Abstract
Pterostilbene (Pter) is reported to exhibit an anticancer effect in hepatocellular carcinoma (HCC). In order to explore the anticancer mechanism in HCC cells, the present study aimed to investigate whether pterostilbene (Pter) may increase phosphatase and tensin homolog (PTEN) expression through targeted downregulation of microRNA (miRNA/miR)-19a in hepatocellular carcinoma (HCC). The proliferation, apoptosis and cell cycle was analyzed in the SMMC-7721 HCC cell line by MTT assays and flow cytometry methods. Cells were divided into five treatment groups: Pter treatment, miR-19a inhibitor transfection, Pter + miR-19a inhibitor, negative control transfection and blank control. The expression of miR-19a and PTEN was detected by reverse transcription-quantitative polymerase chain reaction and western blot analysis following treatment. Furthermore, a luciferase reporter gene assay was performed to determine whether the PTEN gene was a direct target of miR-19a. The results demonstrated that Pter treatment or miR-19a inhibitor transfection downregulated miR-19a and induced PTEN/Akt pathway regulation, which led to proliferation inhibition, cell cycle arrest in the S phase, increased apoptosis and reduced cell invasion. These results indicated that Pter may increase PTEN expression through the direct downregulation of miR-19a in HCC. Therefore, miR-19a may have potential as a novel molecular marker for HCC and Pter may be a promising clinical target with the potential to be developed as a HCC therapy.
Keywords: phosphatase and tensin homolog, Akt, microRNA-19a, pterostilbene, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is among the most common types of malignant tumor. Various genetic and environmental risk factors have been identified for HCC, including aflatoxin, hepatitis, genetic factors (liver cancer of first-degree relatives) and adverse environmental factors (chemical contamination of the water environment) (1). A previous study identified microRNAs (miRNAs/miRs), which are short non-coding RNAs, as potential biomarkers and therapeutic targets in HCC. An increasing amount of research has demonstrated that miRNAs modulate gene expression in cancer and may promote or inhibit tumor growth (2–4). miRNAs affect the stability and transcriptional activity of their target mRNAs by binding to the 3′-untranslated regions (UTRs) of mRNA (5). miRNAs may be employed as biomarkers and have an important role in cancer diagnosis and prognosis. Therefore, miRNAs have the potential to be developed as targets for anti-cancer drugs (6).
miR-19a forms part of the miR-17-92 cluster. Research has revealed that its overexpression occurs in various tumor types including liver (3,4), lung (7), prostate (8) and breast cancer (9), gastric carcinoma (10), colorectal cancer (11) and osteosarcoma (12), and this overexpression may stimulate tumor growth and inhibit tumor cell apoptosis. In the present study, the downregulation of miR-19a prevented proliferation, promoted cell apoptosis and inhibited cell invasion in the SMMC-7721 HCC cell line. Additionally, reduced miR-19a expression led to the upregulation of phosphatase and tensin homolog (PTEN) and suppression of tumor growth, demonstrating that PTEN may exhibit antioncogenic effects in HCC cells. Previous investigation of the molecular biology of HCC has demonstrated that the PTEN/Akt signaling pathway (13–17) is closely associated with the proliferation, apoptosis and cell cycle arrest of HCC cells (18–21). It has also been reported that miR-19a may regulate the PTEN/Akt signaling pathway and subsequently affect the biological behavior of HCC cells (22,23). These discoveries are important for the regulation of cancer cell growth and the development of targeted cancer therapies.
Pterostilbene (Pter) is a derivative of resveratrol, which is an established epigenetic modulator. It is extracted from dietary compounds such as peanuts, grapes, berries and red wine, and is associated with anti-inflammatory, antioxidative, cardioprotective and antioncogenic activity (17,24). Pter may modulate various molecular targets, including miR-19a, to promote apoptosis, cell cycle arrest and cancer growth suppression (25). The present study demonstrated that the antioncogenic effect of Pter may be a result of miR-19a downregulation, which subsequently leads to an increase in PTEN expression.
Materials and methods
Reagents
Pter (purity, >99%; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) was dissolved in dimethyl sulfoxide (DMSO; Sigma-Aldrich; Merck KGaA, Darmstadt, Germany).
Cell culture
The SMMC-7721 HCC cell line (China Center for Type Culture Collection, Wuhan University, Wuhan, China) was cultured and maintained in RPMI-1640 (Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc.) with 100 U/ml penicillin and 100 U/ml streptomycin. Cells were incubated at 37°C with 5% CO2.
Cell transfection
SMMC-7721 cells at a density of 60–70% confluence were divided into the following five groups: Pter treatment group (50 µM Pter), miR-19a inhibitor group (transfected with miR-19a inhibitor 5′-UCAGUUUUGCAUAGAUUUGCACA-3′), Pter + miR19a inhibitor group (50 µM Pter and transfected with miR-19a inhibitor), negative control group (transfected with negative control 5′-UUGUACUACACAAAAGUACUG-3′) and the blank group (DMSO treatment) at 37°C for 48 h. Lipofectamine 2000 (5 µl; Invitrogen; Thermo Fisher Scientific, Inc.) was diluted to 100 µl in serum-free RPMI-1640 medium. miR-19a inhibitor (20 pM) or negative control (20 pM) was also diluted to 50 µl in the serum-free RPMI-1640 medium at room temperature for 5 min. Both of the dilutions were subsequently mixed at room temperature for 20 min prior to transfer into culture plates for cell transfection at 37°C in 5% CO2 for 6 h. The transfected cells were subsequently cultivated in complete medium (RPMI-1640 medium containing 10% FBS with 100 U/ml penicillin and 100 U/ml streptomycin) for 48 h and then the subsequent experiments including qPCR, western blotting, MTT and cell cycle and apoptosis assay were performed.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Following treatment of cells (5×107 cells/well) with Ctrl (DMSO), 5, 25, 50 and 100 µM Pter at 37°C for 24 h, and the establishment of the 5 treatment groups described above, the total RNA was extracted from SMMC-7721 cells using TRIzol (Life Technologies; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. RNA concentration was detected at an absorbance of A260 and A280. cDNA was synthesized by using a PrimeScript RT reagent kit (Takara Bio, Inc., Otsu, Japan). The PCR amplification primer sequences are presented in Table I. qPCR was performed by the MiniOpticon Real-Time PCR system (Bio-Rad Laboratories, Inc., Hercules, CA, USA) using SYBR Premix Ex Taq (Takara Bio, Inc.). Following initial denaturation at 95°C for 30 sec, amplifications were performed for 40 cycles at 95°C for 5 sec and 60°C for 30 sec. miR-19a levels were compared to U6 as an internal reference and PTEN was compared with β-actin. qPCR was repeated 3 times. The relative expression of miRNA and mRNA was calculated by the 2−ΔΔCq method (26).
Table I.
Primer sequences for quantitative polymerase chain reaction amplification.
| Primer sequence | ||
|---|---|---|
| Target | Forward | Reverse |
| miR-19a | 5′-GCGTGTGCAAATCTATGCAA-3′ | 5′-GTGCAGGGTCCGAGGT-3′ |
| U6 | 5′-CTCGCTTCGGCAGCACA-3′ | 5′-AACGCTTCACGAATTTGCGT-3′ |
| PTEN | 5′-CCAAGCTTATGACAGCCATCATC-3′ | 5′-CGCGGATCCTCAGACTTTTGTAA-3′ |
| β-actin | 5′-GAATCAATGCAAGTTCGGTTCC-3′ | 5′-TCATCTCCGCTATTAGCTCCG-3′ |
miR, microRNA; PTEN, phosphatase and tensin homolog.
Western blot analysis
Following treatment of cells (5×104 cells/well) with Ctrl (DMSO), 5, 25, 50 and 100 µM Pter at 37°C for 24 h, and establishment of the 5 treatment groups described above, whole proteins were extracted by RIPA lysis buffer (Beyotime Institute of Biotechnology, Shanghai, China) from SMMC-7721 cells which was quantified by a BCA Protein Assay kit (Beyotime Institute of Biotechnology) and separated at 20 µg/lane by 15% SDS-PAGE prior to transfer onto polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.) using an Electrophoresis Transfer System (Bio-Rad Laboratories, Inc.). The blots blocked with 5% fat-free milk in a Tris-buffered saline with 5% Tween 20 (TBST) for 1 h at room temperature, were probed with anti-PTEN (mouse monoclonal; cat. no. ab79156; 1:500; Abcam, Cambridge, UK), anti-Akt (Rat polyclonal; cat. no. ab8805; 1:500; Abcam), anti-phosphorylated (p)-Akt (Rat polyclonal; cat. no. ab8933; 1:500; Abcam) and anti-β-actin (Rat polyclonal; cat. no. ab8227; 1:2,000; Abcam) primary antibodies overnight at 4°C, and subsequently treated with the respective secondary antibodies (Rat, cat. no. ab218695; 1:4,000 or mouse cat. no. ab131368; 1:4,000 dilution; Abcam) for 1 h at 4°C. The proteins were visualized by using an enhanced chemiluminescence detection kit (Thermo Fisher Scientific, Inc., Waltham, MA, USA). Densitometric analysis was performed to quantify protein expression using ImageJ software version 1.49 (National Institutes of Health, Bethesda, MD, USA).
MTT assay
Following treatment of cells (5×104 cells/well) with Ctrl (DMSO), 20, 40, 60, 80 and 100 µM Pter at 37°C for 24, 48 or 72 h, and establishment of the five treatment groups described in the cell transfection section above, the proliferation of SMMC-7721 was detected by MTT assays (Sigma-Aldrich; Merck KGaA). To investigate cell viability, 0.5 mg/ml MTT solution was added into each well following seeding of the treated cells for 24 h at a density of 3×105 cells/ml per well. Cells were subsequently incubated for 4 h at 37°C with 5% CO2. The supernatant liquid was removed and 150 µl DMSO was added to each well. A microplate reader (BioTek Instruments, Inc., Winooski, VT, USA) was used to determine optical density values at 570 nm after 24, 48 and 72 h. For the experiments that consisted of five different treatment groups, measurements were taken after 24 h.
Cell cycle and apoptosis analysis by flow cytometry
Cell cycle and apoptosis was examined in the five treatment groups by flow cytometry. Annexin V-fluorescein isothiocyanate (10 µl) and propidium iodide (PI; 5 µl, Sigma-Aldrich; Merck KGaA) were incubated with the cells (5×105 cells/well) in each group in the dark at 4°C for 30 min. A flow cytometer (BD Biosciences, San Jose, CA, USA) was used to calculate the percentage of apoptotic cells with FlowJo version 10 software (FlowJo LLC, Ashland, OR, USA).
To analyze the cell cycle, cells (5×105 cells/well) were washed twice with PBS and fixed with 75% ethanol overnight at 4°C. The cells were subsequently stained with 5 µl PI/ribonuclease A (Sigma-Aldrich; Merck KGaA) at 4°C for 30 min in the dark. Data was analyzed with a flow cytometer (BD Biosciences). A total of 14,000 fluorescence signals for each PI-stained sample were collected and calculated by ModFit LT version 3.2 software (Verity Software House, Inc., Topsham, ME, USA).
Cell invasion
SMMC-7721 cell invasion ability was analyzed in Transwell chambers (pore size, 8 µm; Corning Incorporated, Corning, NY, USA) coated with Matrigel in serum-free medium. Following incubation, the 5×104 cells/well were added into the upper chamber at 37°C in a humidified 5% CO2 for 24 h. Then the cells were removed from the upper chamber (medium containing serum-free RPMI-1640) and the invading cells in the lower chamber (medium containing 20% FBS) were fixed with 4% paraformaldehyde at 37°C for 15 min, then stained with 0.1% crystal violet at 37°C for 15 min and counted using a phase-contrast microscope at ×200 magnification of each membrane in 5 representative fields.
Luciferase reporter gene assay
The binding sequence of PTEN 3′UTR and miR-19a was predicted by TargetScanHuman version 7.1 (www.targetscan.org/vert_71/). The pGL3-Luciferase vector with firefly and Renilla luciferase activity (Wuhan Jin Kairui Biological Engineering Co., Ltd, Wuhan, China) was digested with XbaI and the target sequence was inserted. The promoter sequence of PTEN 3′UTR containing the binding site was synthesized as the wild-type (WT) plasmid (PTEN-WT). Similarly, a mutated binding site of PTEN was constructed as the mutant-type (MT) plasmid (PTEN-MT). The cells (5×104 cells/well) were transfected by using Lipofectamine 2000® (5 µl; Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h. The cells were subsequently co-transfected with the luciferase reporter plasmid (20 pM) and the miR-19a inhibitor (20 pM) or negative control (20 pM) prior to treatment with the Dual-Luciferase Reporter Assay System (Promega Corporation, Madison, WI, USA), according to the manufacturer's protocol. Reporter gene activity was detected and normalized with the ratio of Renilla and firefly luciferase activity.
Statistical analysis
Data are presented as the mean ± standard deviation from three independent experiments. All statistical analyses were performed with SPSS version 21.0 software (IBM Corp., Armonk, NY, USA). And unpaired Student's t-tests were employed to analyze the difference between two groups. P<0.05 was considered to indicate a statistically significant difference.
Results
Pter and miR-19a inhibitor downregulate miR-19a expression
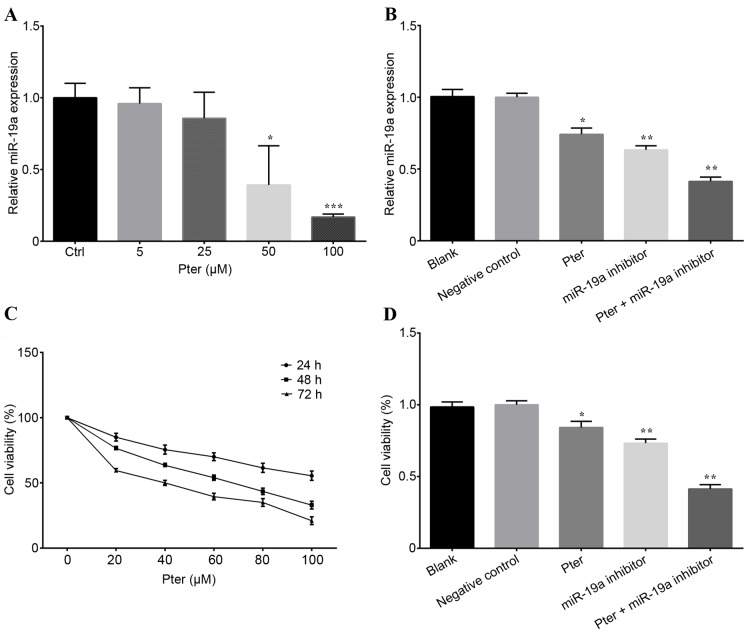
Previous reports have indicated that the modulation of PTEN-targeting miRNAs by Pter in prostate cancer led to the overexpression of PTEN and downregulation of PI3K-Akt pathway activity (17,27). Thus, the present study aimed to confirm this hypothesis in HCC cells. qPCR was performed to measure the relative expression of miR-19a in SMMC-7721 cells. The results demonstrated that miR-19a expression was decreased by Pter treatment in a dose-dependent manner in SMMC-7721 cells, indicating that Pter may exhibit important antioncogenic activity (P<0.05; Fig. 1A). In addition, miR-19a expression was also significantly lower in the Pter, miR-19a inhibitor and Pter + miR-19a inhibitor groups, compared with the blank or negative control groups (P<0.05; Fig. 1B).
Figure 1.
Pter treatment and miR-19a inhibitor transfection affected the miR-19a expression and cell viability. (A) Cells were treated with Pter (0, 5, 25, 50 and 100 µM) for 24 h and miR-19a expression was detected by RT-qPCR. (B) Relative quantification of miR-19a expression was calculated by RT-qPCR for SMMC-7721 cells treated with 50 µM Pter, miR-19a inhibitor, 50 µM Pter + miR-19a inhibitor, negative control and blank. (C) An MTT assay was performed to detect cell viability following treatment with Pter at different concentrations (0, 20, 40, 60, 80 and 100 µM) for 24, 48 and 72 h. (D) Cells were treated with 50 µM Pter, miR-19a inhibitor, 50 µM Pter + miR-19a inhibitor, negative control and blank for 24 h. Cell viability was subsequently assessed by an MTT assay. Each experiment was performed in triplicate. Data are presented as the mean ± standard deviation. For part A, *P<0.05 and ***P<0.001 vs. Ctrl; for parts B and D, *P<0.05 and **P<0.01 vs. blank or negative control group. The Pter-only treatment group was compared with the blank control group, while the miR-19a inhibitor and Pter + miR-19a inhibitor groups were compared with the negative control group. Pter, pterostilbene; miR, microRNA; RT-qPCR, reverse transcription-quantitative polymerase chain reaction; Ctrl, control.
Pter and miR-19a inhibitor treatment represses cell proliferation
The MTT assay revealed that Pter repressed SMMC-7721 cell proliferation in a time- and dose-dependent manner (Fig. 1C). Furthermore, after 24 h, the cell viability in the miR-19a inhibitor and Pter + miR-19a inhibitor groups was markedly reduced compared with the blank or negative control group (P<0.05; Fig. 1D). Additionally, a synergistic effect was observed between Pter and the miR-19a-inhibitor. Both Pter and miR-19a inhibitor treatments significantly repressed SMMC-7721 cell proliferation (Fig. 1D). Pter exerted the most potent inhibitory effect on SMMC-7721 cell proliferation.
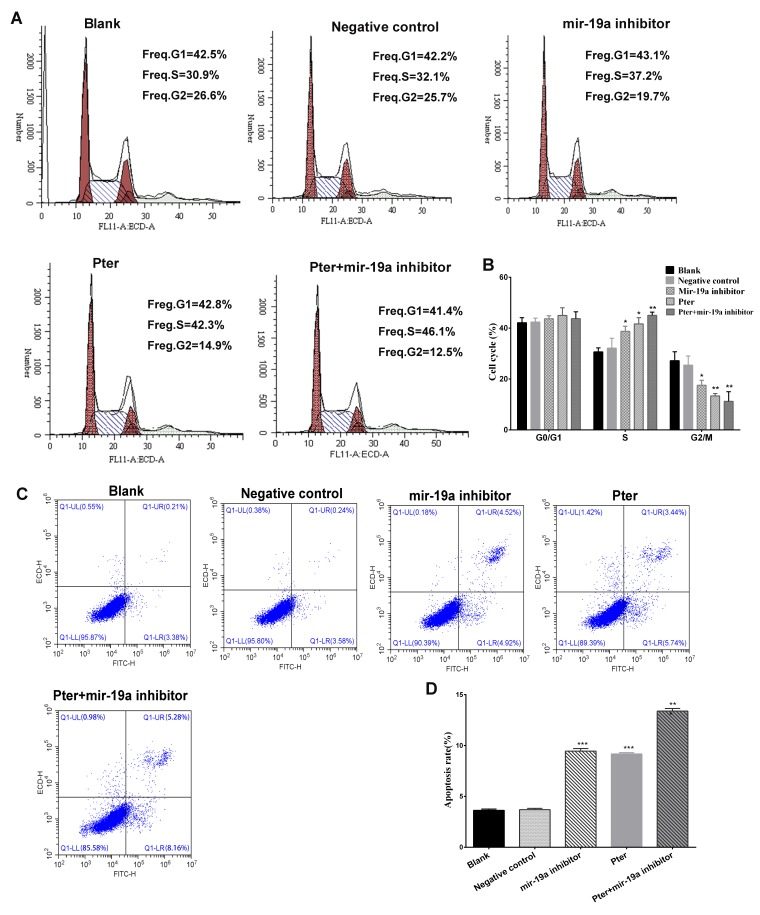
Inhibition of miR-19a reduces cell cycle progression, increases apoptosis and inhibits invasion
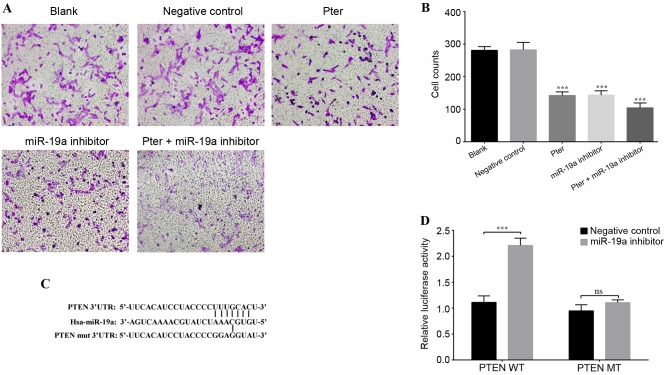
Flow cytometry was performed to analyze the cell cycle of the cells treated with Pter. No statistically significant differences were observed between the cells in the blank and negative control groups in each phase (P>0.05; Fig. 2A and B). However, the Pter treatment and miR-19a inhibitor groups demonstrated a significantly higher number of cells in the S phase, and significantly fewer cells in the G2/M phase, compared with the blank or negative control groups (P<0.05; Fig. 2A and B), indicating increased cell cycle arrest in S phase and reduced cell mitosis following Pter treatment or miR-19a inhibition. Furthermore, the apoptosis of cells was also analyzed; the apoptotic rate of the blank and negative control groups were not significantly different, but Pter treatment or miR-19a inhibition significantly increased the apoptotic rate compared with blank or negative control groups (P<0.05; Fig. 2C and D). Synergy between Pter treatment and miR-19a inhibitor transfection was also observed (Fig. 2C and D). Additionally, cell invasion ability was significantly decreased by Pter and miR-19a inhibitor, compared with blank or negative control groups (Fig. 3A and B).
Figure 2.
Flow cytometry analysis of cell cycle and apoptotic rate. (A) Cell cycle analysis of the Pter, miR-19a inhibitor, Pter + miR-19a inhibitor, negative control and blank control groups. (B) Quantification of the cell cycle analysis for each treatment group. (C) Representative flow cytometry plots indicating the apoptotic rate (the early and late stage apoptosis) within Pter, miR-19a inhibitor, Pter + miR-19a inhibitor, negative control and blank control groups. (D) Quantification of the apoptotic rate for each treatment group. *P<0.05, **P<0.01 and ***P<0.001 vs. blank or negative control groups. The Pter-only treatment group was compared with the blank control group, while the miR-19a inhibitor and Pter + miR-19a inhibitor groups were compared with the negative control group. Pter, pterostilbene; miR, microRNA.
Figure 3.
Cell invasion and luciferase reporter gene assays. (A) Representative images of the lower chamber of the Transwell invasion assay in different treatment groups. (B) Cell counts using a phase-contrast microscope revealed that Pter and miR-19a inhibitor groups exhibited significantly reduced cell invasion ability. This reduction was further enhanced in the Pter + miR-19a group. (C) Binding sequence between miR-19a and WT PTEN 3′-UTR, and the MT binding site of the PTEN 3′-UTR. PTEN was confirmed as a direct downstream target of miR-19a by TargetScanHuman. (D) A luciferase reporter gene activity assay was performed following co-transfection of PTEN 3′UTR MT/WT and miR-19a inhibitor or negative control. miR-19a inhibited the luciferase activity of the WT luciferase plasmid, while there was no significant difference when the miR-19a inhibitor was co-transfected with the MT luciferase plasmid. For part B, ***P<0.001 vs. blank or negative control groups; for part D, ***P<0.001, as indicated. In part B, the Pter-only treatment group was compared with the blank control group, while the miR-19a inhibitor and Pter + miR-19a inhibitor groups were compared with the negative control group. Pter, pterostilbene; miR, microRNA; WT, wild type; phosphatase and tensin homolog; UTR, untranslated region; MT/mut, mutant type; ns, not significant.
Targeted regulation of PTEN 3′UTR by miR-19a
A luciferase reporter gene assay was performed to verify whether PTEN may be a direct target gene of miR-19a. It was demonstrated that the target sequence of miR-19a and PTEN 3′-UTR matched (Fig. 3C). Therefore, the pGL3-Luciferase vector was constructed and the PTEN 3′UTR WT (AGCUUA) or MT (ACACGG) sequence was inserted downstream, and the vector was subsequently co-transfected with the miR-19a inhibitor or negative control. Following PTEN 3′-UTR-WT co-transfection with miR-19a inhibitor, luciferase activity was significantly higher compared with the negative control group; however, no statistically significant difference was observed between the negative control and miR-19a inhibitor group when co-transfected with PTEN 3′UTR-MT (Fig. 3D). There results indicated that the base sequence of miR-19a matched the PTEN mRNA 3′UTR and that PTEN is a target site of miR-19a.
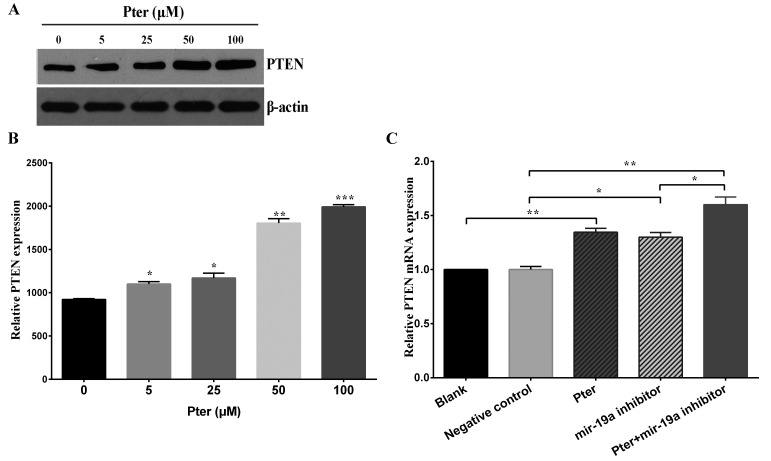
Pter downregulates PTEN mRNA and protein expression through miR-19a
Initially, western blot analysis was performed to determine PTEN protein expression following treatment with various concentrations of Pter. The results demonstrated that Pter increased PTEN protein expression in a dose-dependent manner (Fig. 4A and B). Furthermore, to investigate the association between miR-19a and PTEN expression further, SMMC-7721 cells with low expression levels of miR-19a were established by miR-19a inhibitor transfection. The Pter and miR-19a inhibitor groups exhibited significantly increased expression of PTEN mRNA compared with the blank or negative control groups, respectively (P<0.05; Fig. 4C). A synergistic effect was observed in the Pter + miR-19a inhibitor group (P<0.05; Fig. 4C).
Figure 4.
Pter downregulated PTEN protein and mRNA expression in SMMC-7721 cells. (A) SMMC-7721 cells were treated with increasing concentrations (0, 5, 25, 50 and 100 µM) of Pter for 24 h. The control group was treated with dimethyl sulfoxide. PTEN protein expression was detected by western blot analysis and representative bands are presented. (B) Western blot analysis results were quantified with ImageJ analysis software. (C) PTEN mRNA expression following treatment with 50 µM Pter, miR-19a inhibitor, 50 µM Pter + miR-19a inhibitor, negative control and blank. Cells were harvested and the relative expression of PTEN mRNA was calculated by reverse transcription-quantitative polymerase chain reaction. Data are presented as the mean + standard deviation of the mean of three independent experiments. For part B, *P<0.05, **P<0.01 and ***P<0.001 vs. 0 µM Pter group; for part C, *P<0.05 and **P<0.01, as indicated. Pter, pterostilbene; PTEN, phosphatase and tensin homolog; miR, microRNA.
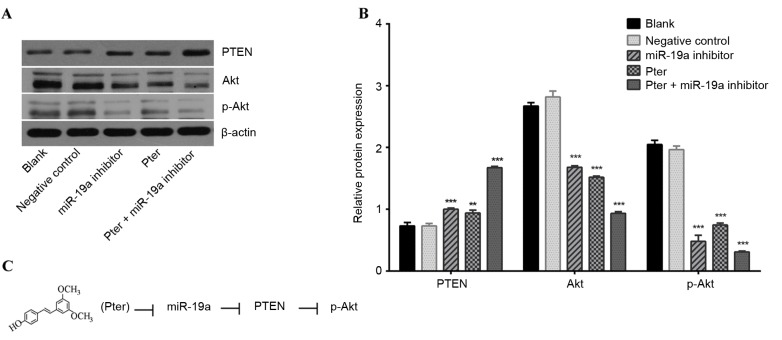
Additionally, reduced miR-19a expression induced a marked upregulation of PTEN protein expression compared with the negative control group (Fig. 5A and B). These results indicated that Pter treatment may enhance PTEN expression via the downregulation of miR-19a.
Figure 5.
PTEN, Akt and p-Akt levels in SMMC-7721 cells following miR-19a inhibitor transfection and Pter treatment for 24 h. (A) PTEN, Akt and p-Akt levels in SMMC-7721 cells were detected by western blot analysis and representative bands are presented. (B) Relative quantification of western blot analysis results by ImageJ analysis software. (C) Schematic representation of the proposed model of Pter mediation of the PTEN/Akt signaling pathway via miR-19a. **P<0.01 and ***P<0.001 vs. blank or negative control groups. The Pter-only treatment group was compared with the blank control group, while the miR-19a inhibitor and Pter + miR-19a inhibitor groups were compared with the negative control group. PTEN, phosphatase and tensin homolog; p-, phosphorylated-; miR, microRNA; Pter, pterostilbene.
Pter treatment and miR-19a inhibitor transfection mediates the PTEN/Akt signaling pathway
PTEN, Akt and p-Akt levels were detected by western blot analysis. Previous research has indicated that PTEN is an inhibitor of the PI3K/Akt pathway (28). The present study revealed a negative association between PTEN and miR-19a expression, and a positive association between PTEN and Akt expression, which indicates an association between the PI3K/Akt pathway and miR-19a. In the current study, the Pter and miR-19a inhibitor groups exhibited significantly upregulated PTEN protein expression and significantly reduced Akt and p-Akt levels (P<0.05; Fig. 5A and B). These results indicate that miR-19a may mediate the downstream targets of the PTEN/Akt signal pathway to affect the biological behavior of HCC (Fig. 5C).
Discussion
Previous research has demonstrated that miR-19a overexpression occurs in various tumor types. Pter was reported to inhibit the overexpression of certain oncogenic miRNAs (oncomiRs), including miR-19a in prostate cancer cells (29). miR-19a has been previously reported to regulate PTEN expression in various types of cancer, including osteosarcoma (18), myeloma (22), HCC (19) and breast cancer (23). Furthermore, the regulation of PTEN by Pter was demonstrated to exert marked anticancer effects (17,29). Therefore, in the present study, the ability of Pter to increase PTEN expression through direct downregulation of miR-19a in HCC was investigated and confirmed. The potential molecular mechanism of this action is presented in Fig. 5C.
An increasing number of studies have demonstrated the anticancer properties of certain dietary agents, indicating potential for their clinical application. The anticancer effects exerted by several of these dietary compounds has been reported to occur through the mediation of abnormal miRNA expression in malignant cells (17,30,31). Therefore, the present study aimed to confirm a direct association between miR-19a and one of its targets, PTEN, as well as the effect of the oncomiR on tumor cell function. The results of the current study demonstrated that Pter may have an important role in reversing the silencing of tumor suppressor PTEN expression by miR-19a. Although several reports have revealed the oncogenic potential of miR-19a in targeting the 3′UTR of PTEN mRNA in malignant cells (15,16,32), the present study focused on the ability of Pter to inhibit miR-19a expression.
In the current study, low miR-19a expression, established by transfection of a miR-19a inhibitor, induced a significant increase in PTEN 3′UTR luciferase activity. Pter also inhibited the expression of miR-19a in a dose-dependent manner, indicating that Pter treatment may directly ameliorate miR-19a overexpression. Overall, the results indicated that Pter downregulated miR-19a expression, which led to increased PTEN expression and subsequent PI3K/Akt signaling pathway regulation.
PTEN has been reported to participate in processes that are involved in basic cell function, including apoptosis, proliferation and cell cycle arrest (33). Chang et al (34) identified that PTEN/p-Akt pathway activation may have prognostic value in HCC. Furthermore, a lack of PTEN was hypothesized to continuously activate signaling pathways to promote uncontrollable cancer cell growth (35). In the present study, miR-19a downregulation by Pter recovered PTEN expression and directly affected the transcriptional activity of the Akt pathway.
The interaction between miR-19a and PTEN in HCC was investigated further. It was demonstrated that PTEN was a target gene of miR-19a through online TargetScan prediction. Increased PTEN expression was previously detected in HCC cells transfected with miR-19a inhibitor, which led to reduced cell growth and invasion and an increase in the apoptotic rate (19,36). Existing data indicates that miR-19a downregulation may reduce HCC occurrence by increasing PTEN expression. Consistently, the present study demonstrated that Pter treatment and miR-19a inhibitor transfection significantly decreased miR-19a expression and enhanced PTEN expression.
Wu et al (32) reported that malignant esophageal cells exhibited excessive Akt expression, which was implicated in uncontrollable cancer cell proliferation, and that PTEN repressed the Akt activity. It was reported that miR-19a and the 3′UTR of PTEN mRNA exhibit homology between their sequences and the interaction of these sequences may lead to PTEN downregulation and subsequent Akt upregulation (37). In the present study, miR-19a inhibitor transfection led to increased PTEN expression, which may be implicated in the cell cycle arrest in the S phase, increased apoptosis, decreased invasion and reduced viability that was observed in SMMC-7721 cells following miR-19a inhibitor transfection and Pter treatment.
In conclusion, the results of the present study demonstrated that Pter inhibited miR-19a expression, which subsequently led to regulation of the PTEN/Akt pathway, reduced cell viability, cell cycle arrest, apoptosis promotion and cell invasion inhibition in HCC cells. The present study may provide a foundation for future research, as miR-19a may have potential as a molecular HCC marker and Pter may be developed as a therapy for patients with HCC.
References
- 1.Schulze K, Imbeaud S, Letouzé E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang B, Pan X, Cobb GP, Anderson TA. microRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 3.Urtasun R, Elizalde M, Azkona M, Latasa MU, García-Irigoyen O, Uriarte I, Fernández-Barrena MG, Vicent S, Alonso MM, Muntané J, et al. Splicing regulator SLU7 preserves survival of hepatocellular carcinoma cells and other solid tumors via oncogenic miR-17-92 cluster expression. Oncogene. 2016;35:4719–4729. doi: 10.1038/onc.2015.517. [DOI] [PubMed] [Google Scholar]
- 4.Zhu H, Han C, Wu T. MiR-17-92 cluster promotes hepatocarcinogenesis. Carcinogenesis. 2015;36:1213–1222. doi: 10.1093/carcin/bgv112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 6.Sapre N, Selth LA. Circulating MicroRNAs as biomarkers of prostate cancer: The state of play. Prostate Cancer. 2013;2013:539680. doi: 10.1155/2013/539680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li L, Song W, Yan X, Li A, Zhang X, Li W, Wen X, Zhou L, Yu D, Hu JF, Cui J. Friend leukemia virus integration 1 promotes tumorigenesis of small cell lung cancer cells by activating the miR-17-92 pathway. Oncotarget. 2017;8:41975–41987. doi: 10.18632/oncotarget.16715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar S, Hicks C, Levenson AS. Resveratrol and prostate cancer: Promising role for microRNAs. Mol Nutr Food Res. 2011;55:1219–1229. doi: 10.1002/mnfr.201100141. [DOI] [PubMed] [Google Scholar]
- 9.Liu F, Zhang F, Li X, Liu Q, Liu W, Song P, Qiu Z, Dong Y, Xiang H. Prognostic role of miR-17-92 family in human cancers: Evaluation of multiple prognostic outcomes. Oncotarget. 2017;8:69125–69138. doi: 10.18632/oncotarget.19096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strickertsson JA, Rasmussen LJ, Friis-Hansen L. Enterococcus faecalis infection and reactive oxygen species Down-regulates the miR-17-92 cluster in gastric adenocarcinoma cell culture. Genes (Basel) 2014;5:726–738. doi: 10.3390/genes5030726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Knudsen KN, Nielsen BS, Lindebjerg J, Hansen TF, Holst R, Sorensen FB. microRNA-17 Is the Most Up-Regulated member of the miR-17-92 cluster during early colon cancer evolution. PLoS One. 2015;10:e0140503. doi: 10.1371/journal.pone.0140503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zou P, Ding J, Fu S. Elevated expression of microRNA-19a predicts a poor prognosis in patients with osteosarcoma. Pathol Res Pract. 2017;213:194–198. doi: 10.1016/j.prp.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 13.Zhou B, Wang J, Zheng G, Qiu Z. Methylated urolithin A, the modified ellagitannin-derived metabolite, suppresses cell viability of DU145 human prostate cancer cells via targeting miR-21. Food Chem Toxicol. 2016;97:375–384. doi: 10.1016/j.fct.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Song L, Liu S, Zhang L, Yao H, Gao F, Xu D, Li Q. MiR-21 modulates radiosensitivity of cervical cancer through inhibiting autophagy via the PTEN/Akt/HIF-1α feedback loop and the Akt-mTOR signaling pathway. Tumour Biol. 2016;37:12161–12168. doi: 10.1007/s13277-016-5073-3. [DOI] [PubMed] [Google Scholar]
- 15.Li C, Song L, Zhang Z, Bai XX, Cui MF, Ma LJ. MicroRNA-21 promotes TGF-β1-induced epithelial-mesenchymal transition in gastric cancer through up-regulating PTEN expression. Oncotarget. 2016;7:66989–67003. doi: 10.18632/oncotarget.11888. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Fang H, Xie J, Zhang M, Zhao Z, Wan Y, Yao Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9:953–961. [PMC free article] [PubMed] [Google Scholar]
- 17.Dhar S, Kumar A, Rimando AM, Zhang X, Levenson AS. Resveratrol and pterostilbene epigenetically restore PTEN expression by targeting oncomiRs of the miR-17 family in prostate cancer. Oncotarget. 2015;6:27214–27226. doi: 10.18632/oncotarget.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao D, Chen Y, Chen S, Zheng C, Hu J, Luo S. MiR-19a regulates the cell growth and apoptosis of osteosarcoma stem cells by targeting PTEN. Tumour Biol. 2017;39:1010428317705341. doi: 10.1177/1010428317705341. [DOI] [PubMed] [Google Scholar]
- 19.Yu G, Chen X, Chen S, Ye W, Hou K, Liang M. MiR-19a, miR-122 and miR-223 are differentially regulated by hepatitis B virus X protein and involve in cell proliferation in hepatoma cells. J Transl Med. 2016;14:122. doi: 10.1186/s12967-016-0888-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ma Q, Peng Z, Wang L, Li Y, Wang K, Zheng J, Liang Z, Liu T. miR-19a correlates with poor prognosis of clear cell renal cell carcinoma patients via promoting cell proliferation and suppressing PTEN/SMAD4 expression. Int J Oncol. 2016;49:2589–2599. doi: 10.3892/ijo.2016.3746. [DOI] [PubMed] [Google Scholar]
- 21.Liu K, Liu S, Zhang W, Jia B, Tan L, Jin Z, Liu Y. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34:1003–1010. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, Chen Y, Zhao P, Zang L, Zhang Z, Wang X. MicroRNA-19a functions as an oncogene by regulating PTEN/AKT/pAKT pathway in myeloma. Leuk Lymphoma. 2017;58:932–940. doi: 10.1080/10428194.2016.1213827. [DOI] [PubMed] [Google Scholar]
- 23.Li X, Xie W, Xie C, Huang C, Zhu J, Liang Z, Deng F, Zhu M, Zhu W, Wu R, et al. Curcumin modulates miR-19/PTEN/AKT/p53 axis to suppress bisphenol A-induced MCF-7 breast cancer cell proliferation. Phytother Res. 2014;28:1553–1560. doi: 10.1002/ptr.5167. [DOI] [PubMed] [Google Scholar]
- 24.Aggarwal BB, Bhardwaj A, Aggarwal RS, Seeram NP, Shishodia S, Takada Y. Role of resveratrol in prevention and therapy of cancer: Preclinical and clinical studies. Anticancer Res. 2004;24:2783–2840. [PubMed] [Google Scholar]
- 25.Wang G, Dai F, Yu K, Jia Z, Zhang A, Huang Q, Kang C, Jiang H, Pu P. Resveratrol inhibits glioma cell growth via targeting oncogenic microRNAs and multiple signaling pathways. Int J Oncol. 2015;46:1739–1747. doi: 10.3892/ijo.2015.2863. [DOI] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Dhar S, Kumar A, Li K, Tzivion G, Levenson AS. Resveratrol regulates PTEN/Akt pathway through inhibition of MTA1/HDAC unit of the NuRD complex in prostate cancer. Biochim Biophys Acta. 2015;1853:265–275. doi: 10.1016/j.bbamcr.2014.11.004. [DOI] [PubMed] [Google Scholar]
- 28.Luo H, Yang Y, Duan J, Wu P, Jiang Q, Xu C. PTEN-regulated AKT/FoxO3a/Bim signaling contributes to reactive oxygen species-mediated apoptosis in selenite-treated colorectal cancer cells. Cell Death Dis. 2013;4:e481. doi: 10.1038/cddis.2013.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A, Rimando AM, Levenson AS. Resveratrol and pterostilbene as a microRNA-mediated chemopreventive and therapeutic strategy in prostate cancer. Ann N Y Acad Sci. 2017;1403:15–26. doi: 10.1111/nyas.13372. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Kong D, Wang Z, Sarkar FH. Regulation of microRNAs by natural agents: An emerging field in chemoprevention and chemotherapy research. Pharm Res. 2010;27:1027–1041. doi: 10.1007/s11095-010-0105-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Milenkovic D, Jude B, Morand C. miRNA as molecular target of polyphenols underlying their biological effects. Free Radic Biol Med. 2013;64:40–51. doi: 10.1016/j.freeradbiomed.2013.05.046. [DOI] [PubMed] [Google Scholar]
- 32.Wu YR, Qi HJ, Deng DF, Luo YY, Yang SL. MicroRNA-21 promotes cell proliferation, migration, and resistance to apoptosis through PTEN/PI3K/AKT signaling pathway in esophageal cancer. Tumour Biol. 2016;37:12061–12070. doi: 10.1007/s13277-016-5074-2. [DOI] [PubMed] [Google Scholar]
- 33.Wang G, Li Y, Wang P, Liang H, Cui M, Zhu M, Guo L, Su Q, Sun Y, McNutt MA, Yin Y. PTEN regulates RPA1 and protects DNA replication forks. Cell Res. 2015;25:1189–1204. doi: 10.1038/cr.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chang RM, Xu JF, Fang F, Yang H, Yang LY. MicroRNA-130b promotes proliferation and EMT-induced metastasis via PTEN/p-AKT/HIF-1α signaling. Tumour Biol. 2016;37:10609–10619. doi: 10.1007/s13277-016-4919-z. [DOI] [PubMed] [Google Scholar]
- 35.Li P, Mao WM, Zheng ZG, Dong ZM, Ling ZQ. Down-regulation of PTEN expression modulated by dysregulated miR-21 contributes to the progression of esophageal cancer. Dig Dis Sci. 2013;58:3483–3493. doi: 10.1007/s10620-013-2854-z. [DOI] [PubMed] [Google Scholar]
- 36.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV, Cheng JQ. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-6426. [DOI] [PubMed] [Google Scholar]