenase-2, multidrug resistance 1, and breast cancer resistance protein gene polymorphisms and inflammatory bowel disease in the Danish population. Scand J Gastroenterol. 2009;44:65–73. doi: 10.1080/00365520802400826. [DOI] [PubMed] [Google Scholar]
- 50.Cox DG, Crusius JB, Peeters PH, Bueno-de-Mesquita HB, Pena AS, Canzian F. Haplotype of prostaglandin synthase 2/cyclooxygenase 2 is involved in the susceptibility to inflammatory bowel disease. World J Gastroenterol. 2005;11:6003–6008. doi: 10.3748/wjg.v11.i38.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garrity-Park MM, Loftus EV, Jr, Bryant SC, Smyrk TC. A biomarker panel to detect synchronous neoplasm in non-neoplastic surveillance biopsies from patients with ulcerative colitis. Inflamm Bowel Dis. 2016;22:1568–1574. doi: 10.1097/MIB.0000000000000789. [DOI] [PubMed] [Google Scholar]